SUMMARY
F-BAR domains control membrane interactions in endocytosis, cytokinesis, and cell signaling. Although generally thought to bind curved membranes containing negatively charged phospholipids, numerous functional studies argue that differences in lipid-binding selectivities of F-BAR domains are functionally important. Here, we compare membrane-binding properties of the S. cerevisiae F-BAR domains in vitro and in vivo. Whereas some F-BAR domains (such as Bzz1p and Hof1p F-BARs) bind equally well to all phospholipids, the F-BAR domain from the RhoGAP Rgd1p preferentially binds phosphoinositides. We determined X-ray crystal structures of F-BAR domains from Hof1p and Rgd1p, the latter bound to an inositol phosphate. The structures explain phospholipid-binding selectivity differences, and reveal an F-BAR phosphoinositide binding site that is fully conserved in a mammalian RhoGAP called Gmip, and is partly retained in certain other F-BAR domains. Our findings reveal previously unappreciated determinants of F-BAR domain lipid-binding specificity, and provide a basis for its prediction from sequence.
INTRODUCTION
Interaction of proteins with cellular membrane surfaces depends on an ever-growing group of phospholipid-binding domains, which recognize specific phospholipid headgroups or a more general property of the membrane such as charge or curvature (Hurley, 2006; Lemmon, 2008; Moravcevic et al., 2012). The BAR (Bin/Amphiphysin/Rvs-like) domain superfamily (Mim and Unger, 2012) exemplifies the second of these groups, comprising ‘banana-shaped’ dimeric helical bundles that appear capable of sensing and/or creating membrane curvature (Qualmann et al., 2011). A structure of the amphiphysin BAR domain (Peter et al., 2004) provided the first clues for how this might be achieved, revealing a concave cationic surface on a crescent-shaped dimer that abuts (and deforms) anionic membranes.
F-BAR domains (Itoh and De Camilli, 2006) represent an important subset within the BAR superfamily. They were first noted in adaptor proteins of the PCH family (Saccharomyces pombe cdc15 homology) involved in endocytosis, cytokinesis, actin nucleation and signaling (Chitu and Stanley, 2007; Lippincott and Li, 2000). The conserved region (a portion of the F-BAR domain) was initially termed an FCH domain (for Fes kinase-CIP4 Homology), and is always followed by a coiled-coil region. The coiled-coil was subsequently included in the definition of the extended FCH (or EFC) domain when structural homology to the BAR domain was predicted (Itoh and De Camilli, 2006; Tsujita et al., 2006) and then confirmed crystallographically (Henne et al., 2007; Shimada et al., 2007). F-BAR domains form crescent-shaped helical dimers that are more elongated than classical BAR domains, and have more shallow curvature. Their membrane association is thought to be driven by nonspecific electrostatic interaction between positively charged residues on the concave face of the crescent and negatively charged membrane surfaces (Itoh and De Camilli, 2006). F-BAR domains also polymerize into helical coats that tubulate membranes (Frost et al., 2008). Differences in their own shapes and modes of polymerization are likely to underlie some of the distinct properties that are now being uncovered for F-BAR domains and F-BAR proteins (Arasada and Pollard, 2011; Coutinho-Budd et al., 2012; Itoh and De Camilli, 2006; Qualmann et al., 2011; Roberts-Galbraith and Gould, 2010). Beyond this ability to recognize and influence mesoscale properties of membranes, however, it now seems clear that phospholipid-binding selectivity itself also plays an important role in defining F-BAR domain function (Wang et al., 2009; Zhao et al., 2013). We identified one F-BAR domain – from S. cerevisiae Rgd1p – in a screen of yeast proteins that specifically recognize phosphoinositides (Moravcevic et al., 2010). Rgd1p is a GTPase-activating protein (GAP) specific for the Rho3 and Rho4 small GTPases, which control actin cytoskeleton organization and stress signaling pathways (Doignon et al., 1999; Lefèbvre et al., 2012; Roumanie et al., 2000).
Combining cellular and in vitro approaches, we compare the phospholipid-binding properties of S. cerevisiae F-BAR domains. We also describe crystal structures of the F-BAR domains from Rgd1p (the only yeast example that selectively binds phosphoinositides) and Hof1p (which binds all phospholipids). Our structures explain the phospholipid specificity differences, and – importantly – reveal an inositol phosphate binding site in the first structure of an F-BAR domain bound to a lipid headgroup. Analyzing which elements of this binding site are conserved in mammalian F-BAR domains provides valuable insight into phospholipid-binding selectivities, and allowed us to identify an F-BAR domain in Gmip, a poorly studied human RhoA-specific GAP that faithfully preserves the Rgd1p phosphoinositide-binding site. Elucidating the binding mode and ligand specificities of these domains is important because F-BAR-containing proteins play key roles as adaptors at the membrane-cytosol interface in numerous fundamental cellular processes, and have also been implicated in cancer, neurological and metabolic disorders (Roberts-Galbraith and Gould, 2010).
RESULTS
Identification of the F-BAR domain from S. cerevisiae Rgd1p as a phosphoinositide-binding domain
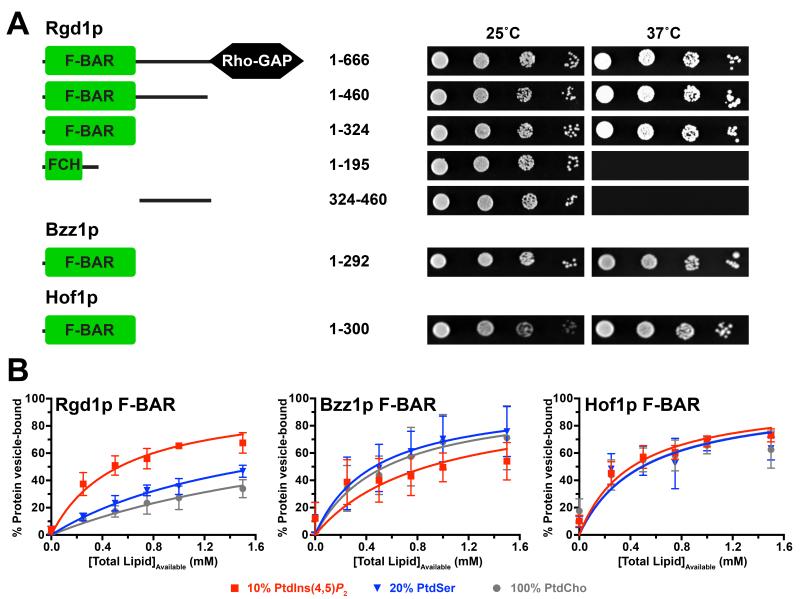
The S. cerevisiae Rho GTPase-activating protein (GAP) Rgd1p (Doignon et al., 1999) was first identified as a potential phosphoinositide-binding protein in a screen of yeast open reading frames that identified 128 yeast proteins with this property (Moravcevic et al., 2010; Zhu et al., 2001). Independent functional studies have also revealed that the subcellular localization and GAP activity of Rgd1p are regulated by phosphoinositides (Prouzet-Mauleon et al., 2008). Using a Ras-rescue assay (Isakoff et al., 1998), we found that fusing full-length Rgd1p to a non-farnesylated, constitutively active (Q61L), Ras variant promotes its recruitment to the membrane to overcome the Ras-activation defect in a cdc25ts cell at the restrictive temperature (Figure 1A). Ras rescue requires the complete F-BAR domain, with neither the FCH domain alone nor the region C-terminal to the F-BAR domain being sufficient to drive Q61L Ras to the membrane (Figure 1A). In vitro binding studies (Figure 1B) further showed that the recombinant Rgd1p F-BAR domain (amino acids 1-324) associates preferentially with vesicles containing phosphatidylinositol-(4,5)-bisphosphate (PtdIns(4,5)P2) over those containing phosphatidylcholine (PtdCho) alone or 20% (mole/mole) phosphatidylserine (PtdSer) in a PtdCho background. We estimate a molar partition coefficient (K) (Kavran et al., 1998; Peitzsch and McLaughlin, 1993) of 1,880 M−1 for this interaction, which would correspond to a dissociation constant (KD) of approximately 53 μM if we assumed that each F-BAR domain protein binds one PtdIns(4,5)P2 molecule.
Figure 1. The Rgd1p F-BAR domain has distinct membrane binding properties.
(A) Ras Rescue (Isakoff et al., 1998) studies show that the N-terminal 324 amino acids (1-324, corresponding to the F-BAR domain) of Rgd1p are necessary and sufficient to recruit non-farnesylated Ha-RasQ61L fusions to the membrane and rescue growth of cdc25ts yeast cells at 37°C. Bzz1p and Hof1p F-BAR domains (residues 1-292 and 1-300 respectively) also show membrane recruitment. Schematic figures of the proteins fused to Q61L Ras are shown at left. On the right, representative results are shown for serial dilutions of yeast cultures expressing the noted fragments spotted in duplicate onto selection plates and incubated at the permissive (25°C) or restrictive (37°C) temperature for 4-5 days.
(B) Vesicle sedimentation studies with histidine-tagged F-BAR domains (10 μM) incubated with increasing concentrations of SUVs containing 20% (mole/mole) PtdSer or 10% (mole/mole) PtdIns(4,5)P2. The percentage of protein pelleting with the vesicles was measured, and data fit as described (Kavran et al., 1998). Mean and standard deviation (SD) are shown for at least three independent experiments.
Phosphoinositides direct subcellular localization of Rgd1p F-BAR
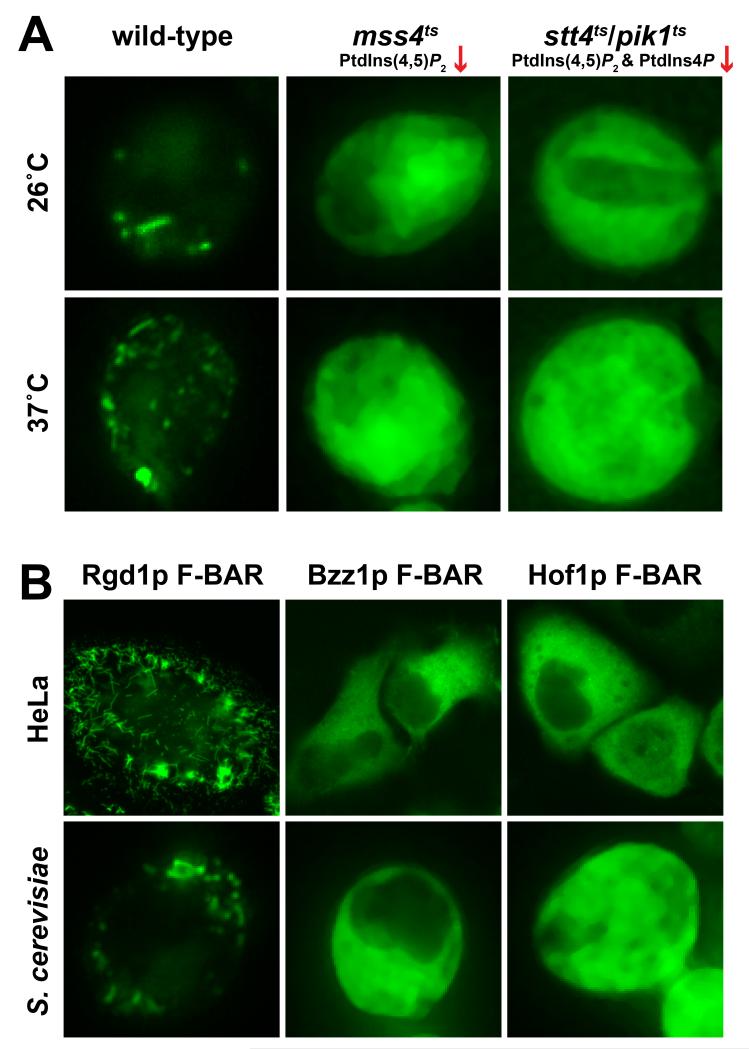
The Rgd1p F-BAR domain displays a punctate distribution when expressed as a GFP fusion protein in yeast (Figure 2A), and clearly tubulates membranes in HeLa cells (Figure 2B), reminiscent of the structures described previously for several mammalian F-BAR domains (Itoh et al., 2005; Tsujita et al., 2006). Importantly, the punctate/tubular distribution of Rgd1p F-BAR was abolished in yeast strains with reduced levels of PtdIns(4,5)P2 (Figure 2A) caused by temperature sensitive mutations in the major PtdIns4P 5-kinase (mss4ts) or in both the Stt4p and Pik1p PtdIns 4-kinases (stt4ts/pik1ts). PtdIns(4,5)P2 levels are reduced even at the permissive temperature (26°C) by ~40% in mss4ts cells, and by over 80% at the restrictive temperature (37°C) (Stefan et al., 2002). In stt4ts/pik1ts cells, PtdIns4P and PtdIns(4,5)P2 levels are approximately 40% and 62% of wild-type levels respectively at 26°C, and fall to less than 10% of wild-type levels at the restrictive temperature (Audhya et al., 2000). The phosphoinositide-dependence of Rgd1p F-BAR localization is also consistent with previous studies in which a genomic copy of GFP-fused full-length Rgd1p showed clear mislocalization in phosphoinositide-deficient strains (Prouzet-Mauleon et al., 2008).
Figure 2. Subcellular localization of GFP-fused S. cerevisiae F-BAR domains in yeast and mammalian cells.
(A) The GFP-fused Rgd1p F-BAR domain was expressed in wild-type yeast cells (left), cells with a temperature sensitive mutation in Mss4p, the major PtdIns4P 5-kinase (mss4ts; middle), or cells with temperature sensitive mutations in both major PtdIns 4-kinases Stt4p and Pik1p (stt4ts/pik1ts; right). Experiments were performed at (37°C) or below (26°C) the restrictive temperature. PtdIns(4,5)P2 levels are greatly reduced in the mutant strains even at 26°C (Audhya et al., 2000; Stefan et al., 2002). Images are representative of >90% of cells observed, with >100 cells observed in each of at least three independent experiments. (B) Comparison of localization of GFP-fused Rgd1p, Bzz1p, and Hof1p F-BAR domains in HeLa cells (upper panels) and wild-type S. cerevisiae cells (lower panels). Representative cells from >300 analyzed are shown.
Distinct lipid selectivities for other S. cerevisiae F-BAR domains
We also assessed the membrane-association abilities of other S. cerevisiae F-BAR domains. Bzz1p is a SH3 domain-containing regulator of actin polymerization (Soulard et al., 2002), and Hof1p is a Src homology 3 (SH3) domain-containing adaptor protein involved in cytokinesis and septum formation (Nishihama et al., 2009; Vallen et al., 2000). The fourth bona fide yeast F-BAR domain is found in the RhoGAP Rgd2p (Roumanie et al., 2001), but we were unable to produce protein of sufficient quality to include this domain in our studies. Like Rgd1p F-BAR, both the Bzz1p and Hof1p F-BAR domains were able to recruit Q61L Ras to the membrane in Ras rescue assays (Figure 1A), and both bound to the PtdIns(4,5)P2-containing vesicles with similar affinities, with K values of 1,140 M−1 and 2,400 M−1 for the Bzz1p and Hof1p F-BAR domains respectively (Figure 1B). Importantly, however, neither the Hof1p nor the Bzz1p F-BAR domains showed the same preference for PtdIns(4,5)P2-containing vesicles that we saw for Rgd1p, consistent with another recent report (Zhao et al., 2013). In fact, the Hof1p and Bzz1p F-BAR domains both bound just as well to vesicles containing 20% (mole/mole) PtdSer or indeed to vesicles containing only PtdCho. The selectivity difference between these two F-BAR domains and Rgd1p F-BAR was further evident in subcellular localization studies (Figure 2B); the Bzz1p and Hof1p F-BAR domains were diffusely localized in HeLa and yeast cells when over-expressed as GFP fusion proteins, with no sign of the tubular/punctate distribution seen with Rgd1p.
The membrane tubulation by the Rgd1p F-BAR domain in HeLa cells resembles that seen for most mammalian F-BAR domains, including those from FBP17, CIP4, FCHo1/2, pacsin 1, nostrin, and PSTPIP1/2 (Henne et al., 2010; Icking et al., 2006; Itoh and De Camilli, 2006; Itoh et al., 2005; Tsujita et al., 2006), which all bind relatively non-specifically to anionic phospholipids. The FBP17, CIP4, and pacsin 1 F-BAR domains preferentially bind PtdSer-containing membranes, and phosphoinositides typically further enhance membrane binding (Henne et al., 2010; Itoh et al., 2005; Tsujita et al., 2006). By contrast, the cellular localization properties of the Bzz1p and Hof1p F-BAR domains more closely resemble the mammalian Fer/Fes F-BAR domain, which shows less clear membrane tubulation in cells, and binds even more promiscuously to phospholipids (McPherson et al., 2009; Tsujita et al., 2006).
Structural comparison of Rgd1p and Hof1p F-BAR domains
To understand the origin of differences in membrane-binding properties of F-BAR domains, which may be crucial for the distinct functions of closely-related proteins that contain them (Arasada and Pollard, 2011; Qualmann et al., 2011; Tanaka-Takiguchi et al., 2013), we determined the X-ray crystal structures of the F-BAR domain from Hof1p (residues 1-300) using multi-wavelength anomalous diffraction (MAD) methods, and the Rgd1p F-BAR domain (residues 24-333) using molecular replacement. Importantly, the Rgd1p F-BAR domain structure was determined in complex with bound myo-inositol-1,2,3,4,5,6-hexakisphosphate (InsP6) as a mimic of a phosphoinositide headgroup – representing to the best of our knowledge the first example of an F-BAR domain structure bound to a lipid headgroup mimetic (see Table 1 for data collection and refinement statistics). Representative electron density, including that around the ligand-binding site of Rgd1p F-BAR, is shown in Figure S1.
Table 1. Data collection and refinement statistics.
| Rgdlp F-BAR InsP6 |
Hof1p F-BAR |
|||
|---|---|---|---|---|
| Data collection | ||||
| Space group | C2 | C2 | ||
| Cell dimensions | ||||
| a, b, c (Å) | 179.0, 74.1, 105.7 | 137.0, 44.0, 95.7 | ||
| α, β, γ (°) | 90, 104.3, 90 | 90, 133.5, 90 | ||
| Peak | Inflection | Remote | ||
|
|
||||
| Resolution (Å)a | 50-3.3 | 50-2.7 | 50-2.7 | 50-2.7 |
| R sym b | 0.14 (0.53) | 0.10 (0.30) | 0.08 (0.39) | 0.07 (0.30) |
| I / σI | 14. 0 (3.2) | 37.2 (5.7) | 32.9 (3.8) | 33.9 (4.9) |
| Completeness (%) | 99.7 (100) | 99.7 (97.2) | 99.4 (96.2) | 98.9 (90.4) |
| Redundancy | 4.2 (4.3) | 7.0 (5.7) | 6.9 (5.1) | 7.2 (6.0) |
| Refinement | ||||
| Resolution (Å) | 31-3.3 | 40-2.7 | ||
| No. reflections | 19341 | 11489 | ||
| Rwork/Rfreec | 26/30 | 20/24 | ||
| No. atoms | 4394 | 2226 | ||
| Protein | Rgd1p | Hof1p | ||
| A:aa 30-326 | aa 2-275 | |||
| B:aa 25-326 | ||||
| Ligand molecules | 2 (InsP6) | |||
| Water molecules | 8 | 40 | ||
| B-factors | ||||
| Protein | 64.4 | 68.6 | ||
| Ligand/ion | 98.8 | |||
| Water | 31.1 | 68.7 | ||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.006 | 0.003 | ||
| Bond angles (°) | 1.09 | 0.61 | ||
See also Figure S1.
Highest-resolution shell data are shown in parentheses. Each dataset was collected from a single crystal.
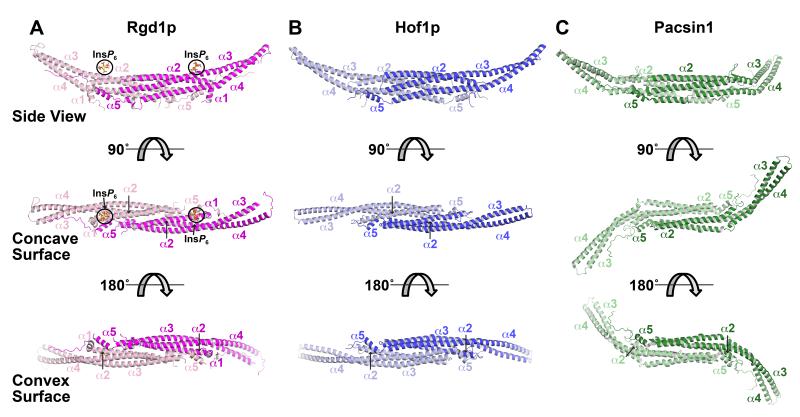
Both domains display the typical F-BAR fold (Figure 3), forming an elongated crescent-shaped dimer. Dissociation constants (KD) for dimerization were measured by sedimentation equilibrium analytical ultracentrifugation to be approximately 3 μM for both F-BAR domains (Figure S2), similar to the value of 2.5 μM reported for the FCHo2 F-BAR domain (Henne et al., 2007). As with other F-BAR domains (Henne et al., 2007; Shimada et al., 2007), the structure is dominated by a core of three long α helices (α2, α3, and α4), with additional shorter helices at the amino (α1) and carboxy (α5) termini. Most of helix α2 from each molecule, and approximately half of each helix α3 and α4 come together to form a closely packed six-helix bundle at the dimer interface. In addition, the amino-terminal α1 helix associates with carboxy-terminal α5 helix of its dimerization partner (only one turn of helix α1 was ordered for Hof1p, presumably because of the boundaries used). An unstructured carboxy-terminal region follows helix α5 in each monomer and also packs against its dimerization partner. The Dali server (Holm and Rosenström, 2010) identified the CIP4 F-BAR domain (PDB entry 2EFK (Shimada et al., 2007)) as the most similar to that from Rgd1p (with FBP17 as the second ranked), and the FBP17 F-BAR domain (PDB entry 2EFL (Shimada et al., 2007)) as most similar to that from Hof1p (with CIP4 as second ranked). The monomers of the Rgd1p and Hof1p F-BAR domains also overlay very well with one another, with a Cα position rmsd of just 2.8 Å – despite sequence identity between the two domains of just 17%. Similarly, Rgd1p F-BAR overlays well with the FBP17 or CIP4 F-BAR domains, with Cα position rmsd values of 2.8 Å and 3.0 Å respectively (corresponding values for Hof1p are 3.1 Å and 2.7 Å). The FCHo2 F-BAR domain is more distinct, with Cα overlay rmsd values of 4.0 Å and 4.4 Å for Rgd1p and Hof1p respectively. There appear to be at least two ‘classes’ of F-BAR domain dimer. Those from Rgd1p, Hof1p, CIP4, and FBP17 (Shimada et al., 2007) all resemble straight round brackets (Figures 3A,B), whereas those from FCHo2 and Pacsin F-BAR domain dimers have a pronounced ‘tilde’ or ‘S’ shape (Henne et al., 2007; Wang et al., 2009) (Figure 3C) that arises because the wings extending beyond the dimer interface core are twisted. Although F-BAR domains differ in their types of curvature, the fact that those from Rgd1p and Hof1p are very similar in this regard argues that this distinction cannot explain their quite different membrane-binding properties.
Figure 3. Structures of the Rgd1p and Hof1p F-BAR domains.
(A) Cartoon of the dimeric Rgd1p F-BAR domain, with one molecule colored magenta (right) and the other pink (left). Three views are shown, with the F-BAR domain progressively rotated around its long axis. The middle (orthogonal) view looks into the concave surface, and the bottom view is 180° rotated so that the view is into the convex surface. The five primary α-helices are labeled. The two bound InsP6 molecules (bound to the concave surface) are labeled and circled in black. See also Figures S2 and S3.
(B) The Hof1p F-BAR domain represented as in (A), but with the two monomers colored dark blue (right) and light blue (left). See also Figures S2 and S3.
(C) Structure of the Pacsin-1 F-BAR domain from PDB entry 3HAI (Wang et al., 2009), illustrating the ‘S’ or tilde shape seen in some F-BAR domains.
A phosphoinositide headgroup-binding site in Rgd1p F-BAR
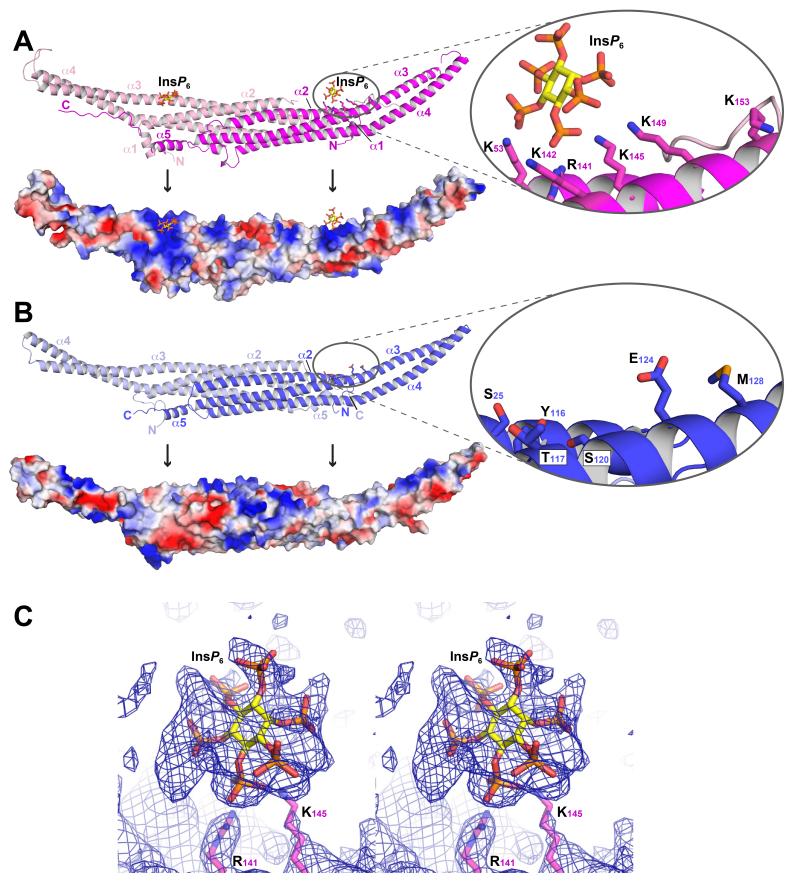
Our ability to visualize InsP6 bound to the Rgd1p F-BAR domain provided one clear explanation for the difference in properties of the Rgd1p and Hof1p F-BAR domains, and yielded the first view of an F-BAR domain bound to a lipid headgroup mimetic (Figure 4A). Several mutational studies have implicated basic residues on the inner part of the concave F-BAR domain in binding to negatively-charged membrane surfaces (Henne et al., 2007; Reider et al., 2009; Shimada et al., 2007; Shimada et al., 2010). Likewise, the Rgd1p and Hof1p F-BAR domains both display clusters of positively-charged lysines and arginines on their concave faces (Figures 4A,B and S3). However, these clusters are not conserved in location – consistent with the considerable sequence divergence between the two domains.
Figure 4. Phosphoinositide binding site specific to Rgd1p F-BAR domain.
(A) The Rgd1p F-BAR domain, labeled as in Figure 3A, with the InsP6 binding site in the right-hand molecule circled and detailed in the inset. In the lower part of the panel, the Rgd1p F-BAR domain is shown with its surface colored according to electrostatic potential (blue is positive; red is negative). See also Figure S4.
(B) The Hof1p F-BAR domain, with the region corresponding to the Rgd1p InsP6 binding site circled and shown in detail in the inset. The lower panel shows the Hof1p F-BAR domain with its surface colored according to electrostatic potential as for (A). Black vertical arrows point to the location of the InsP6 binding site in Rgd1p or its equivalent in Hof1p.
(C) Composite omit map contoured at 1σ for the region around the InsP6 binding site in Rgd1p F-BAR. The electron density indicates that InsP6 binds in multiple different orientations, consistent with the lack of stereospecificity and smearing of density. InsP6 (shown as sticks) was fit in one ‘average’ orientation, and refined as a rigid-body.
The most distinctive basic patch on the concave surface of the Rgd1p F-BAR domain contained clear electron density for InsP6 in crystals that had been soaked with this headgroup mimetic, with two InsP6 molecules bound per dimer. The electron density suggests that InsP6 does not have a highly preferred binding orientation (Figure 4C), and efforts to fit multiple orientations indicated that many are possible – as also reported for InsP6 binding to AP180 (Ford et al., 2001). InsP6 was therefore fit into the density as a rigid body (see Experimental Procedures) to yield an ‘average’ orientation for visual representation. The binding site is formed by a surface-lying “patch” of lysine and arginine side-chains (Figure 4A), reminiscent of similar inositol phosphate binding sites in the AP180 N-terminal homology (ANTH) domain, the amino-terminal part of the AP2 α-subunit, arrestin, and the pleckstrin homology (PH) domain from β-spectrin (Moravcevic et al., 2012). Side-chains in Rgd1p that contact InsP6 most closely (within ~3 Å of one of the InsP6 phosphates) are those from R141 and K145 in helix α3, plus K53 in helix α2 (Figure 4A,C). K142, K149 and K153 (all in helix α3) also contribute to a larger positively-charged region that accommodates the InsP6 molecule. This basic patch is starkly absent from the Hof1p F-BAR domain (Figure 4B), replaced by a neutral or even negatively charged region in which all of the basic residues involved in the Rgd1p/InsP6 interaction have neutral or anionic counterparts in Hof1p: S25; Y116; T117; S120; E124 and M128. The major basic patch on the concave surface of the Hof1p F-BAR domain is approximately half-way along the dimer (Figures 4B and S3), almost 30 Å away from the location of the Rgd1p InsP6 binding site.
Consistent with the surface location of its InsP6 binding site and heterogeneity of InsP6 orientation, Rgd1p shows little stereospecificity in binding to phosphoinositides (Figure S4A), binding with essentially the same affinity in surface plasmon resonance (SPR) studies to vesicles containing PtdIns(4,5)P2, PtdIns(3,4)P2, or PtdIns(3,5)P2 at (10% mole/mole). Moreover, the isolated headgroups of these three phosphoinositides were equally potent in their ability to compete the Rgd1p F-BAR domain off membranes containing 10% (mole/mole) PtdIns(4,5)P2 (Figure S4B). This specificity – or lack thereof – resembles that seen for phosphoinositide binding by ANTH domains (Ford et al., 2001), the AP2 α subunit (Collins et al., 2002; Gaidarov et al., 1996), arrestin (Gaidarov et al., 1999), the β-spectrin PH domain (Hyvönen et al., 1995), and several others (Moravcevic et al., 2012).
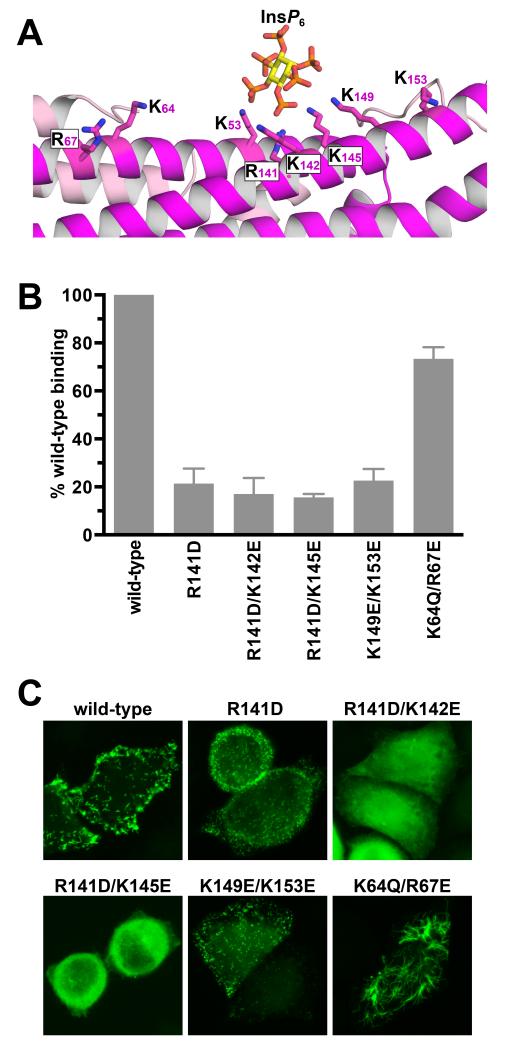
Mutating the crystallographically-observed phosphoinositide binding site alters subcellular localization of the Rgd1p F-BAR domain
To investigate the physiological relevance of the inositol phosphate binding site observed in the Rgd1p F-BAR domain, we mutated the key contributing lysines in various combinations to aspartates or glutamates, and assessed the consequences for both in vitro PtdIns(4,5)P2 binding and subcellular localization (Figure 5). SPR experiments showed that mutating basic residues in the crystallographic InsP6-binding site diminishes Rgd1p binding to membranes that contain 10% (mole/mole) PtdIns(4,5)P2. Mutating R141, in the center of the binding site, reduced binding of 30 μM Rgd1p F-BAR by >75%. Additional mutations reduced PtdIns(4,5)P2 binding further, as seen for R141D/K142E and R141D/K145E doubly-mutated variants (Figure 5B). Glutamate substitutions at K149 and K153 – at the periphery of the binding site – also diminished PtdIns(4,5)P2 binding (Figure 5B). By contrast, mutating K64 and R67, which are more distant from the binding site (Figure 5A), had little effect. The subcellular distribution of overexpressed GFP-fused Rgd1p F-BAR domains in HeLa cells showed similar trends (Figure 5C). The K64Q/R67E variant was indistinguishable from wild-type with essentially no diffuse cytoplasmic fluorescence, and formation of tubule-like structures.
Figure 5. Interrogation of the crystallographically observed phosphoinositide binding in Rgd1p F-BAR by mutagenesis.
(A) Expanded view of the InsP6 binding site in Rgd1p F-BAR, in the same orientation as Figure 4A, showing candidate basic residues involved in InsP6 binding.
(B) Assessment of PtdIns(4,5)P2 binding by Rgd1p F-BAR mutated variants using SPR. Purified proteins (30 μM) were injected onto a sensorchip bearing 10% (mole/mole) PtdIns(4,5)P2 in DOPC, and the binding signal (± SD for at least three experiments) plotted as a function of the value recorded for wild-type protein.
(C) The same mutations were also introduced into the GFP-fused Rgd1p F-BAR domain and expressed in HeLa cells to assess effects on subcellular localization. Over 300 cells were examined for each variant, and representatives are shown.
The R141D and K149E/K153E variants showed significant diffuse cytoplasmic fluorescence, but retained some reticular or punctate (but not tubular) localization. The R141D/K142E and R141D/K145E doubly-mutated variants, which had the lowest in vitro PtdIns(4,5)P2-binding affinities, showed only diffuse cytoplasmic fluorescence (Figure 5C). These findings argue that the crystallographically-observed inositol phosphate binding site in the Rgd1p F-BAR domain contributes significantly to its phosphoinositide-dependent membrane tubulation in cells.
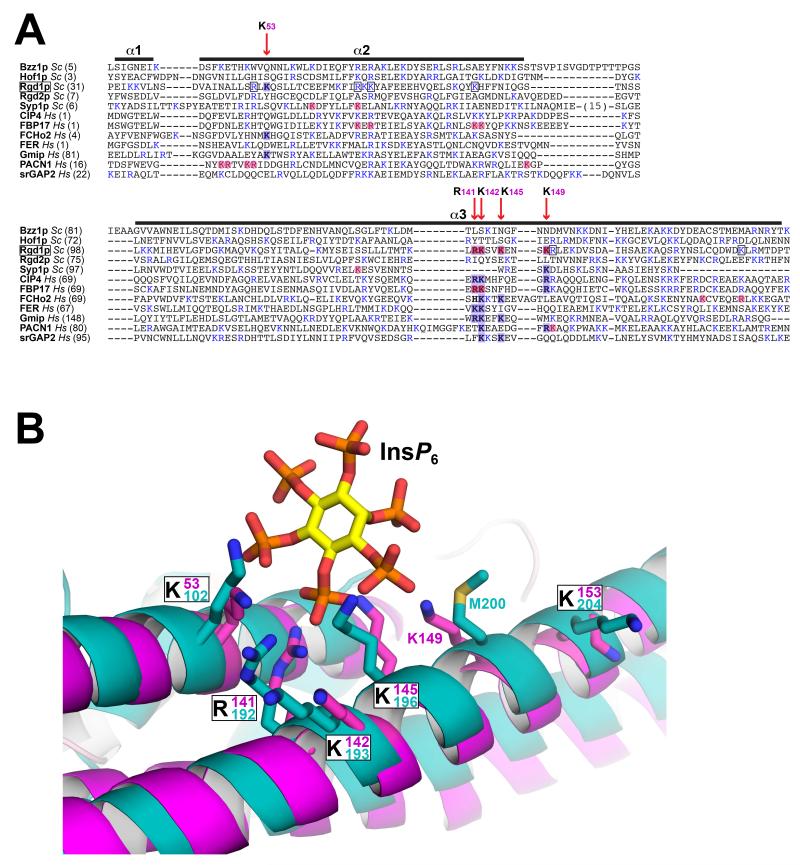
Conservation of elements of the Rgd1p phosphoinositide-binding site in other F-BAR domains: Implications for functional selectivity
Elements of the Rgd1p InsP6-binding site are clearly conserved in some, but not all mammalian F-BAR domains (Figure 6A), but this site is unique to Rgd1p among the S. cerevisiae F-BAR domains. The CIP4 and FBP17 F-BAR domains (which have the same straight bracket shape) retain three of the basic residues in α3, and mutation of these residues in FBP17 impairs binding to negatively-charged membranes (Shimada et al., 2007; Tsujita et al., 2006). The lysine in α2 (K53 in Rgd1p) is not conserved in these two F-BAR domains (Figure 6A), and regions elsewhere in this helix have been implicated in membrane binding (Shimada et al., 2007; Tsujita et al., 2006), implying a much more delocalized binding site (consistent with less phosphoinositide specificity). The FCHo2 F-BAR domain (which has an ‘S’ or tilde-shape) retains the α2 lysine and part of the α3 site, but mutational studies again suggest a more delocalized binding site (Henne et al., 2007). Interestingly, the Pacsin-1 F-BAR domain (PACN1) retains little of this site, but basic residues in broadly the same region contribute to membrane binding that has been shown to also involve an array of other features (Shimada et al., 2010; Wang et al., 2009), including a ‘wedge’ in the middle of α3. Similarly, the membrane-binding properties of the Fer and srGAP2 F-BAR domains are quite different (Coutinho-Budd et al., 2012; Tsujita et al., 2006) – apparently resembling inverted BAR or IBAR domains (Guerrier et al., 2009) – and these lack the inositol phosphate binding site. Taken together, these considerations suggest that multiple headgroup binding sites (and possibly membrane insertion loops) along the length of the F-BAR domain contribute to membrane association and deformation. Superimposed upon this arrangement are more specific binding sites such as the inositol phosphate binding site seen in Rgd1p – which confers the unusual phosphoinositide dependence of this F-BAR domain.
Figure 6. Conservation of InsP6 binding site elements in other F-BAR domains.
(A) The S. cerevisiae and human F-BAR domains listed were aligned using PROMALS3D (Pei and Grishin, 2014), which includes evolutionary and structural information. Only helices α1 to α3 are shown, corresponding to the first half of the F-BAR domain, which contains the Rgd1p InsP6 binding site. Basic residues are colored blue. Positions of key residues in the Rgd1p F-BAR InsP6 binding site are depicted with red vertical arrows, labeled with the Rgd1p residue number – and shaded blue where they occur in Rgd1p or other F-BAR domains. Residues shaded light red correspond to residues implicated in binding of other F-BAR domains to anionic membranes by mutational studies. Where a basic residue is also present at one of these positions in Rgd1p, it is boxed. (B) Close-up view of the Rgd1p F-BAR (magenta) InsP6 binding site overlaid with the structure of the Gmip (GEM interacting protein – a Rho-GAP) F-BAR domain shown in cyan (PDB entry 3QWE). The two F-BAR domain structures are compared in Figure S5. The InsP6 binding site is almost completely conserved in Gmip F-BAR, with the exception of K149 (replaced by M200, but with K201 close by).
Intriguingly, one mammalian F-BAR domain appears to conserve almost all aspects of the Rgp1p inositol phosphate-binding site (Figure 6A) – that from Gem-interacting protein, or Gmip (Aresta et al., 2002). Gmip has not typically been included in lists of F-BAR proteins, but the structure of its amino-terminal domain was deposited in the PDB (entry 3QWE) by the Structural Genomics Consortium, and overlays very well (Cα rmsd of 3.1 Å) with Rgd1p F-BAR (Figure S5). More importantly, the Rgd1p InsP6-binding site is structurally very well conserved in the human Gmip F-BAR domain (Figure 6B). Given this unique resemblance among mammalian F-BAR domains to Rgd1p, it is intriguing that Gmip, like Rgd1p, is also a Rho-GAP, specific for RhoA (Aresta et al., 2002). This correspondence may signal an analogous functional dependence on phosphoinositides in the related functions of these yeast and mammalian Rho-GAPs that have been documented in the secretory pathway (Johnson et al., 2012; Lefèbvre et al., 2012). Gmip has also been reported to play important roles in cortical actin remodeling in early mitosis (Andrieu et al., 2014) and the speed of neuronal migration in the postnatal brain (Ota et al., 2014) – both processes in which phosphoinositides are likely to play an important regulatory role.
DISCUSSION
Despite the fact that F-BAR domains all appear to associate with negatively-charged membrane surfaces in a non-specific manner (Itoh and De Camilli, 2006), there are many studies suggesting that different F-BAR domains are functionally quite distinct (Arasada and Pollard, 2011; Qualmann et al., 2011; Roberts-Galbraith and Gould, 2010). The different shapes of the F-BAR dimers, exemplified in a comparison for Rgd1p and Pacsin1 (Figure 3), will undoubtedly contribute to these differences. It is also possible that some F-BAR domains have protein binding partners – with binding of arfaptin’s BAR domain to small GTPases (Tarricone et al., 2001) providing a precedent for this in the related N-BAR family. Although none of the F-BAR domains may have the high degree of phospholipid-binding specificity described for certain PH and other domains (Lemmon, 2008; Moravcevic et al., 2012), it also seems highly likely that the different lipid specificity profiles of individual F-BAR domains will be important in defining their precise function (Tanaka-Takiguchi et al., 2013; Zhao et al., 2013). Different lipid specificity profiles may alter the locations to which F-BAR proteins are recruited. Alternatively, membrane-associated F-BAR domains may laterally recruit (and possibly cluster) acidic phospholipids such as PtdIns(4,5)P2 and thus stabilize the formation of lipid microdomains or rafts with potentially important functional consequences. As recently suggested for the BAR domain superfamily in general (Tanaka-Takiguchi et al., 2013; Zhao et al., 2013), the distinct lipid selectivities of F-BAR domains may profoundly alter the nature of the lipid microdomains that they stabilize.
Our studies with the S. cerevisiae Rgd1p, Hof1p, and Bzz1p F-BAR domains exemplify such differences. Rgd1p F-BAR showed clear selectivity for phosphoinositides, also manifested for the intact protein in cellular studies (Prouzet-Mauleon et al., 2008) – whereas Hof1p and Bzz1p did not. We were not able to crystallize the Bzz1p F-BAR domain, so our subsequent analysis focused on Rgd1p and Hof1p. The two F-BAR domains appear structurally quite similar, despite low (17%) sequence identity. The sensitivity of Rgd1p F-BAR behavior to phosphoinositides is satisfyingly explained by the inositol phosphate binding site revealed in our structure – the first crystallographic view of ligand binding to an F-BAR domain. A similar site appears to be used in a subset of mammalian F-BAR domains (such as FBP17 and CIP4), but apparently with less selectivity for phosphoinositides (Henne et al., 2007; Shimada et al., 2007; Shimada et al., 2010; Tsujita et al., 2006). Still others appear to have limited interaction with phosphoinositides and show quite distinct membrane tubulation behavior in cells (Coutinho-Budd et al., 2012; Tsujita et al., 2006). It seems likely that these F-BAR domains interact with a different combination of membrane lipids than Rgd1p. Recent studies with the Hof1p F-BAR domain further suggest possible protein interactions. The Hof1p N-terminal F-BAR domain appears to direct interactions of the protein with septin complexes both in vivo and in vitro (Meitinger et al., 2011; Oh et al., 2013), which are responsible for the temporally controlled recruitment of Hof1p to the bud neck. The Hof1p F-BAR domain also appears to bind to a chitin synthase that is required for primary septum formation (Oh et al., 2013). It seems reasonable to suggest that these interactions cooperate with lipid binding in directing Hof1p to the appropriate location at the appropriate time through a coincidence detection mechanism (Lemmon, 2008; Moravcevic et al., 2012). We suggest that the inositol phosphate binding site in the Rgd1p F-BAR domain takes the place of one or more of the (presumably specific) sites for protein binding on the Hof1p F-BAR domain.
Since their initial discovery, F-BAR domains and other members of this broader superfamily have been shown both to associate with membranes and to induce their curvature in key cellular processes. As structural and functional information on these domains has accumulated, it has become increasingly clear that F-BAR domains are quite complex integrators. They must be recruited to the correct membranes in the cell and must also exert the appropriate specific effects – be it stabilizing precise geometry, specific lipid (or protein) composition, or other influences. The binding site for phosphoinositide headgroups seen here for Rgd1p F-BAR is likely to be just one example of a relatively specific site in these domains. It is used in an altered guise in several other F-BAR domains involved in endocytic processes, but is unexpectedly conserved in a mammalian Rho-GAP named Gmip that may have functional parallels to yeast Rgd1p. Future analysis – and integration – of individual binding sites in these complex domains will be required to fully appreciate their range of functions and dysfunction in disease.
EXPERIMENTAL PROCEDURES
Ras rescue assays
Ras rescue assays were performed exactly as described (Yu et al., 2004). Briefly, DNA encoding the noted F-BAR proteins (or respective fragments) was subcloned into the modified p3S0BL2 vector (Isakoff et al., 1998) to generate a plasmid encoding a Ha-Ras Q61L fusion. This plasmid was transformed into cdc25ts yeast cells, and its ability to rescue the growth defect at 37°C was assessed as described (Isakoff et al., 1998).
Microscopy
For analysis of protein localization in yeast, DNA fragments encoding F-BAR domains were subcloned into a modified pGO-GFP vector (Cowles et al., 1997) to generate green fluorescent protein (GFP) fusions, and were transformed into wild-type (BY4741) yeast cells as described (Audhya and Emr, 2002). To assess effects of altered phosphoinositide metabolism, mss4ts-AA107 and stt4ts/pik1ts-AA105 mutant yeast strains were employed (Audhya et al., 2000). To analyze subcellular localization in mammalian cells, F-BAR domains were subcloned into a modified pEGFP-C1 vector (Clontech) to generate the appropriate GFP fusion proteins. HeLa cells were then transiently transfected using Lipofectamine 2000 (Invitrogen). Images of GFP localization in cells were collected at 100X and 40X magnification for yeast and mammalian cells respectively, using a Leica model DMIRBE microscope and images were processed using Volocity deconvolution software (Improvision).
Vesicle sedimentation binding studies and surface plasmon resonance (SPR)
For in vitro binding studies, the F-BAR domains from Rgd1p (residues 1-324), Bzz1p (residues 1-350) and Hof1p (residues 1-300) were expressed in E. coli BL21 (DE3) with an N-terminal hexahistidine tag. Rgd1p and Hof1p F-BAR domains were purified by Ni-NTA affinity chromatography, cation exchange, and gel filtration, and the Bzz1p F-BAR domain by Ni-NTA affinity chromatography and gel filtration only. Vesicle sedimentation binding assays were performed as described (Kavran et al., 1998). F-BAR proteins at 10 μM were mixed with small unilamellar vesicles (SUVs) of defined lipid composition containing 80-100% brominated phosphatidylcholine to increase vesicle density (Tortorella and London, 1994). Vesicles were added at different [Total Lipid]Available concentrations, assuming that 50% of the lipid is accessible on the SUV outer leaflet. Mixtures were centrifuged for 1 h at 80,000 rpm in a Beckman Optima TLX benchtop ultracentrifuge at 25°C, and the percentage of F-BAR protein sedimenting in the pellet was evaluated using the BCA assay (Pierce). Data were fit as described (Kavran et al., 1998).
Surface plasmon resonance (SPR) studies were performed as described (Yu et al., 2004). Vesicles containing dioleoylphosphatidylcholine (DOPC) alone or including the noted percent (mole/mole) of dioleoyl-PtdIns(4,5)P2 or PtdSer were immobilized on L1 sensorchips (BIAcore). Purified test proteins were then flowed over these surfaces at noted concentrations (determined by absorbance at 280 nm). SPR signals were corrected for background (DOPC) binding, and data were analyzed using GraphPad Prism.
Protein preparation, crystallization and data collection
DNA encoding the F-BAR domains from Rgd1p (residues 24-333) and Hof1p (residues 1-300), plus N-terminal hexahistidine tags, was subcloned into pET21a (Novagen) for expression in E. coli BL21 (DE3) cells in LB. For generating selenomethionine (SeMet)-containing protein, the Hof1p F-BAR domain was produced from B834 (DE3) methionine auxotrophs in MOPS-based minimal medium (Neidhardt et al., 1974) supplemented with SeMet. Proteins were purified from cell lysates in three steps, using Ni-NTA resin (Qiagen), cation exchange chromatography, and a Superdex 200 size exclusion column (GE Healthcare). Crystals were grown at 21°C using the hanging drop vapor diffusion method by mixing equal parts of protein (at 300-500 μM) and reservoir solutions. Hof1p F-BAR crystals were obtained from 0.1 M Na citrate, pH 5.5, containing 0.1 M ammonium acetate, and 5-7% (w/v) PEG3350. Rgd1p F-BAR crystals were obtained from 0.1 M citrate, pH 5.5, containing 0.1-0.2 M (NH4)2SO4 plus 10-20% (w/v) PEG3350. Single Rgd1p F-BAR crystals were obtained by micro-seeding into drops with 2-4% lower PEG3350 concentrations and were soaked in reservoir solution with 5 mM InsP6 for 12 h. Crystals were cryo-protected by direct transfer into reservoir solution containing 20% (w/v) glycerol, and were flash frozen in liquid nitrogen. Data were collected at 100K at the Advanced Photon Source (Argonne, IL) beamlines 23ID-D and 23ID-B (for Hof1p), or the Cornell High Energy Synchrotron Source (CHESS) beamline F1 (microbeam) and were processed using HKL2000 (Otwinowski and Minor, 1997). The Rgd1p F-BAR dataset was collected at a wavelength of 0.91790 Å, and Hof1p F-BAR datasets were collected at wavelengths of 0.97949 Å (peak), 0.97965 Å (inflection) and 0.94949 Å (remote).
Structure determination and refinement
Experimental phase information was obtained for Hof1p F-BAR using data collected from the SeMet-containing crystals, with multi-wavelength anomalous diffraction (MAD) methods implemented in the program SHELX C/D/E (Schneider and Sheldrick, 2002). The 8 Se sites found with SHELX were then refined with SOLVE/RESOLVE (Terwilliger, 2000; Terwilliger and Berendzen, 1999). The resulting experimentally phased map was excellent (Figure S1A), and allowed amino acids 2-274 to be traced using the program Coot (Emsley and Cowtan, 2004). For Rgd1p F-BAR domain, the structure was solved by molecular replacement (MR) using Phaser (CCP4, 1994), with a search model based on the Hof1 F-BAR domain structure that retained the core three-helix bundle mutated to poly-ala. For both structures, cycles of manual building/rebuilding using Coot were alternated with rounds of refinement employing REFMAC (CCP4, 1994) and solvent flattening with the program DM (CCP4, 1994), plus composite omit maps calculated with CNS (Brünger et al., 1998). The density for the InsP6 molecule (Figure 4C) suggested that it does not have a highly preferred binding orientation, consistent with the lack of stereospecificity in binding. To avoid distortion of the geometry arising from multiple poses, we refined the InsP6 molecule as a rigid body. TLS refinement (Winn et al., 2001) was employed in REFMAC. PHENIX (Adams et al., 2010) was used for later stages of refinement, and for final model validation. In the final Hof1p and Rgd1p F-BAR models respectively, 0.37% and 0.72% of residues are Ramachandran outliers. Data collection and refinement statistics are presented in Table 1. Structure figures were generated using PyMol (http://www.pymol.org/).
Supplementary Material
HIGHLIGHTS.
F-BAR domains differ in their phospholipid-binding specificity
The S. cerevisiae Rgd1p F-BAR domain selectively binds phosphoinositides
Crystallography identifies a phosphoinositide-binding site in Rgd1p F-BAR domain
This site is partly conserved in other F-BAR domains, but fully in a human RhoGAP
ACKNOWLEDGMENTS
We thank members of the Lemmon laboratory for constructive comments; and Erfei Bi, Scott Emr and Daryll DeWald for yeast strains used in this study. Crystallographic results were obtained in research conducted in part at the GM/CA Collaborative Access Team at the Advanced Photon Source (APS), funded in whole or in part with funds from the National Cancer Institute (Y1-CO-1020) and National Institute of General Medical Science (Y1-GM-1104). Use of APS was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38.
Additional crystallographic data were collected at beamline F1 at the Cornell High Energy Synchrotron Source (CHESS), supported by the NSF and the NIH/NIGMS under NSF award DMR-0225180, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which was supported by award RR-01646 from the NIH, through its National Center for Research Resources. This work was funded in part by NIH grant R01-GM056846 (to M.A.L.), a Postdoctoral Fellowship from the Jane Coffin Childs Memorial Fund for Medical Research (Fund Project No. 61-1316 to J.A.K.), NIH Training Grant in Structural Biology (T32-GM008275 to K.R.S.), and a predoctoral fellowship from the American Heart Association Great Rivers Affiliate (K.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
ACCESSION CODES
Coordinates and structure factors for the two structures have been deposited in the PDB under accession codes 4WPC (Rgd1p F-BAR/InsP6 complex) and 4WPE (Hof1p F-BAR domain).
AUTHOR CONTRIBUTIONS
K.M. and M.A.L. conceived and designed the project, which was supervised by M.A.L. K.M. was responsible for the execution of most experiments, together with J.M.M. in microscopy studies and yeast studies. K.M. performed all cloning, protein purification, binding analysis, crystallization, and crystallographic model building. K.M., D.A., K.R.S., J.A.K., and K.M.F. all contributed to data processing, structure determination, and structural analysis; K.M. and M.A.L. wrote the manuscript. All authors read and approved the manuscript.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrieu G, Quaranta M, Leprince C, Cuvillier O, Hatzoglou A. Gem GTPase acts upstream Gmip/RhoA to regulate cortical actin remodeling and spindle positioning during early mitosis. Carcinogenesis. 2014:pii: bgu185. doi: 10.1093/carcin/bgu185. [DOI] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr. Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aresta S, de Tand-Heim MF, Béranger F, de Gunzburg J. A novel Rho GTPase-activating-protein interacts with Gem, a member of the Ras superfamily of GTPases. Biochem. J. 2002;367:57–65. doi: 10.1042/BJ20020829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chitu V, Stanley ER. Pombe Cdc15 homology (PCH) proteins: coordinators of membrane-cytoskeletal interactions. Trends Cell Biol. 2007;17:145–156. doi: 10.1016/j.tcb.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Coutinho-Budd J, Ghukasyan V, Zylka MJ, Polleux F. The F-BAR domains from srGAP1, srGAP2 and srGAP3 regulate membrane deformation differently. J. Cell Sci. 2012;125:3390–3401. doi: 10.1242/jcs.098962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- Doignon F, Weinachter C, Roumanie O, Crouzet M. The yeast Rgd1p is a GTPase activating protein of the Rho3 and rho4 proteins. FEBS Letts. 1999;459:458–462. doi: 10.1016/s0014-5793(99)01293-4. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidarov I, Chen Q, Falck JR, Reddy KK, Keen JH. A functional phosphatidylinositol 3,4,5-trisphosphate/phosphoinositide binding domain in the clathrin adaptor AP-2 alpha subunit. J. Biol. Chem. 1996;271:20922–20929. doi: 10.1074/jbc.271.34.20922. [DOI] [PubMed] [Google Scholar]
- Gaidarov I, Krupnick JG, Falck JR, Benovic JL, Keen JH. Arrestin function in G protein-coupled receptor endocytosis requires phosphoinositide binding. EMBO J. 1999;18:871–881. doi: 10.1093/emboj/18.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, McMahon HT. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Kent HM, Ford MG, Hegde BG, Daumke O, Butler PJ, Mittal R, Langen R, Evans PR, McMahon HT. Structure and analysis of FCHo2 F-BAR domain: A dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15:839–852. doi: 10.1016/j.str.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nuc. Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. Membrane binding domains. Biochim. Biophys. Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvönen M, Macias MJ, Nilges M, Oschkinat H, Saraste M, Wilmanns M. Structure of the binding site for inositol phosphates in a PH domain. EMBO J. 1995;14:4676–4685. doi: 10.1002/j.1460-2075.1995.tb00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icking A, Schilling K, Wiesenthal A, Opitz N, Müller-Esterl W. FCH/Cdc15 domain determines distinct subcellular localization of NOSTRIN. FEBS Letts. 2006;580:223–228. doi: 10.1016/j.febslet.2005.11.078. [DOI] [PubMed] [Google Scholar]
- Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, Lemmon MA, Aronheim A, Skolnik EY. Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 1998;17:5374–5387. doi: 10.1093/emboj/17.18.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim. Biophys. Acta. 2006;1761:897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Monfregola J, Napolitano G, Kiosses WB, Catz SD. Vesicular trafficking through cortical actin during exocytosis is regulated by the Rab27a effector JFC1/Slp1 and the RhoA-GTPase-activating protein Gem-interacting protein. Mol. Biol. Cell. 2012;23:1902–1916. doi: 10.1091/mbc.E11-12-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- Lefèbvre F, Prouzet-Mauléon V, Hugues M, Crouzet M, Vieillemard A, McCusker D, Thoraval D, Doignon F. Secretory pathway-dependent localization of the Saccharomyces cerevisiae Rho GTPase-activating protein Rgd1p at growth sites. Eukaryot. Cell. 2012;11:590–600. doi: 10.1128/EC.00042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell. Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lippincott J, Li R. Involvement of PCH family proteins in cytokinesis and actin distribution. Microsc. Res. Tech. 2000;49:168–172. doi: 10.1002/(SICI)1097-0029(20000415)49:2<168::AID-JEMT9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- McPherson VA, Everingham S, Karisch R, Smith JA, Udell CM, Zheng J, Jia Z, Craig AW. Contributions of F-BAR and SH2 domains of Fes protein tyrosine kinase for coupling to the FcepsilonRI pathway in mast cells. Mol. Cell. Biol. 2009;29:389–401. doi: 10.1128/MCB.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 2011;25:875–888. doi: 10.1101/gad.622411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mim C, Unger VM. Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci. 2012;37:526–533. doi: 10.1016/j.tibs.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA, Lemmon MA. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: Facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R, Schreiter JH, Onishi M, Vallen EA, Hanna J, Moravcevic K, Lippincott MF, Han H, Lemmon MA, Pringle JR, Bi E. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J. Cell Biol. 2009;185:995–1012. doi: 10.1083/jcb.200903125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Schreiter J, Nishihama R, Wloka C, Bi E. Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol. Biol. Cell. 2013;24:1305–1320. doi: 10.1091/mbc.E12-11-0804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H, Hikita T, Sawada M, Nishioka T, Matsumoto M, Komura M, Ohno A, Kamiya Y, Miyamoto T, Asai N, et al. Speed control for neuronal migration in the postnatal brain by Gmip-mediated local inactivation of RhoA. Nat. Comm. 2014;5:4532. doi: 10.1038/ncomms5532. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol. Biol. 2014;1079:263–271. doi: 10.1007/978-1-62703-646-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitzsch RM, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Prouzet-Mauleon V, Lefebvre F, Thoraval D, Crouzet M, Doignon F. Phosphoinositides affect both the cellular distribution and activity of the F-BAR-containing RhoGAP Rgd1p in yeast. J. Biol. Chem. 2008;283:33249–33257. doi: 10.1074/jbc.M805161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Koch D, Kessels MM. Let’s go bananas: revisiting the endocytic BAR code. EMBO J. 2011;30:3501–3515. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Báez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Gould KL. Setting the F-BAR: functions and regulation of the F-BAR protein family. Cell Cycle. 2010;9:4091–4097. doi: 10.4161/cc.9.20.13587. [DOI] [PubMed] [Google Scholar]
- Roumanie O, Peypouquet MF, Bonneu M, Thoraval D, Doignon F, Crouzet M. Evidence for the genetic interaction between the actin-binding protein Vrp1 and the RhoGAP Rgd1 mediated through Rho3p and Rho4p in Saccharomyces cerevisiae. Mol. Microbiol. 2000;36:1403–1414. doi: 10.1046/j.1365-2958.2000.01958.x. [DOI] [PubMed] [Google Scholar]
- Roumanie O, Weinachter C, Larrieu I, Crouzet M, Doignon F. Functional characterization of the Bag7, Lrg1 and Rgd2 RhoGAP proteins from Saccharomyces cerevisiae. FEBS Letts. 2001;506:149–156. doi: 10.1016/s0014-5793(01)02906-4. [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Shimada A, Takano K, Shirouzu M, Hanawa-Suetsugu K, Terada T, Toyooka K, Umehara T, Yamamoto M, Yokoyama S, Suetsugu S. Mapping of the basic amino-acid residues responsible for tubulation and cellular protrusion by the EFC/F-BAR domain of pacsin2/Syndapin II. FEBS Letts. 2010;584:1111–1118. doi: 10.1016/j.febslet.2010.02.058. [DOI] [PubMed] [Google Scholar]
- Soulard A, Lechler T, Spiridonov V, Shevchenko A, Shevchenko A, Li R, Winsor B. Saccharomyces cerevisiae Bzz1p is implicated with type I myosins in actin patch polarization and is able to recruit actin-polymerizing machinery in vitro. Mol. Cell. Biol. 2002;22:7889–7906. doi: 10.1128/MCB.22.22.7889-7906.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Audhya A, Emr SD. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Takiguchi Y, Itoh T, Tsujita K, Yamada S, Yanagisawa M, Fujiwara K, Yamamoto A, Ichikawa M, Takiguchi K. Physicochemical analysis from real-time imaging of liposome tubulation reveals the characteristics of individual F-BAR domain proteins. Langmuir. 2013;29:328–336. doi: 10.1021/la303902q. [DOI] [PubMed] [Google Scholar]
- Tarricone C, Xiao B, Justin N, Walker PA, Rittinger K, Gamblin SJ, Smerdon SJ. The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature. 2001;411:215–219. doi: 10.1038/35075620. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella D, London E. Method for efficient pelleting of small unilamellar model membrane vesicles. Anal. Biochem. 1994;217:176–180. doi: 10.1006/abio.1994.1106. [DOI] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Navarro MV, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12700–12705. doi: 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- Zhao H, Michelot A, Koskela EV, Tkach V, Stamou D, Drubin DG, Lappalainen P. Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep. 2013;4:1213–1223. doi: 10.1016/j.celrep.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.