Abstract
Synthetic biology is a relatively new field with the key aim of designing and constructing biological systems with novel functionalities. Today, synthetic biology devices are making their first steps in contributing new solutions to a number of biomedical challenges, such as emerging bacterial antibiotic resistance and cancer therapy. This review discusses some synthetic biology approaches and applications that were recently used in disease mechanism investigation and disease modeling, drug discovery and production, as well as vaccine development and treatment of infectious diseases, cancer, and metabolic disorders.
Keywords: synthetic biology, disease mechanism, drug discovery, vaccine development, therapeutic treatment, cancer treatment, infectious diseases, metabolic disorders
1. Introduction
One of the main goals of synthetic biology is to use well-characterized functional modules for engineering biological systems with novel functionalities. These functional modules are usually protein-coding or regulatory DNA parts, which are cloned from various organisms or synthesized, and assembled together into genetic circuits. Since the development of the first engineered gene networks, a toggle switch1 and an oscillator,2 in the beginning of the century, the field has accelerated with the fabrication of even more sophisticated gene circuits such as clocks, counters, logic processors, pattern detectors, and intracellular communication modules.3−9
Today, synthetic biology is still an emerging field and is rapidly expanding. This raises the question of what the unique contributions of synthetic biology are that could not be addressed otherwise. What distinguishes a synthetic biology approach from simply a biological approach is that it seeks to solve problems through the construction of new models rather than through analysis and observation alone. This approach allows a new perspective, where biology, chemistry, and engineering are combined to revisit old biological questions. Being able to construct and emulate a biological system helps in better understanding of relevant biological phenomena as well as allows the development of a more precise, predictable means of biological manipulation in agriculture, bioenergy production, and therapeutics.
One of the scopes of synthetic biology is to contribute solutions to biomedical challenges. These challenges include, among others, growing antibiotic resistance in bacteria,10 accelerated emergence of new infectious diseases,11 and evolving cancer drug resistance.12,13 To address these challenges, synthetic biology envisions the development of custom-designed, easily regulated, and safe devices that would complement human immune defenses and address metabolic abnormalities.
This review focuses on recent advances in synthetic biology that are holding promise for the future development of human therapeutics. First, we will discuss some synthetic biology approaches that were recently used in disease mechanism investigation and disease modeling. Next, we will briefly cover current examples of the field’s contributions in drug discovery and production (Figure 1). Finally, we will review recent progress of synthetic biology strategies in vaccine development, treatment of infections diseases, cancer, and metabolic disorders.
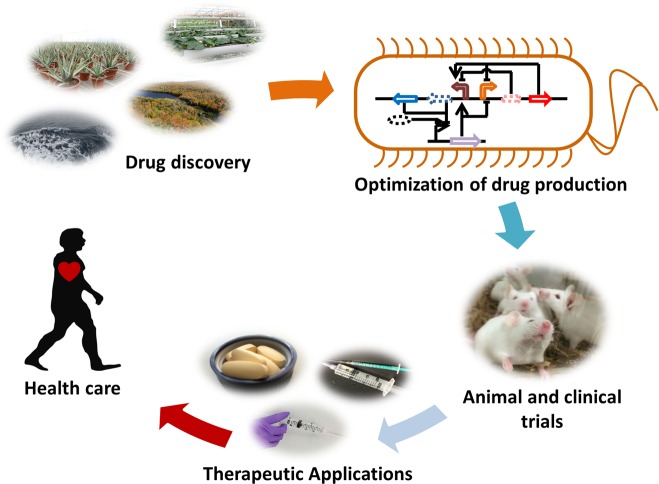
Figure 1.
Outlined scheme of the typical pipeline of drug development consisting of drug discovery, production, animal and clinical trials, and therapeutic administration. Natural resources are often used for novel therapeutic discovery. Once identified, the therapeutic compounds are optimized for bioproduction via refactoring of metabolic pathways, usually in a heterologous host. Next, animal and consequently clinical trials are performed for the assessment of therapeutic efficacy and safety. Finally, novel therapeutics are administered in the clinic to treat human diseases.
2. Disease Mechanism Investigation
2.1. Immunological Disorders
Synthetic biology has been helpful in providing mechanistic insights into certain human disorders. In particular, it provided a framework for generating disease models and discovering new drug targets. For example, the contribution of genetic defects that result in abnormal B cell development in agammaglobulinemia, a primary immunodeficiency, was investigated by reconstituting functional parts of key natural complexes in an orthogonal environment.14 This approach is often implemented to provide an isolated, well-characterized environment. Patients were screened for genes that were expressed in the early stages of B cell differentiation. Once a defect in the immunoglobulin-β (Igβ) encoding gene was identified, the mutant Igβ and other constituents of the human B cell antigen receptor (BCR) were reconstituted on the surface of cultured Drosophila melanogaster cells. Mutant Igβ abolished the assembly of the BCR on the cell surface, and failure to assemble the BCR complex, in turn, caused a complete block of B cell development. The described process was proposed as a likely mechanism of agammaglobulinemia in some patients.14
In another example, synthetic presentation of an entire human peptidome on the surface of T7 phage allowed to discover new self-antigens (autoantigens) that could lead to autoimmune diseases.15 Enrichment of autoantigens was carried out using antibodies from patients with neurological syndromes. The enriched antigens were subjected to high-throughput sequencing, which revealed new antigens that could be used in accurate diagnostic tests and designing new therapeutics.
2.2. Genome Editing Tools for Cancer Study
Disease modeling is another explored avenue that is made possible by utilization of synthetic biology. For example, genome editing tools such as zinc finger nucleases (ZFN), transcription activator-like nucleases (TALEN), and clustered regularly interspaced short palindromic repeats (CRISPR) in combination with the Cas9 nuclease (CRISPR/Cas9 system), are being increasingly used in gene therapy and disease modeling (reviewed in refs (16−18)). These genome editing tools usually function by introducing a sequence-specific double strand break, which is consequently repaired by either error-prone nonhomologous end joining (NHEJ) or homologous recombination (HR) pathways. While the former allows for knocking out the gene of interest, the latter pathway allows gene segment replacement or site-specific gene knock-in. Because of their exceptional precision and relative simplicity in designing, TALENs and CRISPR/Cas9 tools have been important in modeling and drug target discovery, especially for a complex group of diseases such as cancer.
Certain diseases have been linked to chromosomal rearrangements and provide a great challenge for disease modeling. While knocking-in or coexpression of rearranged genes is possible, these models are often unconvincing in pinpointing the exact contribution of the rearrangements. As an example of an alternative approach, TALENs were used to introduce androgen receptor (AR) gene rearrangement to model a castration-resistant prostate cancer (CRPC) phenotype in a human cell line.19 Two pairs of TALENs were cotransfected to introduce a deletion and an inversion of exons 5–7 of the AR gene. The resulting splice variant of the AR gene was discovered to drive its independence from androgen in the genome engineered cell line, which is proposed to be the mechanism of CRPC. ZFN and TALEN technologies have also been implemented in modeling cancer-relevant chromosomal translocations, such as those found in Ewing sarcoma and anaplastic large cell lymphoma.20
Nuclease-assisted genome engineering can also be a powerful tool for generation of genome-scale knockout screening, as was shown using the CRISPR/Cas9 system.21,22 Using lentiviral vectors, libraries of tens of thousands of unique guide RNA sequences, which guide CRISPR/Cas9 specificity, were delivered into human cells. Such screening allowed identification of genes essential for cell viability in cancer and pluripotent stem cells, as well as genes whose loss conferred resistance to chemotherapeutic agent vemurafenib.21 In another work, screening was performed for the identification of members of the DNA mismatch repair pathway and for genes whose loss conferred resistance to a chemotherapeutic agent etoposide.22
3. Drug Discovery and Production
3.1. Discovery
With the rise of multiresistant pathogens, novel antimicrobial compounds are increasingly needed. Since the discovery line of novel drugs has diminished in the recent years, novel approaches, such as those proposed by synthetic biology, are in demand. For example, a synthetic mammalian gene circuit was utilized for the discovery of novel antituberculosis compounds.23 Ethionamide is an antibiotic often used for treatment of tuberculosis; however, ethionamide-based therapies are sometimes unsuccessful due to the development of resistance by Mycobacterium tuberculosis. The resistance develops when M. tuberculosis protein EthR represses the transcription of EthA, which converts ethionamide into a toxic metabolite. A rationally designed chemical library was screened for a compound that would inhibit EthR binding to EthA promoter. The interaction between the latter was assayed in human cells through a reporter gene expression.23 The screening revealed 2-phenylethyl-butyrate as a potent inhibitor of EthR, which dramatically increased the sensitivity of M. tuberculosis to ethionamide. This work was a demonstration of a generic screening platform for the discovery of novel antituberculosis drugs.
Synthetic biology also paved a new road for discovering novel anticancer agents. Cytotoxic anticancer drugs are believed to discriminate between cancerous and normal tissues by preferentially killing actively dividing cells through targeting DNA replication, which makes cytotoxic drugs more generic compared to “targeted” anticancer drugs. For the discovery of novel cytotoxic drugs, a high throughput-compatible mammalian cell based assay was devised.24 CHO-K1 cell line was engineered for tetracycline-responsive overexpression of human cyclin-dependent kinase inhibitor p27Kip1, which is a negative regulator of G1-S transition. The engineered cells proliferate normally in the presence of tetracycline. However, upon withdrawal from the antibiotic, they diverge into a heterogeneous population of growth-arrested and proliferation-competent cells due to a spontaneous loss of p27Kip1 in roughly half of the cells.24 These proliferating cells were assumed to imitate the neoplastic cell characteristics. The assay was validated by scoring the viability of arrested and proliferating cells upon exposure to clinically licensed cytotoxic drugs. It is expected that the assay will be useful for high throughput screening of novel anticancer drugs.
3.2. Drug Production
Natural products have been valuable in therapeutic areas such as infectious diseases and oncology.25 These drugs, however, are produced in small amounts in native hosts, and therefore, extraction of these drugs from native hosts is usually uneconomical or can have a negative impact on the environment. A powerful solution is drug production in metabolically engineered microorganisms or plant cells, which can be made capable of large-scale production (reviewed in refs (26−28)). A prominent recent example is pathway optimization for overproduction of taxadiene, a precursor to an anticancer drug taxol. In this study, Escherichia coli was used as an expression host, where the overall pathway was partitioned into two modules.29 Unlike in previous studies, where these two modules were engineered separately, here, the modules’ expression was varied simultaneously by using various promoters and gene copy numbers. This approach allowed identification of an optimally balanced pathway using a small combinatorial space. The titer of taxadiene was improved 15,000-fold, yielding approximately 1 g/L of taxadiene in fed-batch bioreactor fermentations. Another impactful example was conducted in Saccharomyces cerevisiae for the production of artemisinic acid, a precursor to antimalarial drug. For the first time, artemisinic acid was produced by the expression of a complete biosynthetic pathway, which included a newly discovered plant dehydrogenase and a second cytochrome.30 This development resulted in 10-fold increase in artemisinic acid titers, yielding up to 25 g/L.
Besides microorganisms, plants have also been used for the production of therapeutically valuable plant metabolites. In this approach, natural products are usually produced in plant cell cultures that are induced from either established callus cultures or multipotent cambial meristematic cells.27,31 However, in the past two decades, production of natural products in plant hairy roots has received a lot of attention.32 Hairy roots are a result of genetic transformation of plant cells due to an infection with Agrobacterium rhizogenes. Hairy roots are another attractive system for drug production since in contrast to cell cultures they are phenotypically and genetically stable and can be exploited for a long time. For example, three genes involved in the biosynthesis pathway for tanshinone, a cardiovascular disease agent, were introduced in Salvia miltiorrhiza hairy root cultures, resulting in significantly improved production of tanshinone (4.74-fold) and increased antioxidant activity.33
4. Vaccine Development
In vaccination, the introduced vaccine stimulates the adaptive immunity response to a specific pathogen. Conventional vaccinations are usually delivered by injection; however, the procedure is not without risks or discomfort.34 One example of addressing this issue is engineering of oral plant tissue-based vaccinations, which considerably reduce the risk of contamination with mammalian pathogens and do not require costly purification and downstream processing. Moreover, rigid plant cell walls protect antigen degradation in the acidic environment of the stomach.35 For example, unicellular green alga Chlamydomonas reinhardtii chloroplasts were used for vaccine formulation against Staphylococcus aureus infection.36 Chloroplasts were engineered for stable expression of the D2 fibronectin-binding domain of S. aureus fused with a mucosal adjuvant cholera toxin B subunit, which improves antigen-specific immune responses. Mice treated with transgenic algae had significantly reduced pathogen load in the spleen and the intestine, and 80% of the pretreated mice survived lethal doses of S. aureus, which makes C. reinhardtii an attractive platform for oral vaccine development.36
S. cerevisiae can also be manipulated to express foreign antigens that would stimulate an immunologic response.37 Recombinant yeast vaccines engineered to express viral or tumor antigens have been demonstrated to activate dendritic cells (DCs) and confer protective cell-mediated immunity against tumor cells. One example is S. cerevisiae vector engineered to express a transgene encoding human carcinoembryonic antigen (CEA), which is associated with tumor growth. Human DCs were activated by CEA and subsequently activated cytotoxic T-cells specific for CEA+ human tumor cells.38
Reengineering of viruses for vaccine design is another interesting strategy in vaccine development. Thus, poliovirus was synthetically attenuated by recoding the poliovirus capsid protein with underrepresented codons and synthesizing the recoded DNA de novo.39 Recoding of poliovirus decreased rates of protein translation and resulted in attenuation of the virus in mice. The attenuated virus generated effective immune response in mice, indicating that virus attenuation via codon deoptimization could provide an alternative method of vaccine generation.
5. Treatment of Infectious Diseases
5.1. Treatment of Bacterial Infections by Designer Bacteriophages
With the bacterial antibiotic resistance becoming an increasing concern, synthetic biology is turning back to an almost 100-year-old idea of using bacteriophages to fight bacterial pathogens.40 One source of bacterial resistance to antibiotics and host defenses is the development of biofilms, which are surface-associated communities in a hydrated matrix of extracellular polymers. An example of constructing a bacteriophage with an increased bactericidal ability was engineering of T7, an E. coli-specific phage, to express dispersin B (DspB) enzyme for biofilm degradation.41 Intracellular expression and release of DspB to the environment upon cell lysis allowed biofilm dispersal and a two orders-of-magnitude improvement in decreasing biofilm cell counts compared to a nonenzymatic phage.
In a following study, M13 phage was engineered to enhance the efficacy of antibiotics in a phage–drug combination therapy.42 The phage was modified to overexpress LexA3, a repressor of SOS response in E. coli. It was reasoned that LexA3, which has also been previously shown to inhibit emergence of antibiotic resistance, would enhance bacterial killing by bactericidal antibiotics via disabling of the SOS response. The study showed that the engineered phage improved the bactericidal effect of a quinolone drug by several orders-of-magnitude in vitro and significantly increased survival of infected mice in vivo.
In a different investigation, M13 phage was engineered to inhibit infection of Chlamydia trachomatis, a common cause of sexually transmitted diseases.43 The phage was engineered for enhanced internalization into bacteria-containing parasitophorous vacuoles, also known as inclusions, in the mammalian host cells. The phage capsid proteins were fused with two functional peptides: integrin binding peptide RGD, known to induce integrin mediated endocytosis, and a segment of the polymorphic membrane protein (PmpD), an autotransporter protein from C. trachomatis (Figure 2a). The study showed increased uptake of the phage into the lumen of the inclusions and significant amelioration of bacterial infection in HeLa and primary endocervical cells.
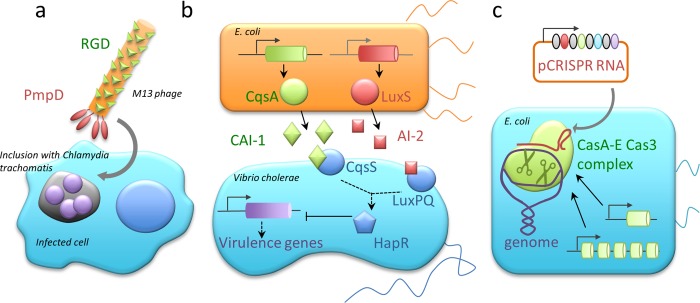
Figure 2.
Biosynthetic devices for the treatment of infectious diseases. (a) Engineering bacteriophages against pathogenic bacteria. M13 phage was engineered for enhanced mammalian host cell internalization by fusing two functional peptides, RGD and PmpD, to the coat proteins of the phage.43 (b) Engineering commensal bacteria against pathogenic bacteria. E. coli strain Nissle 1917 expressing autoinducer 2 (AI-2) was engineered to express cholera autoinducer 1 (CAI-1), both of which are molecules synergistically coordinating quorum sensing in Vibrio cholerae. Through a signal transduction cascade, CAI-1 and AI-2 inhibit the expression of virulence genes in V. cholerae.44 (c) Type I-E CRISPR/Cas endonuclease system was engineered for cytotoxicity against target bacteria via site-specific introduction of a double strand break in the genome. CRISPR RNA was designed for homology to a target gene in a region that is unique to the strain, allowing strain-specific removal of E. coli.46
5.2. Treatment of Bacterial Infections by Commensal Bacteria
Diminishing virulence of pathogenic bacteria by prophylactic consumption of engineered commensal bacteria is another promising approach in synthetic biology. One study exploited the quorum sensing mechanism of Vibrio cholerae in regulating its infection cycle.44 High concentrations of cholera autoinducer 1 (CAI-1) and autoinducer 2 (AI-2) are known to inhibit virulence gene expression in V. cholerae. To interrupt the virulence of V. cholerae, an E. coli strain Nissle 1997, which expresses AI-2 natively, was engineered to express CAI-1 (Figure 2b). When cocultured with the engineered E. coli strain, cholera toxin and toxin-coregulated pilus expression was inhibited in V. cholerae.44 Furthermore, pretreatment of an infant mouse model with engineered commensal bacteria significantly disrupted V. cholerae colonization.45 Thus, engineering commensal bacteria is a promising strategy in the treatment of bacterial infections.
5.3. Sequence-Specific Endonucleases for Disruption of Bacterial and Viral Infections
Another novel approach of developing antibacterial and antiviral agents is exploitation of sequence-specific endonucleases, such as ZFNs, TALENs, or the CRISPR/Cas system. While the NHEJ pathway in most eukaryotes can quickly repair extensive site-specific double strand DNA breaks, poor efficiency or absence of this pathway in many prokaryotes can render the aforesaid endonucleases lethal. Thus, cytotoxicity of the CRISPR/Cas system was exploited for programmable removal of bacterial strains46 (Figure 2c). It was shown that the CRISPR/Cas system targeting endogenous genes at species-specific sites can be employed for the development of species and even strain specific bactericidal agents. This was shown in an experiment with a mixed population of E. coli strains K-12 and BL21, with the extent of removal of >99.999% of the targeted bacterial strain.
Genome editing tools can also be used to establish viral resistance in humans or develop potent virus disrupting agents. For example, HIV-1 resistance in human CD4+ T cells was established using a ZFN targeting human endogenous HIV coreceptor CCR5.47 When targeting directly viral genomes, TALENs and CRISPR/Cas9 were shown to be successful in disrupting hepatitis B virus48 and latent HIV-1 provirus,49 respectively.
6. Cancer Treatment
6.1. Oncolytic Virotherapy
Chemotherapy and radiotherapy are used extensively in the clinic but often target noncancerous tissues and have limited toxicity to cancer cells. More sophisticated technologies capable of discriminating between cancerous and healthy tissues are therefore needed. One of the developing fields that can potentially provide a solution is oncolytic virotherapy, which focuses on engineering viruses capable of infecting and killing cancers. Some viruses are naturally oncotropic, and many also have tissue tropism,50,51 which has been a starting point for engineering tumor-specific oncolytic viruses.
Greater viral specificity for tumor cells has been the preferred direction of recent studies. The specificity for tumors can be engineered at the stage of virus entry via receptor targeting, which requires the modification of receptor binding proteins. This usually is achieved via fusion of single chain antibodies to the attachment proteins displayed on the viral surface (reviewed in ref (52)). However, tumor cells can develop resistance by inhibition or mutation of the target antigen.53 To counteract this process, an oncolytic virus was engineered for bispecific targeting of tumor antigens. This was made possible by using designed ankyrin repeat proteins (DARPins), engineered antibody mimetic proteins that are smaller in size and less prone to aggregation compared with larger single-chain antibodies (Figure 3a). DARPins specific for two different tumor markers were fused with the measles virus attachment protein. As a result, a virus with conserved oncolytic potency and attenuated potency in nontarget tissue was generated.53 The multiplex targeting approach might be helpful in counteracting resistance development in carcinoma cells.54
Figure 3.
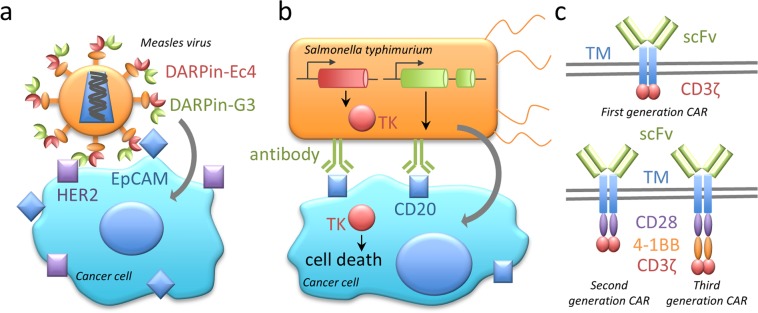
Biosynthetic devices for cancer therapy. (a) Oncolytic virotherapy. To counteract the development of resistance in cancer cells, the measles virus was engineered for multiplex targeting of cancer antigens. To this end, DARPins EC4 and G3 targeting two different tumor markers, EpCAM and HER2, were fused to the virus attachment protein.53 (b) Designer anticancer bacteria. Salmonella typhimurium was engineered to specifically infect CD20+ tumor cells via surface-expression of anti-CD20 antibody. The cytotoxicity was achieved through expression of herpes simplex virus thymidine kinase (TK) prodrug-converting enzyme.57 (c) Structure of chimeric antigen receptors (CARs) for adoptive T cell therapy. First, second, and third generations of CARs differ by the number of intracellular signaling domains. scFv, single fragment length antibody. TM, transmembrane domain. CD28, 4-1BB, and CD3ζ, intracellular signaling domains.61
When it comes to therapeutic efficacy, cytotoxicity of oncolytic viruses can be enhanced through a so-called “arming” strategy, where the virus is engineered to express a protein that sensitizes both infected tumor cells and surrounding uninfected tumor cells. Recent clinical studies demonstrated the increased efficacy of oncolytic viruses when replicative viral oncolysis was combined with stimulation of inflammation and adaptive immunity against tumor antigens. The first report in humans was an oncolytic adenovirus armed with granulocyte macrophage colony-stimulating factor (GMCSF).55 GMCSF-armed virus was reported to mediate antitumor immunologic response by recruiting natural killer cells and inducting of tumor-specific cytotoxic T cells.
6.2. Designer Anticancer Bacteria
Engineering bacteria to invade and kill cancer cells is another promising strategy for cancer treatment. Salmonella, Clostridium, and other genera have been shown to have tumor-tropism and the ability to kill cancer cells, which was exploited for engineering of even more potent anticancer bacterial strains.56 In a recent study, tumor specificity, which is an important attribute of anticancer bacterial therapy, was addressed. To inhibit nonspecific invasiveness of Salmonella, single-domain antibody against human tumor-associated antigen CD20 was expressed on the bacterial cell surface57 (Figure 3b). The engineered Salmonella was found to preferentially invade and destroy CD20+ lymphoma xenografts in mice while significantly minimizing nonspecific cell invasion.
6.3. Chimeric Antigen Receptors
Adoptive T cell therapy has been shown to be effective in initiating lasting antitumor responses.58 In some cases, though, the effectiveness of the technique is limited since the function of redirected T cells relies on the presentation of tumor antigens by the major histocompatibility complexes (MHC), which are often inhibited in cancerous cells. Engineering of T cells to express chimeric antigen receptors (CARs) allows MHC-independent T cell activation and proliferation (reviewed in refs (59−61)). CARs are modular fusion proteins consisting of an extracellular antigen recognition element, usually single chain variable fragment (scFv) antibody, a transmembrane domain, and an intracellular signaling domain, usually from T cell coreceptor CD3ζ or Fc receptor γ. To date, the most encouraging clinical observations were achieved with CARs specific for CD19 antigen, which is expressed in B cell malignancies such as B cell leukemia/lymphoma, but not by normal essential tissues.62,63
To increase signaling strength and persistence, CARs of second and third generations have been developed with two or three different costimulatory signaling domains fused in a single polypeptide chain61−63 (Figure 3c). Engineered T cells with second generation CARs, containing costimulatory domains from both CD3ζ and CD28, have been shown to demonstrate improved expansion and persistence compared to those of the first generation CARs.64 Moreover, the third generation CARs, also containing a third costimulatory domain from the costimulatory molecule 4-1BB, was shown to increase cytotoxicity of engineered T cells compared to the second generation CARs.65
These advancements improved effectiveness of the CAR technology, but the challenge of specificity remains to be addressed. In most of the studies up to date, engineered T cells recognize a single antigen,59−61 which can also be presented by noncancerous tissues, raising the concern of off-tumor cytotoxicity. In a recent approach, a CAR-based AND logic gate was used to create T cells capable of recognizing two antigens, but neither of the antigens alone.66,67 Thus, it was shown that cotransduced T cells administered in mice destroy prostate tumors expressing both tumor antigens PSMA and PSCA, but not tumors expressing either antigen alone.66 Engineering dual-specific CAR-T cells to recognize both mesothelin and a-folate antigens resulted in potent activity against a mouse xenograft model of ovarian cancer.67 These studies showed the efficacy of engineering dual-specific CAR-T cells for minimizing parallel reactivity against normal tissues bearing a single antigen.
7. Treatment of Metabolic Disorders
7.1. Bacterial Devices
Disorders of human metabolism encompass a diverse group of complex diseases that usually result from genetic enzyme deficiency or epigenetic alterations.68 For many metabolic disorders, treatment is currently unavailable, while others are controlled by dietary restriction or supplementation. With the aim of tackling these disorders, the first synthetic biology proof-of-principle studies have been conducted by engineering bacterial circuits capable of restoring normal metabolism. Thus, for treatment of diabetes, bacteria were engineered to stimulate intestinal epithelial cells to secrete insulin in response to glucose.69E. coli strain Nissle 1917 was engineered to express and secrete glucagon-like peptide 1 (GLP-1) as well as pancreatic and duodenal homeobox gene 1 (PDX-1), proteins that are known to stimulate intestinal epithelial cells to synthesize insulin. Cultured epithelial cells grown in cell-free media pretreated with engineered bacteria were stimulated to secrete insulin up to 1 ng/mL.69 However, implementation of the strategy in commensal bacteria cocultured with epithelial cells in vitro or in vivo implementation is yet to be shown.
7.2. Mammalian Pharmaceutically Controlled Open-Loop Circuits
To be more therapeutically relevant, the synthetic biology field is currently developing further by expanding its mammalian gene circuit repertoire.70,71 In particular, for the treatment of metabolic disorders, it is imperative to develop interactive gene networks that would stimulate the expression of therapeutics in a controlled, regulated manner. This can be achieved either by open-loop circuits, where an input signal triggers the output in a linear fashion, or a closed-loop circuit, where the output feeds back on the input signal, the latter giving a more stable output control.72 In a recent study, an open-loop circuit for the treatment of the metabolic syndrome was developed.73 Metabolic syndrome is a co-occurrence of functionally linked health problems, such as hypertension, hyperglycemia, obesity, and dyslipidemia, which are usually treated independently. In this study, a synthetic circuit was devised where a pharmaceutical targeting one of the health conditions, antihypertensive drug guanabenz, is also an input for the signal transduction to express GLP-1 fused to leptin. Both of the latter are therapeutic peptide hormones, GLP-1 stimulating the secretion of insulin and leptin regulating energy intake and expenditure (Figure 4a). Administration of this circuit in mice with the metabolic syndrome phenotype resulted in simultaneous attenuation of hypertension, hyperglycemia, obesity, and dyslipidemia. Thus, this study demonstrated the feasibility of treating a complex metabolic health condition by obtaining a triple output upon the administration of a single input, a pharmacological drug.
Figure 4.

Synthetic circuits for treatment of metabolic diseases. (a) Ligand-controlled open-loop circuits. For the simultaneous treatment of interdependent pathologies comprising the metabolic syndrome, antihypertensive drug guanabenz was used to activate a synthetic signal cascade to stimulate the secretion of metabolically active peptides GLP-1 and leptin, fused to a single polypeptide chain.73 (b) Closed-loop circuits. For the treatment of diet-induced obesity, appetite-suppressive peptide hormone pramlintide was placed under the control of a chimeric transcription regulator TtgR-PPARα. Depending on the presence or absence of fatty acids, human PPARα recruits either endogenous transcription co-activators or endogenous transcription co-repressors, respectively. Thus, a synthetic circuit that constantly senses and regulates the blood fatty acid levels was created.75 (c) Optogenetic open-loop circuits. Electromagnetic waves with radio frequencies were used to activate the production of bioengineered insulin. To this end, antibody-coated iron oxide nanoparticles, which heat up upon exposure to radio waves, were engineered to bind to temperature-sensitive channel protein TRPV1. Upon increase in temperature, TRPV1 activates a signal transduction cascade that in turn activates pro-insulin. The expression of the latter resulted in improved glucose homeostasis.77
7.3. Mammalian Closed-Loop Circuits
Small-molecule drug-based intervention to treat physiological abnormalities in metabolic disorder patients may provide a controlled way of therapeutic delivery, yet prolonged daily administration of a drug can lead to unwanted side effects. To this end, the development of closed-loop circuits would be advantageous since it would remove the reliance on repeated administration of a therapeutic. Implementation of this approach was described using a prosthetic gene network that could sense and restore normal physiological uric acid levels through controlled expression of urate oxidase.74 Urate oxidase was put under direct control of bacterial uric acid sensor HucR, which binds its target DNA motif in the absence of uric acid. The synthetic circuit stabilized the blood urate concentration in urate oxidase-deficient mice with acute hyperuricemia.
In another example, a gene circuit for the treatment of diet-induced obesity in mice was designed.75 To this end, a binary synthetic transcription factor was constructed by fusing peroxisome proliferator-activated receptor-α (PPARα) to bacterial DNA-binding repressor TtgR (Figure 4b). PPARα is a lipid receptor and transcription factor that recruits endogenous co-activators in the presence of fatty acids, but associates with endogenous co-repressors in the absence of fatty acids. TtgR-regulated promoter was used to express an appetite-suppressing peptide hormone pramlintide. Engineered cells were encapsulated and implanted in mice with diet-induced obesity, which consequently showed significant reduction in food consumption and blood lipid levels.
7.4. Mammalian Open-Loop Optogenetic Devices
Switching from drug-dependent gene regulation to molecule-free electromagnetic gene regulation in vivo for therapeutic applications is becoming another appealing approach. Thus, the field of optogenetics is becoming increasingly popular due to anatomical specificity and precise temporal control of gene expression. Recently, the first optogenetic device for the controlled production of a therapeutic protein in an animal disease model was reported.76 Here, shGLP-1 hormone was put under transcriptional control of melanopsin, a blue light sensor protein. Melanopsin belongs to a family of ion channel proteins, such as channelrhodopsin, that transform the light-based energy to ion-based membrane potential and trigger an intracellular signaling cascade. The calcium-dependent signaling cascade eventually activates a transcription factor that controls the expression of the hormone. Encapsulated cultured human cells were implanted in a mouse model of human type II diabetes. Upon illumination with blue light, type II diabetic mice showed improved glucose homeostasis.
However, a major limitation of implementing melanopsin and similar ion channels is that animal tissue is an impenetrable obstacle for light waves, and implanted devices are required for the delivery of the signal. A different solution was proposed, where instead of visible light, radio frequencies, which can penetrate deep tissues with minimal energy loss, were used.77 Unlike tissue, metal nanoparticles strongly absorb radio wave energy and heat up as a result. The antibody-coated nanoparticles were designed to bind TRPV1, a modified temperature sensitive channel in the membrane of the target cells (Figure 4c). Upon exposure to low-frequency electromagnetic energy, the local heating activated TRPV1, which in turn activated the downstream calcium-dependent signaling cascade and expression of modified human insulin gene. Mice with tumor xenografts expressing the bioengineered insulin gene that were exposed to radio waves were shown to have insulin expression activated and blood glucose lowered.
8. Conclusions and Perspectives
Despite being a relatively new discipline, synthetic biology holds a great promise for the development of next generation therapeutics. Although traditional genome engineering strategies can potentially correct some of the genetic disorders, many human disorders are not strictly genetic. Thus, complex ailments such as infectious diseases, cancer, metabolic disorders, and many other known diseases are far more complex to be tackled by a simple genetic correction. For example, infectious diseases, as well as cancer, being agents capable of evolving and adapting, are the most difficult in tackling due to the development of resistance to human interventions. Therapeutic agents with exceptional specificity and efficacy, in combination with agents that inhibit adaptability of infections and malignancies, would be required for tackling these disorders. Many metabolic, immunological, neurological, and other disorders often develop in response to a combination of complex genetic background and variable environmental conditions. These pathologies would require state-of the art devices capable of sensing and self-regulating in response to fluctuating internal and external factors. Thus, sophisticated devices, such as those proposed by synthetic biology, are necessary for development of more potent therapeutic solutions than those currently available in the clinic.
In this review, we have discussed some of the current developments of synthetic biology in addressing human disorders. Synthetic bacterial and viral devices were developed from well studied, model microorganisms with an established genome manipulation and functional toolkit and showed potential for the development of even more complex devices. The first mammalian synthetic circuits for therapeutic applications have also been developed in the past decade; however, their development lagged behind bacterial and viral devices due to the limited number of functional parts, more complex metazoan gene circuitry, and the absence of precise and robust genome engineering strategies. However, with the development of cheaper gene synthesis methods,78 new state of the art genome engineering tools,17 and ongoing development of functional parts,79 this field is continually and increasingly growing.
Despite the current progress, synthetic biology has still a long road for clinical application. Yet, with the current tools and the rate of development, it is easy to envision synthetic biology contributing greatly to faster drug discovery and drug development as well as production of new and more affordable medicines. Among other applications, clinical application of commensal bacteria and bacteriophages against pathogenic bacteria, immune cells that kill metastatic solid tumor cells, and therapeutic sensor-effector devices for personalized medicine can one day become a reality.
Acknowledgments
We gratefully acknowledge financial support from the Energy Biosciences Institute, National Institutes of Health (GM077596), Defense Advanced Research Projects Agency (DARPA), Department of Energy (DE-SC0008743), and National Academies Keck Futures Initiative on Synthetic Biology.
Glossary
Abbreviations
- AI-2
autoinducer 2
- APCs
antigen-presenting cells
- AR
androgen receptor
- BCR
B cell antigen receptor
- CEA
carcinoembryonic antigen
- CAI-1
cholera autoinducer 1
- CARs
chimeric antigen receptors
- CRISPR
clustered regularly interspaced short palindromic repeats
- CRPC
castration-resistant prostate cancer
- DARPins
designed ankyrin repeat proteins
- DCs
dendritic cells
- DspB
dispersin B
- GLP-1
glucagon-like peptide 1
- GMCSF
granulocyte macrophage colony-stimulating factor
- HR
homologous recombination
- Igβ
immunoglobulin-β
- MHC
major histocompatibility complexes
- NHEJ
nonhomologous end joining
- PmpD
polymorphic membrane protein
- PPARα
peroxisome proliferator-activated receptor-α
- scFv
single chain variable fragment
- siRNA
small interfering RNA
- TALEN
transcription activator-like nucleases
- ZFN
zinc finger nucleases
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Gardner T. S.; Cantor C. R.; Collins J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403, 339–42. [DOI] [PubMed] [Google Scholar]
- Elowitz M. B.; Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403, 335–8. [DOI] [PubMed] [Google Scholar]
- You L.; Cox R. S. III; Weiss R.; Arnold F. H. Programmed population control by cell-cell communication and regulated killing. Nature 2004, 428, 868–71. [DOI] [PubMed] [Google Scholar]
- Fung E.; Wong W. W.; Suen J. K.; Bulter T.; Lee S. G.; Liao J. C. A synthetic gene-metabolic oscillator. Nature 2005, 435, 118–22. [DOI] [PubMed] [Google Scholar]
- Rinaudo K.; Bleris L.; Maddamsetti R.; Subramanian S.; Weiss R.; Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nature Biotechnol. 2007, 25, 795–801. [DOI] [PubMed] [Google Scholar]
- Tabor J. J.; Salis H. M.; Simpson Z. B.; Chevalier A. A.; Levskaya A.; Marcotte E. M.; Voigt C. A.; Ellington A. D. A synthetic genetic edge detection program. Cell 2009, 137, 1272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E.; Lu T. K.; Wang X.; Shi D.; Church G.; Collins J. J. Synthetic gene networks that count. Science 2009, 324, 1199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T.; Wang X.; Collins J. J. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009, 27, 465–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino T.; Mondragon-Palomino O.; Tsimring L.; Hasty J. A synchronized quorum of genetic clocks. Nature 2010, 463, 326–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassetti M.; Merelli M.; Temperoni C.; Astilean A. New antibiotics for bad bugs: where are we?. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. E.; Patel N. G.; Levy M. A.; Storeygard A.; Balk D.; Gittleman J. L.; Daszak P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan C.; Van Schaeybroeck S.; Longley D. B.; Johnston P. G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 2013, 13, 714–26. [DOI] [PubMed] [Google Scholar]
- Pritchard J. R.; Lauffenburger D. A.; Hemann M. T. Understanding resistance to combination chemotherapy. Drug Resist. Updates 2012, 15, 249–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S.; Lougaris V.; Caraffi S.; Zuntini R.; Yang J.; Soresina A.; Meini A.; Cazzola G.; Rossi C.; Reth M.; Plebani A. Mutations of the Igbeta gene cause agammaglobulinemia in man. J. Exp. Med. 2007, 204, 2047–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larman H. B.; Zhao Z.; Laserson U.; Li M. Z.; Ciccia A.; Gakidis M. A.; Church G. M.; Kesari S.; Leproust E. M.; Solimini N. L.; Elledge S. J. Autoantigen discovery with a synthetic human peptidome. Nat. Biotechnol. 2011, 29, 535–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M.; Yang Y. Targeted genome editing tools for disease modeling and gene therapy. Curr. Gene Ther. 2014, 14, 2–9. [DOI] [PubMed] [Google Scholar]
- Gaj T.; Gersbach C. A.; Barbas C. F. III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.; Xiao L.; Li A. S.; Zhang X.; Sirois P.; Zhang J.; Li K. Biological and biomedical applications of engineered nucleases. Mol. Biotechnol. 2013, 55, 54–62. [DOI] [PubMed] [Google Scholar]
- Nyquist M. D.; Li Y.; Hwang T. H.; Manlove L. S.; Vessella R. L.; Silverstein K. A.; Voytas D. F.; Dehm S. M. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 17492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau M.; Ghezraoui H.; De Cian A.; Guittat L.; Tomishima M.; Perrouault L.; Rene O.; Katibah G. E.; Zhang L.; Holmes M. C.; Doyon Y.; Concordet J. P.; Giovannangeli C.; Jasin M.; Brunet E. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013, 23, 1182–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O.; Sanjana N. E.; Hartenian E.; Shi X.; Scott D. A.; Mikkelsen T. S.; Heckl D.; Ebert B. L.; Root D. E.; Doench J. G.; Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.; Wei J. J.; Sabatini D. M.; Lander E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014, 343, 80–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber W.; Schoenmakers R.; Keller B.; Gitzinger M.; Grau T.; Daoud-El Baba M.; Sander P.; Fussenegger M. A synthetic mammalian gene circuit reveals antituberculosis compounds. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 9994–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Nicolini V.; Fux C.; Fussenegger M. A novel mammalian cell-based approach for the discovery of anticancer drugs with reduced cytotoxicity on non-dividing cells. Invest. New Drugs 2004, 22, 253–62. [DOI] [PubMed] [Google Scholar]
- Baker D. D.; Chu M.; Oza U.; Rajgarhia V. The value of natural products to future pharmaceutical discovery. Nat. Prod. Rep. 2007, 24, 1225–44. [DOI] [PubMed] [Google Scholar]
- Lee S. Y.; Kim H. U.; Park J. H.; Park J. M.; Kim T. Y. Metabolic engineering of microorganisms: general strategies and drug production. Drug Discovery Today 2009, 14, 78–88. [DOI] [PubMed] [Google Scholar]
- Mora-Pale M.; Sanchez-Rodriguez S. P.; Linhardt R. J.; Dordick J. S.; Koffas M. A. Biochemical strategies for enhancing the in vivo production of natural products with pharmaceutical potential. Curr. Opin. Biotechnol. 2014, 25, 86–94. [DOI] [PubMed] [Google Scholar]
- Keasling J. D. Manufacturing molecules through metabolic engineering. Science 2010, 330, 1355–8. [DOI] [PubMed] [Google Scholar]
- Ajikumar P. K.; Xiao W. H.; Tyo K. E.; Wang Y.; Simeon F.; Leonard E.; Mucha O.; Phon T. H.; Pfeifer B.; Stephanopoulos G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 2010, 330, 70–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C. J.; Westfall P. J.; Pitera D. J.; Benjamin K.; Fisher K.; McPhee D.; Leavell M. D.; Tai A.; Main A.; Eng D.; Polichuk D. R.; Teoh K. H.; Reed D. W.; Treynor T.; Lenihan J.; Fleck M.; Bajad S.; Dang G.; Dengrove D.; Diola D.; Dorin G.; Ellens K. W.; Fickes S.; Galazzo J.; Gaucher S. P.; Geistlinger T.; Henry R.; Hepp M.; Horning T.; Iqbal T.; Jiang H.; Kizer L.; Lieu B.; Melis D.; Moss N.; Regentin R.; Secrest S.; Tsuruta H.; Vazquez R.; Westblade L. F.; Xu L.; Yu M.; Zhang Y.; Zhao L.; Lievense J.; Covello P. S.; Keasling J. D.; Reiling K. K.; Renninger N. S.; Newman J. D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–32. [DOI] [PubMed] [Google Scholar]
- Rischer H.; Hakkinen S. T.; Ritala A.; Seppanen-Laakso T.; Miralpeix B.; Capell T.; Christou P.; Oksman-Caldentey K. M. Plant cells as pharmaceutical factories. Curr. Pharm. Des. 2013, 19, 5640–60. [DOI] [PubMed] [Google Scholar]
- Guillon S.; Tremouillaux-Guiller J.; Pati P. K.; Rideau M.; Gantet P. Harnessing the potential of hairy roots: dawn of a new era. Trends Biotechnol. 2006, 24, 403–9. [DOI] [PubMed] [Google Scholar]
- Kai G.; Xu H.; Zhou C.; Liao P.; Xiao J.; Luo X.; You L.; Zhang L. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 2011, 13, 319–27. [DOI] [PubMed] [Google Scholar]
- Giudice E. L.; Campbell J. D. Needle-free vaccine delivery. Adv. Drug Delivery Rev. 2006, 58, 68–89. [DOI] [PubMed] [Google Scholar]
- Streatfield S. J. Delivery of plant-derived vaccines. Expert Opin. Drug Delivery 2005, 2, 719–28. [DOI] [PubMed] [Google Scholar]
- Dreesen I. A.; Charpin-El Hamri G.; Fussenegger M. Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection. J. Biotechnol. 2010, 145, 273–80. [DOI] [PubMed] [Google Scholar]
- Ardiani A.; Higgins J. P.; Hodge J. W. Vaccines based on whole recombinant Saccharomyces cerevisiae cells. FEMS Yeast Res. 2010, 10, 1060–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remondo C.; Cereda V.; Mostbock S.; Sabzevari H.; Franzusoff A.; Schlom J.; Tsang K. Y. Human dendritic cell maturation and activation by a heat-killed recombinant yeast (Saccharomyces cerevisiae) vector encoding carcinoembryonic antigen. Vaccine 2009, 27, 987–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J. R.; Papamichail D.; Skiena S.; Futcher B.; Wimmer E.; Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science 2008, 320, 1784–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M.; Debarbieux L. Tools from viruses: bacteriophage successes and beyond. Virology 2012, 434, 151–61. [DOI] [PubMed] [Google Scholar]
- Lu T. K.; Collins J. J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 11197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T. K.; Collins J. J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 4629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai S. R.; Yoo S. Y.; Lee S. W.; Dean D. Engineered phage-based therapeutic materials inhibit Chlamydia trachomatis intracellular infection. Biomaterials 2012, 33, 5166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan F.; March J. C. Interrupting Vibrio cholerae infection of human epithelial cells with engineered commensal bacterial signaling. Biotechnol. Bioeng. 2008, 101, 128–34. [DOI] [PubMed] [Google Scholar]
- Duan F.; March J. C. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 11260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa A. A.; Klumpe H. E.; Luo M. L.; Selle K.; Barrangou R.; Beisel C. L. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio 2013, 5, e00928–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez E. E.; Wang J.; Miller J. C.; Jouvenot Y.; Kim K. A.; Liu O.; Wang N.; Lee G.; Bartsevich V. V.; Lee Y. L.; Guschin D. Y.; Rupniewski I.; Waite A. J.; Carpenito C.; Carroll R. G.; Orange J. S.; Urnov F. D.; Rebar E. J.; Ando D.; Gregory P. D.; Riley J. L.; Holmes M. C.; June C. H. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 808–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K.; Ely A.; Mussolino C.; Cathomen T.; Arbuthnot P. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Mol. Ther. 2013, 21, 1889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H.; Misawa N.; Kanemura Y.; Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013, 3, 2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze D. Y.; Reid T. R.; Rose S. C. Oncolytic virotherapy. J. Vasc. Interventional Radiol. 2013, 24, 1115–22. [DOI] [PubMed] [Google Scholar]
- Miest T. S.; Cattaneo R. New viruses for cancer therapy: meeting clinical needs. Nat. Rev. Microbiol. 2014, 12, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M. H.; Rottier P. J. Retargeting of viruses to generate oncolytic agents. Adv. Virol. 2012, 2012, 798526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich K.; Hanauer J. R.; Prufer S.; Munch R. C.; Volker I.; Filippis C.; Jost C.; Hanschmann K. M.; Cattaneo R.; Peng K. W.; Pluckthun A.; Buchholz C. J.; Cichutek K.; Muhlebach M. D. DARPin-targeting of measles virus: unique bispecificity, effective oncolysis, and enhanced safety. Mol. Ther. 2013, 21, 849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluza K. M.; Kottke T.; Diaz R. M.; Rommelfanger D.; Thompson J.; Vile R. Adoptive transfer of cytotoxic T lymphocytes targeting two different antigens limits antigen loss and tumor escape. Hum. Gene Ther. 2012, 23, 1054–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerullo V.; Pesonen S.; Diaconu I.; Escutenaire S.; Arstila P. T.; Ugolini M.; Nokisalmi P.; Raki M.; Laasonen L.; Sarkioja M.; Rajecki M.; Kangasniemi L.; Guse K.; Helminen A.; Ahtiainen L.; Ristimaki A.; Raisanen-Sokolowski A.; Haavisto E.; Oksanen M.; Karli E.; Karioja-Kallio A.; Holm S. L.; Kouri M.; Joensuu T.; Kanerva A.; Hemminki A. Oncolytic adenovirus coding for granulocyte macrophage colony-stimulating factor induces antitumoral immunity in cancer patients. Cancer Res. 2010, 70, 4297–309. [DOI] [PubMed] [Google Scholar]
- Forbes N. S. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 2010, 10, 785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa P. E.; Paniccia A.; Monegal A.; de Marco A.; Rescigno M. Salmonella engineered to express CD20-targeting antibodies and a drug-converting enzyme can eradicate human lymphomas. Blood 2013, 122, 705–14. [DOI] [PubMed] [Google Scholar]
- Pedrazzoli P.; Comoli P.; Montagna D.; Demirer T.; Bregni M.; Ebmt S. Is adoptive T-cell therapy for solid tumors coming of age?. Bone Marrow Transplant. 2012, 47, 1013–9. [DOI] [PubMed] [Google Scholar]
- Han E. Q.; Li X. L.; Wang C. R.; Li T. F.; Han S. Y. Chimeric antigen receptor-engineered T cells for cancer immunotherapy: progress and challenges. J. Hematol. Oncol. 2013, 6, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielewski M.; Hombach A. A.; Abken H. Antigen-specific T-cell activation independently of the MHC: Chimeric antigen receptor-redirected T cells. Front. Immunol. 2013, 4, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadelain M.; Brentjens R.; Riviere I. The promise and potential pitfalls of chimeric antigen receptors. Curr. Opin. Immunol. 2009, 21, 215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentjens R. J.; Riviere I.; Park J. H.; Davila M. L.; Wang X.; Stefanski J.; Taylor C.; Yeh R.; Bartido S.; Borquez-Ojeda O.; Olszewska M.; Bernal Y.; Pegram H.; Przybylowski M.; Hollyman D.; Usachenko Y.; Pirraglia D.; Hosey J.; Santos E.; Halton E.; Maslak P.; Scheinberg D.; Jurcic J.; Heaney M.; Heller G.; Frattini M.; Sadelain M. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood 2011, 118, 4817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D. L.; Levine B. L.; Kalos M.; Bagg A.; June C. H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 2011, 365, 725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoldo B.; Ramos C. A.; Liu E.; Mims M. P.; Keating M. J.; Carrum G.; Kamble R. T.; Bollard C. M.; Gee A. P.; Mei Z.; Liu H.; Grilley B.; Rooney C. M.; Heslop H. E.; Brenner M. K.; Dotti G. CD28 costimulation improves expansion and persistence of chimeric antigen receptor-modified T cells in lymphoma patients. J. Clin. Invest. 2011, 121, 1822–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammana S.; Huang X.; Wong M.; Milone M. C.; Ma L.; Levine B. L.; June C. H.; Wagner J. E.; Blazar B. R.; Zhou X. 4-1BB and CD28 signaling plays a synergistic role in redirecting umbilical cord blood T cells against B-cell malignancies. Hum. Gene Ther. 2010, 21, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss C. C.; Condomines M.; Cartellieri M.; Bachmann M.; Sadelain M. Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol. 2013, 31, 71–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanitis E.; Poussin M.; Klattenhoff A. W.; Song D.; Sandaltzopoulos R.; June C. H.; Powell D. J. Chimeric antigen receptor T cells with dissociated signaling domains exhibit focused anti-tumor activity with reduced potential for toxicity. Cancer immunology research 2013, 10.1158/2326-6066.CIR-13-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk R. W.; Vogel H.; Schurmann A. Genetic and epigenetic control of metabolic health. Mol. Metab. 2013, 2, 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan F.; Curtis K. L.; March J. C. Secretion of insulinotropic proteins by commensal bacteria: rewiring the gut to treat diabetes. Appl. Environ. Microbiol. 2008, 74, 7437–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H.; Aubel D.; Fussenegger M. Synthetic mammalian gene circuits for biomedical applications. Curr. Opin. Chem. Biol. 2013, 17, 910–7. [DOI] [PubMed] [Google Scholar]
- Lienert F.; Lohmueller J. J.; Garg A.; Silver P. A. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol. 2014, 15, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M.; Weber W. Therapeutic synthetic gene networks. Curr. Opin. Biotechnol. 2012, 23, 703–11. [DOI] [PubMed] [Google Scholar]
- Ye H.; Charpin-El Hamri G.; Zwicky K.; Christen M.; Folcher M.; Fussenegger M. Pharmaceutically controlled designer circuit for the treatment of the metabolic syndrome. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmer C.; Gitzinger M.; Daoud-El Baba M.; Djonov V.; Stelling J.; Fussenegger M. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotechnol. 2010, 28, 355–60. [DOI] [PubMed] [Google Scholar]
- Rossger K.; Charpin-El-Hamri G.; Fussenegger M. A closed-loop synthetic gene circuit for the treatment of diet-induced obesity in mice. Nat. Commun. 2013, 4, 2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H.; Daoud-El Baba M.; Peng R. W.; Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science 2011, 332, 1565–8. [DOI] [PubMed] [Google Scholar]
- Stanley S. A.; Gagner J. E.; Damanpour S.; Yoshida M.; Dordick J. S.; Friedman J. M. Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science 2012, 336, 604–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T.; Bao X.; Piao W.; Peng J.; Li W.; Yang C.; Xing M.; Zhang Y.; Qi J.; Xu L.; Xu L.; Liu Q. Recent patents on oligonucleotide synthesis and gene synthesis. Recent Pat. DNA Gene Sequences 2012, 6, 10–21. [DOI] [PubMed] [Google Scholar]
- Smolke C. D. Building outside of the box: iGEM and the BioBricks Foundation. Nat. Biotechnol. 2009, 27, 1099–102. [DOI] [PubMed] [Google Scholar]