Abstract
Background
Mouse hepatocarcinogenesis is associated with mutations in Hras but infrequently in Kras. The effect on carcinogenesis of developmental age at the time of ras mutation is unknown.
Aim
We sought to compare quantitatively the effects of expressing mutant H- or Kras genes in fetal versus adult mouse liver.
Methods
We established an inducible system of gene expression in mouse liver to define disease pathogenesis associated with activation of oncogene expression.
Results
Diffuse expression of either oncogene in fetal or adult hepatocytes caused hepatomegaly. For mutant HrasG12V, this phenotype was almost fully reversible and accompanied by apoptosis, indicating that maintenance of hepatomegaly requires continuous HrasG12V expression. We also examined the effect of ras expression on growth of transplanted hepatocytes in an in vivo system that allows us to quantify hepatocyte growth effects in both permissive and restrictive hepatic growth environments. Mutant KrasG12D had no effect on hepatocyte growth in this sysstem. In contrast, HrasG12V induced increased hepatocyte focus growth in quiescent liver, the hallmark of a cell autonomous growth stimulus. HrasG12V also increased the fraction of donor hepatocyte foci that displayed extreme growth, a characteristic of preneoplastic lesions.
Conclusions
The primary effect of diffuse, whole-liver expression of either mutant ras gene in fetal or adult mouse liver is diffuse and progressive hepatic growth. HrasG12V mutation influences hepatocarcinogenesis by conferring cell autonomous growth potential upon foci of expressing cells and by increasing the risk of neoplastic progression. KrasG12D does not share these latter carcinogenic effects in mouse liver.
Keywords: hepatocellular carcinoma, hepatocyte transplantation, oncogene, HrasG12V, KrasG12D
Introduction
The 21 kD ras proteins are central components of cellular signaling pathways that control cell proliferation, differentiation, and survival (1). The mammalian ras genes encode three closely related GTPases, Hras, Kras, and Nras. Mutations in ras genes have been identified in 30% of cancers in humans and also in many rodent spontaneous and chemically-induced tumors (2, 3). Ras genes are oncogenically activated via single point mutations that cluster at codons 12, 13, 61 or 117. These mutations create amino acid changes in the GTP-binding pocket domain that render proteins constitutively active. Hras is the most commonly identified activated oncogene in mouse hepatocellular carcinomas (HCC) (4-6). K- and Nras mutations occur less frequently (7, 8). Ras mutations are identified infrequently in human HCC. However, all human HCCs display elevated ras signaling activity compared to adjacent normal liver in association with decreased expression of ras pathway inhibitors (9-11), supporting a role for ras signaling in these tumors.
Transgenic mice have been developed that target expression of mutant HrasG12V specifically to hepatocytes (12-14). These studies provide hints about the biological effects of mutant ras in liver. Initial targeting used an albumin enhancer/promoter-mutant HrasG12V transgene (AL-HrasG12V) (13). In that study, the majority of founder mice died 1 to 3 days after birth with liver hyperplasia. In a smaller group of founder mice with no evidence of perinatal liver enlargement there was a mild adult hepatic phenotype, including occasional development of HCC. However, these mice developed lung neoplasia at 1-12 months of age. Later, Wang and colleagues also used the mouse albumin enhancer/promoter to target the expression of mutant HrasG12V to hepatocytes in mice (14). By nine months of age, about half the mice developed HCC. Lack of perinatal hepatic hyperplasia suggests that expression was absent, reduced, or not fully penetrant in young mouse liver. Harada and colleagues utilized the cre-recombinase system to induce expression of a mutant HrasG12V in hepatocytes (12). In their system, an HrasG12V coding sequence was cloned downstream from a lox-flanked stop codon plus polyadenylation sequence and introduced into one endogenous Hras locus using gene targeting. In the absence of cre-mediated recombination the HrasG12V sequence was not transcribed. When cre-recombinase was targeted to hepatocytes using an adenoviral vector, hepatocytes that underwent cre-mediated deletion between the loxP sites deleted the stop codon and polyadenylation sequence and expressed the mutant HrasG12V. Transgenic mice were inoculated with adenoviral vectors expressing cre-recombinase at seven weeks of age. By four weeks post-inoculation dysplastic hepatocytes were apparent microscopically, but they were not associated with changes in cell proliferation and there was no progression to dysplatic nodules or hepatocellular carcinoma through seventeen months of age (12). In contrast, when HrasG12V and mutant β-catenin were coactivated in hepatocytes using this approach, focal neoplasia developed.
Collectively, these studies demonstrate diffuse disruption of normal liver growth by mutant HrasG12V when it is expressed in fetal liver, and variable increase in risk for HrasG12V-mediated neoplastic transformation in adult liver. However, they leave unanswered two important mechanistic questions about the effects of ras mutations in rodent liver. First, are the effects of ras mutants different when expression is restricted to adult versus fetal hepatocytes? Second, do these different activated ras genes have different biological effects during hepatocarcinogenesis? If so, what are the differences? The more frequent detection of mutations in Hras than in other ras family members in HCC could be explained if Hras preferentially causes cancer in this organ. We address these questions in this report using the tetracycline inducible system (15, 16) to regulate hepatocyte expression of transgenes encoding mutant HrasG12V or mutant KrasG12D. Expression of mutant ras can be turned on or off in hepatocytes of any age, allowing us to identify phenotypic consequences of adult versus fetal/perinatal mutant ras expression. We then employed these mice together with the newly-developed comparative hepatocyte growth assay to quantify hepatocyte growth characteristics and neoplastic progression frequency associated with each mutant ras. These studies define mechanisms by which mutant ras genes influence hepatocarcinogenesis.
Materials and Methods
Animals
Mice were maintained according to The Guide for the Care and Use of Laboratory Animals in AAALAC-accredited facilities. All experimental procedures were approved by the University of Wisconsin-Madison Animal Care and Use Committee. The transgenic lines used in these studies have been assigned the following genetic designations: MUP-muPA line 350-2, TgN(MupPlau)1Eps; MT-nLacZ line 379-4, TgN(Mt1nLacZ)4Eps; LAP-tTA, TgN(tTALap)5Uh; tetO-hPAP line 1391-4, TgN(tetOpALPP)11Eps; tetO-HrasG12V line 1562-1, TgN(tetOpHRASG12V)15Eps; AL-tetO-HrasG12V line 1480-3, TgN(tetOpHRASG12V)16Eps; tetO-KrasG12D line 1575-11, TgN(tetOpKRASG12D)13Eps.
Transgene construction and identification of transgenic mice
Mice carrying the metallothionein (MT)-nLacZ marker transgene, encoding nuclear-localized ß-galactosidase, have been described (17). MUP-uPA transgenic mice were used as hepatocyte transplant recipients in these studies, and have been described (18). LAP-tTA mice (19) were obtained from Jackson Laboratories (Bar Harbor, Maine) and backcrossed into FVB to establish a uniform strain background for these studies. Data from multiple backcross generations (FVB-3 to FVB-7) were controlled for by simultaneous evaluation of littermate controls.
To construct tetO-hPAP, the human placental alkaline phosphatase (hPAP) coding region with an attached 950 base pair (bp) Simian Virus 40 polyadenylation signal was cloned downstream of DNA containing seven tandem copies of the E.coli bacterial tetracycline operator (tetO) followed by the human cytomegalovirus (CMV) minimal promoter (pmin) region (provided by H. Bujard). tetO-hPAP transgenic mice were identified by PCR. To construct tetO- KrasG12D, the KrasG12D coding sequence (provided by Manual Perucho) with hGH polyadenylation sequence was inserted downstream of the tetOCMVpmin sequence described above. tetOKrasG12D mice were identified by PCR, using the forward primer 5’ TAGGCGTGTACGGTGGGAGG 3’ and the reverse primer 5’ CTACGCCACAAGCTCCAACTACCAC 3’. Twenty-five µl of reaction mixture containing genomic DNA purified from tail biopsies was subjected to the following conditions: (1) 92°C for 2 minutes; (2) 35 cycles of: 45 seconds at 92°C, 90 seconds at 60°C, and 110 seconds at 72°C; and (3) 72°C for 7 minutes. To construct the tetO-HrasG12V transgene, a human activated H-rasG12V gene (provided by R. Palmiter) was cloned downstream of the tetOCMVpmin sequence described above. To construct the ALtetO-HrasG12V transgene, the human activated HrasG12V coding sequence was cloned downstream of a different tetOpmin, which contained two tetO repeats downstream of a minimal promoter from the mouse albumin gene (Alpmin, 300 bp) (pAlbtetO2; provided by William Wilkison). HrasG12V transgenic mice were identified by PCR as described for KrasG12D, using the forward primer 5’ CCTGTCTCCTGCTTCCTCTAG 3’ and the reverse primer 5’ CTCACGGGGTTCACCTGTACTG 3’. All transgenes were generated and maintained on the FVB strain background except MUP-uPA, which was in the (FVB6)F1 strain background.
Comparative hepatocyte growth assay (CHeGA)
Hepatocytes were isolated from LacZ- or hPAP-marked transgenic mice to facilitate identification of donor hepatocytes in recipient mice. Donor hepatocytes were isolated using a modified two-step ethylenediaminetetraacetic acid (EDTA)/collagenase A protocol (18). Approximately 80% of the cells isolated using this method are hepatocytes (20). The concentration of viable hepatocytes was determined by trypan blue exclusion using a hemacytometer. Cells were maintained at 4°C until transplantation. Hepatocytes were transplanted as a 1:1 mixture in 10 ul via intrasplenic injection within 5 hours of isolation, as described (21).
Recipient mice initiate expression of the hepatotoxic mouse major urinary protein-urokinase type plasminogen activator (MUP-uPA) transgene at 3 to 4 weeks of age. The resulting liver disease provides a growth-permissive environment for the healthy donor cells. Donor hepatocytes travel from spleen then proliferate and expand as clonal foci within recipient livers for approximately 4 weeks, at which time the liver is repopulated by a mixture of donor cell clones and healthy endogenous hepatocyte clones derived from progenitors that stably inactivated transgene expression. Recipients are euthanized at 4 or 12 weeks post-transplant. To detect hPAP-marked donor hepatocyte foci in liver, recipient livers are fixed in 4% paraformaldehyde for 1 to 2 hours at 4°C, and samples are treated to detect hPAP activity by inucbation with the hPAP substrate 5-bromo-4-chloro-3-indolyl phosphate (BCIP, Sigma) as described (18). The hPAP-expressing donor hepatocyte foci are visible on the surface of the liver lobes, and display a blue color. To detect LacZ-marked donor hepatocyte foci in liver samples, recipient mice are administered 0.1 mg/kg cadmium sulfate intraperitoneally to induce expression of the MT-nLacZ transgene 16 to 24 hours prior to euthanasia. Livers are fixed in 4% paraformaldehyde for 1-2 hours at 4°C, and samples are treated to detect β–galactosidase activity by incubation with the substrate 5-bromo-4-chlor-3-indolyl-B-D-galactoside (X-gal, United States Biological), as described (17). The β–gal-expressing donor hepatocyte foci are visible on the surface of the liver lobes, and display a blue color product as a result of the β-galactosidase enzymatic reaction.
Cross-sectional surface areas occupied by single blue-staining donor clones are determined on the liver surface under low-power magnification by measuring the major axes (a, b) of each stained focus, then calculating the area of an oval using the formula A = π[1/4 (a+b)]2 (18). For each recipient liver, we measure 50 foci of each donor type.
Quantification of hepatocyte proliferation and apoptosis
One hour prior to euthanasia, all mice were administered 200 mg/kg bromodeoxyuridine (BrdU), a nucleotide analog incorporated into the DNA of cells in the synthesis phase of cellular replication. Cells undergoing replication were identified using immunohistochemistry to detect BrdU incorporation as described (18). The BrdU labeling index was determined by expressing the number of BrdU-positive hepatocyte nuclei as a percentage of the total number of hepatocyte nuclei counted. Apoptotic indices in foci were calculated similarly, using morphological criteria to identify apoptotic cells: (i) chromatin condensation and nuclear fragmentation into apoptotic bodies, (ii) eosinophilic cytoplasm, and (iii) cell shrinkage.
Evaluation of gene expression
Total RNA was isolated from approximately 30 mg of mouse liver, quantified, and subjected to cDNA synthesis as described (22). Fitness of cDNA templates was determined using the SPUD assay (23) and the 3’:5’ GAPDH assay (24). GAPDH, HPRT1, and TBP were selected for mouse liver from a group of 10 commonly used housekeeping genes (25). RT-qPCR reactions were performed using the Biorad iQ5, and data quantification used iQ5 software. Primers used were as follows (5’ to 3’): Hras mutant (human)
F: ggacgaatacgaccccactatagag, R: acacaggaagccctccccggtg; Hras total F: tttgccatcaacaacaccaagtc, R: gcagccaggtcacacttgtt; Kras mutant (human) F: ggacgaatatgatccaacaatagag, R: acaaagaaagccctccccagtc; Kras wildtype (mouse) F: ggatgagtacgaccctacgatagag, R: acaaagaaagccctccccagtt; Kras total F: acacagcaggtcaagaggagtacag, R: cacaaagaaagccctccccagt; Gs F: gtgagcccaagtgtgtggaa, R: gaaggggtctcgaaacatggc; Glt1 F: tgctcatcctccctcttatcatct, R: ggccgctggctttagcat; Oat F: gccctttctggcggtttatac, R: tggtttaatggtcagcattatctca; Otc F: acccttcctttcttaccacacaa, R: ttgagagagccatagtgttcctg; Ass1 F: ctgaaggaacaaggctatgatgt, R: gagagaggtgcccaggagatag; Asl F: tgttttgggcttctctgtgtatg, R: aggctgtctgggtttttcttct; Cps F: ctacccctgcttactgagacctt, R: ccaacagcgtccatctctactt; Arg1 F: ctccaagccaaagtccttagag, R: aggagctgtcattagggacatc; G6p F: cgactcgctatctccaagtga; R: gttgaaccagtctccgacca; Khk F: atgtggtggacaaatacccaga, R: caagcaaggaaaggacagtgc; Acaca F: attgaggatgacaggcttgc, R: tgccatgctcaaccaaagta; Alb F: agataagttgtgtgccattccaa, R: tgtgttgcaggaaacattcgt; Afp F: aactctggcgatgggtgttta, R: acactgatgtctttccactcca; Cyp2e1 F: ggctgtcaaggaggtgctac, R: ctcattaccctgtttcccca; Cyp7a1 F: atgtccacttcatcacaaactcc, R: ctgtgtccaaatgccttcgc; CyclinD1 F: ccctgacaccaatctcctcaac, R: gcatggatggcacaatctcct; Ccnnb1 F: cgttgagttgacaagctctctc, R: cctgtctgtagccaagaggttc; Bax F: ttgctacagggtttcatc, R: tgtccagttcatctccaa.
Evaluation of serum chemistries
Serum chemistries were performed to determine if hepatocyte-targeted expression of activated H- or Kras had an identifiable effect on overall liver function or health. Serum was transported on ice and evaluated using a Hitachi 912 Chemistry Analyzer (Roche Diagnostics).
Results
Regulated gene expression in mouse liver
The tet-inducible system uses two transgenes. LAP-tTA targets expression of the tetracycline transactivator (tTA) to hepatocytes via the liver-enriched activating protein promoter (19). tTA is a chimeric protein containing the herpes simplex virus viral protein 16 transactivator domain and the tet repressor (tetR) moiety from the tet operon of Escherichia coli. The tetR portion contains a DNA-binding domain that allows the protein to bind to tet operator (tetO) sequences in target genes, thereby activating transcription. Target transgenes contain tandem repeats of the 19 base pair tetO sequence adjacent to a minimal promoter (Pmin) from cytomegalovirus (CMV) or the mouse albumin gene, cloned upstream of hPAP, mutant HrasG12V, or mutant KrasG12D gene coding sequences. When both tTA and target transgenes are present in the same mouse, tTA binds to tetO as a dimer and transactivates the expression of hPAP and/or ras genes in hepatocytes. However, tetracycline or more potent analogs such as doxycycline (Dox) bind to the tetR moiety, thereby inhibiting binding of tTA to the tetO sequences and eliminating expression of target genes. For these studies, DOX was administered in the drinking water at 5 mg/ml at the schedules noted in text and tables.
To evaluate the kinetics of target transgene activation and silencing, LAP-tTA/tetOCMV-hPAP bitransgenic mice were subjected to different Dox treatment regimens, and multiple tissues evaluated for hPAP expression (data not shown). In the absence of Dox, liver stained uniformly blue following incubation in the hPAP substrate BCIP. Some staining also was observed in the renal medulla. Monotransgenic LAP-tTA or tetOCMV-hPAP mice displayed no liver staining. Additional animals were administered Dox in drinking water until adulthood, and then Dox was removed. By one week post-Dox, faint staining was present in liver, and by two weeks staining was intense, indicating that Dox clearance and activation of target transgenes occurs within one week. In the converse experiment, livers of initially untreated mice administered Dox for one week stained a faint blue, and after two weeks of treatment were unstained, indicating that Dox suppressed most hPAP activity by one week after drug administration.
Mutant ras expression in hepatocytes reduces mouse longevity, increases liver weight, and alters hepatic gene expression
Non-transgenic mice and mice carrying a single transgene displayed normal liver weight as a percent of body weight both in the presence and absence of Dox (data not shown). To identify the effects of HrasG12V and KrasG12D on liver growth in vivo, we established bitransgenic mice carrying LAP-tTA and an inducible mutant ras transgene (Table 1). Without Dox treatment (target gene on), survival of bitransgenic mice was reduced. The most extreme effect was observed in animals never exposed to Dox. For the KrasG12D line, only one bitransgenic animal could be obtained, and it required euthanasia one day after birth. Onset of lesions in this mouse resembled perinatal disease in AL-HrasG12V transgenic mice (13). Bitransgenic mice for both HrasG12V lines displayed mean survival ranging from 6.4 to 9.5 weeks after birth. Providing Dox to pregnant and lactating mothers of bitransgenic mice blocked ras expression. Following removal of Dox at birth, average mouse survival for H or Kras lines ranged from 5 to 9 weeks. Removal of Dox from adults (6 weeks of age) permitted average survival for an additional 13-15 weeks after initiation of ras expression regardless of the age at transgene initiation. Length of survival typically reflected the animal's condition: mice were euthanized when they became lethargic and lost body mass such that the caudal spine could be palpated easily.
Table 1.
Effect on liver growth of hepatocyte-specific expression of mutant H- or Kras
| Transgene line | Ras expressiona (# mice) | Endstage, weeks post-induction, mean ± SD | Liver/body weight %, mean ± SD |
|---|---|---|---|
| Non-transgenic | None (16) | - | 5.3±0.6 |
| tTA/Hras 1562-1 | Continuous (15) | 6.4±4.2b | 15.6±2.0 |
| On at birth (4) | 6.0±1.0 | 18.4±4.6 | |
| On at 6 weeks (13) | 13±6 | 16.7±3.6 | |
| tTA/Hras 1480-3 | Continuous (17) | 9.5±4.8a | 17.4±3.6 |
| On at birth (7) | 8.9±2.0 | 16.2±2.8 | |
| On at 6 weeks (5) | 15±4 | 14.7±0.6 | |
| tTA/Kras 1575-11 | Continuous (1) | 0.14a | - |
| On at birth (4) | 4.6±1.0 | 33±13 | |
| On at 6 weeks (10) | 13±7 | 16.6±5.1 |
Ras expression was initiated in mice by removing Dox at the stated age. Ras expression was continuous during development in mice that did not receive Dox in utero or after birth.
Age in weeks.
For all bitransgenic mice removed from Dox, liver weights increased significantly compared to non-transgenic or single transgenic control mouse livers, irrespective of age at Dox removal (Table 1). Grossly, 3 of 13 bitransgenic mice (23%) older than 24 weeks of age, two expressing HrasG12V and one expressing KrasG12D, displayed large hepatic masses confirmed microscopically to be hepatocellular trabecular or cystic carcinomas. Hepatic architecture remained normal in the enlarged livers, although two types of hepatocytes were identified (data not shown). Periportal hepatocytes were smaller and more uniform in size than pericentral hepatocytes. At the time of euthanasia, most bitransgenic mice also displayed renal tubule dilatation.
Clinical chemical evaluation of mouse serum identified physical hepatocyte damage (increased ALT) in one of two tTA/ KrasG12D mice, increased blood ammonia in tTA/HrasG12V mice, and elevated albumin and total protein in both (Table 2).
Table 2.
Clinical chemistrya
| Serum parameter | Genotype |
||
|---|---|---|---|
| Controlb | tTA/Hras | tTA/Kras | |
| Alanine amino transferase (ALT) (U/L) | 39±9 (9) | 48±17 (8) | 329±350 (2)* |
| Total protein (g/dL) | 3.9±0.4 (13) | 6.5±1.3 (10)*** | 7.3±1.5 (2)** |
| Albumin (g/dL) | 1.9±0.5 (13) | 2.7±0.4 (9)** | 2.8±0.9 (2)p=006 |
| Glucose (mg/dL) | 258±48 (14) | 254±102 (8) | 190±58 (2) |
| Ammonia (umol/L) | 43±17 (7) | 132±61 (4)* | 46±4 (2) |
Data expressed as mean ± SD (n). Some control and tTA/Hras mice in this study were in the FVB6F1 strain background. All tTA/Hras mice were from line 1562-1, Mutant ras-expressing mice were not receiving Dox, and were approaching end stage disease as noted in Table 1.
Controls were nontransgenic and single transgenic mice that did not express mutant H- or Kras.
p<0.05
p<0.01
p<0.001 versus non-transgenic samples using two-sided Wilcoxon rank sum test.
We also measured ras expression, and examined the effects of HrasG12V on expression of a panel of genes normally active in liver by real time qPCR. For Hras, we measured mutant (human only) and total (mouse plus human) Hras mRNA; we could not develop primers specific for mouse mRNA that did not also cross react with the human mRNA. For Kras, we measured mutant (human only), endogenous wildtype (mouse only), and total Kras mRNA. Compared to nontransgenic control liver, Hras total mRNA increased 2.2-fold and Kras total mRNA increased 2.6-fold (Table 3). Thus, transgene expression for both groups of transgenic mice was close to the physiological range of endogenous ras expression. Kras wildtype mRNA expression also increased, by 1.9-fold, in transgenic mice. Comparing fold increases for wildtype versus total Kras mRNA indicates that about 27% of total expression in transgenic livers was from the transgene. HrasG12V altered expression of nitrogen metabolizing enzymes, and genes reflective of cellular differentiation (Table 4). In particular, alpha-fetoprotein expression was increased 200-fold and Cyp expression was decreased, reflective of a less well-differentiated liver state. Paradoxically, albumin gene expression also was elevated. Albumin typically is expressed at the highest level in well-differentiated hepatocytes.
Table 3.
Relative transgene expressiona
| qPCR target | Genotype (n) |
||
|---|---|---|---|
| Nontransgenic (4) | tTA/Hras (5) | tTA/Kras (3-4) | |
| HrasG12V (human) | 0.013±0.006 | 1.00±0.37** | - |
| Hras total | 1.00±0.04 | 2.18±0.27** | - |
| KrasG12D (human) | 0.003±0.002n=3 | - | 1.00±0.07* |
| Kras wildtype (mouse) | 1.00±0.14 | - | 1.90±0.28* |
| Kras total | 1.00±0.23 | - | 2.62±0.20* |
Data expressed as mean ± SD (n).
p<0.05
p=0.01 versus nontransgenic samples using one-sided Wilcoxon rank sum test.
Wildtype and total ras mRNAs were normalized to equal 1.00 in nontransgenic control mice. Human transgene mRNA was normalized to equal 1.00 in tTA/Hras line 1562-1 and tTA/Kras mice. These mice were not being administered Dox, so mutant ras transgene expression was induced.
Table 4.
Relative hepatic gene expressiona
| Gene | Genotype (n) |
|
|---|---|---|
| Nontransgenic (4) | tTA/Hras (6) | |
| Nitrogen Metabolism | ||
| Glutamine synthase (GS) | 1.00±0.47 | 0.23±0.12** |
| Transporter of glutamate (Glt 1) | 1.00±0.65 | 0.20±0.22* |
| Ornithine aminotransferase (Oat) | 1.00±0.79 | 0.11±0.12** |
| Ornithine carbamylase (Otc) | 1.00±0.71 | 0.87±0.26 |
| Argininosuccinate synthase (Ass1) | 1.00±0.24 | 0.36±0.19** |
| Argininosuccinate lyase (Asl) | 1.00±0.61 | 0.56±0.27 |
| Carbamoylphosphatase synthetase I (Cps) | 1.00±0.12 | 0.76±0.30 |
| Arginase (Arg 1) | 1.00±0.52 | 1.32±0.60* |
| Carbohydrate and Lipid Metabolism | ||
| Glucose-6-phosphatase (G6p) | 1.00±0.96 | 0.74±0.33 |
| Ketohexokinase (Khk) | 1.00±0.70 | 1.69±0.55 |
| Acetyl CoA carboxylase a (Acaca) | 1.00±0.34 | 0.80±0.53 |
| Cellular Differentiation/Turnover | ||
| Albumin (Al) | 1.00±0.23 | 2.56±1.09* |
| Alphafetoprotein (Afp) | 1.00±0.99 | 208±131** |
| Cytochrome p450 2e1 (Cyp2e1) | 1.00±0.27 | 0.28±0.18** |
| Cytochrome p450 7a1 (Cyp7a1) | 1.00±0.02 | 0.30±0.24** |
| Cyclin D1 (Ccnd1) | 1.00±0.71 | 0.45±0.16 |
| Cyclin B1 (Ccnb1) | 1.00±0.21 | 4.50±3.09* |
| Bax (Bax) | 1.00±0.03 | 1.93±0.48** |
Data expressed as mean ± SD. Hepatic gene mRNAs were normalized to equal 1.00 in nontransgenic control mice. Transgenic line 1562-1 mice were not being administed Dox, so HrasG12V expression was induced.
p<0.05
p<0.01 versus non-transgenic samples using two-sided Wilcoxon rank sum test.
Interrupting mutant HrasG12V expression in adult liver reverses hyperplasia
To determine whether HrasG12V-mediated growth effects were reversible, we removed Dox from 6- to 8-week-old bitransgenic line 1562-1 mice, then readministered Dox beginning 12 to 13 weeks later. After Dox removal, bitransgenic mouse livers increased in mass as a percent of body weight (Table 5). However, after 2 weeks of Dox readministration to turn off transgene expression, liver weights decreased significantly to only 1.5-fold above the control values, and remained constant (8.0±1.3% bw at 5 weeks post-Dox readministration, n=10). BrdU labeling index was elevated in mouse liver expressing HrasG12V, and returned to normal after Dox readministration. Finally, the apoptotic index was increased significantly by HrasG12V, and increased even more when HrasG12V expression was turned off by Dox. These findings indicated that mutant ras-induced liver hyperplasia is mostly though not completely reversible once transgene expression is interrupted in adult liver via a mechanism consistent with apoptosis.
Table 5.
Reversibility of mutant Hras-induced liver phenotype in adult line 1562-1 micea
| Genotype (n): | Control (10-11) | tTA/Hras (4-5) | tTA/Hras (7-8) |
|---|---|---|---|
| DOX Status: | - | No DOX | 14 day DOX |
| Transgene status: | none | ON | 14 day OFF |
| Li wt/body wt | 5.1+1.0 | 17.8+3.5** | 8 0+1 5 b *** c |
| BrdU index | 0.66+0.68 | 1.7+1.2* | 1.0+0.6 |
| Apoptosis index | 0.19+0.17 | 0.69+0.45** | 2.0+1.0*** c |
Data expressed as mean ± SD.
p<0.05
p<0.01
p<0.001 versus control samples using two-sided Wilcoxon rank sum test. Experimental mice were removed from Dox when they were 6- to 8-weeks-old, then 12 to 13 weeks later were euthanized (No DOX group) or readministered Dox for 2 weeks prior to euthanization (14 day DOX group).
One mouse with Li wt/body wt of 21% was excluded from this calculation.
p<0.01 versus tTA/Hras No DOX samples using two-sided Wilcoxon rank sum test.
Mutant HrasG12V but not KrasG12D alters transplanted hepatocyte growth and transformation frequency
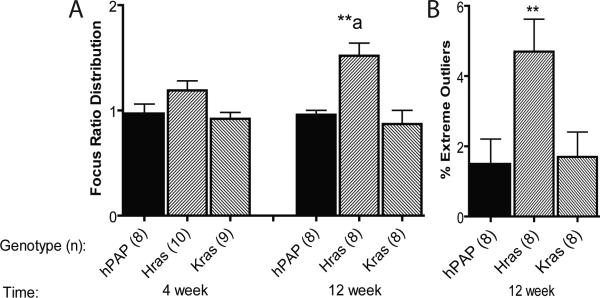
We used CheGA to compare growth of mutant ras-expressing and control donor hepatocyte foci following transplantation. For all recipient animals at each timepoint post-transplant, we measured cross-sectional areas of 50 foci expressing hPAP and 50 foci expressing LacZ. To address inter-recipient focus growth variability, we normalized hPAP focus size distributions by dividing each hPAP focus cross-sectional area by the median lacZ (control) focus cross-sectional area from the same recipient. The median of this distribution should equal one if the lacZ and hPAP hepatocyte foci grow at the same rate; for normal hPAP-marked hepatocytes, all values were close to one (Figure 1A).
Figure 1.
Focus ratio distribution medians (A), and outlier frequency (B) of transplanted hepatocyte foci. **p<0.01 versus hPAP at the same time point using one-sided Wilcoxon rank sum test.
aStatistically different (p=0.015) than HrasG12V 4 week time point using one-sided Wilcoxon rank sum test.
Following transplantation of normal hepatocytes into 4-week-old MUP-uPA mouse livers, there is a permissive or “growth phase” through 4 weeks post-transplant, during which transplanted cells expand as clonal foci. This is followed by a “quiescence phase” that spans weeks 4 to 12 post-transplant, during which normal transplant foci do not grow further (18, 21). KrasG12D expression had no effect on hepatocyte growth at any time post-transplant (Figure 1A). HrasG12V expression did not consistently affect hepatocyte growth in a growth-permissive liver environment, as determined by 4 week focus ratio distribution, but it was sufficient to induce sustained growth in quiescent liver (comparing 4 to 12 weeks post-transplant; Figure 1A).
When examining focus ratio distributions, we observed occasional foci that were much larger than others in the distribution. We used the method of Tukey (26) to quantitate the fraction of “extreme outliers” in each 12 week focus ratio distribution (Figure 1B). Tukey defined interquartile range (IQR) as the distance between the 1st and 3rd quartile in a distribution and extreme outliers as values lying 3xIQR or more above the 3rd quartile. There was no increase in extreme growth outliers detected in KrasG12D foci. However, for HrasG12V, 4.7% of foci fell into the outlier class. These foci likely developed from transplanted cells that had accumulated additional genetic or epigenetic alterations that permitted excessive growth (21).
To identify cell turnover characteristics in transplant foci, we determined hepatocyte DNA synthesis (BrdU labeling) and apoptotic indices in foci during the growth (samples collected 2 weeks post-transplant, n=4-6) and quiescent (samples collected 8-12 weeks post-transplant, n=5-6) phases post-transplant. Relative to control foci, HrasG12V foci displayed slight but significant (p<0.05) increases in apoptosis at 2 weeks (1.1±0.2 versus 0.67±0.10) and BrdU labeling at 8-12 weeks (2.1±0.4versus 1.1±0.6), the latter consistent with the continued growth of these foci. No other differences were observed.
Discussion
We designed these experiments to test two hypotheses. First, we hypothesized that HrasG12V and KrasG12D would have different effects depending on whether expression was initiated in fetal or adult liver. Previously, in the AL- HrasG12V model, we observed that diffuse expression of HrasG12V in fetal hepatocytes caused liver hyperplasia leading to perinatal mouse death (13). Mice from lines surviving to adulthood had normal liver size but developed hepatocellular carcinoma or lung neoplasia at 3 to 12 months of age (13, 27). In the present model, initiating expression of mutant H- or Kras caused similar progressive hepatomegaly and decreased animal survival regardless of whether expression was initiated in fetal or adult liver. These changes were observed with near-physiological levels of mutant ras expression. Our observations indicate that, following diffuse hepatocyte-targeted expression, the primary effect of either mutant ras is diffuse liver enlargement regardless of animal age, disproving our hypothesis. Diffuse growth could represent a direct effect of mutant ras expression, or a compensatory increase in liver mass due to alterations in liver function induced by ras expression in hepatocytes.
Rosseland and colleagues identified different effects of Hras and Kras on proliferation and survival of cultured rat hepatocytes (28). Both HrasG12V and KrasG12V promoted hepatocyte survival (anti-apoptosis), Hras through ERK and PI3K and Kras only through PI3K. However, only HrasG12V enhanced ERK-mediated hepatocyte proliferation. These findings would explain our results if the chronic anti-apoptotic signals of either mutant ras render the liver non-responsive to normal signals regulating livers mass, thereby permitting hepatomegaly
We indirectly evaluated liver function in bitransgenic mice by examining mouse serum clinical chemistry and hepatic gene expression. That increased ALT was found in only one tTA/ KrasG12D mouse indicates that physical hepatocyte injury is not a general cause of the liver hyperplasia. However, HrasG12V generally decreased expression of hepatic genes involved in nitrogen metabolism and hepatocyte differentiation, and was accompanied by a 3-fold elevation in serum ammonia. Changes in the differentiation-specific genes Afp, Cyp2e1, and Cyp7a1 resembled changes observed in mice with hepatomegally secondary to diffuse expression of the Wnt pathway protein β-catenin (22). The 200-fold increase in Afp expression and the increased albumin expression and serum concentration could account for the increased serum total protein. Proapoptotic Bax gene expression also was increased by HrasG12V (this study) and β-catenin (22). Neither changed expression of cyclinD1, but HrasG12V significantly increased cyclinB1 expression. Interestingly, changes in nitrogen metabolizing genes were opposite to those identified in β-catenin mice, demonstrating that the patterns of gene expression changes are regulated differently by the different oncogenes. Overall, the changes observed following HrasG12V expression suggest induction of a periportal hepatocyte metabolic phenotype, whereas β-catenin induces a pericentral hepatocyte metabolic phenotype (22, 29).
Finally, we demonstrated that the HrasG12V-induced growth phenotype is almost fully reversible via apoptosis, indicating that maintenance of increased liver weight requires the continuous presence of mutant ras, and consistent with the reported association of mutant ras expression with hepatocyte survival (28). Loss of HrasG12V anti-apoptotic activity promotes liver mass normalization via hepatocyte death. Regarding the association of ras expression with neoplasia, only a minority of ras-expressing animals (23%) developed hepatocellular neoplasms. Incomplete penetrance likely reflects animal mortality secondary to hepatomegaly.
Compare these results with those of Schuhmacher and colleagues (30) and Chen and colleagues (31). Their models activated expression of HrasG12V in all tissues beginning in fetal development from an endogenous Hras allele. Schuhmacher et al. (30) developed an Hras locus knock-in that created a polycistronic mRNA with HrasG12V coding sequence-IRES-β-galactosidase coding sequence. They identified no liver phenotype, and could not identify any downstream evidence of active HrasG12V signaling, which they attributed to negative feedback.
Chen et al. (31) described different results. They identified a severe phenotype in mice expressing an otherwise unmodified HrasG12V allele from the endogenous Hras locus. These mice had 80% perinatal lethality and, in survivors, a highly penetrant neoplastic phenotype in skin, stomach, and endothelium. Cause of death of neonates was not determined, and, in particular, the liver was not described. Liver in surviving mice was not altered in size, but displayed elevated pMEK and pERK. They did not quantify HrasG12V expression in liver. They addressed the stark difference between their model and Schuhmacher's by suggesting that the IRES-β-galactosidase cassette interfered with expression of HrasG12V such that a threshold for tumorigenesis was not reached.
Neither of those studies definitively addressed the hepatic consequences of physiological levels of HrasG12V expression. Schuhmacher did not precisely quantify HrasG12V expression or identify the pattern of expression in liver, and, as noted by Chen, the HrasG12V expression may be altered by the targeting strategy, which resulted in a bicistronic mRNA. Chen did not quantify or determine the pattern of hepatic HrasG12V expression. They also determined the phenotype in only the 20% minority of mice that survived beyond 14 days of age. Something must have been different about this subset of mice for the animals to have been selected for survival. A likely candidate would be a different pattern or level of expression/effects in survivors than in the majority that did not survive. By using the transgenic approach in our studies, we restricted expression to hepatocytes, thus eliminating the possibility that secondary effects on another organ may influence liver phenotype, or that primary effects on other organs may overshadow a liver phenotype. We also could turn on transgene expression at a time of our choosing, rather than being limited to examining the effects of altered genes throughout development. To address the concern that transgene expression could be at a non-physiological level, thereby inducing diffuse hepatic toxicity, we measured transgene expression using RT-PCR. We identified near-physiological levels of transgene expression. Measurement of serum ALT also indicated that liver toxicity was not a common feature of these mice.
Next, we questioned whether different ras family members could have different biological effects on hepatocarcinogenesis; recall that, when expressed diffusely in mouse liver in our model, their effects appeared to be the same. To examine this question in more detail, we employed our hepatocyte transplantation assay. Following transplantation, normal hepatocytes expand as foci of cells up to 4 weeks post-transplant (growth phase), while no significant additional expansion occurs between 4 and 12 weeks following transplantation (quiescence phase). Neither HrasG12V nor KrasG12D induced consistently increased growth during the growth-stimulatory phase compared to normal cells (assessed at 4 weeks). However, HrasG12V but not KrasG12D induced continuous increased growth during the quiescent phase (weeks 4 through 12 post transplant). We included in donor cells a regulated marker transgene (tetO-hPAP) that, like the ras transgenes, is tTA-responsive. All hepatocyte foci displayed the blue stain, indicating that tTA activity was present in both HrasG12V and KrasG12D foci, ruling out failure of expression of KrasG12D as a cause of the different focus growth characteristics. These findings conform to expectations based on the report of Rosseland and colleagues: whereas both mutant Hras and Kras have anti-apoptotitc properties, only HrasG12V can enhance hepatocyte proliferation via the ERK pathway (28).
The expansion of focus size in quiescent liver identifies hepatic growth as a cell autonomous effect of HrasG12V. This finding also confirms that HrasG12V expression in our model is not directly hepatotoxic. Drinkwater and colleagues correlated increased carcinogen sensitivity and more rapid growth of altered hepatic foci with increased frequency of Hras mutations in different mouse strains following administration of the hepatocarcinogen diethylnitrosamine (32). Our data indicate that HrasG12V expression is sufficient for this growth effect in FVB strain mice, identifying one important mechanistic feature of HrasG12V-mediated transformation that is not shared by KrasG12D. When Harada and colleagues initiated focal hepatocyte expression of HrasG12V in adult liver using retrovirally-delivered cre recombinase, altered foci and neoplasms were not observed (12). In that system, HrasG12V expression is likely activated independently in single hepatocytes, whereas our transplant system involves mutant HrasG12V expression in foci of 2,000-3,000 hepatocytes (18, 21). Thus, HrasG12V expression appears to be insufficient to induce cell autonomous growth in single initiated hepatocytes. Apparently, additional replication of HrasG12V-expressing cells into foci is necessary to permit HrasG12V-mediated focus-autonomous growth.
We identified a second important difference between effects of mutant H- and Kras. Transplant foci expressing HrasG12V displayed a significantly increased outlier fraction (4.7%). KrasG12D and mutant β-catenin did not have this effect (22). This finding indicates that HrasG12V is associated with a higher frequency of cells and foci with extreme growth characteristics. These large foci resemble preneoplastic foci, which have accumulated additional genetic and/or epigenetic changes (21). Thus, in this context, HrasG12V is associated with enhanced neoplastic progression. A differential rate of conversion of foci to neoplasms may explain in part the more frequent identification of Hras than Kras mutations in spontaneous and induced mouse liver tumors. Our data identifies mechanistic roles for HrasG12V mutation in the stages of promotion and progression. KrasG12D does not support these mechanistic effects in mouse liver.
In summary, diffuse hepatic expression of either HrasG12V or KrasG12D, initiated in fetal or adult liver, results in hepatomegaly and reduced mouse survival. However, when expressed in focal collections of hepatocytes, their effects are markedly different. Only HrasG12V increases the likelihood of progression to cancer, likely via the mechanism of induction of cell-autonomous hepatocyte focus growth in an otherwise quiescent liver, and increased incidence of outlier focus development. These results confirm in vivo that mutant Hras and mutant Kras can have distinct growth-related effects on hepatocytes, and my explain the preponderance of Hras mutations versus Kras mutations in mouse liver neoplasia.
ACKNOWLEDGMENTS
We thank Meg Bowden, Renee Szakaly, and Kristen Wentworth for technical assistance. This work was supported by the National Institutes of Health (RO1-DK49787, RO1-ES07671, and PO1-CA022484 to EPS, KO1-RR020033 to TJS, and 5T32-GM07215 to MLF).
REFERENCES
- 1.BAR-SAGI D. A Ras by any other name. Mol Cell Biol. 2001;21(5):1441–3. doi: 10.1128/MCB.21.5.1441-1443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.SILLS RC, BOORMAN GA, NEAL JE, HONG HL, DEVEREUX TR. Mutations in ras genes in experimental tumours of rodents. IARC Sci Publ. 1999;(146):55–86. [PubMed] [Google Scholar]
- 3.ADJEI AA. Blockin1g oncogenic Ras signaling for cancer therapy. J Natl Cancer Inst. 2001;93(14):1062–74. doi: 10.1093/jnci/93.14.1062. [DOI] [PubMed] [Google Scholar]
- 4.REYNOLDS SH, STOWERS SJ, MARONPOT RR, ANDERSON MW, AARONSON SA. Detection and identification of activated oncogenes in spontaneously occurring benign and malignant hepatocellular tumors of the B6C3F1 mouse. Proc Natl Acad Sci U S A. 1986;83(1):33–7. doi: 10.1073/pnas.83.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.REYNOLDS SH, STOWERS SJ, PATTERSON RM, MARONPOT RR, AARONSON SA, ANDERSON MW. Activated oncogenes in B6C3F1 mouse liver tumors: implications for risk assessment. Science. 1987;237(4820):1309–16. doi: 10.1126/science.3629242. [DOI] [PubMed] [Google Scholar]
- 6.WISEMAN RW, STOWERS SJ, MILLER EC, ANDERSON MW, MILLER JA. Activating mutations of the c-Ha-ras protooncogene in chemically induced hepatomas of the male B6C3 F1 mouse. Proc Natl Acad Sci U S A. 1986;83(16):5825–9. doi: 10.1073/pnas.83.16.5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MANAM S, STORER RD, PRAHALADA S, LEANDER KR, KRAYNAK AR, LEDWITH BJ, et al. Activation of the Ha-, Ki-, and N-ras genes in chemically induced liver tumors from CD-1 mice. Cancer Res. 1992;52(12):3347–52. [PubMed] [Google Scholar]
- 8.MARONPOT RR, FOX T, MALARKEY DE, GOLDSWORTHY TL. Mutations in the ras proto-oncogene: clues to etiology and molecular pathogenesis of mouse liver tumors. Toxicology. 1995;101(3):125–56. doi: 10.1016/0300-483x(95)03112-s. [DOI] [PubMed] [Google Scholar]
- 9.CALVISI DF, LADU S, GORDEN A, FARINA M, CONNER EA, LEE JS, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterol. 2006;130(4):1117–28. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 10.CALVISI DF, LADU S, GORDEN A, FARINA M, LEE JS, CONNER EA, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117(9):2713–22. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.YOSHIDA T, HISAMOTO T, AKIBA J, KOGA H, NAKAMURA K, TOKUNAGA Y, et al. Spreds, inhibitors of the Ras/ERK signal transduction, are dysregulated in human hepatocellular carcinoma and linked to the malignant phenotype of tumors. Oncogene. 2006;25(45):6056–66. doi: 10.1038/sj.onc.1209635. [DOI] [PubMed] [Google Scholar]
- 12.HARADA N, OSHIMA H, KATOH M, TAMAI Y, OSHIMA M, TAKETO MM. Hepatocarcinogenesis in mice with beta-catenin and Ha-ras gene mutations. Cancer Res. 2004;64(1):48–54. doi: 10.1158/0008-5472.can-03-2123. [DOI] [PubMed] [Google Scholar]
- 13.SANDGREN EP, QUAIFE CJ, PINKERT CA, PALMITER RD, BRINSTER RL. Oncogene-induced liver neoplasia in transgenic mice. Oncogene. 1989;4(6):715–24. [PubMed] [Google Scholar]
- 14.WANG AG, MOON HB, LEE MR, HWANG CY, KWON KS, YU SL, et al. Gender-dependent hepatic alterations in H-ras12V transgenic mice. J Hepatol. 2005;43(5):836–44. doi: 10.1016/j.jhep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 15.FURTH PA, ST ONGE L, BOGER H, GRUSS P, GOSSEN M, KISTNER A, et al. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc Natl Acad Sci U S A. 1994;91(20):9302–6. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.GOSSEN M, BUJARD H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89(12):5547–51. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.RHIM JA, SANDGREN EP, DEGEN JL, PALMITER RD, BRINSTER RL. Replacement of diseased mouse liver by hepatic cell transplantation. Science. 1994;263(5150):1149–52. doi: 10.1126/science.8108734. [DOI] [PubMed] [Google Scholar]
- 18.WEGLARZ TC, DEGEN JL, SANDGREN EP. Hepatocyte transplantation into diseased mouse liver. Kinetics of parenchymal repopulation and identification of the proliferative capacity of tetraploid and octaploid hepatocytes. Am J Pathol. 2000;157(6):1963–74. doi: 10.1016/S0002-9440(10)64835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.KISTNER A, GOSSEN M, ZIMMERMANN F, JERECIC J, ULLMER C, LUBBERT H, et al. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci USA. 1996;93(20):10933–8. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.BRAUN KM, SANDGREN EP. Cellular origin of regenerating parenchyma in a mouse model of severe hepatic injury. Am J Pathol. 2000;157(2):561–9. doi: 10.1016/S0002-9440(10)64566-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.FIGUEIREDO ML, WENTWORTH KM, SANDGREN EP. Quantifying growth and transformation frequency of oncogene-expressing mouse hepatocytes in vivo. Hepatology. 2010;52(2):634–43. doi: 10.1002/hep.23682. [DOI] [PubMed] [Google Scholar]
- 22.STEIN TJ, JOCHEM A, HOLMES KE, SANDGREN EP. Effect of mutant b-catenin on liver growth homeostasis and hepatocarcinogenesis in transgenic mice. Liver International. 2011;31(3):303–12. doi: 10.1111/j.1478-3231.2010.02430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.NOLAN T, HANDS RE, OGUNKOLADE W, BUSTIN SA. SPUD: a quantitative PCR assay for the detection of inhibitors in nucleic acid preparations. Anal Biochem. 2006;351(2):308–10. doi: 10.1016/j.ab.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 24.NOLAN T, HANDS RE, BUSTIN SA. Quantification of mRNA using real-time RTPCR. Nat Protoc. 2006;1(3):1559–82. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 25.VANDESOMPELE J, DE PRETER K, PATTYN F, POPPE B, VAN ROY N, DE PAEPE A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034.1–11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.TUKEY J. Exploratory Data Analysis. 2nd ed. Addison-Wesley; Reading, MA: 1977. [Google Scholar]
- 27.MARONPOT RR, PALMITER RD, BRINSTER RL, SANDGREN EP. Pulmonary carcinogenesis in transgenic mice. Exp Lung Res. 1991;17(2):305–20. doi: 10.3109/01902149109064420. [DOI] [PubMed] [Google Scholar]
- 28.ROSSELAND CM, WIEROD L, FLINDER LI, OKSVOLD MP, SKARPEN E, HUITFELDT HS. Distinct functions of H-Ras and K-Ras in proliferation and survival of primary hepatocytes due to selective activation of ERK and PI3K. J Cell Physiol. 2008;215(3):818–26. doi: 10.1002/jcp.21367. [DOI] [PubMed] [Google Scholar]
- 29.BRAEUNING A, MENZEL M, KLEINSCHNITZ EM, HARADA N, TAMAI Y, KOHLE C, et al. Serum components and activated Ha-ras antagonize expression of perivenous marker genes stimulated by beta-catenin signaling in mouse hepatocytes. FEBS J. 2007;274(18):4766–77. doi: 10.1111/j.1742-4658.2007.06002.x. [DOI] [PubMed] [Google Scholar]
- 30.SCHUHMACHER AJ, GUERRA C, SAUZEAU V, CANAMERO M, BUSTELO XR, BARBACID M. A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. J Clin Invest. 2008;118(6):2169–79. doi: 10.1172/JCI34385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CHEN X, MITSUTAKE N, LAPERLE K, AKENO N, ZANZONICO P, LONGO VA, et al. Endogenous expression of Hras(G12V) induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proc Natl Acad Sci USA. 2009;106(19):7979–84. doi: 10.1073/pnas.0900343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.BENNETT LM, FARNHAM PJ, DRINKWATER NR. Strain-dependent differences in DNA synthesis and gene expression in the regenerating livers of CB57BL/6J and C3H/HeJ mice. Mol Carcinog. 1995;14(1):46–52. doi: 10.1002/mc.2940140109. [DOI] [PubMed] [Google Scholar]