ColoPrint is a gene expression classifier that distinguishes patients with low or high risk of disease relapse. Results of this study showed that this classifier significantly improved prognostic accuracy, independent of microsatellite status or clinical variables, to facilitate the identification of patients with stage II colorectal cancer at higher risk of recurrence who might be considered for additional treatment.
Keywords: Stage II colon cancer, Risk prediction, Gene expression signature, Risk classification
Abstract
Background.
Approximately 20% of patients with stage II colorectal cancer will experience a relapse. Current clinical-pathologic stratification factors do not allow clear identification of these high-risk patients. ColoPrint (Agendia, Amsterdam, The Netherlands, http://www.agendia.com) is a gene expression classifier that distinguishes patients with low or high risk of disease relapse.
Methods.
ColoPrint was developed using whole-genome expression data and validated in several independent validation cohorts. Stage II patients from these studies were pooled (n = 416), and ColoPrint was compared with clinical risk factors described in the National Comprehensive Cancer Network (NCCN) 2013 Guidelines for Colon Cancer. Median follow-up was 81 months. Most patients (70%) did not receive adjuvant chemotherapy. Risk of relapse (ROR) was defined as survival until first event of recurrence or death from cancer.
Results.
In the pooled stage II data set, ColoPrint identified 63% of patients as low risk with a 5-year ROR of 10%, whereas high-risk patients (37%) had a 5-year ROR of 21%, with a hazard ratio (HR) of 2.16 (p = .004). This remained significant in a multivariate model that included number of lymph nodes retrieved and microsatellite instability. In the T3 microsatellite-stable subgroup (n = 301), ColoPrint classified 59% of patients as low risk with a 5-year ROR of 9.9%. High-risk patients (31%) had a 22.4% ROR (HR: 2.41; p = .005). In contrast, the NCCN clinical high-risk factors were unable to distinguish high- and low-risk patients (15% vs. 13% ROR; p = .55).
Conclusion.
ColoPrint significantly improved prognostic accuracy independent of microsatellite status or clinical variables, facilitating the identification of patients at higher risk who might be considered for additional treatment.
Implications for Practice:
Patients with stage II colon cancer have a low rate of recurrence after surgery. A modest benefit may be derived from chemotherapy after surgery, but this approach requires treating many patients who are already cured and who do not need treatment. A gene expression signature, ColoPrint, is able to distinguish patients with the highest risk of recurrence, who may derive greater benefit from further chemotherapy, and to reassure doctors and patients about the overall excellent prognosis of patients with a low-risk ColoPrint result. This study confirmed the performance of this test in a large cohort of patients across many different centers. This test is currently available commercially for patient testing.
Introduction
Colorectal cancer is the third most common cancer and a leading cause of cancer death worldwide [1, 2]. If this cancer is detected early, many patients can be cured by surgery. Patients with stage II colorectal cancer have an ∼80% chance to stay disease-free even without adjuvant therapy [3]. The challenge is to identify those ∼20% of patients in whom the disease will recur locally or at distant sites. The early identification of these high-risk patients would allow more informed discussions about the risks and benefits of adjuvant therapy and, potentially, more risk-adapted surveillance plans [4]. Much effort has been put into identifying reliable risk factors to assess the individual risk of stage II patients. A number of poor prognostic clinical factors have been identified—low number of assessed lymph nodes (<12); T4 tumors; obstruction or perforation at diagnosis; vascular, lymphatic, or perineural invasion; poor histological differentiation (high grade); and the presence of positive resection margins [5, 6]—but no clinical marker has been shown to improve the selection of patients who benefit from treatment [7]. High microsatellite instability (MSI-H) is the most reliable factor used in the clinic to identify patients who have a good prognosis and who may derive no benefit from adjuvant therapy [8, 9]. Only ∼15% of patients are MSI-H, leaving the majority of patients with indeterminate risk.
Multi-index assays like gene expression profiling or the combination of multiple factors and technologies are likely to provide more robust information regarding individual prognosis. The successful development and wide clinical use of such tests for breast cancer patients show that this approach might provide more information than other individual clinical or molecular factors [10, 11]. Similar gene expression profiles have been developed for colorectal cancer in the past 10 years [12–17], but only a few profiles have been validated in independent studies [18] and have been shown to be technically robust. ColoPrint (Agendia, Amsterdam, The Netherlands, http://www.agendia.com) is an 18-gene expression signature that was developed based on unbiased gene selection, searching the whole genome for genes that have the highest correlation to a tumor-relapse event [19]. The signature was translated into a diagnostic test, validated in three independent studies [19–21], and shown to be technically reproducible and robust [20]. In this study, we described the pooled analysis of all stage II patients from the independent cohorts of patients treated in the U.S., Spain, Germany, Italy, and Austria.
Methods
Patients and Tumor Samples
Frozen tumor samples from patients with stage II colorectal cancer who underwent curative tumor resection were collected prospectively between 1987 and 2009 at six institutes in Spain, Germany, Austria, Italy, and the U.S. (supplemental online Table 1).
Clinical and histopathological data of all patients were collected. All patients were staged according to the seventh edition of the Union for International Cancer Control and American Joint Committee on Cancer tumor staging system [22]. The performance of the ColoPrint classifier has been described previously for several of the cohorts, but they were individually underpowered to detect the performance of the classifier in the clinically relevant stage II population and thus were used for the pooled analysis. The median follow-up time of patients was 81 months (range: 56–178 months).
Gene Expression Analysis
The 18-gene ColoPrint gene expression profile was assessed at Agendia’s laboratories (ISO17025 certified and Clinical Laboratory Improvement Amendments accredited) based on a previously established signature. None of the data sets included in this analysis were part of this initial discovery set. The assessment was made by laboratory staff who were blinded to clinical data, as described previously [19, 20]. Briefly, frozen sections were stained with hematoxylin and eosin; only samples that contained at least 30% tumor cells, as reviewed by a central certified pathologist, were used for RNA isolation. RNAs of adequate quality were amplified, labeled, and hybridized to the custom-designed ColoPrint microarray (Agilent Technologies, Santa Clara, CA, http://www.agilent.com). Approximately 10% of samples submitted for analysis had insufficient tumor cellularity or inadequate RNA quality and were not able to be analyzed. The correlation of the sample expression profile to a template (the mean expression profile of tumors with a known clinical outcome) was calculated (ColoPrint index), and the molecular profile of the sample was determined and categorized as either low risk or high risk [19, 20].
Microsatellite Instability Analysis
MSI status for most patients (n = 306) was determined at local hospitals using their preferred methods. MSI status for patients from the Institut Català d’Oncologia hospital (Barcelona, Spain) was determined by polymerase chain reaction (PCR) amplification of six microsatellite DNA regions (D21S415, D21S1235, D12S95, D4S2948, SIT2, BAT26) or five microsatellite DNA regions (BAT-25, BAT-26, NR-21, NR-24, Mono-27) from paired normal and tumor tissues (MSI analysis system, version 1.2; Promega, Madison, WI, https://www.promega.com). A tumor with only normal markers was defined as microsatellite stable (MSS).
For all patients from Munich, Germany, microsatellite instability was determined using the Qiagen Type-it Microsatellite PCR Kit (Qiagen, Hilden, Germany, http://www.qiagen.com). Two mononucleotide and three dinucleotide Bethesda markers (BAT25, BAT26, D2S123, D5S346, D17S250) were investigated. A tumor with five normal markers was defined as MSS. Irregularity in one marker was defined as low-grade microsatellite instability and as MSS in our binary analysis. MSI status in the patients at MD Anderson Cancer Center (Houston, TX) was determined by immunohistochemistry for MHS2, MSH6, MLH1, and PMS2, with loss of any one protein defining MSI-H. For all patients who had no MSI status determined locally, we used a previously described MSI signature for classification that identifies patients with MSI with high sensitivity and specificity [23].
Statistical Analyses
The primary endpoint was risk of relapse (ROR), which was defined as the probability that patients developed a recurrence (locoregional or metastatic) or did not die of cancer as the first event; data on all other patients were censored on the date of the last follow-up visit or date of death. Deaths not attributed to cancer were censored to evaluate true prognostic prediction. A secondary analysis of disease-free survival was performed, and all deaths were coded as events. Data were analyzed from the date of surgery to the time of the first event or the date on which data were censored, according to the Kaplan-Meier method, and the curves were compared with the log-rank test.
In order to determine the independence of ColoPrint from clinicopathological variables in predicting an individual’s risk of relapse, we used univariate and multivariate analysis. Variables included sex; localization of the tumor; T stage; number of lymph nodes assessed; histological grade; adjuvant chemotherapy administration; MSI status; ColoPrint and the combined National Comprehensive Cancer Network (NCCN) high-risk score, including T4, high grade, lymphovascular or perineural invasion, perforation or obstruction, <12 nodes examined, and positive margins, in which any one positive factor represented a high-risk NCCN score (NCCN guidelines version 3.2013) [24]. Variables with p < .1 in the univariate analysis were entered into the multivariate model with forward entry at p < .05.
Log-rank tests were used in the univariate analysis, and a multivariate Cox model was built. Given prior distribution of high- and low-risk scores and a median follow-up of 81 months, a sample size of 400 patients in this pooled analysis was sufficient to detect a clinically meaningful hazard ratio (HR) of 1.6 for the high-risk cohort, with 90% power and two-sided α of 0.05. All calculations were performed with SPSS version 16.0 (IBM Corp, Armonk, NY, http://www-01.ibm.com/software/analytics/spss/).
Results
Patient Characteristics
The clinical characteristics of the 416 patients (median age: 67 years [range: 33–94 years]) are shown in Table 1. Patients in this pooled analysis were treated at five different hospitals in Europe and one hospital in the U.S. Patient characteristics varied across the studies in age and grade (supplemental online Table 1). Importantly, all patients had large numbers of lymph nodes assessed (median: 19 [range, among hospitals: 13–21.5]), indirectly indicating a high quality of surgery and pathology at all hospitals. A total of 124 patients (29.8%) received adjuvant treatment, with fewer patients treated in Munich than at the other hospitals; this can be partially explained by the fact that these patients were diagnosed earlier in time. Adjuvant therapy was administered as 5-fluorouracil (5-FU) in 94% or 5-FU and oxaliplatin in 6%.
Table 1.
Patient characteristics and ColoPrint results
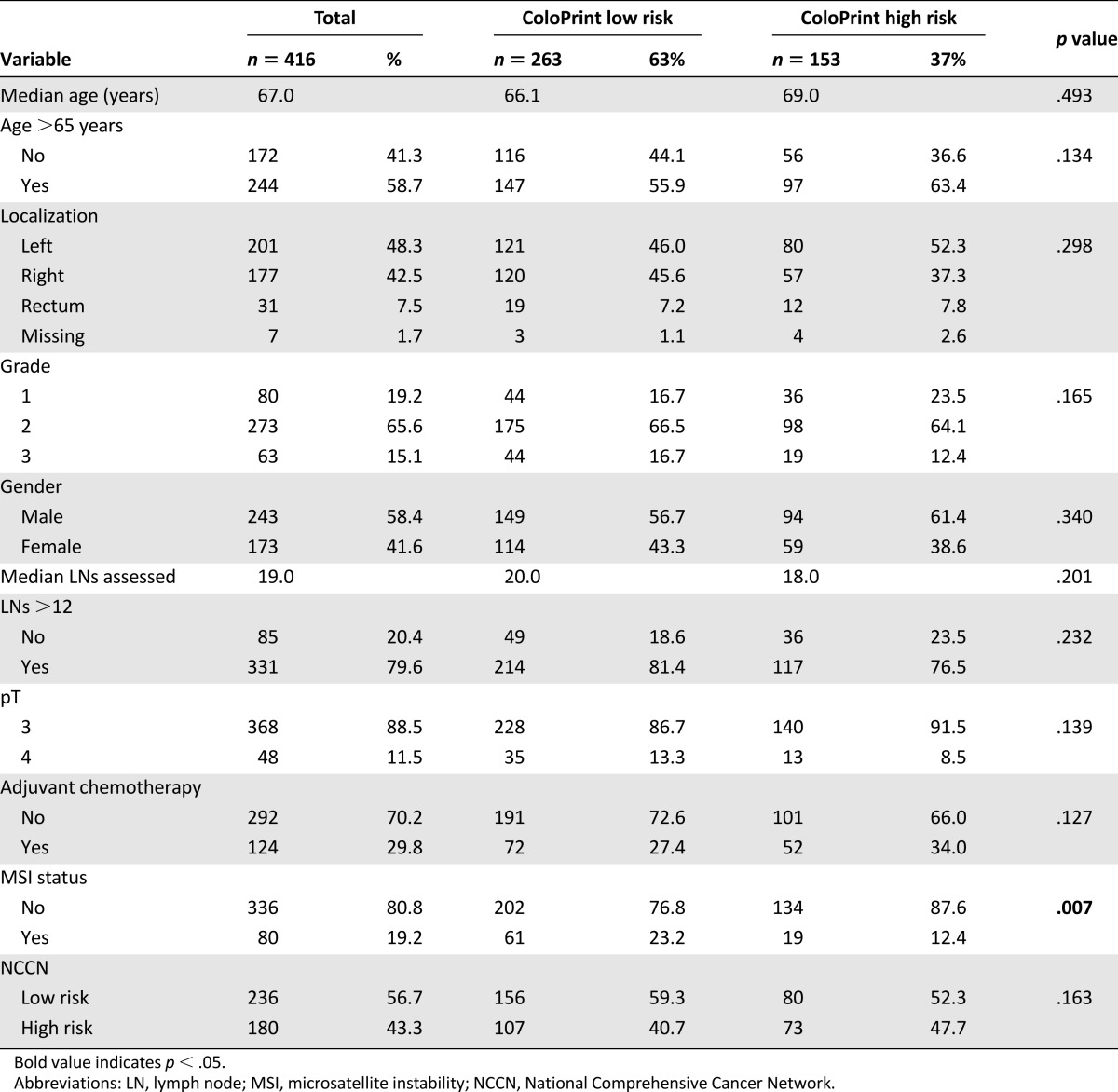
ColoPrint classified 263 patients (63%) as low risk and 153 (37%) as high risk. ColoPrint assessment was not correlated to age, sex, localization of tumor, grade, number of assessed lymph nodes, or T stage. ColoPrint classification also correlated with MSI status because most, but not all, MSI-H patients were classified as low risk (76.8%).
Risk Assessment by ColoPrint and NCCN
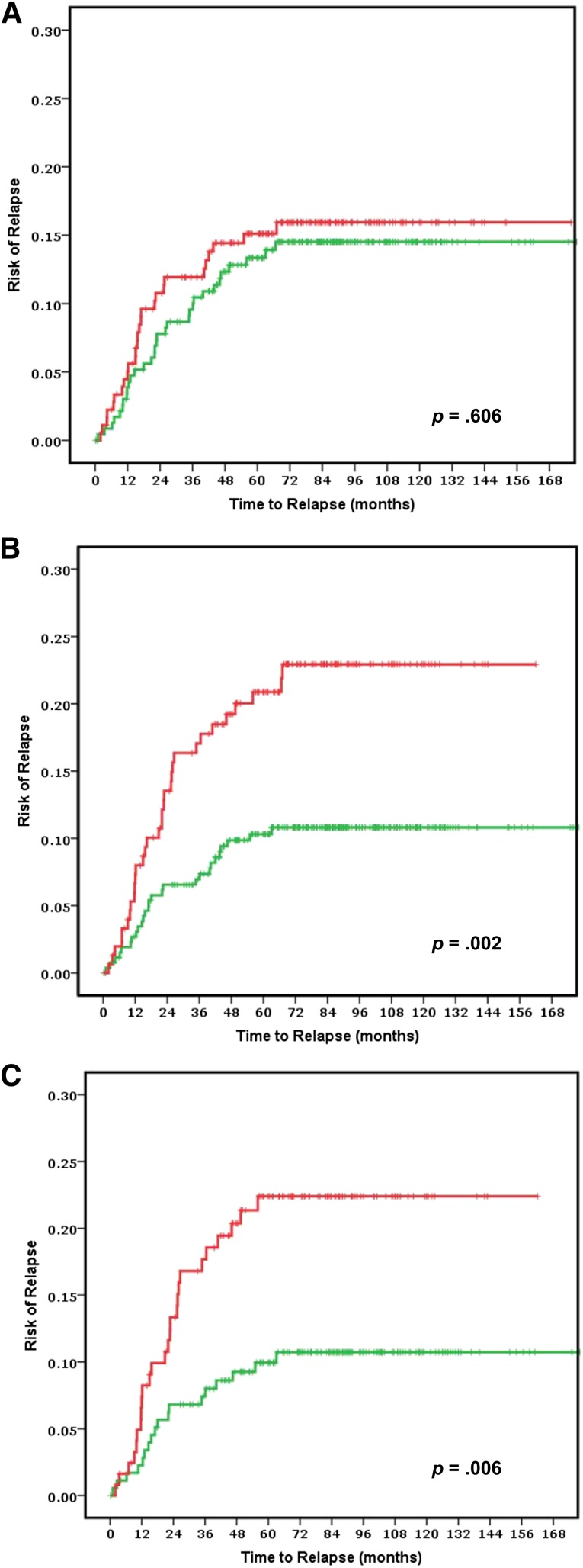
The risk of relapse curves show that ColoPrint high-risk patients had a 5-year ROR of 20.9% (95% confidence interval [CI]: 14.2%–27.6%), whereas patients with a low-risk ColoPrint result had a 5-year ROR of 10.3% (95% CI: 6.6%–14%; p = .004) (Fig. 1). This corresponds to a 2.16-fold higher HR for recurrence in the univariate analysis (95% CI: 1.28–3.65; p = .004). Similarly, the 3-year ROR is 7% for ColoPrint low-risk patients and 17.1% for ColoPrint high-risk patients (HR: 2.55; 95% CI: 1.39–4.68; p = .002).
Figure 1.
Clinical assessment using National Comprehensive Cancer Network guidelines to separate low (green line) and high (red line) risk groups in all stage II patients (n = 416) (A) and ColoPrint assessment in all stage II patients (n = 416) (B) and in the T3-Microsatellite-stable subgroup (n = 301) (C). Results for patients with rectal cancer are shown in supplemental online Figure 1.
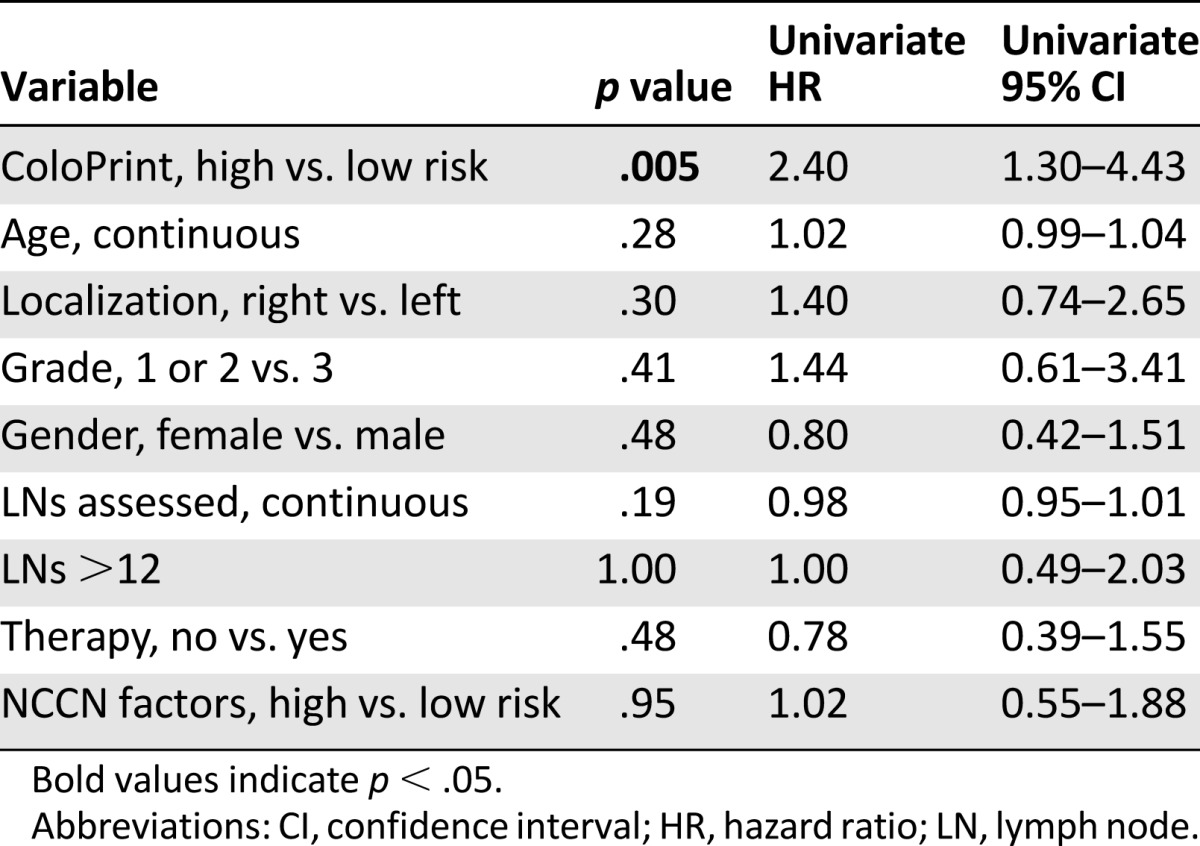
Of the clinical variables, only number of lymph nodes as a continuous variable reached near significance for 5-year ROR (HR: 0.97; p = .053). The MSI status was prognostic with patients with MSI-H having a better prognosis (HR: 0.39; p = .046). Age, localization, grade, sex, T stage (3 vs. 4), therapy, and NCCN risk were not significantly correlated with the time to recurrence (Table 2). The discordance between clinical risk classification using NCCN guidelines and ColoPrint was 45%, in part because ColoPrint identified many patients with T4, high grade, or low number of assessed lymph nodes as low risk (Table 3). Patients classified as clinically high risk using the NCCN guidelines had a 5-year ROR of 15.1% (95% CI: 9.8%–20.4%) compared with 13.3% for NCCN low-risk patients (95% CI: 8.8%–17.8%; p = .55). This clinical risk score was not significant in the univariate analysis (HR: 1.17; p = .6). A multivariate model was constructed with the number of assessed lymph nodes, MSI status, and ColoPrint classification. The number of assessed lymph nodes and MSI status lost their significance in the multivariate analysis, and only ColoPrint remained prognostic (HR: 2.16; p = .004) (Table 2).
Table 2.
Univariate and multivariate analysis of risk of recurrence
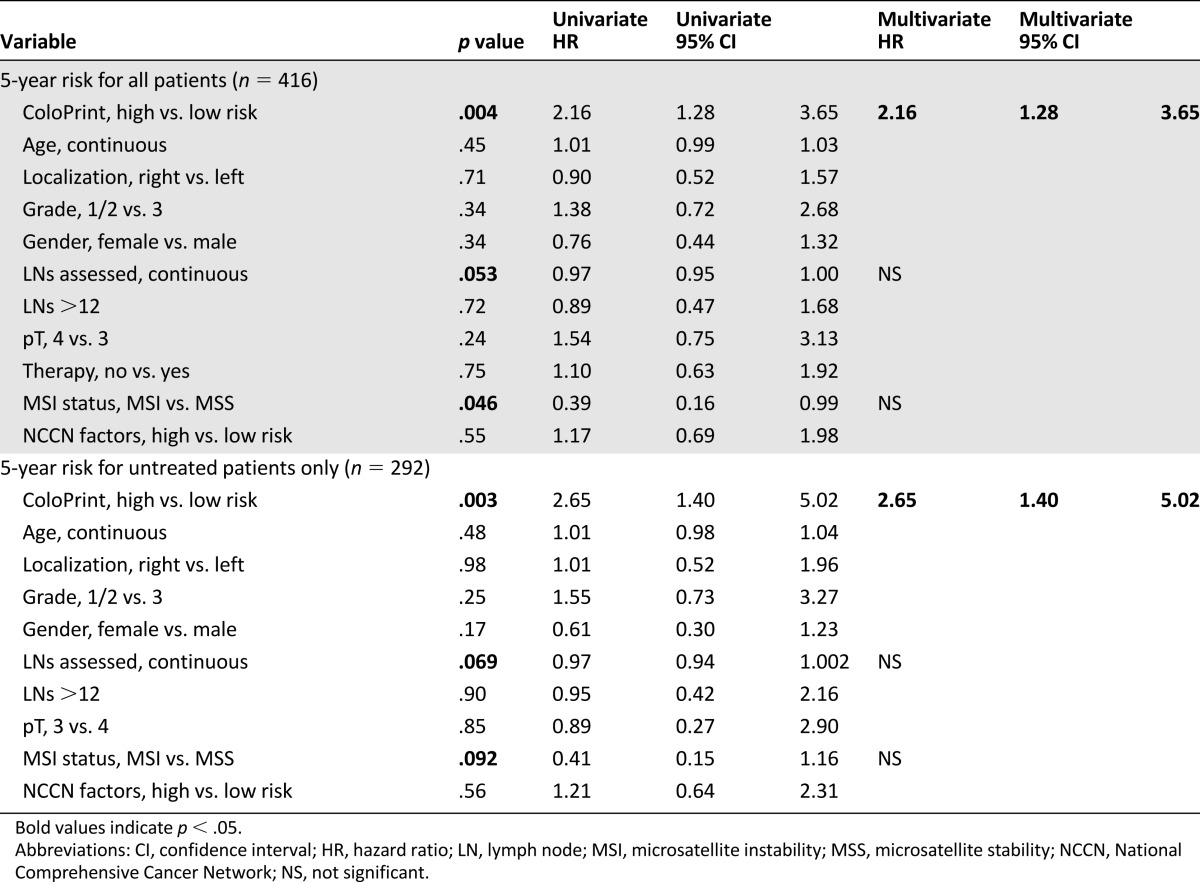
Table 3.
Risk assessment by ColoPrint and NCCN Guidelines

In our data set, 30% of patients received adjuvant 5-FU-based chemotherapy. The decision to administer adjuvant chemotherapy was correlated with cohort site, year, and clinical risk factors but was not correlated with ColoPrint results, which were not available to the treating physicians. The outcome of patients was not improved by chemotherapy (p = .88). Patients who did not receive chemotherapy had a 5-year ROR of 13.8% (95% CI: 9.7%–17.9%), whereas patients who received therapy had a 5-year ROR of 14.8% (95% CI: 8.5%–21.1%). The analysis of ColoPrint in patients who did not receive any adjuvant treatment resulted in the same prognostic power as the analysis of all patients (HR: 2.38; p = .008). Neither the clinical factors nor MSI status were significantly prognostic in this subset of patients (Table 2).
In an analysis of the secondary endpoint of disease-free survival, using any recurrence or any cause of death as an event, ColoPrint also significantly distinguished patients with high risk from those with low risk (HR: 1.86; p = .003) (data not shown).
ColoPrint Analysis in Stage II T3-MSS
MSI and T4 are prognostic indicators currently used in the clinical setting to identify low-risk (MSI-H) and high-risk (T4) stage II patients. In our data set, patients with MSI-H had a low 5-year ROR of 6.6% (95% CI: 0.1%–12.1%), whereas T4 patients had a high 21% ROR (95% CI: 8%–31%). As such, the greatest clinical uncertainty was present for patients with T3 and microsatellite-stable tumors. A subset analyses was performed for patients with T3 and MSS colorectal cancer. ColoPrint was still significantly prognostic in the T3-MSS subgroup (n = 301) with excellent separation. Of the T3-MSS patients, 59% had ColoPrint low-risk results, which resulted in a 5-year ROR of 9.9% (95% CI: 5.4%–14.4%), whereas patients with a high-risk result had a 5-year ROR of 22.4% (95% CI: 14.8%–30%) (Fig. 1C; Table 4). This corresponds to a hazard of relapse of 2.4 (95% CI: 1.3–4.4), with a p value of .005. Analysis of disease-free survival also showed separation of high-risk and low-risk patients with an HR of 1.93 (p = .005) at 5 years (data not shown).
Table 4.
Five-year risk of relapse for T3 microsatellite-stable patients only (n = 301)
Discussion
Risk stratification in stage II colon cancer provides an opportunity for improved delivery of care in a setting of modest adjuvant therapy benefit, competing risks, and necessity to integrate patient preferences. A quarter of colon cancers are diagnosed at stage II, and it has been estimated that 30% of patients subsequently receive adjuvant therapy, with significant variation in practice patterns between providers and healthcare systems. Current guidelines suggest use of clinical or pathological high-risk features and testing for microsatellite instability.
Two of the strongest prognostic factors, the poor-prognosis T4 invasion and good-prognosis mismatch repair deficiency, are commonly used to justify administration and withholding, respectively, of chemotherapy. Tumors that penetrate to the visceral peritoneal surface (T4a) or invade into adjacent structures (T4b) have high rates of recurrence even in the absence of nodal involvement, with higher rates of recurrence than T1–2N1 stage III tumors, and are commonly treated with adjuvant therapy. Conversely, adjuvant therapy is routinely withheld from patients with deficient mismatch repair because of their good prognosis and the accumulating data in some but not all studies suggesting lack of benefit when these patients are treated with 5-FU adjuvant therapy [9, 25]. For the ∼75% remaining stage II patients (the T3N0 MSS population), there is a need for a prognostic model that can be used for clinical decision making.
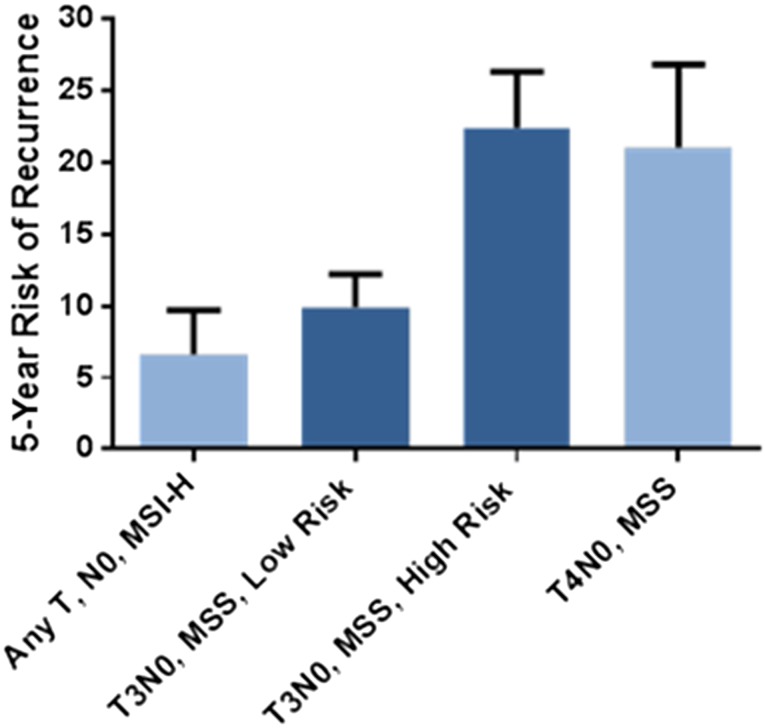
Current clinical and pathological risk factors, as defined by the NCCN Guidelines, poorly predicted outcome in our cohort (HR: 1.3; p = .4), consistent with other analyses. Within the T3N0 MSS population, the additional variables in the NCCN Guidelines likewise did not correlate with outcomes (HR: 1.01; p = .9). In contrast, ColoPrint classification works independently of these clinical-pathological factors, with no statistically significant correlation of the two (odds ratio for ColoPrint [low vs. high risk]: 1.33; 95% CI: 0.890–1.988). The model identifies fewer high-risk patients than the clinical-pathological high-risk guidelines (∼7% less) with a higher risk of relapse (21% in ColoPrint high-risk vs. 15% in clinically high-risk patients). This dichotomization of risk is clinically useful, and it is notable that within the T3N0 MSS population, a low-risk result provides a risk of recurrence equivalent to that of the MSI-H population, whereas a high-risk result indicates a recurrence risk equivalent to T4N0 and some node-positive (stage III) patients (Fig. 2).
Figure 2.
Five-year risk of recurrence by ColoPrint risk groups in patients with T3N0 microsatellite stable colon cancer compared with microsatellite unstable and T4N0 colon cancers.
Abbreviations: MSI-H, high microsatellite instability; MSS, microsatellite stability.
Although there was no difference in the outcomes of patients treated or not treated with adjuvant therapy, we are limited in drawing conclusions based on this finding because patients were not treated within a randomized clinical trial and the potential benefit of chemotherapy may be too small (∼3%–5%) to be detected in this limited data set [26]. A more recent publication [27] also questions the efficacy of 5-FU-based adjuvant treatment in stage II because earlier beneficial results might conceivably be compromised by a proportion of patients with low numbers of assessed lymph nodes and thus with undiagnosed stage III disease. Because the number of assessed lymph nodes in this data set was very high, we can hypothesize that most patients were correctly staged.
As prognostic models improve, it may be possible to re-evaluate existing classification strategies. Although ColoPrint classification also correlates with MSI status, a minority of MSI-H patients are still classified as high risk (24%), suggesting that additional molecular factors may be useful to better segregate this good-prognosis group. Conversely, a subset of stage III colon cancer patients may be overtreated with combination chemotherapy, and improved prognostic models may inform the risk-benefit discussion for these patients. An ideal prognostic model would provide the ability to discriminate between risk groups with clinically meaningful and reproducibly validated differences; to allow testing from limited amounts of formalin-fixed, paraffin-embedded (FFPE) samples; and to be paired with appropriate educational material to facilitate discussion of the results with patients. A variety of assays have been developed that have individual strengths and weaknesses, as recently reviewed [28]. Although ColoPrint provides clinically relevant magnitude of benefit (as noted by the 2.4-fold hazard of relapse between low- and high-risk results for T3-MSS patients), it is currently performed using fresh or frozen tumor samples, requiring prospective planning for fresh tissue collection or mechanisms to routinely collect frozen specimens. Because current clinical practice relies heavily on FFPE samples, this signature is being further developed in FFPE to improve feasibility in clinical practice. Risk models are most useful for patient care when integrated with patient education tools, and numeric literacy allows optimal communication of risk to patients. Several such models are available but are hampered by the limiting discriminatory ability of the existing clinical and pathological risk factors [29, 30].
Collectively, these results further validate the ColoPrint risk index and provide reproducible, clinically meaningful segregation of risk that can be used for patient care. A prospective study has completed enrollment to confirm the clinical utility of these results (ClinicalTrials.gov identifier NCT00903565). By defining a high-risk subset, this advance provides opportunities to study optimal treatment strategies for reduction of risk in this subgroup.
Supplementary Material
Acknowledgments
This study was supported by NIH Grants CA95060 and CA16672. Sample collection and annotation of the Institut Català d’Oncologia study were funded by Instituto de Salud Carlos III, FIS Grant PI11-01439 and CIBERESP CB07/02/2005.
Author Contributions
Conception/Design: Scott Kopetz, Josep Tabernero, Iris Simon, Ramon Salazar
Provision of study material or patients: Scott Kopetz, Josep Tabernero, Robert Rosenberg, Zhi-Qin Jiang, Víctor Moreno, Thomas Bachleitner-Hofmann, Giovanni Lanza, Lisette Stork-Sloots, Dipen Maru, Iris Simon, Gabriel Capellà, Ramon Salazar
Collection and/or assembly of data: Scott Kopetz, Josep Tabernero, Zhi-Qin Jiang, Víctor Moreno, Thomas Bachleitner-Hofmann, Giovanni Lanza, Lisette Stork-Sloots, Dipen Maru, Iris Simon, Gabriel Capellà, Ramon Salazar
Data analysis and interpretation: Scott Kopetz, Josep Tabernero, Robert Rosenberg, Zhi-Qin Jiang, Víctor Moreno, Thomas Bachleitner-Hofmann, Giovanni Lanza, Lisette Stork-Sloots, Dipen Maru, Iris Simon, Gabriel Capellà, Ramon Salazar
Manuscript writing: Scott Kopetz, Josep Tabernero, Robert Rosenberg, Lisette Stork-Sloots, Dipen Maru, Iris Simon, Ramon Salazar
Final approval of manuscript: Scott Kopetz, Josep Tabernero, Robert Rosenberg, Zhi-Qin Jiang, Víctor Moreno, Thomas Bachleitner-Hofmann, Giovanni Lanza, Lisette Stork-Sloots, Dipen Maru, Iris Simon, Gabriel Capellà, Ramon Salazar
Disclosures
Lisette Stork-Sloots: Agendia NV (E); Iris Simon: Agendia (E, IP). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.World Cancer Research Fund International. Colorectal cancer statistics. Available at http://www.wcrf.org/int/cancer-facts-figures/data-specific-cancers/colorectal-cancer-statistics. Accessed December 21, 2014
- 3.Nitsche U, Maak M, Schuster T, et al. Prediction of prognosis is not improved by the seventh and latest edition of the TNM classification for colorectal cancer in a single-center collective. Ann Surg. 2011;254:793–800; discussion 800–801. doi: 10.1097/SLA.0b013e3182369101. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham D, Atkin W, Lenz HJ, et al. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 5.Benson AB, III, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–3419. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 6.Church D, Midgley R, Kerr D. Biomarkers in early-stage colorectal cancer: Ready for prime time? Dig Dis. 2012;30(suppl 2):27–33. doi: 10.1159/000341890. [DOI] [PubMed] [Google Scholar]
- 7.O’Connor ES, Greenblatt DY, LoConte NK, et al. Adjuvant chemotherapy for stage II colon cancer with poor prognostic features. J Clin Oncol. 2011;29:3381–3388. doi: 10.1200/JCO.2010.34.3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087, e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van ‘t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 11.de Snoo F, Bender R, Glas A, et al. Gene expression profiling: Decoding breast cancer. Surg Oncol. 2009;18:366–378. doi: 10.1016/j.suronc.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Jatkoe T, Zhang Y, et al. Gene expression profiles and molecular markers to predict recurrence of Dukes’ B colon cancer. J Clin Oncol. 2004;22:1564–1571. doi: 10.1200/JCO.2004.08.186. [DOI] [PubMed] [Google Scholar]
- 13.Eschrich S, Yang I, Bloom G, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23:3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 14.Barrier A, Boelle PY, Roser F, et al. Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol. 2006;24:4685–4691. doi: 10.1200/JCO.2005.05.0229. [DOI] [PubMed] [Google Scholar]
- 15.Lin YH, Friederichs J, Black MA, et al. Multiple gene expression classifiers from different array platforms predict poor prognosis of colorectal cancer. Clin Cancer Res. 2007;13:498–507. doi: 10.1158/1078-0432.CCR-05-2734. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Y, Casey G, Lavery IC, et al. Development of a clinically feasible molecular assay to predict recurrence of stage II colon cancer. J Mol Diagn. 2008;10:346–354. doi: 10.2353/jmoldx.2008.080011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorissen RN, Gibbs P, Christie M, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park YY, Lee SS, Lim JY, et al. Comparison of prognostic genomic predictors in colorectal cancer. PLoS One. 2013;8:e60778. doi: 10.1371/journal.pone.0060778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salazar R, Roepman P, Capella G, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29:17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- 20.Maak M, Simon I, Nitsche U, et al. Independent validation of a prognostic genomic signature (ColoPrint) for patients with stage II colon cancer. Ann Surg. 2013;257:1053–1058. doi: 10.1097/SLA.0b013e31827c1180. [DOI] [PubMed] [Google Scholar]
- 21.Kopetz S, Jiang ZQ, Overman M, et al. Genomic classifier (ColoPrint) predicts outcome and chemotherapy benefit in stage II and III colon cancer patients. J Clin Oncol. 2013;31(suppl):3612a. [Google Scholar]
- 22.Edge SB, Byrd DR, Compton CC, editors. Colon and rectum. In: Compton CC, Byrd DR, Garcia-Aguilar J et al., eds. AJCC Cancer Staging Manual. 7th ed. New York, NY: Springer; 2010. pp. 143–164. [Google Scholar]
- 23.Tian S, Roepman P, Popovici V, et al. A robust genomic signature for the detection of colorectal cancer patients with microsatellite instability phenotype and high mutation frequency. J Pathol. 2012;228:586–595. doi: 10.1002/path.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines). Colon cancer [version 3, 2013]. Available at http://www.nccn.org/professionals/physician_gls/f_guidelines.asp#site. Accessed December 21, 2014.
- 25.Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 26.Sargent D, Sobrero A, Grothey A, et al. Evidence for cure by adjuvant therapy in colon cancer: Observations based on individual patient data from 20,898 patients on 18 randomized trials. J Clin Oncol. 2009;27:872–877. doi: 10.1200/JCO.2008.19.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Q, Andre T, Grothey A, et al. Comparison of outcomes after fluorouracil-based adjuvant therapy for stages II and III colon cancer between 1978 to 1995 and 1996 to 2007: Evidence of stage migration from the ACCENT database. J Clin Oncol. 2013;31:3656–3663. doi: 10.1200/JCO.2013.49.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharif S, O’Connell MJ. Gene signatures in stage II colon cancer: A clinical review. Curr Colorectal Cancer Rep. 2012;8:225–231. doi: 10.1007/s11888-012-0132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adjuvant! online. Available at http://www.adjuvantonline.com. Accessed December 21, 2014.
- 30.Stage III colon cancer calculator. Available at http://www.mayoclinic.org/medical-professionals/adjuvant-systemic-therapy-tools/colon-cancer.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.