Summary
Cells of Flavobacterium johnsoniae, a rod-shaped bacterium devoid of pili or flagella, glide over glass at speeds of 2–4 μm/s [1]. Gliding is powered by a protonmotive force [2], but the machinery required for this motion is not known. Usually, cells move along straight paths, but sometimes they exhibit a reciprocal motion, attach near one pole and flip end-over-end, or rotate. This behavior is similar to that of a Cytophaga species described earlier [3]. Development of genetic tools for F. johnsoniae led to discovery of proteins involved in gliding [4]. These include the surface adhesin SprB that forms filaments about 160 nm long by 6 nm in diameter, which, when labeled with a fluorescent antibody [2] or a latex bead [5], are seen to move longitudinally down the length of a cell, occasionally shifting positions to the right or the left. Evidently, interaction of these filaments with a surface produces gliding. To learn more about the gliding motor, we sheared cells to reduce the number and size of SprB filaments and tethered cells to glass by adding anti-SprB antibody. Cells spun about fixed points, mostly counterclockwise, rotating at speeds of 1 Hz or more. The torques required to sustain such speeds were large, comparable to those generated by the flagellar rotary motor. However, we found that a gliding motor runs at constant speed rather than constant torque. Now there are three rotary motors powered by protonmotive force: the bacterial flagellar motor, the Fo ATP synthase, and the gliding motor.
Keywords: bacterial motility, torque-speed, type-9 secretion
Results and Discussion
The gliding motor rotates in place
We developed a method for tethering F. johnsoniae to a glass surface using anti-SprB antibody (Figure 1A). This method is similar to the procedure for shearing and tethering cells of Escherichia coli [6, 7], a technique used extensively in studies of chemotaxis of flagellated bacteria. Tethered F. johnsoniae cells rotated about a fixed point, as shown in Movie S1. Tracks of their center of mass were circular (Figure 1B). We tracked the tethering point and found its displacement to be within ~5 nm, which is negligible compared to the μm-sized circular trajectory (Figure 1C). This argues that the SprB filament is connected to a rotary motor that stays in place. To further test for evidence of rotation, we attached a polystyrene bead to a sheared cell and tracked rotation of the bead (Figure 1D, 1E, Movie S2).
Figure 1.
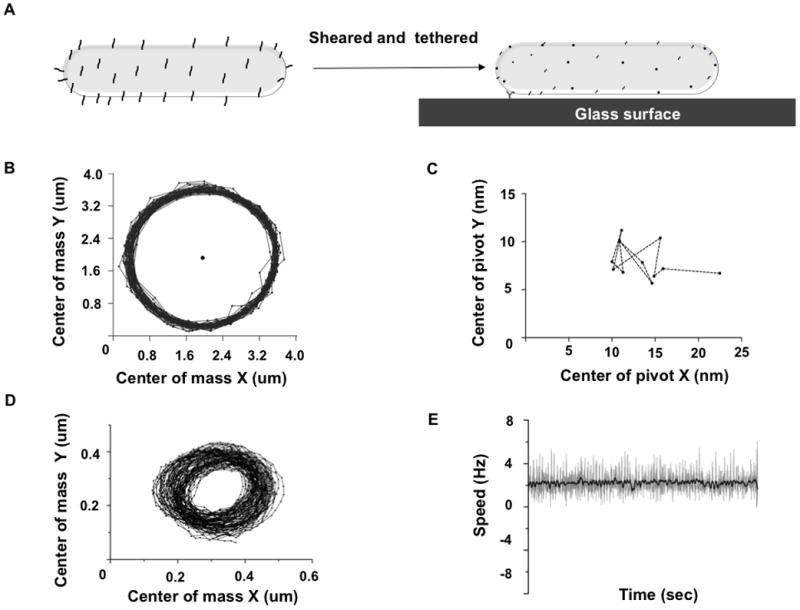
Evidence for a rotary motor. (A) The F. johnsoniae adhesin SprB is present on the cell surface as ~160-nm long filaments. SprB was sheared off and anti-SprB antibody was used to tether F. johnsoniae to a glass surface. (B) The trajectory of the center of mass of a tethered cell plotted over 1000 frames with the center of rotation plotted as a black circle (C) The position of the center of rotation was averaged over 100 frames and plotted as black circles for a movie spanning 1200 frames; the drift of the center of rotation shown with a dotted line was negligible (< 5 nm). (D) Center of mass of a 0.5 μm polystyrene bead tethered onto a sheared cell was tracked over 2192 frames. (E) Speed of rotation is plotted in grey, average speed was calculated every 10 frames and plotted in black.
Most gliding motors rotate counterclockwise
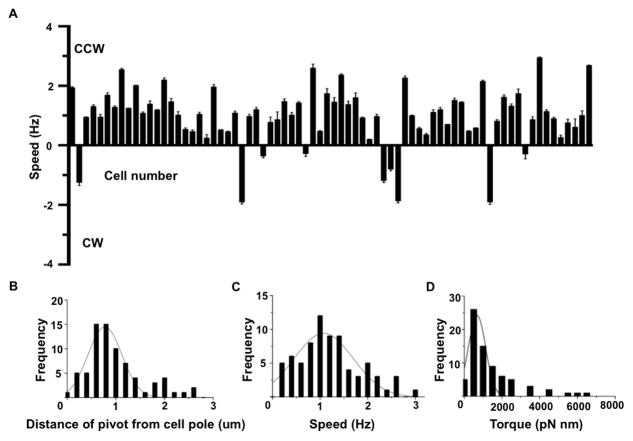
92% of motors rotated counterclockwise and 8% clockwise (Figure 2A). Changes in the direction of rotation were not observed. Presumably, the direction of rotation of motors observed on tethering determines the direction of translation of SprB filaments when cells glide. Fluorescently-labeled SprB has been reported to move along a left-handed closed helical loop [2], while labeling with latex beads has shown that SprB molecules move in different directions, often crossing paths while moving on a single cell [5]. We envision that gliding motors are present along multiple looped tracks and that these tracks intersect each other. Analysis of a population of tethered cells showed that the position of the pivot varied from near the pole to near the middle of the cell, but in a majority of cells, the pivot was near the pole (Figure 2B). This suggests that multiple tracks intersect near the pole.
Figure 2.
Speeds and torques recorded for 74 tethered cells. (A) 92% of cells rotated counterclockwise and 8% clockwise. Speed was calculated by recording cell rotation twice for 1-minute periods. Average speed for each recording was calculated. Error bars represent variation in speed of the same cell between the two recordings. Changes in direction of rotation were not seen. (B) Frequency distribution of pivot position for 74 cells normalized for an average cell length of 6 um. Most cells tethered at a distance of ~1 um from the cell pole. (C) Speeds ranged from 0.2–3 Hz, with a majority of cells rotating with a speed ~1 Hz. (D) The torque varied from ~200 to ~6000 pN nm with the majority of cells at a torque ~1000 pN nm. For torque calculations, see Materials and Methods.
Torque generated by the gliding motor is large
Speeds of rotation were calculated from the center of mass trajectories using custom MATLB codes. The cells rotated with an average angular speed of ~1Hz (Figure 2C). Torque generated by each gliding motor was calculated using a formula described in Materials and Methods [8], based upon measurements of angular speed (Figure 2C), cell length, cell width and trajectory radius (Figure S1). Torque ranged from 200–6000 pN nm, with most cells running at ~1000 pN nm (Figure 2D). Torques measured with motors of E. coli spinning latex beads (~1 μm dia.) averaged ~1300 pN nm [9, 10], so the torques generated by the gliding motor are comparable to those generated by a flagellar motor. Stator elements formed by MotA and MotB proteins act as force-generating units that generate torque for rotation of flagellar motors. It is likely that similar stator elements, albeit made up of different protein subunits, harvest protonmotive force to power rotation of the gliding motor.
The gliding motor runs at constant speed
F. johnsoniae cells tethered in a flow cell were exposed to 8% w/v solutions of Ficoll in motility medium (MM). Ficoll is a viscous agent commonly used to alter the load on bacterial flagellar motors [11]. Rotation speeds of single cells were measured. Speeds did not change significantly (Figures 3A, B). However, the torque generated by the motors, equal to the viscosity times the viscous drag coefficient times the speed, increased dramatically (Figures 3C, D). A gliding cell has multiple moving SprB filaments. It is reasonable for them to move at the same rather than at different speeds. Otherwise, if more than one filament adhered to the substratum, the motors would not work synchronously. In our experiment, speed remains constant but torque increases with increase in load (viscosity). When attached to a bead, which represents a low load compared with that of a tethered cell, the gliding motor rotated the bead at a speed comparable to that of the tethered cell (Figure 1E). We do not know whether speed is an intrinsic property of the motor or whether a cellular mechanism exists that coordinates speeds of different motors.
Figure 3.
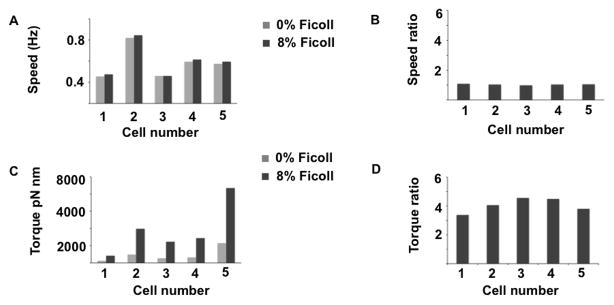
Measured speeds and computed torques for cells in 0% and 8% Ficoll. (A) Speed at 0% and 8% Ficoll. (B) The ratio of speeds at 8% and 0% Ficoll are close to 1. (C) Torque at 0% and 8% Ficoll. (D) The ratio of torques at 8% and 0% Ficoll approximate the ratio of viscosities, 3.91.
The gliding motor is novel
Genome sequencing has shown that F. johnsoniae lacks proteins similar to components of the bacterial flagellar motor [12]. GldJ is a putative component of the gliding motor. Presumably GldJ interacts with the Type-IX protein secretion system (T9SS) and is important for the movement of cell-surface adhesins. GldK, GldL, GldM and GldN are core T9SS proteins and cells lacking these proteins do not exhibit motility. The macromolecular structure of gliding motor and its exact interaction with T9SS is unclear. GldL localizes to the cytoplasmic membrane and it might act as an anchor for the gliding motor [13]. Besides the core T9SS proteins, other Gld and Spr proteins might associate with this motor. The gliding motor appears to associate with T9SS in a manner analogous to the association of the bacterial flagellar motor with the Type-III secretion system (T3SS). In flagellated bacteria, T3SS is required for secretion of axial components of the flagellum. In F. johnsoniae, T9SS is required for secretion of the SprB filament and a mobile adhesin, RemA [13, 14].
Model for Flavobacterium gliding
A model for Flavobacterium gliding was proposed recently [15] in which rotary motors drive baseplates, to which SprB filaments are attached. The baseplates were visualized by cryo-electron tomography [16]. In our model, gliding motors form complexes with T9SS, which span the inner and outer membranes, harvesting protonmotive force to power SprB rotation. The baseplates move along the inner surface of the outer membrane (Figure 4). If this is correct, then shearing breaks filaments and fragments baseplates, allowing a motor to spin a fragment together with one or more of its filaments that are adsorbed to the substratum. To explain movement of sprB along tracks, cells must contain substantial numbers of gliding motors. If there is a molecular rack and pinion that converts rotation to translation, and the pinion rotates, say, 10 Hz, it would have to be 100 nm in diameter to drive the cell 3 μm/s. So, gliding motors could be as large as bacterial flagellar motors. Attempts were made to isolate gliding motors with Cytophaga [3], using the methods developed for flagellar motors, but without success. In an alternative model, SprB filaments might be attached to rotary motors directly, with an unknown mechanism that passes filaments from one motor to the next. Advanced microscopic tools might shed light on motor structure and interactions between motors and baseplates. How the gliding motor generates torque and manages to run at a constant speed are interesting questions that beg for answers. We now know of three rotary motors powered by protonmotive force: the bacterial flagellar motor, the Fo ATP synthase, and the gliding motor. The bacterial motors generate about 25 times more torque than the F1 ATPase.
Figure 4.
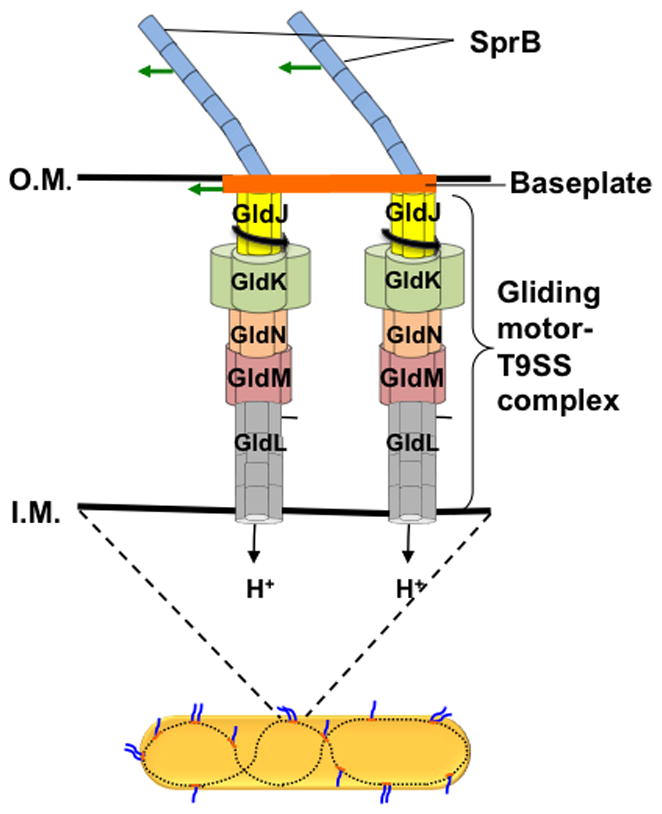
Flavobacterium gliding model. A Flavobacterium cell with two gliding motors attached to a baseplate mounted on a looped track (bottom). Two SprB filaments are attached to the baseplate and move with it. If either of these filaments adheres to the substratum, the cell glides. Shearing shortens the filaments and disrupts the baseplate, so that each filament is driven by a different motor. If one filament adheres to the substratum, the cell body spins about the axis of the motor.
Experimental Procedures
Cell tethering
Cells of wild-type F. johnsoniae CJ1827 were grown overnight at 25°C in motility medium (MM: per liter, 1.1 g Casitone, 0.55 g yeast extract, 1.1 mM Tris, pH 7.5) with shaking at 50 rpm. These cells were inoculated in fresh MM and were grown in the same way to OD600 0.4. Then 500 μL of the culture was passed 50 times through polyethylene tubing of inner diameter 0.58 mm between 1mL syringes equipped with 23-gauge stub adapters, a procedure similar to that used for shearing E. coli. The sheared cells were washed with 500 μL MM. Anti-SprB antibody [5] was purified using Melon Gel IgG Spin Purification Kit (Product # 45206, Thermo Scientific) and preadsorbed against an F. johnsoniae ΔsprB mutant. 40 uL of the suspension of sheared cells was incubated for 20 min with the purified antibody diluted 1:10. After incubation, the cells were washed and resuspended in 40 uL MM. The cells were added to a tunnel slide and incubated for 5 min. The slide was washed 3 times with 200 uL MM.
Imaging and image analysis
Movies of tethered cells were recorded using a phase contrast microscope with a digital camera running at a frame rate of 62 frames per second (Thorlabs, DCC1545M-GL). Custom MATLAB codes were used to analyze cell rotation and bead tracking [17]. The center of mass of the cell was tracked to calculate speed and direction of rotation. Cell length, width and distance of the center of mass from the center of rotation were calculated.
Attachment of beads to sheared cells and measurment of bead rotation
5 μL polystyrene beads (0.5 μm dia., Polysciences Inc.) with 5μL anti-SprB antibody were added to 50 μL of cells and incubated for 5 min. If attached to unsheared cells, the beads traveled down the length of a cell, sometimes moving from side to side, indicative of the translation of SprB. If attached to sheared cells, the beads rotated in place, as shown in Figure 1 D, E. Data were collected for 3 beads rotating at average speeds of 2.19 Hz, 0.71 Hz and 0.38 Hz. The rotating beads were imaged using a phase contrast microscope with a digital camera running at a frame rate of 62 frames per second (Thorlabs, DCC1545M-GL). Movies were analyzed using custom MATLAB codes [17].
Torque calculation
Torques generated by motors spinning tethered cells were calculated for each cell separately using the formula given in [8], Nr = (Cr+ r2Ct)2πf, where r = distance between the center of rotation and the center of mass of a cell, f= rotation rate, and Cr, Ct are rotational and translational frictional drag coefficients respectively. With the cell approximated as a prolate ellipsoid, Cr= (8πηa3/3)/(ln2a/b-0.5), Ct= 8πη a/(ln2a/b+0.5), a = cell length/2, b = cell width/2.
Ficoll experiments
F. johnsoniae cells were grown and sheared as described above. Cells were tethered onto a coverglass attached to a flow cell. Motility medium (MM) was added at the rate of 50 uL/min using a syringe pump (Harvard Apparatus 22). The rotation of a cell was recorded at 0% Ficoll and then a solution at higher concentration was pumped through for a period of 5 min. The rotation rate of the same cell was than recorded. Torque was calculated as described above, using the viscosities measured previously [11]: in cp, 0% 0.986 and 8% 3.86.
Supplementary Material
Distributions for cell length, cell width and radius of trajectory for the tethered cells described in Figure 2 of the main text. Individual values of cell length, cell width and radius of trajectory were used to calculate motor torque shown in Figure 2D in the main text.
Acknowledgments
We thank Mark J. McBride at The University of Wisconsin-Milwaukee for providing the antibody used in this study. This work was supported by National Institutes of Health Grant AI016478.
Footnotes
Author contributions. AS and HCB planned the experiments and wrote the paper. AS performed the experiments. AS and PPL analyzed the data.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6:466–476. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 2.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A. 2013;110:11145–11150. doi: 10.1073/pnas.1219753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lapidus IR, Berg HC. Gliding motility of Cytophaga sp. strain U67. J Bacteriol. 1982;151:384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun TF, Khubbar MK, Saffarini DA, McBride MJ. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J Bacteriol. 2005;187:6943–6952. doi: 10.1128/JB.187.20.6943-6952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson SS, Bollampalli S, McBride MJ. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. Journal of Bacteriology. 2008;190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Block SM, Segall JE, Berg HC. Impulse responses in bacterial chemotaxis. Cell. 1982;31:215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- 7.Silverman M, Simon M. Flagellar rotation and the mechanism of bacterial motility. Nature. 1974;249:73–74. doi: 10.1038/249073a0. [DOI] [PubMed] [Google Scholar]
- 8.Meister M, Berg HC. The stall torque of the bacterial flagellar motor. Biophys J. 1987;52:413–419. doi: 10.1016/S0006-3495(87)83230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahrner KA, Ryu WS, Berg HC. Biomechanics: bacterial flagellar switching under load. Nature. 2003;423:938. doi: 10.1038/423938a. [DOI] [PubMed] [Google Scholar]
- 10.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A. 2006;103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Berg HC. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys J. 2000;78:1036–1041. doi: 10.1016/S0006-3495(00)76662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol. 2009;75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. Journal of bacteriology. 2013;195:3201–3212. doi: 10.1128/JB.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ. Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. Journal of bacteriology. 2012;194:3678–3688. doi: 10.1128/JB.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nan B, McBride MJ, Chen J, Zusman DR, Oster G. Bacteria that glide with helical tracks. Current biology : CB. 2014;24:R169–173. doi: 10.1016/j.cub.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, McBride MJ, Subramaniam S. Cell-surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol. 2007;189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lele PP, Hosu BG, Berg HC. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci U S A. 2013;110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distributions for cell length, cell width and radius of trajectory for the tethered cells described in Figure 2 of the main text. Individual values of cell length, cell width and radius of trajectory were used to calculate motor torque shown in Figure 2D in the main text.