SUMMARY
To provide a temporal framework for the genoarchitecture of brain development, in situ hybridization data were generated for embryonic and postnatal mouse brain at 7 developmental stages for ~2100 genes, processed with an automated informatics pipeline and manually annotated. This resource comprises 434,946 images, 7 reference atlases, an ontogenetic ontology, and tools to explore co-expression of genes across neurodevelopment. Gene sets coinciding with developmental phenomena were identified. A temporal shift in the principles governing the molecular organization of the brain was detected, with transient neuromeric, plate-based organization of the brain present at E11.5 and E13.5. Finally, these data provided a transcription factor code that discriminates brain structures and identifies the developmental age of a tissue, providing a foundation for eventual genetic manipulation or tracking of specific brain structures over development. The resource is available as the Allen Developing Mouse Brain Atlas (developingmouse.brain-map.org).
INTRODUCTION
The diversity of cell types in the brain presents an immense challenge towards understanding cellular organization, connectivity, and function of this organ. The objective definition of cell type remains elusive, but should integrate molecular, anatomic, morphological, and physiological parameters. At both a large and small scale, neuroscientists have flocked to genetic strategies that depend upon known molecular markers to label adult cell types for the purpose of isolating or manipulating specific populations (Siegert et al., 2012; Sugino et al., 2006). However, achieving a fine resolution of cell subtypes will likely require combinatory or intersectional strategies due to the lack of absolute specificity of any single gene marker for a given cell type. Developmental neurobiologists have used careful descriptive analysis and genetic fate-mapping for over a decade to specify the developmental origin of cell types, and typically utilizing an intersectional strategy to map the fate of cells produced at a specified time from a particular anatomic domain (Joyner and Zervas, 2006). In the retina, a transcription factor (TF) code has been deduced for each branch of the retinal cell lineage (Agathocleous and Harris, 2009; Livesey and Cepko, 2001) and this code is evident even in the adult differentiated neurons (Siegert et al., 2012). The success of creating meaningful definitions of cell types may ultimately rely on a combination of classification metrics that include both terminal molecular characteristics as well as their topological developmental origin.
Morphogenesis and functional development of the mammalian central nervous system (CNS) occur via mechanisms regulated by the interaction of genes expressed at specific times and locations during development (Rubenstein and Rakic, 2013; Sanes et al., 2012). Understanding this temporal and regional complexity of gene expression over brain development will be critical to provide a framework to define neuroanatomical subdivisions and the component cell types. To this end, we have generated an extensive dataset and resource that provides spatial and temporal profiling of ~2100 genes across mouse C57Bl/6J embryonic and postnatal development with cellular-level resolution (http://developingmouse.brain-map.org/). Genes were surveyed by high-throughput ISH across seven embryonic and postnatal ages (E11.5, E13.5, E15.5, E18.5, P4, P14 and P28), in addition to P56 data available in the Allen Mouse Brain Atlas. This developmental survey comprises 18,358 sagittal and 1913 coronal ISH experiments, displayed online at 10X resolution and are downloadable via XML. From a neuroanatomical perspective the Allen Developing Mouse Brain Atlas defines a number of CNS subdivisions (described in 2D atlas plates and 3D structural models) based on an updated version of the prosomeric model of the vertebrate brain (Puelles et al., 2012; Puelles and Rubenstein, 2003). Furthermore, a novel informatics framework enables navigation of expression data within and across time points. In addition to stage-specific novel reference atlases, the resource provides an innovative ontogenetic ontology of the full brain with over 2500 hierarchically organized names and definitions, and 434,946 sections of high resolution spatially and temporally linked ISH data, offering rapid access and a range of visualization and analysis tools.
The chosen stages were intended to survey diverse developmental mechanisms, including regional specification, proliferation, neurogenesis, gliogenesis, migration, axon pathfinding, synaptogenesis, cortical plasticity and puberty. The genes selected include: 1) ~800 TFs representing 40% of total TFs, with nearly complete coverage of homeobox, basic helix-loop-helix, forkhead, nuclear receptor, high mobility group and POU domain genes; 2) neurotransmitters and their receptors, with extensive coverage of genes related to dopaminergic, serotonergic, glutamatergic, and GABA-ergic signaling, as well as of neuropeptides and their receptors; 3) neuroanatomical marker genes delineating regions or cell-types throughout development; 4) genes associated with signaling pathways relevant to brain development including axon guidance (~80% coverage), receptor tyrosine kinases and their ligands, and Wnt and Notch signaling pathways; 5) a category of highly studied genes coding for common drug targets, ion channels (~37% coverage), G-protein-coupled receptors (GPCRs; ~7% coverage), cell adhesion genes (~32% coverage), and genes involved neurodevelopmental diseases, which were expected to be expressed in the adult brain or during development (Table S1). A smaller set of genes was surveyed in “old age” (18–33 months).
Analyses of these data identified molecular signatures associated with key developmental events with precise spatiotemporal regulation. These signatures revealed a shift in the organizing principles governing the molecular profiles of brain regions over development, with the coexistence of both dorsoventral (DV) or longitudinal plate-based, and anteroposterior (AP) or neuromere-based organization strongest at E13.5 leading to areal (progenitor domain)-based organization. Finally, by focusing on TFs, unique combinatorial codes were found that precisely define most brain structures and even pinpoint developmental age, a potential starting point to investigate both how regions are specified as well as how they acquire unique functional properties.
RESULTS
New Developmental Reference Atlases
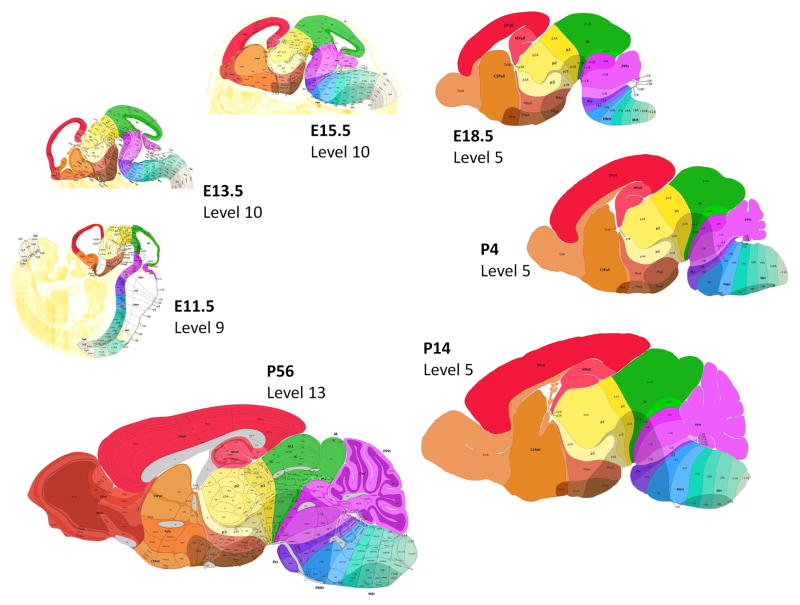
To provide a consistent anatomical context for analysis of ISH data, seven reference atlases were created spanning E11.5 to P56 (161 annotated plates and 1898 supporting reference images; Fig. 1). The reference atlases used a novel ontological approach that classifies brain structures based upon their orthogonal areal neuroepithelial origin in the wall of the neural tube (intersection of fundamental neuromeric and longitudinal zonal units), employing a topological and ontogenetic viewpoint to register the emergence of both transient and definitive nuclear or cortical cell populations in the mantle layer, the final location of postmitotic, terminally differentiated neurons (Puelles and Ferran, 2012; Puelles et al., 2012). Tangentially migrated structures (e.g., pontine nuclei) are classified by postmigratory position. Thus, the reference atlas drawn for the adult contains developmental and morphological concepts that make it distinct, in terms of nomenclature and classification, from that drawn for the Allen Mouse Brain Atlas (Dong, 2008).
Figure 1. Reference framework for the Allen Developing Mouse Brain Atlas.
Representative reference atlas plates from seven developmental ages surveyed in the project are shown. Because P28 and P56 time points are indistinguishable from a neuroanatomic standpoint, the P56 Nissl images used in the reference atlas for the Allen Mouse Brain Atlas were also annotated using the developmental ontology and are supplied as a reference for both P28 and P56 ISH data.
The present ontogenetic ontology (Puelles et al., 2013) has 13 levels of anatomic classification. Early levels 1–3 include definition of protosegments (e.g., forebrain), followed by neuromeric AP subdivisions (e.g., prosomeres). Basic DV subdivisions are defined next, including the alar-basal boundary, plus roof and floor plates (levels 4, 5). Levels 6–8 cover finer areal regionalization into realistic progenitor domains with known differential fates. Stratification refers first to the distinction between ventricular and mantle zones (level 9), and secondly to superficial, intermediate, and periventricular transient strata (level 10) of the mantle zone. Adult brain nuclei and other associated structures (tracts, commissures, circumventricular organs, and glands) largely reside in the mantle zone and are represented at levels 11–13.
Automated and Manual Annotation of the Data Facilitates Navigation of Spatial and Temporal Information
For ISH experiments, the sampling density of tissue sections was scaled by specimen size and age, ranging from 80 μm to 200 μm (Supplemental Experimental Procedures). Most genes (63%) were expressed at all ages and 10% were not expressed in the brain at any stage. The remaining genes (27%) were temporally specific, with 19% exhibiting delayed activation across this time course, potentially associated with terminally differentiated cellular phenotypes (e.g., GPCRs and ion channels; Fig. S1 E,F) and 4% of the genes expressed only at early stages (Fig. S1 A,B; Table S2).
Events that shape the development of the brain from an undifferentiated neuroepithelium populated by neural precursors to a mature, functioning organ occur at different times in different regions (Rubenstein and Rakic, 2013; Sanes et al., 2012), and the ability to parse out specific spatial and temporal patterns of gene expression is highly desirable. The standardized generation of ISH data supported the development of a systematic and automated informatics-based data processing pipeline (Ng et al., 2007) for navigation and analysis of this large and complex dataset, shown in Figure 2(A–C). Tissue sections from each ISH experiment were aligned to age-matched 3D brain models assembled from 2D reference atlas annotation, and ISH signal was quantified across a voxel grid whose dimensions corresponded to the sampling density of the ISH. The ISH data for each gene can be analyzed in a 3D context as a pure voxel grid, or can be contextualized with the neuroanatomic reference atlas. In the online application, this processing supports expression summary statistics, anatomic and temporal-based search, and other advanced search options, all freely available. Using Anatomic Search, for a given age, a user can identify genes enriched within a selected brain structure. Results are rank-ordered based upon their selectivity for that structure by comparing expression in the target brain structure to expression in adjacent brain structures. With Temporal Search, users find genes showing temporal enrichment for a given structure. In this case, results are rank-ordered based upon their selectivity for expression at a given age in comparison to all other ages. These two search options are orthogonal in that Anatomic Search ignores temporal enrichment and Temporal Search ignores anatomic enrichment.
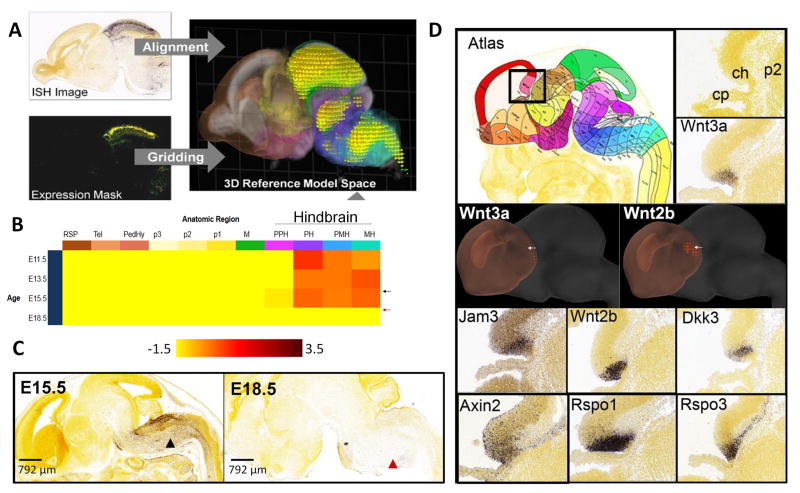
Figure 2. Automated informatics-based pipeline for ISH image analysis.
(A) Image pre-processing, alignment, signal quantification, and summary are provided by a suite of automated modules. An “Alignment” module registers ISH images to the common coordinates of a 3D reference space (Supplemental Experimental Procedures). The “Gridding” module produces an expression summary in 3D for computational expression analysis. The “Unionize” module generates anatomic structure-based statistics by combining grid voxels with the same 3D structural label. In (A), ISH for Tcfap2b is shown at E18.5 with its expression mask and 3D expression summary. (B) Expression summary and (C) ISH for Hoxa2. PH, pontine hindbrain; PMH, pontomedullary hindbrain; and MH, medullary hindbrain (last three columns in Expression Summary in B). (D) Wnt3a was used as a seed gene in NeuroBlast to find other genes in the cortical hem at E13.5. The E13.5 reference atlas is shown; the black box indicates the areas shown in the histology images. The area containing the cortical hem is labeled in a reference HP Yellow stained image (ch, cortical hem; p2, prosomere 2; cp, choroid plexus). 3D images of the atlas structures overlaid with gene expression are shown using the Brain Explorer® 2 3D viewer, where grey represents the entire brain, and orange represents the telencephalic vesicle (Tel) which was used to constrain the search. Voxels found to have gene expression are highlighted, appearing as “bubbles”. Arrows point to the cortical hem. ISH for genes identified by NeuroBlast are shown (sagittal plane; see also Figures S2, S3, and Table S1).
Gene co-expression can imply shared gene function (Hughes et al., 2000; Nayak et al., 2009), protein interactions (Jansen et al., 2002) and common regulatory pathways (Allocco et al., 2004; Segal et al., 2003). We have previously demonstrated that gene-to-gene spatial correlations in adult mouse brain can identify genes belonging to specific functional classes (Hawrylycz et al., 2011) and cell types, such as astrocytes or oligodendrocytes (Lein et al., 2007). An online tool (NeuroBlast) allows identification of genes whose spatial expression patterns are correlated to that of a given gene of interest. The expression pattern of each gene is summarized by a voxel grid encompassing the brain. Pearson’s correlation coefficient is calculated between pairs of genes, over the corresponding voxel sets across the neural primordium. Correlation can also be restricted to a pre-defined anatomic structure. For example, Wnt3a, a ligand in the Wnt signaling pathway, is selectively expressed in the E13.5 cortical hem, a transiently identifiable brain structure that regulates hippocampal development; Wnt3a mutant mice fail to generate a recognizable hippocampus (Lee et al., 2000). Pearson’s correlation coefficient between Wnt3a and the entire gene set across all voxels in the telencephalic vesicle was used to identify genes with spatially comparable expression. The top search returns include eight Wnt signaling genes: Wnt3a, Wnt2b, Dkk3, Axin2, Rspo1, Rspo3, Nkd1, and Rspo2 (DAVID (Huang da et al., 2009a, b)). Other highly correlated genes, Jam3, Dmrt3, Lmx1a, Foxj1, and Id3, could represent candidates for interactions with Wnt signaling or pallial patterning (Fig. 2D).
The NeuroBlast tool can also identify co-expression relationships between a TF and potential downstream targets. In the simplest scenario, a TF would be activated in a cell type and then collaborate with other TFs to activate given enhancer/repressor DNA sequences of its target genes. Positively-regulated target genes should be expressed shortly after, and over time, the spatial expression of the TF should partly match the expression of its downstream targets. We identified a set of 22 genes highly correlated with the TF Pou4f1 which is expressed in the habenula (Fig. S2). Seven of the top genes are presumed to be downstream of Pou4f1 as shown by altered expression levels in a knockout model of Pou4f1 (Efcbp2, Etv1, Chrna3, Nr4a2, Dcc, Sncg, Wif1 (Quina et al., 2009)). These methods can be used to identify and establish a temporal hierarchy of expression of genes activated downstream of any TF.
Although sophisticated image processing tools were developed to annotate ISH expression data, the small size of some brain structures relative to adjacent large empty ventricles as occurs in the E11.5 brain presents challenges for automated tissue registration and analysis. Therefore, expert-guided manual annotation of the ISH data was performed on the four prenatal ages (E11.5 through E18.5) to accurately assign gene expression calls and metrics to specific atlas-defined brain structures and is available online (Fig. S3).
Mapping gene expression to developmental phenomena
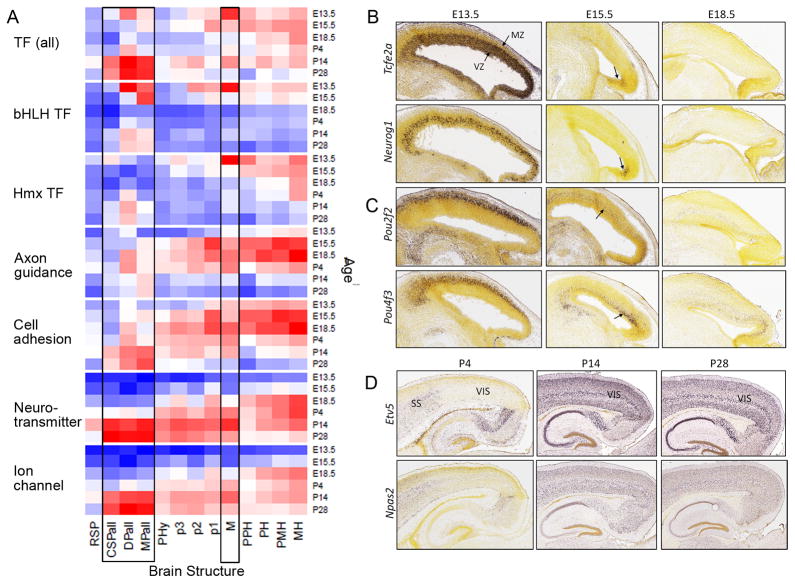
Analyzing temporal peaks of gene expression over development could identify major developmental phenomena associated with a specific brain structure or age. RNA expression levels were investigated for seven functional gene categories across 13 brain regions at 6 ages (E13.5-P28; Fig. 3A). These categories relate to key developmental events such as regional patterning, neurogenesis, differentiation, migration, axogenesis and synaptogenesis, in which developmental timing may vary throughout the CNS (Rubenstein and Rakic, 2013; Sanes et al., 2012). For TFs, two primary peaks are evident, with one peak in the E13.5 midbrain. The Temporal Search tool identified 5 bHLH genes (Tal2, Mxd3, Tcfe2a, Nhlh2 and Neurog1), and 4 homeobox genes (Pou3f3, Lhx1, Pou2f2, and Pou4f3) within the top 20 returns for genes enriched at E13.5 in the midbrain. Most of these bHLH genes were expressed specifically in the ventricular or periventricular strata of the midbrain wall (Fig. 3B), coincident with the timing of peak neurogenesis (Clancy et al., 2001), suggesting a role in growth and generation of neurons in this region. Tcfe2a, for example, maintains stem cells in an undifferentiated state (Nguyen et al., 2006) and is essential for midbrain development in zebrafish (Kim et al., 2000). Neurog1 marks initiation of neurogenesis and promotes cell cycle exit (Bertrand et al., 2002), consistent with its expression in the periventricular zone, where postmitotic neurons exit into the mantle zone. In contrast, homeobox genes that peak in the E13.5 midbrain were primarily enriched in mantle zone, which contains postmitotic maturing neurons, suggesting a role for these genes in differentiation or layering (Fig. 3C). Consistent with this hypothesis, Pou2f2 (Oct2) is known to induce neuronal differentiation (Theodorou et al., 2009) and a close family member of Pou4f3 (Brn3c), regulates the transition from neurogenesis to terminal differentiation (Lanier et al., 2009). The distinct stratification of bHLH and homeobox genes suggest that these TF classes are utilized in the same manner as in the retina, in which the bHLH activators regulate layer specificity of retinal cell types but not neuronal fate, but the homeobox genes regulate neuronal subtype specification (Hatakeyama and Kageyama, 2004). Later, the the midbrain shows peak expression of axon guidance and cell adhesion genes around birth, followed by expression of neurotransmitter-related genes and ion channels in postnatal ages, consistent with the expected progression of neural development.
Figure 3. Anatomic and temporal expression by gene class.
(A) Normalized average expression level for gene classes by age and anatomic region. Expression level is calculated as in Methods and normalized across gene class with higher expression levels in red, lower in blue. Abbreviations: Genes: bHLH, basic helix loop helix; Hmx, homeobox. Structures: RSP, rostral secondary prosencephalon; CSPall, central subpallium; DPall, dorsal pallium/isocortex; MPall, medial pallium; PHy, peduncular hypothalamus; p3, prosomere 3 (prethalamus and prethalamic tegmentum); p2, prosomere 2 (thalamus and thalamic tegmentum); p1, prosomere 1 (pretectum and pretectal tegmentum); M, midbrain; PPH, prepontine hindbrain; PH, pontine hindbrain; PMH, pontomedullary hindbrain; MH, medullary hindbrain. (B–D) Genes identified using online Temporal Search feature. (B, C) Temporal Search for genes enriched in E13.5 midbrain identified bHLH genes expressed in ventricular (VZ) and periventricular zones (B), and homeobox genes in mantle zone (MZ) (C). (D) Temporal Search for genes enriched at P28 in the telencephalic vesicle. Although these genes are expressed in the P4 somatosensory cortex (SS), they exhibit striking lack of expression in visual cortex (VIS). These genes are expressed throughout neocortex after eye opening (P14 and P28; see also Figure S1 and Table S2).
A second expression peak for TFs was identified in dorsal pallium (isocortex), medial pallium (hippocampus) and central subpallium (striatum/pallidum) at P14 and P28, the period when activity-dependent processes are sculpting the brain’s wiring diagrams. A Temporal Search for genes enriched at P28 in the telencephalic vesicle (inclusive of these regions) reveals enrichment for immediate-early genes (Fos, Egr1, Homer1, Arc, Ets2, Dusp14, Hlf, Bcl6, Etv5 and Per1), many of which are TFs. Immediate-early genes are rapidly induced following stimuli, believed to reflect neuronal activation. A subset of these genes is induced in the visual cortex and striatum by sleep deprivation (Thompson et al., 2010) presumably due to increased visual stimulation during sleep deprivation in the light phase. Many immediate-early genes appear to be strongly enriched in visual cortex starting at P14. For instance, expression of Etv5 and Npas2 is not detected in the P4 visual cortex (note expression in surrounding cortical regions in Fig. 3D), whereas by P14 and P28 visual cortex expression is prominent. Thus, Etv5 and Npas2 expression may reflect activity-induced transcription resulting from accrued visual input to the visual cortex after eye opening.
For other gene categories, brainstem exhibits peak expression at mid-late embryonic stages, whereas telencephalic regions exhibit late postnatal peak expression. This trend is observed for the axon guidance, cell adhesion, neurotransmitter and ion channel gene categories, and parallels the timing of maturation of these regions. Neurotransmitter and ion channel classes represent late differentiation variables of the neuronal phenotypes; genes in these categories exhibit very low expression at the earliest age, E13.5, across all brain regions.
To illustrate clusters of genes with temporal co-expression patterns, we focused on the diencephalon. We clustered genes based on their co-expression patterns in voxels annotated for the diencephalon. However, when genes were clustered at each age, no significant coherence was observed across the entire time period (data not shown), although clear differences were observed between embryonic and postnatal ages. This observation led us to group the time points into 3 periods, for independent analysis of co-expression: “embryonic” (E13.5, E15.5, and E18.5), “postnatal” (P4, P14, and P28), and “all” (E13.5, E15.5, E18.5, P4, P14, and P28). In order to extract expression trends over these time periods, weighted gene co-expression network analysis (WGCNA) was used to group genes into clusters with co-expression patterns across the data set (Zhang and Horvath, 2005). The eigengene of each cluster, a measure of the average expression of all the genes within a cluster, represents an expression trend over time observed for the diencephalon (Horvath, 2011).
In most cases, the clusters were comprised of genes delineating particular spatially discrete, contiguous sets of voxels. Example modules are shown for “embryonic” period clustering (Fig. S4). The temporal pattern of expression in the diencephalon is plotted ordering the module eigengenes from E13.5 to E18.5 (Fig. S4 B–K). Because the expression data is comprised of voxels with known anatomic location, the average expression pattern of the cluster can also be plotted back into a 3D model to determine the spatial expression pattern of each cluster. Clustering results are available for “embryonic”, “postnatal” and “all” (Figs. S4–6) and gene ontology results for a subset of modules (Tables S3–S5, respectively). The two most frequent anatomic expression patterns in the diencephalon identified by WGCNA clustering across any timeframe were expression in the thalamus (Figs. S4 B–D and S5 B–F) and subsets of diencephalic regions that specifically exclude the thalamus (Figs. S4 E,F and S5 G). The thalamus clusters were enriched in metabotropic glutamate receptor group I pathways, ion transport, and synaptic transmission genes. In some cases, specific nuclei of the thalamus were identified, e.g., the parafascicular nucleus or the posterior ventromedial nucleus (Fig. S4 C,D, and 6E).
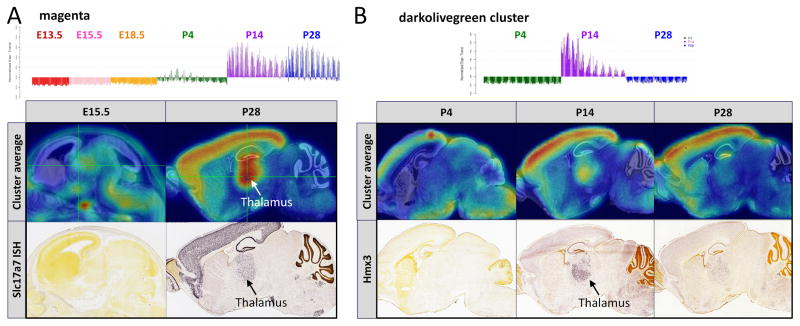
Temporal expression patterns can also be identified using the WGCNA approach. When examining the “all” period that spans E13.5 to P28, two clusters are identified in which genes have strong upregulation of expression in the diencephalon at P14 and P28 (Fig. S6). In the magenta cluster (Fig. 4A), GO analysis identifies enrichment of genes in the metabotropic glutamate receptor group III pathway (p = 0.028; e.g., Slc17a7, Grm4, Slc17a6, Grin2b, Grin2c, Grm1, Slc1a1). Examining the postnatal (P4, P14, and P28) cluster identified a set of genes (Plp1, Cnp, Mbp, Mog, Mobp, and Olig1) strongly upregulated at P14 and P28, including genes heavily enriched in oligodendrocytes (Fig. S4, cluster grey60). Although oligodendrocytes are produced as early as E18.5 (Hardy and Friedrich, 1996), these data show that several well-known oligodendrocyte genes do not exhibit widespread distribution in the diencephalon until P14. A particularly intriguing temporal expression pattern is the occurrence of strong, thalamus-specific expression of predominantly TF genes at P14 (Fig. 5B), a phenomenon that is either weak or undetectable at P4 and P28. The timing may coincide with eye opening and the initial reception of visual stimulation by the thalamus, occuring around P12–13, or with other delayed synaptogenesis-related developmental event. Note that thalamic nuclei corresponding to visual, somatosensitive, somatomotor, and auditory systems are represented in this cluster.
Figure 4. Temporal expression patterns in the diencephalon identified by WGCNA.
(A) Voxelized expression data from 6 ages were used to cluster genes by WGCNA; the magenta cluster is a temporally regulated cluster. The plot (top) shows the eigengene for the cluster across individual voxels at each age. Underneath, the top panels illustrate average expression levels at the indicated stages. The ISH for a gene example is shown at the bottom panels. (B) Voxelized expression data from postnatal ages were used to cluster genes by WGCNA. The darkolivegreen cluster shows strong upregulation at P14 (see also Figures S4, S5, S6 and Tables S3–S5).
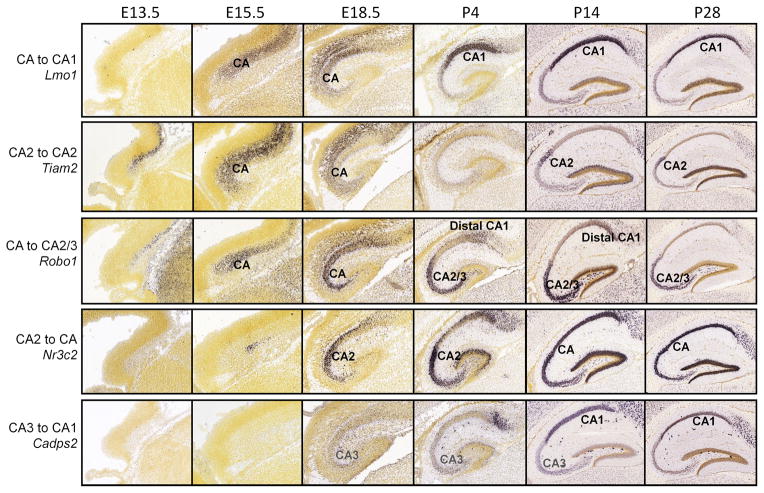
Figure 5. Changes in specificity of gene markers for hippocampal fields.
The top three genes are expressed initially in the entire CA pyramidal layer in the embryo, and eventually display specificity in only one CA field by P28. Nr3c2 is expressed in a subset of cells at E15.5, with enrichment in CA2 around birth, but is expressed throughout CA by the adult. Finally, Cadps2 exhibits transient weak expression in CA3 prior to strong CA1 staining in the adult.
Molecular cohesion of anatomic regions over development
An obvious application of this dataset is to find gene markers selective for specific structures over time, to assess the earliest appearance of that structure in the embryo as well as to characterize how sets of developmentally important genes may change over time. Numerous markers were identified from the Allen Mouse Brain Atlas that subdivide the hippocampus CA region into fields CA1, CA2, and CA3. These genes exhibited complex spatiotemporal expression. First, many markers were not apparent before P14 (e.g., CA2 markers Sostdc1, Stard5, and Fgf5; CA1 markers Plekhg1, SStr4, Htr1a, and Igfbp4; available online) and may relate to terminal differential functions of these CA fields rather than to developmental identity. Other markers are expressed in the full CA pyramidal layer at earlier ages, becoming regionalized at later stages (Fig. 5), or are regionally restricted at E18.5/P4, and become widely expressed across the CA by P28 (e.g., Nr3c2; Fig. 5). Other genes show changing specificity, such as Cadps2 which is expressed in CA3 at age P4, in both CA1 and CA3 at P14, and is CA1 specific at P28. Within a brain region, a variety of events may drive dynamic or transient gene expression and could provide intriguing clues about the process of development within a given region. In order to provide users with another mode of navigating spatiotemporal gene expression, we created a new version of Anatomic Gene Expression Atlas (AGEA) that incorporates developmental age.
Gene expression profiling has been invaluable for refining our understanding of neuroanatomy and development, insofar as gene expression correlations can recapitulate known functional divisions of the brain, provide a hint of their embryological origin (Ng et al., 2009; Zapala et al., 2005) and serve as fiducials to compare particular brain structures across species and time (Puelles et al., 2000). The original AGEA released as part of the Allen Mouse Brain Atlas was a powerful tool to identify correlated voxels at age P56 and find corresponding genes. In the Allen Developing Mouse Brain Atlas, AGEA has undergone a significant advance to allow users to explore spatiotemporal genetic relationships and identify voxels in the brain that show highly correlated gene expression across different ages. Thus, the molecular signatures of brain regions (Puelles and Ferran, 2012) can be used to follow the progressive development of anatomic domains as a surrogate for actual fate-mapping experiments.
A correlation map for each fixed age (Ng et al., 2009) is generated by evaluating each seed voxel against every other target voxel in the 3D reference model. The values obtained across the voxels of each map represent the Pearson correlation coefficients between the seed voxel and every other location over the set of 2,000 genes. Correlations are also calculated between each seed voxel and target voxels of adjacent ages, resulting in a combined total of 265,621 online 3D browsable maps. These correlation maps allow visualization of voxels that share a correlated transcriptome profile, and typically identify adjacent voxels that reflect local neuroanatomy.
Furthermore, the user can view correlation maps that “walk” across the different ages. In this technique, the highest correlates of a chosen voxel are identified at adjacent ages thereby enabling a type of anatomic “virtual molecular fatemap.” By selecting an initial seed voxel at P28, the user can navigate across time to find the highest correlated voxel at P14, subsequently P4, then E18.5, and so on to provide a “reverse molecular fatemap”. A “forward molecular fatemap” is similarly constructed by selecting an initial seed voxel at E13.5 and moving forward in time. The thalamus, the olfactory bulb, and cortex each exhibit coherent and identifiable anatomic precursors as shown in reverse correlation maps traced from P28 to E13.5, highlighting the molecularly consistent anatomic origin of these structures (Fig. 6A). Once such spatiotemporal correlations are established, the AGEA application lists the most significant correlated genes.
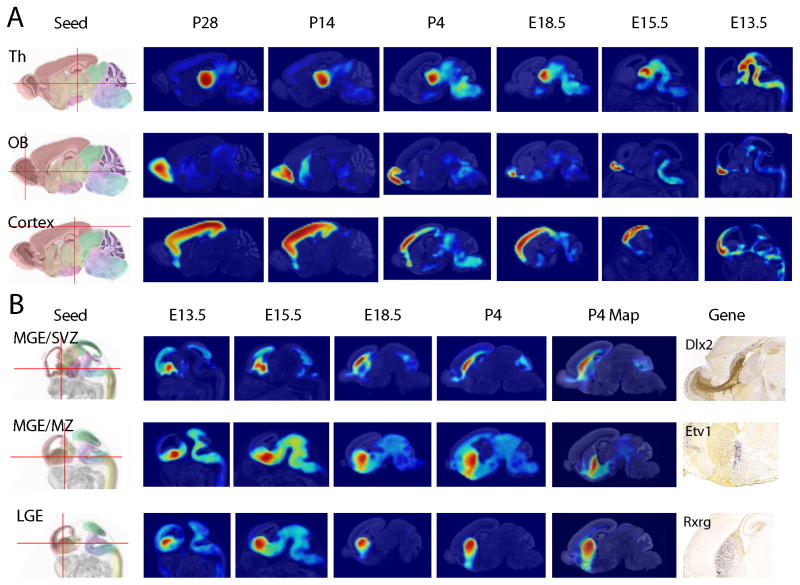
Figure 6. Virtual fatemaps using AGEA.
(A) Virtual (reverse) fate mapping is constructed starting with an initial seed voxel selected at P28. The highest correlated voxel at the next youngest age is calculated in stepwise fashion iteratively until E13.5, and a correlation map is generated at each age. Method is shown for thalamus (Th), olfactory bulb (OB), and cortex. (B) Virtual (forward) fate map of the ganglionic eminences. The initial seed voxel was selected manually at E13.5, and the highest correlated voxel at the next oldest age was automatically selected in stepwise fashion until P28. ISH data at P4 for a supporting gene is shown for each example: Dlx2 for MGE/SVZ; Etv1 for MGE/MZ; and Rxrg for LGE).
To illustrate a virtual forward molecular fate map we selected an initial seed voxel in the E13.5 ganglionic eminences. The highest correlated voxel at the next oldest age was calculated and automatically selected in stepwise fashion. The lateral ganglionic eminence (LGE) is a source of striatal projection neurons, and the medial ganglionic eminence (MGE) is a source of pallidal, diagonal and preoptic projection neurons, as well as of striatal and cortical interneurons; the latter migrate tangentially from the MGE to the cortex and intersperse across the cortical layers amongst the glutamatergic neurons. In the forward map, a seed point chosen in the subventricular zone (SVZ) of the MGE at E13.5 correlates highly to the P4 cortical SVZ and rostral migratory stream, both of which undergo late neurogenesis and tangential migration; they likely share part of their transcriptomic profiles with the neurogenic subpallial SVZ. A seed point in the E13.5 LGE results in a set of highly correlated voxels in the striatum by P4, consistent with current knowledge about the origin and local radial layering of these neurons. These techniques provide a novel method for understanding the molecularly-defined precursor domains and the development of anatomic structures; its results serve also as tests of the structural interpretations introduced in the reference atlases. These “virtual fate maps” are based on the assimilation of data from over 2000 genes, and are geared to identify the best temporal match for the correlates of any structure recognizable at the given magnification. While this works easily for broad definitions of structures (e.g., olfactory bulb), it does not necessarily work for finer subdivisions of the brain. In practice, a limit is imposed by the level of neuroanatomical knowledge of the user (aided by the reference atlases). Anatomically expert users may guess where new interesting seed voxels can be found. Exploring true cell fate specification would require two things: 1) analysis with cellular level resolution to discriminate diverse cell types present in the brain; and 2) using genes or methods to consistently label cell types over time rather than rely on transiently expressed genes. However, the use of many genes at once provides a measure of relatedness that can inform novel insights about the development of the brain.
Molecular principles of brain organization
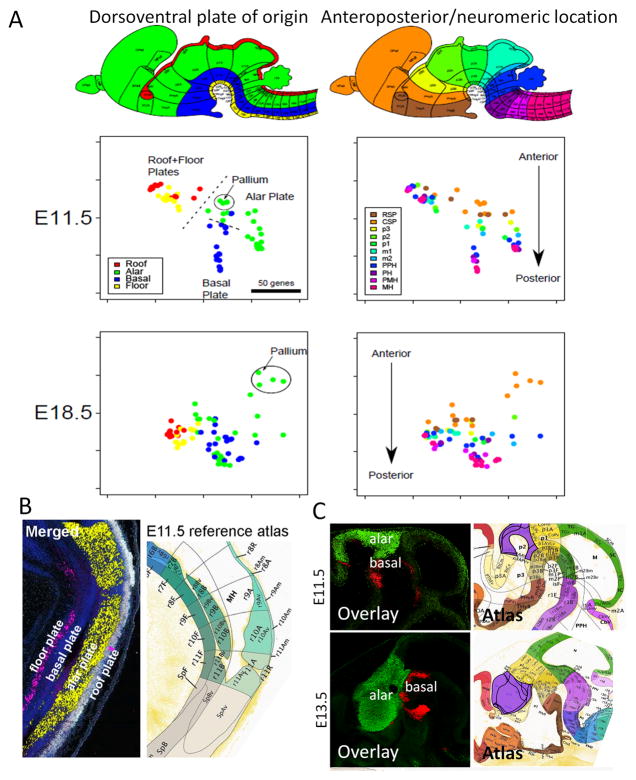
TFs are key regulators for the specification of cell fate during neural development and thus the profiling of ~800 TFs with a relatively fine spatiotemporal sampling may reveal organizing principles of the brain. Multidimensional scaling (MDS) was applied to the binarized (on-off) manual annotation expression data. The MDS visualization allows for qualitative comparision of the relationship of gene expression and structural development. Points represent anatomic structures at a given developmental age, and the distance between them represents proximity on the basis of gene expression. A progressive change was observed in how TF expression correlates with progressive brain regionalization from E11.5 to E18.5 (Fig. 7). At E11.5, brain structures clustered primarily by their longitudinal zonal origin within the major DV columns or ‘plates’ (roof, alar, basal, floor), and secondarily by neuromeric location along the AP axis, jointly defining a checker-board pattern of primary histogenetic areas (Fig. 7). This implies that gene functions shared along the longitudinal dimension of the whole neural tube – underpinning subsequent segmental serial similarity, known as metamery (Puelles and Rubenstein, 2003) - are activated earlier and more distinctly than differential AP molecular patterning of the neuromeric domains. In Figure 7, panels B and C, gene expression patterns were overlaid to demonstrate the clear plate-based (DV) and neuromeric (AP) organization. Between E11.5 and E18.5, a gradual shift occurs in the molecular organization of the brain, resulting in the emergence of a secondary organization with mixed DV and AP features, appearing areal by E18.5 (as shown by the stronger AP organization). By E18.5 structures derived from alar and basal plates are no longer demarcated easily on the sole basis of their TF expression, possibly the result of DV tangential migrations (data not shown). The same is true for floor and roof plate-derived structures, although a distinction remains between alar-basal and roof-floor. Therefore, by late prenatal stages, brain regional identity is defined areally, instead of by plate-of-origin or neuromere; this switch occurs between E15.5 and E18.5, as shown by TF expression.
Figure 7. Multidimensional scaling shows a shift from dorsoventral (plate)-based to anteroposterior neuromere-based organization of the embryonic brain.
(A) Two-dimensional visualization of regions characterized by differences in TF expression, using standard MDS for two embryonic ages. The brain schematic on the top shows brain structures color-coded by DV plates or AP/neuromeric position. The distance between any two regions (dots) represents the number of genes that are differentially expressed between them, as determined by “expressed” versus “undetected” calls in the manual annotation. Left, structures are colored by DV location (roof, red; alar, green; basal, blue; yellow, floor); right, regions are colored by AP location, divided into the following gross categories: rostral secondary prosencephalon (RSP), caudal secondary prosencephalon (CSP), prosomeres 1–3 (p1, p2, p3), mesomeres 1–2 (m1, m2), prepontine hindbrain (PPH), pontine hindbrain (PH), pontomedullary hindbrain (PMH) and medullary hindbrain (MH). (B, C) Examples of genes showing DV organization at E11.5 in the hindbrain (B) and in the diencephalon (C). Genes in (B) are: floor plate, Arx; alar plate, Ascl1; roof plate, Msx1. Genes in (C) are: alar plate, Tcf7l2 and basal plate, Foxa1.
In general, alar-derived structures in the forebrain and midbrain show the largest variation over time, followed by basal-derived structures in the same two brain parts. The roof and floor plate-derived structures brain-wide, as well as the alar and basal-derived hindbrain structures, show the least variable expression over time. Some subregions of the alar telencephalon, including neocortex, hippocampus, and olfactory bulb (red samples in left MDS plots), follow a unique trajectory separate from other alar plate-derived structures. These samples reasonably cluster with other alar structures at E11.5, when the plate-of-origin dominates region identity, but as they differentiate they become increasingly distinct from all other brain regions. One caveat is that TF expression is not necessarily linked to mechanisms of anatomic regionalization (boundary building), since other functions exist (e.g., control of proliferation and neurogenesis). These analyses are intended to assess the most evident principles of organization based upon a broad sampling of genes, acknowledging that selected functionally relevant markers can be used for more precise investigation of longitudinal or transverse boundaries.
The change from plate and neuromeric organization to largely areal organization reflects an acquisition of mature properties and a loss of early patterning cues. Finer subdivisions emerge as distinct structures over this period of embryonic development, lending to the dominance of areal and even strata-related identity by E18.5. We used the binarized TF data to assess the emergence of complexity over this time period, defined as the number of distinct binarized spatial expression patterns exhibited by the TFs within a given brain structure. For example, there are 12 distinct level 5 structures in the diencephalon in the reference atlas; a given gene can be “on” (detected) or “off” (undetected) in each structure, resulting in one of 4096 (212) possible combinations. Taking all the TFs into account, the complexity of a region is the total number of distinct spatial expression patterns observed within that region. Based upon independent analysis of four brain regions (secondary prosencephalon, diencephalon, midbrain, and hindbrain), the number of distinct expression patterns increased from E11.5 to E13.5, with a twofold increase in secondary prosencephalon. We detected no significant increase in the diversity of patterns after E13.5 (Fig. S7). Based on the spatial patterns shared by the largest number of genes, it appears the common expression modes at E11.5 were defined by expression throughout a large DV/AP region (e.g., Hox genes in the hindbrain and spinal cord) or by genes restricted to a longitudinal plate (e.g., Shh and Pax7). In the older embryo, however, the most frequent spatial expression patterns were restricted to individual brain structures (e.g., pallium or olfactory bulb). The peak in expression patterns at E13.5 could be due to the temporary coexistence of both DV (plate)-based and neuromeric/AP-based patterning.
The TFs were further analyzed to determine if brain regions (defined as atlas ontology level 7 for pallium and level 5 for other brain structures) can be distinguished by a binary pattern of TF expression at each age across embryonic development (E11.5–E18.5); basically, we sought unique combinatorial expression patterns to define each age by brain structure combination. In order to identify putative genes that are involved in structural identity, we used a criterion that a gene must be expressed in all descendants of a given atlas structure down to level 10, the deepest level of the ontology short of individual nuclei or layers, in order to be called “widely expressed” for that level 5/7 brain structure (as opposed to “locally expressed” or “not expressed”).
To find a binarized TF code, for each structure a unique set of widely expressed and not expressed genes was identified. Several pairs of regions cannot be distinguished based on this criterion; these pairs of regions show widespread expression of the same genes and not expression of the same genes. Although differences noted in locally expressed genes imply that the expression patterns in such brain structures are not identical, they cannot be definitively distinguished with any combination of TFs. In addition, the “locally expressed” characterization means that transitivity of distinction between gene pairs is not preserved: if regions A and B cannot be distinguished, and regions B and C cannot be distinguished, it does not necessarily follow that regions A and C cannot be distinguished, because there might be a gene with widespread expression in A that shows local expression in B and no expression in C.
We identified a minimal set of ~80 TFs that provide a unique signature for every “distinguishable” region over four ages; 830 out of 13,944 total possible structure and region-pairs cannot be distinguished, the vast majority of which are pairs of regions at E18.5 (Fig. S8B). For the remaining regions with distinct signatures, Figure S8A shows a spatiotemporal TF code at key prenatal stages in development. These genes include known region-specific markers such as Foxg1, a marker of telencephalic development, or the set of Hox genes, known to be involved in hindbrain and spinal cord patterning. The list also includes genes involved in reprogramming to stem cells, or in vitro transdifferentiation. However, some of the selected genes have not been as widely studied, and are thus potentially interesting candidates for further analysis for their role in structural identity along both the spatial and temporal axes.
This minimal TF code is not unique, and alternative or complementary codes could exist. Indeed, the full set of ~800 TFs itself forms a comprehensive code, although it provides no more information than the minimal 83 gene set. The majority of genes in the minimal TF code presented here are necessary (i.e., some pairs of anatomic structures are distinguished by a single gene). The remaining genes may still be biologically relevant, in that they distinguish particular subregions from each other. Overall, this analysis shows that a reduced set of less than 100 TFs is sufficient to generate a unique spatiotemporal code for all distinguishable primary/secondary brain structures at a medium-scale partitioning of the developing mouse brain wall. A simpler example of how the TF code can distinguish six structures at 4 ages is shown (Fig. 8A).
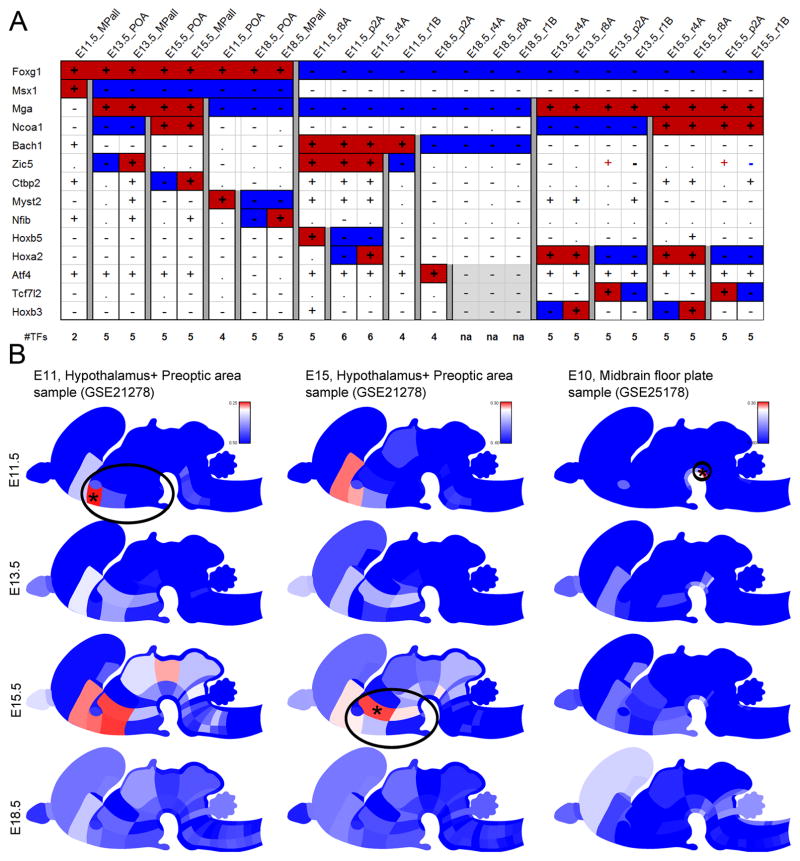
Figure 8. A transcription factor code can uniquely identify the developmental age and anatomic structure in a sample profiled by microarray.
(A) 14 genes can distinguish six brain structures at 4 ages; in this example, three atlas structures at E18.5 (gray shade) remain indistinguishable with this code. (B) Identifying the anatomic region and biological age of a microarray sample based upon the TF code. For each sample, the GEO ID is given; the best match to a given age x region combination in the ADMBA is color-coded (red, high correlation; blue, low correlation; asterisk, best match). In each case, the TF code accurately identifies the closest age x brain structure. Note the anatomic criteria used for obtaining the microarray samples may have differed in part with our criteria, leading to the dispersion of the correlative results (see also Figures S7 and S8).
To demonstrate the utility of this TF code for cross-platform comparisons of developmental time and region, we used published microarray datasets for mouse embryonic hypothalamus and preoptic area sampled from E11 to E18. Cross-platform comparison is compounded by the underappreciated challenges of converting a scale of microarray expression values into a thresholded, binarized expression call comparable to our manual annotation data; thus a perfect match was not anticipated. A mismatch score was calculated between the microarray set and the age x anatomic structure-specific TF code, and using this score, the appropriate age and anatomic structure for each microarray sample could be identified based upon the best match of each sample to the TF code (Fig. 8B).
Discussion
The Allen Developing Mouse Brain Atlas uses histological and molecular profiling to provide a window into the temporal dynamics of over 2100 genes over neural development in the mouse. Due to compromises of scale, a number of key genes surely are not represented in this Atlas. The gene set was selected to survey key functional classes and categories based on known pathways important for development. 90% of these genes were detected in brain at some stage of development, as compared to the Allen Mouse Brain Atlas encompassing more than 20,000 genes in the C57Bl/6J P56 mouse, of which 78.8% are expressed at some level in the adult murine brain (Lein et al., 2007). It is notable that even using a pre-selected set of roughly 2000 genes, representing 10% of the genome, the analyses of the resulting dataset provided great insight into the organization of the brain, underpinning significantly our novel ontology (Puelles et al., 2013) and reference atlases.
While neuroanatomists have long used expression of key genes to guide their understanding of brain architecture only more recently have integrated studies over genome-scale datasets been possible (Bota et al., 2003; Diez-Roux et al., 2011; Dong et al., 2009; Hawrylycz et al., 2010; Lein et al., 2007; Ng et al., 2009; Ng et al., 2010; Puelles and Ferran, 2012; Swanson, 2003; Thompson et al., 2008). In this resource, we provided a temporal framework to understand the genoarchitecture of brain development, and new tools for the community to access these data. The manual annotation data that interprets expression patterns based on the ontology, and the seven reference atlases provides support for users unfamiliar with neuroanatomy, aiding them to assign observed ISH signal to atlas structures; discovery tools such as NeuroBlast and AGEA enable users to achieve explicit identification of new genes of interest. Furthermore, in the three youngest embryonic ages, ISH, 3D models and AGEA tools are available for the entire embryo, encompassing not only spinal cord and peripheral nervous system, but also organs such as lung, heart, and kidney.
The temporal resolution of these data provided several major findings. First, gene expression exhibits complex dynamics over development; a set of marker genes at one stage may not necessarily define the same brain structure at a distant stage of development. However, by integrating the data of ~2000 genes, large brain areas (i.e., at the level of thalamus, cortex, or striatum) and relatively smaller subregions can be tracked in a stepwise fashion from embryonic to postnatal ages, demonstrating their molecular coherence across development, irrespective of emergent changes. Over the course of embryonic development, we observed that the organizing principles for the brain shift from a largely DV or longitudinal, plate-based organization of the brain (classic columns) to an AP, neuromeric or transversally-delimited organization of the brain, that eventually transforms by orthogonal intersection of DV and AP units into the areal organization of individual histogenetic or progenitor domains, key for understanding the production of differential cell types. This order (AP to plate to areal) is consistent with the purposeful ordering of the reference atlas ontology, reflecting the order of key stages of developmental patterning. Although the hallmarks of AP patterning remain through the time course (previously observed in gene expression from adult tissues (Zapala et al., 2005)), the molecular signature of the major longitudinal plates appears to be transient; alar or basal plate signatures become indistinguishable by E18.5, and discrete late neuronal populations or complexes more closely identify with their final areal position/context.
Due to the complexity of developmental gene expression, it would be useful to have a molecular signature, or “barcode” that identifies a particular brain structure at a given stage of brain development. This barcode could provide enable the development of intersectional strategies to target and manipulate cells at a precise stage of development, and could also help identify the developmental age of cells generated from pluripotent stem cells by directed differentiation in vitro. The developmental phenomena that underlie brain development in tetrapods and possibly in all vertebrates have striking similarities in the types of genes and networks activated to govern the precise development of each brain region, though the timing of individual regions may vary; several neuroanatomists are developing pan-mammalian ontologies that assume a common developmental progression underlies this process in humans, non-human primates and mice, hopefully without undermining the future pan-vertebrate developmental and adult brain ontology predicted by evolutionary theory and genomics. Thus the identification of a perfected TF code that could potentially align homologous structures along comparable developmental stages across different vertebrate species is highly possible, irrespective of predictable variations. The TF code introduced in this paper is a humble beginning to the deduction of a molecular signature that describes brain regionalization in its entirety, as an extension of the codes previously deduced for simpler systems such as retinal development (Hatakeyama and Kageyama, 2004), and should illuminate hypercomplex systems such as human brain development. However, in our approach a shared code may not necessarily consist of, or contain, all factors that are causative for the specification of cell types. The TF code presented includes genes known to be key factors in direct reprogramming to specific cell types in culture (e.g., Ascl1, Pou3f2, Sox2, Gata2, Nr2f1, Foxg1), and it probably also includes TFs that are involved in more downstream developmental differentiation processes such as axogenesis or dendritic maturation, providing a hallmark of the developmental age of the region. Future efforts could be targeted to refine the TF code to find causative genes and pan-mammalian or pan-vertebrate genes.
EXPERIMENTAL PROCEDURES
ISH
A high-throughput ISH platform described previously (Lein et al., 2007) generated ISH data for ~2,000 genes across 7 ages including four embryonic (days post conception: E11.5, E13.5, E15.5, and E18.5) and three postnatal ages (P4, P14, and P28 days after birth, where day of birth is P0), with the addition of a yellow nuclear counterstain, and modified protocols optimized for each age. Full methodological details are supplied (Supplemental Experimental Procdures).
Reference Atlases
For each reference atlas, tissue sections were stained by Nissl/cresyl violet or a nuclear HP Yellow stain to aid identification of anatomic structures for expert delineation (done by L. Puelles on the basis of a novel ontogenetic ontology based upon the prosomeric model (Puelles et al., 2012). High resolution images of tissue sections were obtained from automated microphotographic digitalizing systems and processed through our standard image pipeline, then exported to Adobe Illustrator CS graphics software for delineation of brain structures. Line drawings were converted to polygons corresponding to individual structures which were named systematically according to the ontology in the Illustrator file, converted to scalable vector graphics (SVG), databased, and lofted into 3D for use in the informatics pipeline.
Informatics Processing and Data Analysis
Full methodological details on the pipeline including development of NeuroBlast and AGEA are supplied in Supplemental Experimental Procedures. Pearson correlation was used to compare expression profiles. The statistical package R (http://www.r-project.org/) was used for data analysis and visualization. Expression clusters were visualized by projecting voxel expression data into a plane of section. Using expression values for the voxels of the diencephalon, we created co-expression gene networks using WGCNA (Zhang and Horvath, 2005). Gene ontology analysis was performed using DAVID (Huang da et al., 2009a, b). For the MDS analysis, all manually annotated data was binarized to expressed (=1) and not expressed (=0) calls for each anatomic structure at level 5 of the ontology (level 7 for pallial substructures). The distance between each pair of structures was calculated as the number of genes expressed in one structure that were not expressed in the other; this is equivalent to the Manhattan distance between the structures’ expression vectors. This distance matrix was then projected onto 3 dimensions using the classical MDS package cmdscale in R. For visualization in two dimensions, the first two coordinates were chosen to plot the structure labels. In all plots shown, the eigenvalue-based goodness of fit measure as reported by the cmdscale package was at least 0.65.
Manual annotation
ISH experiments were annotated by expert developmental neuroanatomists. Complete sets of image series of E11.5, E13.5, E15.5, and E18.5 experiments were manually annotated. Three metrics were used: intensity, density, and pattern. These metrics were scored for each brain structure according to a standard scheme (Fig. S3) and entered into the hierarchically organized ontology of anatomical structures. At each developmental stage, annotation was performed for anatomic structures belonging to the most detailed level of the ontology (down to Level 10) that were identifiable as exhibiting differential expression. For example, if the pallium exhibited a homogeneous pattern but the subpallium exhibited a different pattern, annotation would be recorded for each of these structures individually. When a given brain structure was annotated, that annotation data was intended to represent the complete set of “child” or “descendent” structures of this level in the hierarchical tree (corresponding to an anatomical region), such that the expression call for pallium would then apply to its children: medial pallium, lateral pallium, ventral pallium, and dorsal pallium.
Manual annotation was not performed for every structure at every level of the ontology, which amounts to over 1,500 brain structures. Instead, the annotation strategy ensured that every “branch” of the ontological tree was annotated. For example, for the four major parts of the brain: forebrain, midbrain, hindbrain, and spinal cord, a given gene may have expression in only diencephalon. Therefore, midbrain, hindbrain, and spinal cord would be annotated as “undetected”, and the forebrain expression may be addressed by providing the actual expression information for diencephalon, while producing an “undetected” call for the sibling structure, secondary prosencephalon.
Transcription factor code
For every pair of anatomic structures, TFs were identified that show widespread expression in one structure and no expression in the other, generating a combinatorial set of structure-pairs, each linked to a set of TFs. Widespread expression was defined as expression in all children of the structure to level 10, the deepest level of the ontology. Next, we identified all pairs of brain structures that could be distinguished only by a single gene. All of these genes were included in the final set. For the remaining structure-pairs, identifying a minimal set of TFs to distinguish each brain region is equivalent to the set cover problem (an NP-hard problem). We used a heuristic pruning approach to approximate a minimal set: starting with the full set of unselected genes, we randomly removed one, and re-examined the remaining data to identify structure-pairs that now had a single gene distinguishing the pair members. These genes were added to the final list, and the pruning process continued until all remaining genes were crucial to distinguish at least one structure-pair. An exhaustive search over every possible selection path was not feasible, so this process was repeated 100 times and the gene set with the fewest members was selected.
The TF code was applied to three Affymetrix mouse genome microarray data sets from the Gene Expression Omnibus (GEO) with IDs GSE21278 and GSE25178. Because the tissues profiled in these data sets do not correspond exactly to specific anatomic structures defined in the atlas ontology described here, we compared the thresholded expression profile from the GEO datasets to the full TF code for every time point and structure in the ontology, and ranked the matches using the following metric:
where FN = number of genes called “present” in the GEO set but “undetected” in our code, FP = number of genes called “absent” in the GEO set but called “widely expressed” in our code, TN = number of genes called “absent” in the GEO set and “undetected” in our code, and TP = number of genes called “present” in the GEO set and “widely expressed” in our code. For each GEO sample dataset we ranked all the structures by how well they scored according to this match score. The brain structure with the best match score for each of the GEO datasets is starred (Fig. 8).
Supplementary Material
Figure S1. Temporal Gene Expression in the Brain by Class, related to Figure 3. (A) Percentage of genes by expression trend in brain: ON, on at all stages; OFF, off at all stages; OFF-ON, genes not expressed at E11.5 but eventually turned on; ON-OFF, genes that are expressed at E11.5 but eventually turned off; Complex patterns, e.g. OFF-ON-OFF. Note that transcription factors account for 75% of the genes in the ON-OFF category. (B–F), Distribution of OFF-ON (purple) and ON-OFF (green) genes shown as percentages from five gene functional categories or pathways over seven stages. (B) Transcription Factors, (C) Wnt Signaling, and (D) Receptor Tyrosine Kinases (RTKs) and ligands all exhibit both ON-OFF and OFF-ON trends of expression, whereas GPCRs (E) and Ion Channels (F), tend to be expressed largely later in development (OFF-ON).
Figure S2. Using NeuroBlast tool to identify genes based on spatial correlation of expression, related to Figure 2. NeuroBlast was used to identify top search returns in the diencephalon (yellow 3D structure) at the indicated ages for the seed gene Pou4f1/Brn3a, a transcription factor. Expression of Efcbp2, Etv1, and Chrna3 are highly correlated with Pou4f1 across most ages. Insets show ISH in the sagittal plane for these genes in the habenula. Etv1 at E13.5 and Chrna3 at E13.5 and E15.5 are weakly expressed (arrow indicates area of incipient expression). The correlation between each gene and Pou4f1/Brn3a is given in the lower left.
Figure S3. Manual annotation of embryonic gene expression data, related to Figure 2. (A) Metrics used to annotate expression patterns included density, intensity, and pattern. (B) The annotation process began with viewing all images in a series, and opening a separate window to record metrics for each structure in the ontology. The “2D annotation” refers to an initial assignment of expression to a structure, and the “3D annotation” refers to further assignment of expression to strata within that region (e.g., ventricular zone, mantle zone).
Figure S4. Spatial and Temporal Expression Profiles across E13.5, E15.5, and E18.5, related to Figure 4. (A) Cluster dendrogram groups genes into distinct modules using a vector of expression energy across all diencephalon voxels spanning the time window E13.5, E15.5, and E18.5 with the y-axis corresponding to co-expression distance between genes and the x-axis to genes. Two colorbars label the modules assigned by dynamic tree cutting (top) and by dynamic tree cutting followed by merging close modules (bottom), which is used in analyses. For the early period, expression levels for the diencephalon voxels at E13.5, E15.5, and E18.5 time points were concatenated as a vector for each gene. (B–K) Spatial realization of examples of clusters, showing the eigengene of voxels over time (top, E13.5 (red), E15.5 (pink), and E18.5 (orange)) and the plot of the average spatial expression of the cluster genes on the Nissl atlas (bottom, ordered E13.5, E15.5, and E18.5). (B–D) p2/thalamus modules violet, darkgreen, and red are shown. Not thalamus modules light green and salmon are shown in E and F. p3/prethalamus modules pale turquoise and royal blue are shown in G and H. A ventricular zone module (orange) is shown in I. Midnight blue and pink are modules with expression in the roof, choroid, and habenula (J and K).
Figure S5. Spatial and Temporal Expression Profiles across P4, P14 and P28, Related to Figure 4. (A) Cluster dendrogram groups genes into distinct modules using a vector of expression energy across all diencephalon voxels spanning the time window P4, P14, and P28 with the y-axis corresponding to co-expression distance between genes and the x-axis to genes. Two colorbars label the modules assigned by dynamic tree cutting (top) and by dynamic tree cutting followed by merging close modules (bottom), which is used in analyses. (B–J) Spatial realization of examples of clusters, showing the eigengene of voxels over time (top, P4 (green), P14 (purple), and P28 (blue)) and the plot of the average spatial expression of the cluster genes on the Nissl atlas (bottom, ordered P4, P14, and P28). (B–F) p2/thalamus modules dark orange, dark olive green, sienna 3, pale turquoise, and violet are shown. Not thalamus module orange is shown in G. Substantia nigra module dark grey is shown in (H). Roof, choroid plexus, and habenula modules yellow green and light green are shown in (I) and (J). A glial module grey60 is shown in (K).
Figure S6. Spatial and Temporal Expression Profiles across “all” timepoints, related to Figure 4. (A) Cluster dendrogram groups genes into distinct modules using a vector of expression energy across all diencephalon voxels spanning the time window P4, P14, and P28 with the y-axis corresponding to co-expression distance between genes and the x-axis to genes. Two colorbars label the modules assigned by dynamic tree cutting (top) and by dynamic tree cutting followed by merging close modules (bottom), which is used in analyses. (B–F) Spatial realization of examples of clusters, showing the eigengene of voxels over time (top, E13.5 (red), E15.5 (pink), E18.5 (yellow), P4 (green), P14 (purple), and P28 (blue)) and the plot of the average spatial expression of the cluster genes on the Nissl atlas (bottom, same order as top panel).
Figure S7. Expression patterns in three brain regions, related to Figure 8. (A–C) Most common patterns in Secondary Prosencephalon, Diencephalon, and Hindbrain using manual annotation data; number of subregions per structure was 16, 12, and 16, respectively. Brain schematics indicate expression pattern (red, expressed; blue, not expressed). Circles show number of genes per pattern. (D) Number of patterns observed; note that patterns that differ by expression in a single subregion are grouped together as a single pattern to allow for possible error.
Figure S8. A minimal transcription factor code with maximal discrimination between age-specific brain regions, related to Figure 8. (A) Brain regions are labeled on the right and the color bar provides the atlas-region color (also demonstrated in schematic at the top). Royal blue denotes a gene is “widely expressed” and white denotes “not expressed”. Cyan indicates “locally expressed” within a structure. Only genes that are widely expressed or not expressed can be used to discriminate structures. (B) Brain region pairs for each age that are indistinguishable based upon transcription factor expression.
Genes selected for in situ hybridization probes are listed. The table shows the gene symbol, gene name, Entrez Gene ID, gene classification, and weight gene co-expression analysis (WGCNA) module for the early (E13.5, E15.5, and E18.5), middle (E18.5 and P4), and late (P4, P14, and P28) periods as well as the correlation to the eigengene for each gene within each period. Gene class categories (column D) include axon guidance pathway, cell adhesion, ion channel, neurotransmitter pathway, Notch signaling, RTKs and RTKs ligands (receptor tyrosine kinase), transcription factor activity, bHLH TF (basic helix loop helix transcription factor), forkhead TF, homeobox TF, nuclear receptor, POU domain genes, and Wnt signaling.
Genes were evaluated for expression over development in brain: ON, on at all stages; OFF, off at all stages; OFF-ON, genes not expressed at E11.5 but eventually turn on; ON-OFF, genes that are expressed at E11.5 but eventually turn off; Complex patterns, e.g. OFF-ON-OFF. Gene symbol, Entrez Gene ID, expression (on or off for E11.5, E13.5, E15.5, E18.5, P4, P14, and P28), class (on, off, off-on, etc.), and classification of gene as a transcription factor, Wnt Signaling, receptor tyrosine kinase (RTK) and ligand, ion channel or GPCR are provided.
Gene ontology (GO) analysis was carried out on individual modules using DAVID with GO biological process 5, GO molecular function 5, PANTHER biological process all, PANTHER molecular function all, KEGG pathway, and PANTHER pathway with the entire early gene list as the background gene list. A GO significant summary is provided containing any DAVID result with BH (Benjamini-Hochberg) p-value < 0.1. For each individual module, all DAVID results are shown separately.
Gene ontology (GO) analysis was carried out on individual modules using DAVID with GO biological process 5, GO molecular function 5, PANTHER biological process all, PANTHER molecular function all, KEGG pathway, and PANTHER pathway with the entire late gene list as the background gene list. A GO significant summary is provided containing any DAVID result with BH (Benjamini-Hochberg) p-value < 0.1. For each individual module, all DAVID results are shown separately.
Gene ontology (GO) analysis was carried out on individual modules using DAVID with GO biological process 5, GO molecular function 5, PANTHER biological process all, PANTHER molecular function all, KEGG pathway, and PANTHER pathway with the entire middle gene list as the background gene list. A GO significant summary is provided containing any DAVID result with BH (Benjamini-Hochberg) p-value < 0.1. For each individual module, all DAVID results are shown separately.
Highlights.
We generated a survey ~2100 genes over 7 stages of mouse brain development.
Automated and manual image analysis allows discovery of genoarchitecture.
Transcription factors exhibit temporal shifts in molecular organizing principles.
A transcription factor code of 83 genes uniquely identifies age and brain region.
Acknowledgments
We wish to thank the Allen Institute founders, Paul G. Allen and Jody Allen, for their vision, encouragement, and support. We express our gratitude to past and present Allen Institute staff members from the Structured Science and Technology teams for their technical assistance and to Conor Kelly for assistance with reference atlas production, and to Sara Ball for manuscript edits. We also wish to thank the Allen Developing Mouse Brain Atlas Advisory Council Members Gregor Eichele, Josh Huang, Alexandra Joyner, Marc Tessier-Lavigne, Joseph Takahashi, and Phyllis Wise. We wish to thank Eric Turner for discussion on the habenula.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annual review of cell and developmental biology. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- Allocco DJ, Kohane IS, Butte AJ. Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinformatics. 2004;5:18. doi: 10.1186/1471-2105-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bota M, Dong HW, Swanson LW. From gene networks to brain networks. Nat Neurosci. 2003;6:795–799. doi: 10.1038/nn1096. [DOI] [PubMed] [Google Scholar]
- Clancy B, Darlington RB, Finlay BL. Translating developmental time across mammalian species. Neuroscience. 2001;105:7–17. doi: 10.1016/s0306-4522(01)00171-3. [DOI] [PubMed] [Google Scholar]
- Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HW. The Allen Reference Atlas: A Digital Color Brain Atlas of the C57BL/6J Male Mouse. 1. Hoboken, NJ: John Wiley & Sons, Inc; 2008. [Google Scholar]
- Dong HW, Swanson LW, Chen L, Fanselow MS, Toga AW. Genomic-anatomic evidence for distinct functional domains in hippocampal field CA1. Proc Natl Acad Sci U S A. 2009;106:11794–11799. doi: 10.1073/pnas.0812608106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RJ, Friedrich VL., Jr Oligodendrocyte progenitors are generated throughout the embryonic mouse brain, but differentiate in restricted foci. Development. 1996;122:2059–2069. doi: 10.1242/dev.122.7.2059. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hawrylycz M, Bernard A, Lau C, Sunkin SM, Chakravarty MM, Lein ES, Jones AR, Ng L. Areal and laminar differentiation in the mouse neocortex using large scale gene expression data. Methods. 2010;50:113–121. doi: 10.1016/j.ymeth.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Hawrylycz M, Ng L, Page D, Morris J, Lau C, Faber S, Faber V, Sunkin S, Menon V, Lein E, et al. Multi-scale correlation structure of gene expression in the brain. Neural Netw. 2011;24:933–942. doi: 10.1016/j.neunet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Horvath S. Weighted Network Analysis: Applications in Genomics and Systems Biology. 1. New York: Springer; 2011. [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;235:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- Kim CH, Oda T, Itoh M, Jiang D, Artinger KB, Chandrasekharappa SC, Driever W, Chitnis AB. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature. 2000;407:913–916. doi: 10.1038/35038097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier J, Dykes IM, Nissen S, Eng SR, Turner EE. Brn3a regulates the transition from neurogenesis to terminal differentiation and represses non-neural gene expression in the trigeminal ganglion. Dev Dyn. 2009;238:3065–3079. doi: 10.1002/dvdy.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Nayak RR, Kearns M, Spielman RS, Cheung VG. Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. Genome Res. 2009;19:1953–1962. doi: 10.1101/gr.097600.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Bernard A, Lau C, Overly CC, Dong H, Kuan C, Pathak S, Sunkin SM, Dang C, Bohland JW, et al. An Anatomic Gene Expression Atlas of the Adult Mouse. Brain Nature Neuroscience. 2009;12:356–362. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- Ng L, Lau C, Sunkin SM, Bernard A, Chakravarty MM, Lein ES, Jones AR, Hawrylycz M. Surface-based mapping of gene expression and probabilistic expression maps in the mouse cortex. Methods. 2010;50:55–62. doi: 10.1016/j.ymeth.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Ng L, Pathak SD, Kuan C, Lau C, Dong H, Sodt A, Dang C, Avants B, Yushkevich P, Gee JC, et al. Neuroinformatics for genome-wide 3D gene expression mapping in the mouse brain. IEEE/ACM Trans Comput Biol Bioinform. 2007;4:382–393. doi: 10.1109/tcbb.2007.1035. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- Puelles L, Ferran JL. Concept of neural genoarchitecture and its genomic fundament. Frontiers in neuroanatomy. 2012;6:47. doi: 10.3389/fnana.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L, Harrison M, Paxinos G, Watson C. A developmental ontology for the mammalian brain based on the prosomeric model. Trends Neurosci. 2013;36:570–578. doi: 10.1016/j.tins.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Puelles L, Kuwana E, Puelles E, Bulfone A, Shimamura K, Keleher J, Smiga S, Rubenstein JL. Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol. 2000;424:409–438. doi: 10.1002/1096-9861(20000828)424:3<409::aid-cne3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Puelles L, Martinez-de-la-Torre M, Bardet S, Rubenstein JL. The Hypothalamus. In: Watson C, Paxinos G, Puelles L, editors. The Mouse Nervous System. Waltham: Elsevier; 2012. pp. 221–312. [Google Scholar]
- Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- Quina LA, Wang S, Ng L, Turner EE. Brn3a and Nurr1 mediate a gene regulatory pathway for habenula development. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29:14309–14322. doi: 10.1523/JNEUROSCI.2430-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Rakic P, editors. Comprehensive Developmental Neuroscience. Oxford: Academic Press; 2013. [Google Scholar]
- Sanes DH, Reh TA, Harris WA. Development of the Nervous System. 3. New York: Elsevier, Inc; 2012. [Google Scholar]
- Segal E, Shapira M, Regev A, Pe’er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Siegert S, Cabuy E, Scherf BG, Kohler H, Panda S, Le YZ, Fehling HJ, Gaidatzis D, Stadler MB, Roska B. Transcriptional code and disease map for adult retinal cell types. Nat Neurosci. 2012;15:487–495. S481–482. doi: 10.1038/nn.3032. [DOI] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Swanson LW. Brain Architecture: Understanding the Basic Plan. Oxford: Oxford University Press; 2003. [Google Scholar]
- Theodorou E, Dalembert G, Heffelfinger C, White E, Weissman S, Corcoran L, Snyder M. A high throughput embryonic stem cell screen identifies Oct-2 as a bifunctional regulator of neuronal differentiation. Genes Dev. 2009;23:575–588. doi: 10.1101/gad.1772509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, Pathak SD, Jeromin A, Ng LL, MacPherson CR, Mortrud MT, Cusick A, Riley ZL, Sunkin SM, Bernard A, et al. Genomic anatomy of the hippocampus. Neuron. 2008;60:1010–1021. doi: 10.1016/j.neuron.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Wisor JP, Lee CK, Pathak SD, Gerashchenko D, Smith KA, Fischer SR, Kuan CL, Sunkin SM, Ng LL, et al. Molecular and anatomical signatures of sleep deprivation in the mouse brain. Front Neurosci. 2010;4:165. doi: 10.3389/fnins.2010.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, Tynan W, Broide RS, Helton R, Stoveken BS, et al. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Temporal Gene Expression in the Brain by Class, related to Figure 3. (A) Percentage of genes by expression trend in brain: ON, on at all stages; OFF, off at all stages; OFF-ON, genes not expressed at E11.5 but eventually turned on; ON-OFF, genes that are expressed at E11.5 but eventually turned off; Complex patterns, e.g. OFF-ON-OFF. Note that transcription factors account for 75% of the genes in the ON-OFF category. (B–F), Distribution of OFF-ON (purple) and ON-OFF (green) genes shown as percentages from five gene functional categories or pathways over seven stages. (B) Transcription Factors, (C) Wnt Signaling, and (D) Receptor Tyrosine Kinases (RTKs) and ligands all exhibit both ON-OFF and OFF-ON trends of expression, whereas GPCRs (E) and Ion Channels (F), tend to be expressed largely later in development (OFF-ON).
Figure S2. Using NeuroBlast tool to identify genes based on spatial correlation of expression, related to Figure 2. NeuroBlast was used to identify top search returns in the diencephalon (yellow 3D structure) at the indicated ages for the seed gene Pou4f1/Brn3a, a transcription factor. Expression of Efcbp2, Etv1, and Chrna3 are highly correlated with Pou4f1 across most ages. Insets show ISH in the sagittal plane for these genes in the habenula. Etv1 at E13.5 and Chrna3 at E13.5 and E15.5 are weakly expressed (arrow indicates area of incipient expression). The correlation between each gene and Pou4f1/Brn3a is given in the lower left.
Figure S3. Manual annotation of embryonic gene expression data, related to Figure 2. (A) Metrics used to annotate expression patterns included density, intensity, and pattern. (B) The annotation process began with viewing all images in a series, and opening a separate window to record metrics for each structure in the ontology. The “2D annotation” refers to an initial assignment of expression to a structure, and the “3D annotation” refers to further assignment of expression to strata within that region (e.g., ventricular zone, mantle zone).
Figure S4. Spatial and Temporal Expression Profiles across E13.5, E15.5, and E18.5, related to Figure 4. (A) Cluster dendrogram groups genes into distinct modules using a vector of expression energy across all diencephalon voxels spanning the time window E13.5, E15.5, and E18.5 with the y-axis corresponding to co-expression distance between genes and the x-axis to genes. Two colorbars label the modules assigned by dynamic tree cutting (top) and by dynamic tree cutting followed by merging close modules (bottom), which is used in analyses. For the early period, expression levels for the diencephalon voxels at E13.5, E15.5, and E18.5 time points were concatenated as a vector for each gene. (B–K) Spatial realization of examples of clusters, showing the eigengene of voxels over time (top, E13.5 (red), E15.5 (pink), and E18.5 (orange)) and the plot of the average spatial expression of the cluster genes on the Nissl atlas (bottom, ordered E13.5, E15.5, and E18.5). (B–D) p2/thalamus modules violet, darkgreen, and red are shown. Not thalamus modules light green and salmon are shown in E and F. p3/prethalamus modules pale turquoise and royal blue are shown in G and H. A ventricular zone module (orange) is shown in I. Midnight blue and pink are modules with expression in the roof, choroid, and habenula (J and K).
Figure S5. Spatial and Temporal Expression Profiles across P4, P14 and P28, Related to Figure 4. (A) Cluster dendrogram groups genes into distinct modules using a vector of expression energy across all diencephalon voxels spanning the time window P4, P14, and P28 with the y-axis corresponding to co-expression distance between genes and the x-axis to genes. Two colorbars label the modules assigned by dynamic tree cutting (top) and by dynamic tree cutting followed by merging close modules (bottom), which is used in analyses. (B–J) Spatial realization of examples of clusters, showing the eigengene of voxels over time (top, P4 (green), P14 (purple), and P28 (blue)) and the plot of the average spatial expression of the cluster genes on the Nissl atlas (bottom, ordered P4, P14, and P28). (B–F) p2/thalamus modules dark orange, dark olive green, sienna 3, pale turquoise, and violet are shown. Not thalamus module orange is shown in G. Substantia nigra module dark grey is shown in (H). Roof, choroid plexus, and habenula modules yellow green and light green are shown in (I) and (J). A glial module grey60 is shown in (K).
Figure S6. Spatial and Temporal Expression Profiles across “all” timepoints, related to Figure 4. (A) Cluster dendrogram groups genes into distinct modules using a vector of expression energy across all diencephalon voxels spanning the time window P4, P14, and P28 with the y-axis corresponding to co-expression distance between genes and the x-axis to genes. Two colorbars label the modules assigned by dynamic tree cutting (top) and by dynamic tree cutting followed by merging close modules (bottom), which is used in analyses. (B–F) Spatial realization of examples of clusters, showing the eigengene of voxels over time (top, E13.5 (red), E15.5 (pink), E18.5 (yellow), P4 (green), P14 (purple), and P28 (blue)) and the plot of the average spatial expression of the cluster genes on the Nissl atlas (bottom, same order as top panel).
Figure S7. Expression patterns in three brain regions, related to Figure 8. (A–C) Most common patterns in Secondary Prosencephalon, Diencephalon, and Hindbrain using manual annotation data; number of subregions per structure was 16, 12, and 16, respectively. Brain schematics indicate expression pattern (red, expressed; blue, not expressed). Circles show number of genes per pattern. (D) Number of patterns observed; note that patterns that differ by expression in a single subregion are grouped together as a single pattern to allow for possible error.
Figure S8. A minimal transcription factor code with maximal discrimination between age-specific brain regions, related to Figure 8. (A) Brain regions are labeled on the right and the color bar provides the atlas-region color (also demonstrated in schematic at the top). Royal blue denotes a gene is “widely expressed” and white denotes “not expressed”. Cyan indicates “locally expressed” within a structure. Only genes that are widely expressed or not expressed can be used to discriminate structures. (B) Brain region pairs for each age that are indistinguishable based upon transcription factor expression.
Genes selected for in situ hybridization probes are listed. The table shows the gene symbol, gene name, Entrez Gene ID, gene classification, and weight gene co-expression analysis (WGCNA) module for the early (E13.5, E15.5, and E18.5), middle (E18.5 and P4), and late (P4, P14, and P28) periods as well as the correlation to the eigengene for each gene within each period. Gene class categories (column D) include axon guidance pathway, cell adhesion, ion channel, neurotransmitter pathway, Notch signaling, RTKs and RTKs ligands (receptor tyrosine kinase), transcription factor activity, bHLH TF (basic helix loop helix transcription factor), forkhead TF, homeobox TF, nuclear receptor, POU domain genes, and Wnt signaling.
Genes were evaluated for expression over development in brain: ON, on at all stages; OFF, off at all stages; OFF-ON, genes not expressed at E11.5 but eventually turn on; ON-OFF, genes that are expressed at E11.5 but eventually turn off; Complex patterns, e.g. OFF-ON-OFF. Gene symbol, Entrez Gene ID, expression (on or off for E11.5, E13.5, E15.5, E18.5, P4, P14, and P28), class (on, off, off-on, etc.), and classification of gene as a transcription factor, Wnt Signaling, receptor tyrosine kinase (RTK) and ligand, ion channel or GPCR are provided.
Gene ontology (GO) analysis was carried out on individual modules using DAVID with GO biological process 5, GO molecular function 5, PANTHER biological process all, PANTHER molecular function all, KEGG pathway, and PANTHER pathway with the entire early gene list as the background gene list. A GO significant summary is provided containing any DAVID result with BH (Benjamini-Hochberg) p-value < 0.1. For each individual module, all DAVID results are shown separately.
Gene ontology (GO) analysis was carried out on individual modules using DAVID with GO biological process 5, GO molecular function 5, PANTHER biological process all, PANTHER molecular function all, KEGG pathway, and PANTHER pathway with the entire late gene list as the background gene list. A GO significant summary is provided containing any DAVID result with BH (Benjamini-Hochberg) p-value < 0.1. For each individual module, all DAVID results are shown separately.
Gene ontology (GO) analysis was carried out on individual modules using DAVID with GO biological process 5, GO molecular function 5, PANTHER biological process all, PANTHER molecular function all, KEGG pathway, and PANTHER pathway with the entire middle gene list as the background gene list. A GO significant summary is provided containing any DAVID result with BH (Benjamini-Hochberg) p-value < 0.1. For each individual module, all DAVID results are shown separately.