Summary
Chemical shifts are obtained at the first stage of any protein structural study by NMR spectroscopy. Chemical shifts are known to be impacted by a wide range of structural factors and the artificial neural network based TALOS-N program has been trained to extract backbone and sidechain torsion angles from 1H, 15N and 13C shifts. The program is quite robust, and typically yields backbone torsion angles for more than 90% of the residues, and sidechain χ1 rotamer information for about half of these, in addition to reliably predicting secondary structure. The use of TALOS-N is illustrated for the protein DinI, and torsion angles obtained by TALOS-N analysis from the measured chemical shifts of its backbone and 13Cβ nuclei are compared to those seen in a prior, experimentally determined structure. The program is also particularly useful for generating torsion angle restraints, which then can be used during standard NMR protein structure calculations.
Keywords: NMR, Chemical shifts, Protein structure, Side-chain conformation, Artificial neural network, Secondary structure, Backbone torsion angle
1 Introduction
1.1 Relations between chemical shifts and protein structure
The first step of any protein structural study by NMR spectroscopy typically involves assignment of the multitude of NMR resonances to individual nuclei. Originally, for proteins extracted from natural sources, this only involved assignment of the hydrogen NMR spectra (1, 2). However, due to extensive resonance overlap in 1H NMR spectra, this technology was restricted to relatively small proteins. With advances in molecular biology, the vast majority of today’s structural studies focus on cloned proteins, typically overexpressed in Escherichia coli (3–5). By using suitable isotopically enriched growth media, it then is readily feasible to obtain essentially full incorporation of the NMR-observable stable isotopes 13C and 15N. These nuclei not only are key to dispersing the crowded NMR spectra in three or four orthogonal frequency dimensions, dramatically reducing the resonance overlap problem, the 13C and 15N chemical shifts themselves have proven to be important reporters on local backbone conformation (6–8). NMR chemical shifts in proteins are exquisitely sensitive to local conformation. However, they depend on many different factors, including backbone and side-chain torsion angles, neighboring residues, ring currents caused by nearby aromatic groups, hydrogen bonding, electric fields, local strain and geometric distortions, as well as solvent exposure (9–15). This not only has made it difficult to separately quantify the relation between each of these parameters and the chemical shift, it also makes it impossible to uniquely attribute such a structural parameter to any individual chemical shift.
For protein NMR spectroscopy, triple resonance correlation experiments, which link the resonances of directly bonded 1H, 13C and 15N nuclei are commonly used to assign the chemical shifts of 1H, 13C, and 15N nuclei in proteins (16–18). The chemical shift assignment procedure usually consists of two steps: (1) sequence-specific assignment of the backbone atoms, and (2) side-chain assignments. Nearly complete chemical shift assignments for backbone and side-chain atoms are commonly required to assign nuclear Overhauser enhancement (NOE) spectra, which classically are used to derive interproton distances that serve as the primary experimental restraints for calculating the protein structure. The backbone (1Hα, 13C′, 13Cα, 15N and 1HN) and 13Cβ chemical shifts, which are generally obtained in the earliest stage of any protein NMR study, are particularly useful reporters on local conformation. Their link to secondary structure, as well as to hydrogen bonding and χ1 sidechain torsion angles, has been long recognized and has been the focus of both empirical studies as well as quantum-chemical calculations (11–15, 19, 20).
1.2 Protein backbone and side-chain conformation from NMR chemical shifts
The rapid increase in the number of proteins for which both high-resolution structural coordinates have been deposited in the Protein Data Bank (PDB) (21) and NMR chemical shift assignments are available in the BioMagResBank (BRMB) (22), have stimulated the development of quantitative empirical methods to study the relation between protein structure and chemical shifts (23). Among the wide array of empirical methods, TALOS (20) and its two successors TALOS+ (24) and TALOS-N (25) have become particularly widely used for making accurate ϕ/ψ backbone torsion angle predictions on the basis of the backbone (13Cα, 13C′, 15N, 1Hα and 1HN) and 13Cβ chemical shift assignments. These ϕ/ψ predictions can be used to validate NOE-derived NMR structures that didn’t use chemical shift derived input parameters or, conversely, to generate additional restraints as input to the protein structure calculation and refinement protocols.
The original TALOS program (Torsion Angle Likeliness Obtained from Shifts) searches a protein database, consisting originally of only 20 proteins, but later expanded to ca 200 proteins, with both high-resolution X-ray coordinates and NMR chemical shift assignments. TALOS identifies the ten tripeptide fragments that represent the best match in terms of chemical shifts and residue types to those of a tripeptide segment whose assignments are known and whose structure is under study (the “target protein”). The assumption underlying TALOS is that fragments with similar chemical shifts and residue type typically have similar backbone conformations. Thus, if these ten best matched fragments have consistent, narrowly clustered values for the ϕ/ψ angles of their center residue, their averages and standard deviations are used as a prediction for the ϕ/ψ angles of the center residue of the target protein tripeptide. If the ϕ/ψ angles of the center residue of these ten best matched tripeptides fall in different regions of the Ramachandran map, the matches are declared ambiguous, and no prediction is made for the central residue. With this quality control criterion, TALOS predicted ϕ/ψ torsion angles for, on average, ca 72% of the residues in any given target protein. For TALOS validation proteins, where the true ϕ/ψ angles are known, only about 1.8% of the predictions were inconsistent with crystallographically determined ϕ/ψ torsion angles. Excluding these 1.8% erroneous predictions, a root mean square (RMS) difference of ca 13° is observed between predicted and crystallographically observed ϕ/ψ torsion angles.
Although rather robust, the original TALOS program was unable to make definitive predictions for about 28% of the residues in any given protein. Most of these 28% are located outside regular secondary structure, exactly those regions where backbone torsion angle information is most needed. TALOS+ was developed to address this shortcoming, and to extend the coverage of the program {Shen, 2009 #4712}. For a given residue in the target protein, TALOS+ first uses an artificial neural network (ANN) module to predict its three-state distribution in the Ramachandran map, i.e., α, β, and positive-ϕ. This three-state distribution is subsequently used to guide the database search procedure for the 10 best matches. With the incorporation of the ANN, TALOS+ is able to increase its coverage to ca 88%, without sacrificing accuracy. Thus, compared to the original TALOS program, the fraction of residues whose backbone angles cannot be predicted is reduced from ~28% to ~12%. Importantly, most of the additional ϕ/ψ torsion angle predictions are made for residues in loop or turn regions, where this information is needed most.
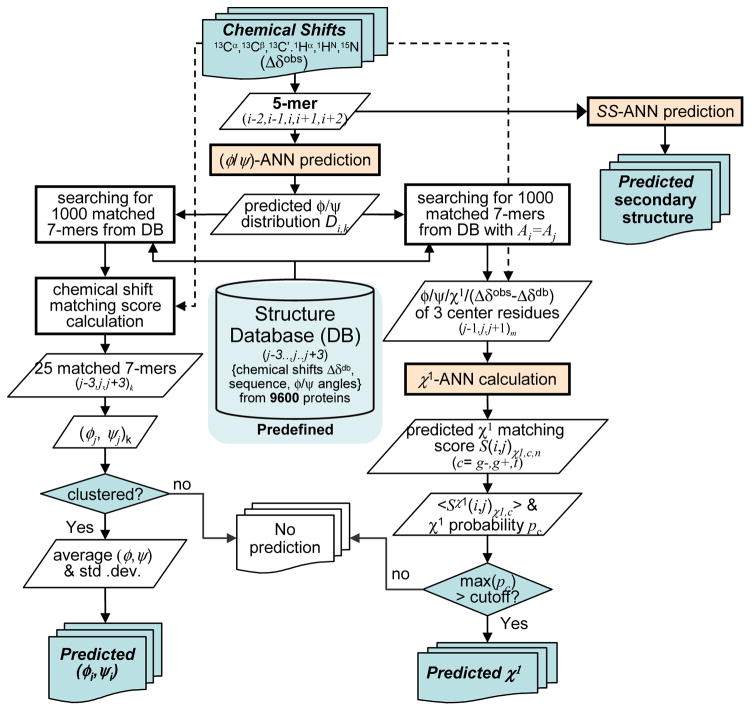
The recently introduced TALOS-N program relies far more extensively on neural network analysis of the input chemical shift data than TALOS+, thereby further increasing coverage, accuracy and reliability. In addition, TALOS-N is the first program to generate quite accurate predictions for the side-chain χ1 torsion angles (Fig. 1).
Fig. 1.
Flowchart of the TALOS-N program (reproduced from (25) with permission from Springer).
For the ϕ/ψ torsion angle prediction of a given residue i in the target protein, a well-trained two-level feed-forward multilayer ANN, referred to as a (ϕ,ψ)-ANN, is first used by TALOS-N to predict the 324-state ϕ/ψ distribution of residue i on the basis of the NMR chemical shifts and residue type of itself and its adjacent residues (i−2 to i+2). Here, the 324-state ϕ/ψ distribution corresponds to the likelihood that residue i adopts torsion angles that fall in any of the 324 voxels, of 20°×20° each, that make up the Ramachandran map. The ANN-predicted ϕ/ψ distribution is then used solely to search a large crystallographic database (containing 9523 proteins, with chemical shifts added by a computational method (26)), for a pool of 1000 heptapeptide fragments with ϕ/ψ angles that best match the 324-state ϕ/ψ distribution. These top 1000 fragments then are further evaluated for the agreement between their computed chemical shifts and experimental values of the corresponding heptapeptide segment (i−3 to i+3) in the target protein. The 25 best-matched database heptapeptides are retained, and the ϕ/ψ angles of their center residues are inspected by using an advanced clustering analysis, and subsequently used to make a prediction for the ϕ/ψ angles of the query residue. Validation on an independent set of proteins indicates that backbone torsion angles can be predicted for a larger, ≥ 90% fraction of the residues, with an error rate of ca 3.5% when using an acceptance criterion that is nearly two-fold tighter than that used previously by TALOS and TALOS+. The RMS difference between predicted and crystallographically observed ϕ/ψ torsion angles is ca 12°, also slightly better than what was obtained with the earlier versions of the program.
To predict the χ1 rotameric state (g−, g+ or t) for a given residue i (of residue type a) in the target protein, TALOS-N uses another set of ANNs, referred to as (χ1)a-ANNs. The (χ1)a-ANN has been trained to correlate the center residue likelihood of adopting each of the three χ1 rotameric states to the differences between its observed chemical shifts and those expected on the basis of its backbone conformation. A separate database search procedure is subsequently used to estimate the three-state probability of residue i to adopt the three χ1 rotameric states. With an optimized error control criterion, TALOS-N predicts χ1 rotameric states for ca 50% of the residues, with an “error rate” of ca 10% when comparing the predicted χ1 rotameric state to that of any given reference structure. However, we note that the true error is likely to be much lower, as for proteins that have multiple available independently solved X-ray structures, the χ1 rotameric states of any “erroneous” χ1 prediction is typically in agreement with that of another X-ray structure (25).
Similar to TALOS+, TALOS-N is also implemented with an ANN-based module for predicting secondary structure (SS) from the NMR chemical shifts. For this purpose, TALOS-N uses two separate ANNs, referred to as SS-ANN and SSseq-ANN, which are trained to correlate the three-state secondary structure classification (helix, sheet, and coil) of a residue to both the chemical shifts and amino acid sequence, or to amino acid sequence alone, respectively. The output of these two ANNs is used in a hybrid manner to predict secondary structure for any residue in a protein, regardless of the completeness of chemical shift assignments. The overall correctness of the SS prediction is ca 88% when NMR chemical shifts are available, dropping to ca 81% when no chemical shifts are available. In the absence of chemical shifts, TALOS-N matches the accuracy of the best sequence-only secondary structure prediction programs (27, 28).
2 Materials
In this chapter, we use the protein DinI (29) to illustrate the use of TALOS-N for predicting its backbone ϕ/ψ and side-chain χ1 torsion angles, as well as its secondary structure classification. To follow the examples, both the TALOS-N software package and an input file with correctly formatted chemical shift assignments are needed.
2.1 Software Requirements
The TALOS-N software package, including the required binaries for three of the most common operating systems, Linux, Mac OS X and Windows, as well as the requisite protein database and scripts can be downloaded from http://spin.niddk.nih.gov/bax/software/TALOS-N/ and installed straightforwardly (see Note 1). Alternatively, a server version of TALOS-N can be used directly, without installing the TALOS-N software (http://spin.niddk.nih.gov/bax/nmrserver/talosn/).
2.2 Data Requirements
An input table containing both the full amino acid sequence and the NMR chemical shift assignments is required, to be prepared with a specific data format (general purpose NMRPipe table format). As an example, an excerpt of such a file is shown below for the protein DinI:
DATA FIRST_RESID 2
DATA SEQUENCE RIEVTIAKT SPLPAGAIDA LAGELSRRIQ YAFPDNEGHV SVRYAAANNL
DATA SEQUENCE SVIGATKEDK QRISEILQET WESADDWFVS E
VARS RESID RESNAME ATOMNAME SHIFT
FORMAT %4d %1s %4s %8.3f
2 R C 174.123
2 R CA 55.537
2 R CB 32.786
2 R H 8.772
2 R HA 4.994
2 R N 123.394
3 I C 173.941
3 I CA 60.986
The protein’s amino acid sequence should be provided in one or more lines starting with the tag “DATA SEQUENCE”. Only the one-character residue name is allowed (see Note 2) and space characters in the sequence are ignored. An optional line beginning with a tag of “DATA FIRST_RESID” is needed to specify the first residue number of the amino acid sequence listed in the “DATA SEQUENCE” line if the first residue listed is not residue number 1. For the chemical shift table, columns for residue number, one-character residue type (see Note 2), atom name (see Note 3) and chemical shift value must be included, and their definitions (“RESID”, “RESNAME”, “ATOMNAME”, and “SHIFT” respectively) must be pre-declared in a line beginning with a “VARS” tag; a line beginning with a “FORMAT” tag is also required (immediately after the “VARS” line) to define the data type of each corresponding column of the table.
Note that all chemical shifts used as input for TALOS-N are required to be properly referenced (see Note 4) to ensure the accuracy and reliability of the predictions. If the protein sample used to collect the NMR chemical shift data is perdeuterated, 2H isotope corrections (30) need to be applied for 13Cα and 13Cβ chemical shifts (see Note 5).
Other standard chemical shift formats, such as the NMR-Star format used by the BMRB database, can also be used as input after a format conversion. A conversion script is provided in the TALOS-N package for this purpose (see Note 6). The server version of TALOS-N includes automated chemical shift format identification and can use the NMR-Star format chemical shift file directly as input, without requiring prior format conversion.
3. Methods
3.1 TALOS-N prediction
The TALOS-N prediction can be performed for DinI with an input chemical shift file of name “inCS.tab” by typing the command:
talosn -in inCS.tab
The program first converts the chemical shifts (δ) of each query residue to its corresponding secondary chemical shifts (Δδ) by subtracting a residue-type dependent random coil value, as well as corrections to account for the residue types of its two immediate neighbors. The converted secondary chemical shifts are stored in a file named “predAdjCS.tab” (in the “SHIFT” column), together with the original chemical shifts (“CS_OBS”) and the corresponding corrections (“CS_ADJ”, which is the random coil value including nearest neighbor (i±1) residue type correction) used to calculate the secondary chemical shifts. To make a ϕ/ψ angle prediction, the converted secondary chemical shifts together with the amino acid type information are used as inputs for the (ϕ/ψ)-ANN to calculate the 324-state ϕ/ψ distribution for each predictable residue (see Note 7), with the output stored in a file named “predANN.tab”. A database search step is then performed to search a 9523-protein database for the 25 best matched heptapeptides in terms of the 324-state ϕ/ψ angle distribution, the secondary chemical shifts, and the amino acid type. A single file, “predAll.tab”, is generated in this step to store the information of those best database matches for each of the residues in the target protein. A final summarization and quality control step is performed to identify outliers in the 25 best-matching heptapeptides by evaluating the clustering of the ϕ/ψ angles of their center resides in the Ramachandran map, or by using the observed ϕ/ψ of a reference structure if such a structure is available (this requires an additional option “-ref ref.pdb” in the command line, where “ref.pdb” is the name for the reference structure). A summary file “pred.tab” is then created, displaying the average ϕ and ψ values (in the PHI and PSI columns) and their respective standard deviations (DPHI and DPSI), as well as an aggregate, weighted χ2 score (DIST, see eq 12 of reference (25)), reflecting how well the target protein chemical shifts match those of the database fragments. An excerpt of this file for DinI is shown below:
VARS RESID RESNAME PHI PSI DPHI DPSI DIST S2 COUNT CS_COUNT CLASS
FORMAT %4d %s %8.3f %8.3f %8.3f %8.3f %8.3f %5.3f %2d %2d %s
2 R -107.206 129.115 9.502 7.843 0.293 0.873 25 16 Strong
3 I -117.237 126.352 6.691 6.523 0.180 0.883 25 18 Strong
For each predictable residue, or residue with sufficient input chemical shifts (see Note 7), a final classification is made (listed as the last “CLASS” column in the “pred.tab” file) for its ϕ/ψ angle prediction by a summarization step, detailed below. Prior to making this final classification, the program calculates the predicted backbone rigidity as reflected in the “random coil index” order parameter, RCI-S2 (31), which scales between 0 (total disorder) and 1 (fully rigid). Its values are included under the “S2” column in the “pred.tab” file. Residues below the threshold RCI-S2 ≤0.6 are assigned as dynamic (receiving a “Dyn” classification) in “pred.tab”. For other residues, a classification of strongly unambiguous is assigned (with a “Strong” tag) if the center residues of all 25 best matching heptapeptides locate in a consistent ϕ/ψ region in the Ramachandran map. A generously unambiguous classification is assigned (with a “Generous” tag) if the center residues of only the top 10 best matches cluster in a consistent ϕ/ψ region. All other cases are considered ambiguous (classified with a “Warn” tag), even though inspection of their Ramachandran map population may contain very useful information. For example, often the ambiguous residues will cluster in two distinct regions of the Ramachandran map, and the investigator can explore both options during structure calculations.
For the predictable residues, the ϕ/ψ angles are calculated by averaging the ϕ/ψ angles of the center residues of all 25 best matches (for residues classified as “Strong”) or from the top 10 best matches (for a “Generous” prediction), and shown in the “PHI”/“PSI” columns. The estimated uncertainties in the predicted ϕ/ψ angles are calculated from their standard deviations from these averages and listed in the “DPHI” and “DPSI” columns. Only when a known reference structure is provided as input to the program, the predicted ϕ/ψ values will be compared to the observed ϕ/ψ angles in this reference structure for all unambiguously predicted (“Strong” and “Generous”) residues. A prediction is labeled as “Bad” if the predicted and the observed ϕ/ψ angles are not consistent (see Note 8).
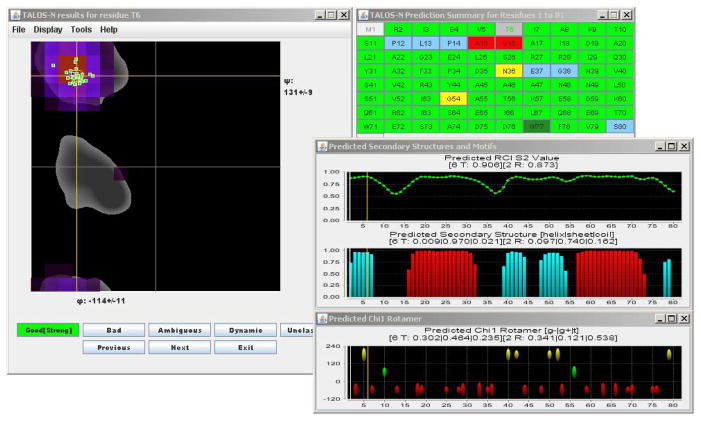
For DinI, 71 residues (out of a total of 81) are obtained with unambiguous ϕ/ψ angles prediction, 2 have an ambiguous ϕ/ψ angle prediction, and 6 are predicted as dynamic (Fig. 2). Among those 71 unambiguous predictions, 70 are classified as “Strong” and two (Ala15 and Gly16) are designated “Bad” after inspecting their consistency relative to the ϕ/ψ angles observed in the reference NMR structure. It is worth noting that in the reference structure, these latter two residues (with ϕ/ψ angles of −57°/49° and −176°/−18°, respectively) are located in very lowly populated regions of the Ramachandran map, i.e., they are statistically unlikely to occur. Without further experimental data, it is not possible to decide whether the “Bad” classification refers to the reference structure, or to the quality of the prediction.
Fig. 2.
Graphic TALOS-N inspection interface for protein DinI. (For details, see 3.2, or the TALOS-N webpage http://spin.niddk.nih.gov/bax/software/TALOS-N/)
After TALOS-N prediction of ϕ/ψ angles has been completed, another database search and ANN-based procedure is performed to predict the χ1 rotameric states. A χ1 rotamer prediction summary file “predChi1.tab” is created with an excerpt of this file shown below for DinI:
VARS RESID RESNAME CS_COUNT CHI1_OBS Q_Gm Q_Gp Q_T CLASS
FORMAT %4d %s %2d %8.3f %5.3f %5.3f %5.3f %s
2 R 16 -69.938 0.341 0.121 0.538 na
3 I 18 -62.494 0.873 0.063 0.063 g-
4 E 18 -61.087 0.312 0.093 0.595 na
5 V 18 178.554 0.073 0.055 0.872 t
6 T 18 66.182 0.302 0.464 0.235 na
7 I 18 -75.725 0.713 0.143 0.143 g-
For a query residue of residue type a (excluding Gly, Pro, and Ala) with sufficient input chemical shifts (see Note 7), TALOS-N first searches the database for the 1000 best-matched heptapeptides in terms of the backbone torsion angles and residue types. It then uses a trained (χ1)a-ANN to calculate a χ1 matching score for each database match, which measures the likelihood of the center residue of the database heptapeptide to adopt the same χ1 rotameric state as the query residue. The program then derives a three-state probability score, Pc, for the query residue to adopt each of the three χ1 rotameric states (c = g−, g+ and t, stored in the columns “Q_Gm”, “Q_Gp” and “Q_T”, respectively, in “predChi1.tab”). TALOS-N then classifies the prediction for the query residue to adopt χ1-rotamer state c (g−, g+ or t, as listed in the last column of “CLASS” in the “predChi1.tab” file) only when the predicted probability for state c is significantly higher than that for the other two states, by default Pc > 0.6. Otherwise, an ambiguous classification is assigned (with a “na” tag). Details of other contents in “predChi1.tab” are as follows: the column of “CS_COUNT” is for the count of the experimental chemical shifts of the target residue itself and its two neighbors; when a reference structure is provided, a “CHI1_OBS” column is provided to display the χ1 angle observed in the reference structure. For DinI, TALOS-N makes χ1 rotameric state predictions for 30 out of a total of 61 (non-Gly/-Pro/-Ala) residues, among which three (Asp35, Asn48 and Asp75) differ in their predicted χ1 rotameric state from the reference NMR structure (PDB entry 1GHH).
Next to predicting ϕ, ψ and χ1 torsion angles, TALOS-N also predicts the protein’s secondary structure. For residues with chemical shift assignments, a two-level neural network, SS-ANN, is trained to make a three-state secondary structure prediction (H, E or L, representing for α-helix, β-sheet and loop, respectively) on the basis of both the chemical shifts and the amino acid sequence information. In addition, another two-level ANN, referred to as SSseq-ANN, is trained by using solely the amino acid sequence information. It can be used to make predictions for residues that lack chemical shift information. However, this SSseq-ANN is used more generally by TALOS-N in a hybrid manner with the SS-ANN to make secondary structure prediction for proteins when chemical shift assignments are incomplete. TALOS-N generates an output file “predSS.tab” to store the predicted secondary structure. An excerpt of this file is shown below for DinI:
VARS RESID RESNAME CS_CNT CS_CNT_R2 Q_H Q_E Q_L CONFIDENCE SS_CLASS
FORMAT %4d %1s %2d %2d %8.3f %8.3f %8.3f %4.2f %s
1 M 10 4 0.333 0.333 0.333 0.00 L
2 R 16 6 0.097 0.740 0.162 0.58 E
3 I 18 6 0.027 0.970 0.003 0.94 E
4 E 18 6 0.006 0.968 0.026 0.94 E
5 V 18 6 0.004 0.963 0.033 0.93 E
6 T 18 6 0.009 0.970 0.021 0.95 E
Details of its contents are as follows: the column of “CS_CNT_R2” lists the number of experimental chemical shifts of the target residue; “CS_CNT” contains the count of experimental chemical shifts of the target residue plus its two immediate neighbors; the columns “Q_H”, “Q_E” and “Q_L” list the SS-ANN (or SSseq-ANN) predicted probability for the target residue to be of secondary structure type “H”, “E” and “L”, respectively; the values shown in the “CONFIDENCE” column represent the confidence of the 3-state secondary structure prediction for a given target residue, calculated from the difference of maximal and median values of “Q_H”, “Q_E” and “Q_L”; the text listed in the “SS_CLASS” column shows the final secondary structure classification assigned by the program, i.e., one of the three states with the maximal predicted probability.
For DinI, when comparing to the output of the DSSP program (32) for the reference structure (PDB entry 1GHH), the overall correctness ratio of the TALOS-N predicted secondary structure is 70/81. In this respect, it is important to note that even for proteins of known structure, secondary structure assignment can be ambiguous, as reflected in only ca 90% agreement among the output of some of the most popular programs (23).
As mentioned above, TALOS-N predictions can either be made locally by downloading the requisite programs, or can be performed via the TALOS-N server (http://spin.niddk.nih.gov/bax/nmrserver/talosn/), which requires a chemical shift file as input, and an e-mail address to send back the prediction results, including all abovementioned output files, such as “pred.tab”, “predChi1.tab”, “predSS.tab”, “predS2.tab”, “predAll.tab”, “predAdjCS.tab” and “predANN.tab”.
3.2 Manual inspection and adjustment
The TALOS-N predictions can be inspected and further adjusted by using a Java graphic program, jrama. Two examples of command line calls of this program are:
jrama -in pred.tab jrama -in pred.tab -ref DinI.pdb
Fig. 2 shows the jrama graphic interface, loaded with the TALOS-N predicted results for DinI. The left panel of the graphic interface shows a map of the ϕ/ψ angles of the center residues of the 25 best matched heptapeptides in the database (green squares) and the query residue Thr-6 (blue, depicting the angles observed in the NMR-derived PDB entry 1GHH), superimposed on a Ramachandran map depicting in gray the “most favorable” ϕ/ψ angles for Thr, i.e., those most commonly observed in high-resolution crystal structures of a very large array of proteins. The 324 (ϕ/ψ)-ANN predicted scores for Thr-6 are shown as colored voxels, but only for those that are populated at least one standard deviation above the average predicted voxel density. The top right panel displays the amino acid sequence of DinI, with residues colored according to their ϕ/ψ prediction classification. Missing predictions (e.g., residue M1) are shown in light grey, consistent predictions in light or dark green (for “Strong” and “Generous” predictions, respectively), ambiguous predictions in yellow, and dynamic residues in blue. Three other panels correspond to the RCI-S2 value, the predicted secondary structure (red, αhelix; aqua, β-sheet), with the height of the bars reflecting the probability assigned by the SS-ANN secondary structure prediction. The bottom right panel depicts the χ1 rotamer predictions (red oval: g−; green: g+; yellow: t), with the height of the ovals corresponding to the probability assigned by the χ1 rotameric state prediction.
The TALOS-N prediction (including the summary of TALOS-N predicted ϕ/ψ angles) is normally performed with the default parameters and settings. However, the left panel also can be used to manually adjust the prediction classification of a given query residue according to a user’s preference. The prediction files then will be overwritten to reflect any changes made interactively.
3.3 Generation of angular restraints
The TALOS-N output can be converted into ϕ and ψ torsion angle restraints that then can be used directly as input for a conventional protein NMR structure calculation procedure (33, 34). Two convenient scripts, “talos2dyana.com” and “talos2xplor.com”, are included in the TALOS-N software package for this purpose. These scripts read predicted ϕ and ψ angles from the TALOS-N prediction summary file “pred.tab”, and generate for each residue with an unambiguous TALOS-N prediction (classified as “Strong” or “Generous”) a ϕ and a ψ torsion angle restraint (see Note 9). These torsion angle restraints can be stored in either CYANA format, as shown below for residues 2 and 3 of DinI:
2 ARG PHI -127.2 -87.2 2 ARG PSI 109.1 149.1 3 ILE PHI -137.2 -97.2 3 ILE PSI 106.4 146.4
or in XPLOR format:
assign (resid 1 and name C ) (resid 2 and name N )
(resid 2 and name CA ) (resid 2 and name C ) 1.0 -107.2 20.0 2
assign (resid 2 and name N ) ( resid 2 and name CA )
(resid 2 and name C ) (resid 3 and name N ) 1.0 129.1 20.0 2
assign (resid 2 and name C ) (resid 3 and name N )
(resid 3 and name CA ) (resid 3 and name C ) 1.0 -117.2 20.0 2
assign (resid 3 and name N ) (resid 3 and name CA )
(resid 3 and name C ) (resid 4 and name N ) 1.0 126.4 20.0 2
These input restraints then can be used for protein structure calculations as a complement to the conventional NOE distance restraints. Note that such chemical shift derived torsion angle restraints alone are typically insufficient to reach a converged protein structure as each torsion angle contains a substantial uncertainty (±20°, in the above example), and these uncertainties rapidly accumulate when building the protein chain. Moreover, as mentioned above, predictions are generally only about 90% complete and may contain errors.
Acknowledgments
This work was funded by the Intramural Research Program of the NIDDK, NIH.
Footnotes
An installation shell script “install.com” is provided with the TALOS-N software package, which can be use for installing and configuring the TALOS-N program on a Linux or a Mac OS X system. After the installation, two starting shell scripts “talosn” and “jrama” are generated with properly configured installation paths for the system-specific binary and all required databases. For a Windows system, TALOS-N can be installed by simply uncompressing the package. However, when running the TALOS-N program, the TALOS-N installation path ($talosnDir”) must be specified on the fly, for example with the command (see Methods 3.1) of “$talosnDir/bin/TALOS.exe –in inCS –talosnDir $talosnDir”.
In both the sequence header and the chemical shift data table, the lower case “c” must be used for oxidized Cys (δ13Cβ ~ 42.5 ppm) and upper case “C” for reduced Cys (δ13Cβ ~ 28 ppm); “h” for protonated His and “H” for deprotonated His.
Atom names should be given exactly as: “HA” for Hα atoms of all non-Gly residues; “HA2” for the first Hα atom of a Gly residues and “HA3” for the second; “C” for C′ (CO) atoms; “CA” for Cα atoms; “CB” for Cβ atoms; “N” for amide nitrogen atoms, “HN” for amide hydrogens. Data for all other atom types will be ignored.
All 13C chemical shifts (including δ13Cα, δ13Cβ, and δ13C′) should be referenced relative to the methyl groups of 4,4-dimethyl-4-silapentane-1-sulfonic acid, or DSS (35). The 15N chemical shifts used as input for TALOS-N should be referenced relative to liquid ammonia at 25 °C (35). A pre-check module in TALOS-N will be used to identify possible referencing problems with the δ13Cα, δ13Cβ, δ13C′ and δ1Hα chemical shift inputs (36) when running a typical TALOS-N command with an additional “-check” option, for example by using the command line input argument “talosn -in inCS.tab -check”. This module first converts the chemical shifts (δ) of each residue to secondary chemical shifts (Δδ; see Methods 3.1), and subsequently evaluates these by correlating Δδ13Cα, Δδ13Cβ, Δδ13C′ and Δδ1Hα to the reference-free entity, Δδ13Cα - Δδ13Cβ (36). The estimated chemical shift referencing offsets, as well as their corresponding fitting error, will be printed for δ13Cα, δ13Cβ, δ13C′ and δ1Hα. An offset correction generally is only needed when the estimated referencing offset exceeds the average fitting error by more than about five standard deviations. This pre-check module will also identify residues with unusual chemical shifts, for which secondary chemical shifts fall outside the expected range. Such chemical shift outliers, especially those with highly unusual chemical shifts, for which secondary chemical shifts deviate from the expected range by more than 2 times of the normal range of secondary chemical shifts, may correspond to experimental errors, and need to be inspected carefully prior to using them for making torsion angle predictions.
2H isotope chemical shift corrections for 13Cα and 13Cβ (30) can be applied before starting the TALOS-N prediction, i.e., when generating the secondary chemical shifts. To do this, an additional option “-iso” must be added when running a TALOS-N prediction; for example by using a command line argument of the form “talosn -in inCS.tab -iso”.
A conversion Unix shell script, bmrb2talos.com, is included with the TALOS-N package and can be used to convert a NMR-Star format chemical shift table, used by the BMRB database, to TALOS format. An example command line for using this script is “bmrb2talos.com bmrb.str > inCS.tab”.
To ensure the prediction accuracy and reliability for a given query residue, the chemical shift sufficiency is first inspected by the program for the residue itself and its two immediate neighbors. If at least two of the three residues have at least three chemical shifts, the center residue is considered to be predictable.
The consistency between the predicted ϕ/ψ values (ϕpred/ψpred) and the observed ϕ/ψ angles (ϕobs/ψobs) is defined by .
For a residue with a “Strong” classification of its prediction, the φ and ψ angle restraints are set to <ϕ> ± 2σ and <ψ> ± 2σ, where <ϕ> and <ψ> are the averaged TALOS-N predictions, and 2σ is the larger of 20° or two standard deviations of the TALOS-N prediction. For a residue classified with a “Generous” prediction, the ϕ and ψ angle restraints are less tight, <ϕ> ± 3σ and <ψ> ± 3σ, with an allowed range of the larger of 30° or three standard deviations of the TALOS-N prediction.
References
- 1.Wüthrich K. NMR of Proteins and Nucleic Acids. John Wiley & Sons; New York: 1986. [Google Scholar]
- 2.Englander SW, Wand AJ. Main-chain-directed strategy for the assignment of 1H NMR spectra of proteins. Biochemistry. 1987;26:5953–5958. doi: 10.1021/bi00393a001. [DOI] [PubMed] [Google Scholar]
- 3.Oh BH, Westler WM, Darba P, Markley JL. Protein 13C spin systems by a single two-dimensional nuclear magnetic resonance experiment. Science. 1988;240:908–911. doi: 10.1126/science.3129784. [DOI] [PubMed] [Google Scholar]
- 4.Ikura M, Kay LE, Bax A. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of larger proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. application to calmodulin. Biochemistry. 1990;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 5.Wagner G. Prospects for NMR of large proteins. J Biomol NMR. 1993;3:375–385. doi: 10.1007/BF00176005. [DOI] [PubMed] [Google Scholar]
- 6.Saito H. Conformation-dependent C13 chemical shifts - A new means of conformational characterization as obtained by high resolution solid state C13 NMR. Magn Reson Chem. 1986;24:835–852. [Google Scholar]
- 7.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 8.Spera S, Bax A. Empirical correlation between protein backbone conformation and Ca and Cb 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- 9.Haigh CW, Mallion RB. Ring current theories in nuclear magnetic resonance. Prog Nucl Magn Reson Spectrosc. 1979;13:303–344. [Google Scholar]
- 10.Avbelj F, Kocjan D, Baldwin RL. Protein chemical shifts arising from alpha-helices and beta-sheets depend on solvent exposure. Proc Natl Acad Sci U S A. 2004;101:17394–17397. doi: 10.1073/pnas.0407969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Dios AC, Pearson JG, Oldfield E. Secondary and tertiary structural effects on protein NMR chemical shifts - an ab initio approach. Science. 1993;260:1491–1496. doi: 10.1126/science.8502992. [DOI] [PubMed] [Google Scholar]
- 12.Case DA. The use of chemical shifts and their anisotropies in biomolecular structure determination. Curr Opin Struct Biol. 1998;8:624–630. doi: 10.1016/s0959-440x(98)80155-3. [DOI] [PubMed] [Google Scholar]
- 13.Vila JA, Aramini JM, Rossi P, Kuzin A, Su M, Seetharaman J, Xiao R, Tong L, Montelione GT, Scheraga HA. Quantum chemical C-13(alpha) chemical shift calculations for protein NMR structure determination, refinement, and validation. Proc Natl Acad Sci U S A. 2008;105:14389–14394. doi: 10.1073/pnas.0807105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohlhoff KJ, Robustelli P, Cavalli A, Salvatella X, Vendruscolo M. Fast and Accurate Predictions of Protein NMR Chemical Shifts from Interatomic Distances. J Am Chem Soc. 2009;131:13894–13895. doi: 10.1021/ja903772t. [DOI] [PubMed] [Google Scholar]
- 15.Asakura T, Taoka K, Demura M, Williamson MP. The relationship between amide proton chemical shifts and secondary structure in proteins. J Biomol NMR. 1995;6:227–236. doi: 10.1007/BF00197804. [DOI] [PubMed] [Google Scholar]
- 16.Bax A, Grzesiek S. Methodological advances in protein NMR. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 17.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog NMR Spectrosc. 1999;34:93–158. [Google Scholar]
- 18.Salzmann M, Wider G, Pervushin K, Senn H, Wuthrich K. TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J Am Chem Soc. 1999;121:844–848. [Google Scholar]
- 19.Wagner G, Pardi A, Wuthrich K. Hydrogen-Bond Length And H-1-Nmr Chemical-Shifts In Proteins. J Am Chem Soc. 1983;105:5948–5949. [Google Scholar]
- 20.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 21.Berman HM, Kleywegt GJ, Nakamura H, Markley JL. The Protein Data Bank at 40: Reflecting on the Past to Prepare for the Future. Structure. 2012;20:391–396. doi: 10.1016/j.str.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markley JL, Ulrich EL, Berman HM, Henrick K, Nakamura H, Akutsu H. BioMagResBank (BMRB) as a partner in the Worldwide Protein Data Bank (wwPDB): new policies affecting biomolecular NMR depositions. J Biomol NMR. 2008;40:153–155. doi: 10.1007/s10858-008-9221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wishart DS. Interpreting protein chemical shift data. Prog Nucl Magn Reson Spectrosc. 2011;58:62–87. doi: 10.1016/j.pnmrs.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y, Bax A. SPARTA plus: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 28.Rost B, Sander C. Prediction of protein secondary structure at better than 70 percent accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 29.Ramirez BE, Voloshin ON, Camerini-Otero RD, Bax A. Solution structure of DinI provides insight into its mode of RecA inactivation. Protein Sci. 2000;9:2161–2169. doi: 10.1110/ps.9.11.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maltsev AS, Ying JF, Bax A. Deuterium isotope shifts for backbone 1H, 15N and 13C nuclei in intrinsically disordered protein alpha-synuclein. J Biomol NMR. 2012;54:181–191. doi: 10.1007/s10858-012-9666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 33.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 34.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 35.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wuthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids (Reprinted from Pure and Applied Chemistry, vol 70, pgs 117–142, 1998) J Mol Biol. 1998;280:933–952. doi: 10.1006/jmbi.1998.1852. [DOI] [PubMed] [Google Scholar]
- 36.Wang LY, Eghbalnia HR, Bahrami A, Markley JL. Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J Biomol NMR. 2005;32:13–22. doi: 10.1007/s10858-005-1717-0. [DOI] [PubMed] [Google Scholar]