Abstract
Multivariate statistical techniques are used extensively in metabolomics studies, ranging from biomarker selection to model building and validation. Two model independent variable selection techniques, principal component analysis and two sample t-tests are discussed in this chapter, as well as classification and regression models and model related variable selection techniques, including partial least squares, logistic regression, support vector machine, and random forest. Model evaluation and validation methods, such as leave-one-out cross-validation, Monte Carlo cross-validation, and receiver operating characteristic analysis, are introduced with an emphasis to avoid over-fitting the data. The advantages and the limitations of the statistical techniques are also discussed in this chapter.
Keywords: Metabolomics, Mass spectrometry, Multivariate statistics, Classification
1 Introduction
In many typical metabolomics studies, a large number of metabolite molecular signatures are measured quantitatively from biofluids, such as blood and urine, from either animals or humans. Metabolomics data provide important information about biomarkers and metabolic pathways related to disease, gender, diet, etc. Since metabolites are the downstream products of genes and gene expression, and thus are very sensitive to various biological states, they can potentially be more readily used for early disease detection than other molecular information, as well as for providing contemporaneous information for a variety of other studies. Liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) are analytical techniques that have often been used in metabolomics studies to generate the high content data. Another important component is to apply multivariate statistical techniques to analyze the MS-based metabolomics data.
There are multiple statistical techniques available for every data analysis task, including many powerful classification and regression models, and multiple model validation methods. Biomarker selection can be either an independent step or a byproduct of a model. Various statistical techniques can be combined in a proper way to identify potential biomarkers and demonstrate how useful the selected biomarkers are. This article discusses four models, partial least squares, logistic regression, support vector machine, and random forest, and the related model based variable selection techniques. Two model independent variable selection techniques, principal component analysis and two sample t-tests are also introduced.
A key issue for metabolomics studies is to avoid over-fitting the data. Because of the large number of metabolites and the relatively small sample size, a complex model can over-utilize (over-fit) the data specific information and show very good performance, but that good result is useless if it cannot be duplicated using a new set of test data. Proper model evaluation and validation is therefore a necessary step to understand the true performance of a model and the potential biomarkers. Cross-validation, Monte Carlo cross-validation (MCCV), and receiver operating characteristic (ROC) analysis are discussed in this article.
2 Data Processing and Metabolite Identification
2.1 Data Extraction
Liquid chromatography (LC) and gas chromatography (GC) are the two most commonly used separation methods prior to mass spectrometric (MS) analysis. Generally, the instrument manufacturer will provide software to process raw data and generate a list of ion intensities/areas for detected peaks after peak picking, peak de-convolution, peak alignment, etc. For example, Agilent’s MassHunter Qualitative Analysis software can be used for peak identification and Agilent’s Mass Profiler Professional software also has functions for metabolite identification. The compound identification can be assisted by various databases containing the information of retention time, accurate mass, and tandem mass spectrometry. The NIST MS database [1] is often used for the data from electron ionization (EI) that is typically utilized in GC-MS. The Metlin Database [2] is especially suitable for the identification of metabolites in LC-MS data sets as it contains the MS/MS spectra of more than ten thousand standard compounds.
2.2 Preprocessing
Prior to the actual multivariate statistical analysis, some steps are necessary to be performed on spectral data in order to obtain meaningful information. Useful preprocessing methods mainly include normalization, mean-centering, and scaling. These steps can be used individually or in combination. It should be noted that the reproducibility of MS can be a concern (i.e., operated without internal standards for each compound of interest), and thus quality controls are normally included in experiments to compensate instrument drift.
2.2.1 Normalization
For biofluids, especially urine samples, the concentrations of metabolites are highly dependent on factors that are generally not of interest to metabolomics studies, such as the amount of consumed water. Normalization is an important and effective method to exclude or reduce the unwanted overall variations in spectral data.
There are three commonly used approaches to normalize data sets in metabolomics. The first approach is total signal normalization, which calculates the total intensity of the whole spectrum or total ion count and then sets it to a constant value. The second method is vector length normalization. Each spectrum containing many variables can be regarded as a point in a high dimensional space. The vector length normalization sets the Euclidean distance in the multidimensional space to be constant. The last approach is to divide the whole spectrum by a certain peak intensity (typically the largest peak). In the medical community, creatinine is often used as an intensity reference for urine samples. This is due to the underlying assumption of constant excretion of creatinine into urine, which generally holds except for some diseases that affect kidney function. Creatinine levels in urine also show a slight age dependence [3].
2.2.2 Mean-Centering
Mean-centering is often carried out to center the data distribution at the origin in the multidimensional space. In a typical data matrix used for multivariate statistical analysis, each row represents a different sample while the metabolite identities, m/z, or peak variables are aligned into specific columns. Mean-centering is performed by subtracting the mean value of each peak (column) from the corresponding variable in each sample (row).
2.2.3 Scaling
Multivariate statistical analysis tends to focus on metabolites with high intensities. However, low-concentration metabolites may also play important roles in the biological processes. Scaling is often used in metabolomics to change the emphasis from metabolites with high concentrations to those with moderate or small abundances. Variance scaling calculates the standard deviation of each variable (column) and then divides each column by this value. The combination of mean-centering and variance scaling is termed auto-scaling. Auto-scaling sets all variables to unit variance. Auto-scaling is highly recommended when variables have different units. Nevertheless, one drawback of this operation is that it may increase the contribution of noise variables to the analysis. To minimize the undesirable noise effect, Pareto scaling, which uses the square root of the standard deviation as the scaling parameter, can be used. Pareto scaling falls in between no-scaling and auto-scaling, and thus has become more popular in metabolomics to avoid over-manipulating the data. In addition, log scaling can also be used to reduce the effect of large peaks in data analysis and make the data more normal-distributed (many statistical methods were developed assuming that the data follow a normal distribution). However, a drawback of log scaling is that it is unable to deal with zero/negative values.
3 Variable Selection (See Note 1)
After the metabolomics data are created and organized into matrix format, multivariate statistical analysis is performed to identify bio-marker candidates and to examine how well the different groups in the data set are separated using those biomarker candidates through a classification model. Variable selection is a step to identify relevant and putative biomarkers. Variable selection can be performed prior to building a classification model or as a byproduct of a classification model. GC-MS data and LC-MS data typically contain signals from several hundred to over a thousand metabolites. Their peak intensities are collected for every sample and become candidates for variable selection. The number of candidate variables is typically relatively large compared with the sample size, and sometimes much larger than the sample size. Because many of the candidate variables are not related to the study, variable selection is a crucial step for metabolomics data analysis and modeling.
There are multiple popular variable selection techniques with slightly different objectives. Some variable selection techniques directly identify relevant metabolites, while other techniques serve as a screening process to eliminate irrelevant metabolites. However, none of the techniques can claim that its results are consistently better than others. Furthermore, biological and analytical knowledge must be combined with statistical techniques for biomarker selection; e.g., certain metabolites have been previously shown to be related to a disease. Metabolic pathways also provide important information about disease related biomarkers.
In this section, we discuss principal component analysis (PCA) and two sample t-tests. Neither of these techniques depends on the choice of the classification model. Model based variable selection procedures through penalized logistic regression and partial least squares discriminant analysis (PLS-DA) are discussed in Subheading 4.
3.1 Principal Component Analysis
PCA can be described as follows. Assume there are p metabolites, X=(X1, X2,…, Xp), with a variance-covariance matrix Σ. Let (λk, ek), k =1,2,…, p, be the eigenvalue and eigenvector pairs of Σ. Arrange the eigenvalues such that λ1 ≥ λ2 ≥ … ≥ λp. The kth principal component (PC) is Wk = ekTX, and its variance is equal to λk. ek is the loading of the kth PC. The principal components, W1, W2,…, Wp, are formed by an orthogonal transformation of the metabolites X. The first PC W1 follows the direction of the maximum variance. The kth PC, Wk, follows the direction of the largest variance in the subspace that is perpendicular to the first k−1 PCs. The PCs thus form another set of orthogonal axes in the multidimensional space. The sum of the variances of the PCs equals to the sum of the variances of the metabolites X.
When the metabolites X are correlated, some smaller eigenvalues, λK+1,…, λp, are close to 0. Hence, the first k PCs, W1,…, Wk, can capture a large portion of the total variance in the original data. In subsequent analysis, the metabolites X can be replaced by the first k PCs, W1,…, Wk, without much loss of information [4].
It is a common practice to keep the first two or three PCs and examine the score plots. There is no guarantee that the different groups will be well-separated on the PC score plots, since PCA is not designed for classification purposes. However, when the groups are well separated, which happens in many studies, the metabolites that have large loadings in the first two or three PCs can be selected as potential biomarkers. PCA thus serves for a useful variable selection purpose (e.g., see refs. 5–8).
PCA can be applied to the scaled data instead of the original data. Several scaling options are discussed in Subheading 2. PCs from the unscaled data are dominated by a few metabolites with the largest peak intensities. Scaling (partially) solves this problem, but the PC scores and loadings from the scaled data are more difficult to interpret. Also notice that there is no apparent relationship between the PC scores and loadings from the original and the scaled data.
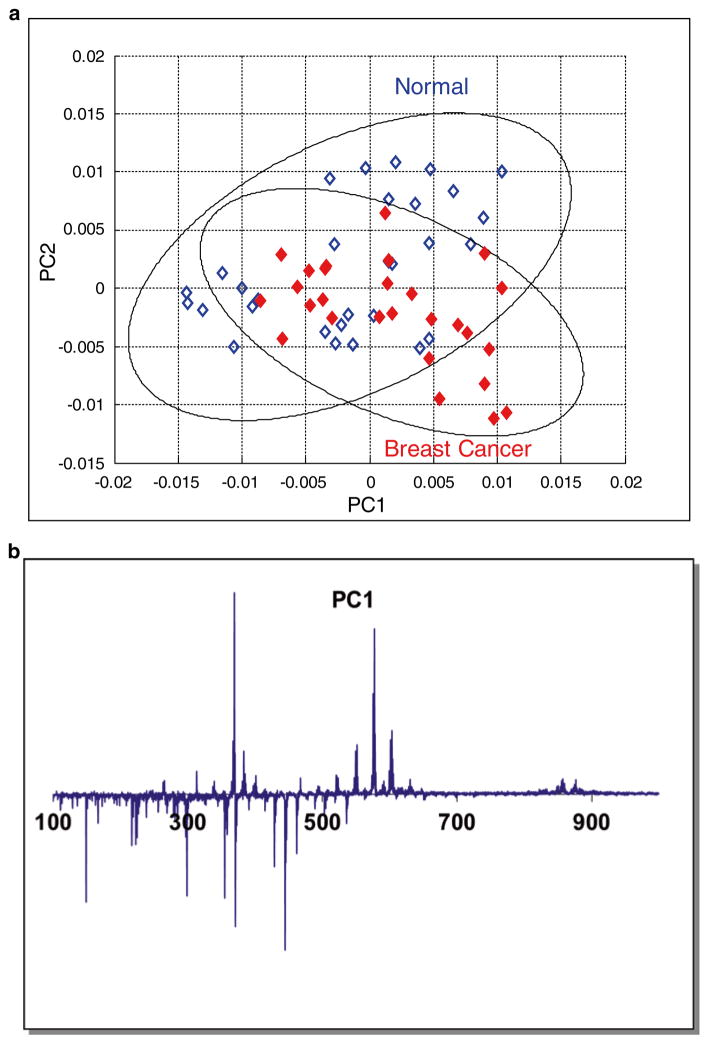
In a breast cancer study [9], PCA was applied to the mean-centered DART-MS spectra of 57 serum samples. There were 30 healthy controls and 27 breast cancer patients. Figure 1 shows the PC score plot and the PC loading plot. DART-MS PC scores alone cannot separate the two groups, providing a counter example to show that PCA is not designed for classification. Hence, a hybrid method, the principal component directed partial least squares model was proposed [9] to combine the NMR and DART-MS spectra. Using both spectra the proposed classification model then successfully separated the two groups and identified breast cancer related biomarker candidates.
Fig. 1.
(a) PCA score plot of the DART-MS spectra in a breast cancer study. Ellipses show the 95 % confidence regions of the two groups; (b) PC1 loading plot of the DART-MS spectra in the breast cancer study. Reproduced from ref. 9 with permission
3.2 Two Sample T-Tests and Multiple Comparisons
Two sample t-tests are used to show which metabolites have the power to differentiate the different groups in the data set (e.g., ref. 8). There are a variety of t-tests: the original Student’s t-test assumes normally distributed data with equal group variances; Welch’s t-test allows for unequal variances; the Wilcoxon-Mann-Whitney test uses a ranked set of values and thus allows for non-normally distributed data sets; and several other variants. The two sample t-test is applied to one metabolite at a time (i.e., a univariate analysis) to determine whether the mean values of the two groups are different. The null hypothesis for the test is H0 : μgroup1 = μgroup2, and the alternative hypothesis is Ha : μgroup1 ≠ μgroup2. If the p-value for the test is smaller than a cutoff value, typically 0.05, the null hypothesis is rejected. If the p-value is large, there is no significant difference between the mean values for the two groups, indicating the metabolite has little power to separate them. On the other hand, a small p-value does not guarantee that the metabolite has sufficient power to separate the two groups in classification. Even if the mean values are different for the two groups, the samples from the two groups may still show large overlap. Hence the metabolites with small p-values must be further evaluated when building a classification model.
The 0.05 cutoff value is often used when the t-test for a metabolite is examined individually, without considering the tests for other metabolites. A multiple comparison procedure can be employed, in which a smaller cutoff value is used, to control the overall error caused by using all the t-tests together. Because of the large number of metabolites in the data, simple multiple comparison procedures such as the Bonferroni correction are too conservative and do not work well [10]; they set a cutoff value that is too close to 0 as the number of tests becomes large.
Multiple comparison procedures have a history dating back to the 1950s, and there are now many proposed procedures. The Benjamini–Hochberg procedure [11] controls the false discovery rate at level α (e.g., α = 0.05) as follows. Assume there are m metabolites being examined. It is a step-up procedure:
Order the p-values as p(1) ≤ p(2) ≤ … ≤ p(m);
Find the largest k such that , and reject the null hypotheses for the corresponding tests.
Additional methods for false discovery rate are also commonly used, such as the q-value [12] and the Benjamini–Hochberg–Yekutieli procedure under dependence assumptions [13].
However, since the metabolites with small p-values are further examined when building a classification model, and the final decision regarding which biomarkers are related to the study is not based on the t-tests alone, a simple 0.05 cutoff can often be used instead of a multiple comparison procedure [14].
3.3 Example Analysis
As an example, we apply several of the statistical techniques described in this chapter to a liver cancer LC-MS dataset [15] for demonstration purposes. Hepatitis C virus (HCV) infection of the liver is a major risk factor for the development of hepatocellular carcinoma (HCC). Serum samples (30 HCC patients with underlying HCV and 22 HCV patients without HCC) were obtained from the Indiana University/Lilly tissue bank, and targeted LC-MS/MS data were obtained and used in the study. 73 targeted metabolites were detected in both the HCC patients and the HCV patients, of which 39 were detected in negative ionization mode and 34 were detected in positive ionization mode. 16 of the 73 metabolites had p-values less than 0.05, as shown in Table 1. The four metabolites with the smallest p-values were used in constructing classification models later. Despite the small number of samples available, this data set is useful to provide examples of the different analysis approaches.
Table 1.
List of metabolites with significant mean changes between HCC and HCV patients
| Metabolite | a FC | p-Value |
|---|---|---|
| Tyrosine | 0.8 | 0.016 |
| Phenylalanine | 0.9 | 0.013 |
| Glycerol | 0.8 | 0.018 |
| Methionine | 0.7 | 0.0032 |
| Creatine | 2.1 | 0.029 |
| Homocysteine | 0.8 | 0.036 |
| 2-Deoxyguanosine | 0.3 | 0.015 |
| Xanthine | 0.8 | 0.011 |
| 1-Methyladenosine | 1.4 | 0.011 |
| N2,N2-Dimethylguanosine | 0.5 | 0.0018 |
| 5-Hydroxymethyl-2′-deoxyuridine | 1.5 | 0.00088 |
| 1-Methylinosine | 0.5 | 0.0075 |
| 1-Methylguanosine | 0.6 | 0.0078 |
| N-Carbamoyl β-alanine | 0.7 | 0.016 |
| Aconitic acid | 0.7 | 0.029 |
| Uric acid | 0.7 | 0.0069 |
FC: mean fold change (HCC/HCV)
4 Classification Models
Classification models can be constructed in a variety of ways, such as using all the metabolite signals in the data, followed by the selection of biomarkers from the model results. Another option is to employ variable selection as the first step, where several metabolites are identified as either potentially correlated with the group differences, or having little power to separate the groups (after which they could then be eliminated). Following the variable selection step, classification models can be constructed based on the combined pool of candidate metabolites, which will be further examined in this process. The final classification model may use either all the candidate variables or just a subset of them for the best classification performance. Because the sample size is often not large, the final classification model needs to be validated to avoid over-fitting the data.
Classification models utilize two matrices, an X matrix for the metabolite peak intensities and a Y matrix for the class labels. There are multiple powerful classification methods and numerous variations of them. This section introduces several popular methods used in the metabolomics field: PLS-DA, logistic regression, support vector machine (SVM), and random forest.
4.1 Partial Least Squares Discriminant Analysis
The selection of axes in general PLS models is based on the regression of X against Y, and thus PLS can express the maximum variance from both X and Y matrices. As a bilinear model, PLS fits the data and recasts them as score plots, loading plots, and weight plots. While loading plots summarize the observations in the X matrix, weight plots express the correlation between the X matrix and score values. The PLS score plot is generated by projection of the original spectra onto the new coordinate system. Each orthogonal axis in the score plot is called a latent variable (LV), similar to a PC in PCA. Corresponding loadings or weights contain information about the importance of each variable in the model.
For classification purposes, Y is a dummy matrix, i.e., 0s and 1s are often used to represent the group assignment of samples. With such a Y matrix, PLS is referred to PLS-DA. PLS-DA is a typical supervised method in that it requires the class membership knowledge of biological specimens. If PCA is not successful in showing the subtle difference among the sample groups, PLS-DA modeling can be used to maximize the separation among the sample groups and target putative biomarkers for metabolomics studies. Notably, variable importance in projection (VIP) values estimate the importance of each variable in the projection used in a PLS model and are often used for variable selection.
4.1.1 PLS Analysis Example
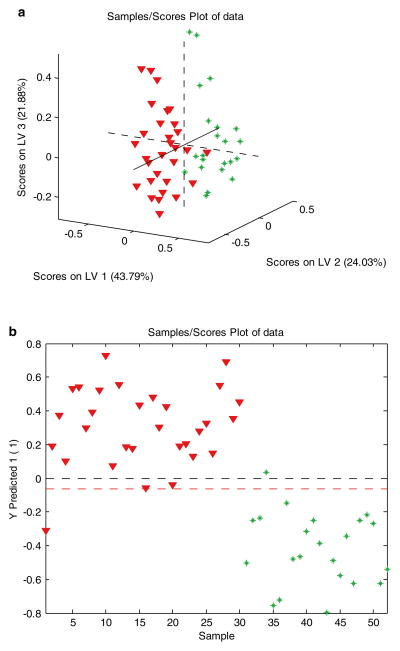
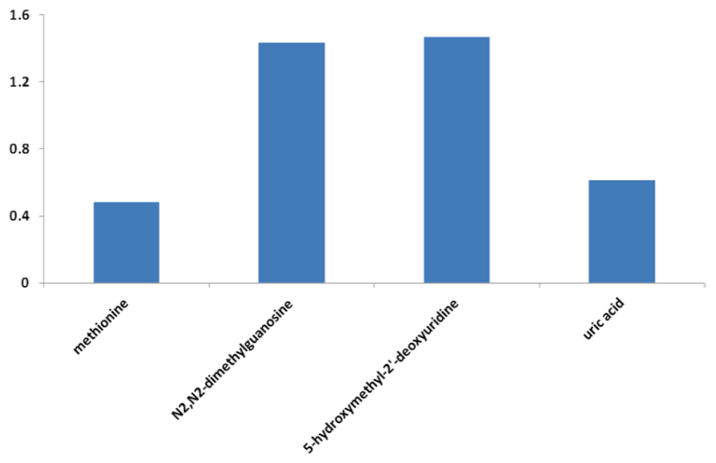
A PLS-DA model was developed for the liver cancer data based on four metabolites with the lowest p-values [15]. Figure 2 shows the three dimensional PLS-DA score plot and the predicted Y values of the liver cancer data. The two groups, HCC and HCV are well separated, with only a small overlap. Figure 3 shows the PLS-DA VIP plot of the liver cancer data, indicating the relative contributions of each of the four metabolites to the overall model.
Fig. 2.
(a) PLS-DA three-dimensional score plot of the liver cancer data; (b) PLS-DA model predicted Y values of the liver cancer data
Fig. 3.
PLS-DA VIP plot of the liver cancer data
A challenge in using multivariate supervised methods is that they may over-fit the data and give too optimistic results. A strict cross-validation step is necessary before drawing a reliable conclusion. Generally speaking, there are “internal” and “external” cross-validations. The leave-one-out cross-validation procedure, a typical internal cross-validation approach, is commonly employed to select the number of LVs and to find an optimal PLS-DA model. From the internal cross-validation step, the root-mean-square error of cross-validation (RMSECV) curve is created, representing the accuracy of prediction, and is used to choose the number of LVs. Afterwards, external cross-validation is employed to measure the model performance. There are several options for external cross-validation, such as splitting data into two halves, k-fold cross-validation, and MCCV, as described in Subheading 5. If a test set of samples acquired from one or more different locations is available, it offers a more accurate validation result for a set of putative biomarkers.
4.1.2 Orthogonal Signal Correction
It is well known that metabolic profiles of biological samples provide a fingerprint of endogenous markers that can be correlated to a number of factors such as disease, diet, toxicity, medication intake, etc. Because these factors can cause significant changes in metabolic profiles, the analysis of MS data is often aided by the use of statistical methods that can de-convolute the metabolite contributions from specific factors. For this purpose, orthogonal signal correction (OSC) was developed and introduced to the metabolomics field to remove chemical and thermal noise, as well as other variables that are not of interest to the study [16]. OSC is a PLS based data filtering technique. There are two matrices involved, the X matrix (spectral data) and the Y matrix (variables of interest). In the OSC step, the structure in the X matrix that is mathematically orthogonal to the Y matrix data is subtracted. Hence the corrected X matrix contains the Y matrix related variation. A PLS model based on the corrected X matrix now focuses more on the variables of interest.
4.2 Logistic Regression
The standard logistic regression model predicts the probabilities of a sample being a member of either of two groups for a set of metabolite peak intensities. Mathematically, let p(1|X) and p(2|X) be the probabilities that a sample belongs to group 1 and 2, respectively, given the metabolite peak intensities X (note that p(1|X)+p(2|X) = 1). The probabilities are modeled as a function of X as follows in Eq. (1):
| (1) |
The response variable of the i-th sample for the logistic regression model, Yi, is binary, 1 or 0, corresponding to the two groups. The coefficients in the above model are estimated by maximizing the log likelihood function Σ(Yilog(p(1|Xi))+(1−Yi) log(p(2|Xi))), where Xi contains the metabolite peak intensities of the i-th sample. With the estimated coefficients from a training set of data, the probabilities can be computed for a test set. A test sample is classified into a group with a probability larger than 0.5. As a classification model, standard logistic regression can be used in place of PLS-DA (e.g., ref. 17).
There is one major difference between the two models. PLS-DA can be constructed using all the metabolites in the data, without any prior variable selection step, whereas the standard logistic regression model has difficulty handling a large number of variables. Typically, therefore, variable selection is performed before fitting a standard logistic regression model [18]. The standard logistic regression model is then constructed using the selected metabolites from the previous variable selection step. In general PLS-DA can handle high dimensional, correlated data better than logistic regression, which can become an issue for highly correlated metabolites.
4.2.1 Penalized Logistic Regression
Penalized logistic regression, a variant of the logistic regression model [19], can handle a large number of variables and has a built-in stepwise variable selection process. For penalized logistic regression, the probabilities are also modeled as a function of the metabolite intensities, according to Eq. (1). To handle a large number of variables, with p potentially much larger than n (the number of samples), the penalized logistic regression model estimates the coefficients by maximizing a penalized log likelihood function, , where λ is a constant prespecified by users, often chosen from cross-validation. A stepwise variable selection process is combined with penalized logistic regression to eliminate the variables that are powerless in terms of classifying the two groups. Penalized logistic regression itself is a classification model that uses all the variables. Another option is to combine the metabolites selected by penalized logistic regression with selected biomarkers from other variable selection techniques, and build a standard logistic regression model. The penalized logistic regression and the stepwise variable selection can be performed using the “stepPlr” package in R [20].
4.3 Support Vector Machine
Support vector machine (SVM) is a robust classification technique, initially proposed to use a linear decision boundary to separate the two classes [21]. Since then, there have been extensive studies of SVM in the machine learning and statistics communities. Many variants of SVM have been proposed. Now SVM can perform nonlinear classification using a kernel function. Steinwart and Christmann [22] provide a comprehensive and in-depth discussion of the various approaches for SVM.
A general description of SVM is as follows. Let the response variable for a sample Yi be +1 or −1, indicating two groups. Let f(Xi) be a decision function that computes the estimated response for a sample given the metabolite intensities Xi. Let L(Y, f(X)) be a classification loss function. Consider a set of decision functions in a reproducing kernel Hilbert space (RKHS) H. The best decision function f minimizes .
The above target function has two components. measures the classification loss of n data points n using the decision function f. λ is a positive constant prespecified by users, often chosen through experiments. measures the complexity of the decision function f. While the best decision function needs to minimize the classification loss, there is also a penalty for a complex decision function through the term .
A straightforward loss function is a 0–1 loss, i.e., L=1 if the sign of f(Xi) is not equal to the sign of Yi, and L=0 otherwise. The simple 0–1 loss is not a convex function and makes the optimization problem much harder. It is often replaced by a convex loss function, which provides a unique minimizer for the optimization problem. The best decision function has the general form fλ=Σαik(Xi, ●), where k(Xi, ●) is the kernel that belongs to the RKHS, and the coefficients αi depend on the data.
Although certain variants of SVM can handle data with redundant noise variables, and thus are suitable tools for metabolomics studies (e.g., refs. 23, 24), using a subset of metabolites selected from the variable selection step in SVM will remove the unpredictable effects of such noise variables. There are a number of software tools and packages that can build an SVM model, for example, libsvm [25], Weka [26], and several R packages, e.g., package “e1071” [27].
Standard SVM output class labels are +1 or −1. Standard SVM does not compute the probabilities p(1|X) or p(2|X) as does logistic regression. However, probabilistic scores are needed later for ROC analysis. Various approaches have been proposed to return probabilistic scores for a sample (e.g., ref.28). The above-mentioned software tools have options to output probabilistic scores instead of class labels.
4.4 Random Forest
The random forest approach consists of a collection of classification trees. A single classification tree is an unstable classification model. The entire tree structure can change significantly because of some (small) perturbation in the data. Formed as an ensemble of classification trees, however, random forest becomes a robust classifier [29, 30].
A classification tree is formed by binary recursive partitioning [31]. In the first step, all samples from both groups are combined in one node. An optimal partition in the shape of Xk > C is used to divide the node into two “branching” nodes based on the value of one variable Xk. This process is repeated for each of the branching nodes. A tree grows until a stopping criterion is satisfied. The variable Xk and the cutoff value C are chosen to split a node to minimize the sum of the impurity of the two branching nodes. Let p1 and p2 be the proportions of group 1 and 2 samples in a node, respectively. There are multiple ways to define the impurity of a node (e.g., misclassification error 1 − max(p1, p2), Gini index 2p1p2, entropy - p1 log p1 - p2 log p2, etc.). A typical approach is to grow a large tree and then prune it back. A sample point, based on the variable values, travels down the tree and falls into one of the terminal nodes. It is classified into the majority class in that terminal node.
The random forest is then formed by a set of de-correlated trees. The random forest is constructed as follows [30]: first, create B bootstrap samples from the training data; then grow a tree from every bootstrap sample; finally, return the collection of trees. A sample point is classified into a class by taking the majority vote of the trees. Random forest can return probabilistic scores for a sample point instead, by using the proportion of trees voting for each class. R has a package “randomForest” [32] that is useful to build the model.
For computation efficiency and to de-correlate the trees (a tree grown from a bootstrap sample is different from a standard classification tree), users can prespecify the number of variables m to be evaluated at each partition and the minimum node size s. Often m can be set to two or three with good results. Then at each partition, a bootstrap tree randomly selects m variables and choses the optimal partition based on the m variables. Random forest grows every bootstrap tree to full size without pruning.
4.4.1 Random Forest Analysis Example
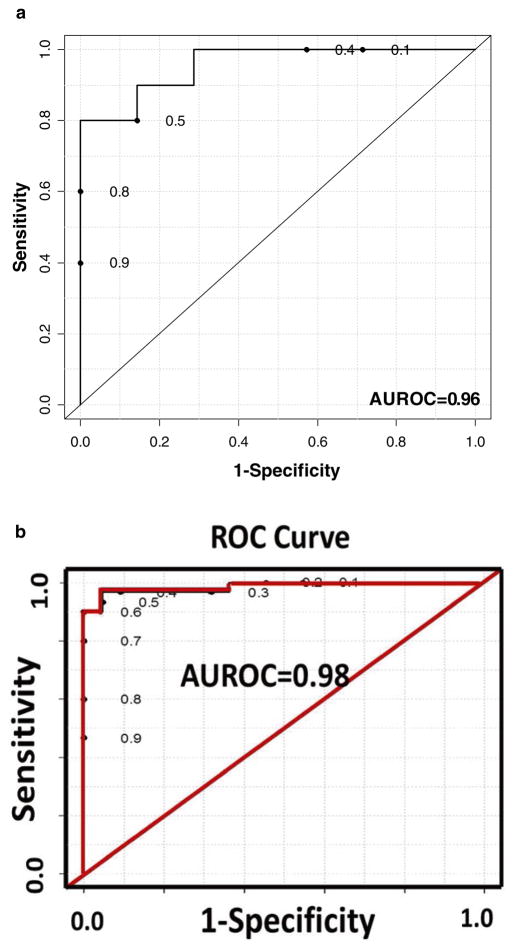
Using the four selected metabolites in the liver cancer study [15], we built a random forest model. The 52 samples were randomly split into a training data (20 HCC samples and 15 HCV samples) and a test data (10 HCC samples and 7 HCV samples). In Fig. 4, the liver cancer results from PLS-DA and random forest are compared using ROC curves. The area under the ROC curve (AUROC, introduced in Subheading 5) is 0.96 for the random forest model (see Fig. 4a), which is similar, but not quite as good as the model built using PLS-DA (see Fig. 4b).
Fig. 4.
(a) ROC curve generated by the Random Forest statistical model using four selected metabolites in a liver cancer study; (b) ROC curve generated from the PLS-DA model constructed using the same four selected metabolites in the liver cancer study. Reproduced from ref. 15 with permission
5 Model Performance Evaluation and Validation
Accurately assessing the performance of a classification model is a critical issue. A successful classification model confirms that the selected biomarkers have sufficient power to separate the groups and thus are useful in the study. Because of the relatively small sample size of typical metabolomics data sets, the proper evaluation of a chosen classification model is an issue that requires attention.
The classification error rate is one measure of model performance. If a model is constructed using a training set of samples and the error rate is computed using the same training set, the resulted re-substitution error rate is highly optimistically biased, i.e., much smaller than the error rate from a future test data. One simple method to estimate the classification error rate without such bias is to first divide the sample set into two subsets, build the model using one subset of the data, and then test the performance using the other subset. If a dataset with class labels from a different location or a later time period is available, this is a very natural choice for estimating the classification error rate. However, this simple method has drawbacks for a small dataset, and also does not fully utilize all the information available. Although the estimated error rate is theoretically unbiased in this approach, it has a large variance; the estimated error rates using this approach vary a lot from one test data set to another. Another approach to obtain an unbiased error rate estimate is to use cross-validation, discussed later in this section.
Besides the classification error rate, which gives a single number that summarizes the model performance, the ROC curve is another method to demonstrate the classification model performance. Furthermore, MCCV and bootstrapping, two strong model validation methods, can be employed to show the variation of the model performance.
5.1 Receiver Operating Characteristic Curve
ROC analysis is a graphical tool that shows how the true positive rate (i.e., sensitivity) changes with the false positive rate (i.e., 1−specificity). To perform ROC analysis, a classification model needs to output probabilistic scores of different classes for every test data point, not simply class labels. At different threshold values, a classification model that is better than a random guess will have the true positive rate increase faster than the false positive rate. Plotting the true positive rate versus false positive rate creates an ROC curve. The area under the ROC curve (AUROC) is a measure of classification model performance. AUROC close to 1 indicates a successful classification model. The shape of the ROC curve is also an important indicator of model performance. A sharp increase in true positive rate with minor increase in false positive rate is most desirable.
In the liver cancer study [15], a PLS-DA model was constructed using the four selected metabolites with the smallest p-values. The ROC curve for the PLS-DA model is shown in Fig. 4a. The AUROC is 0.98. The HCC and HCV groups are well separated under the PLS-DA model using the four selected biomarkers. Figures 2, 3, and 4 show that the four selected biomarkers have robust, good performance in different classification models.
5.2 Cross-Validation
Cross-validation is a technique that can be applied to different types of models, including classification models. Leave-one-out cross-validation and k-fold cross-validation are popular choices. As an example, assume the sample size is n. For leave-one-out cross-validation, every time n−1 samples are used as a training set to fit a classification model, and the remaining sample that is left out is used for testing. This process is repeated n times, and every sample serves as a test data once and only once. A model that is built on n−1 samples is nearly as accurate as the model built on all n samples. The classification error rate is estimated as the proportion of misclassified test data points.
As leave-one-out cross-validation fits the classification model n times, it is computationally demanding. K-fold cross-validation simplifies this process. The whole dataset is divided into k equal size subsets (e.g., k=5 or k = 10). For each iteration, k−1 subsets are combined and serve as a training set, and the one remaining subset serves as the test set. Again, every sample serves as a test data point once and only once. Both the leave-one-out and k-fold cross-validation error rate estimates are unbiased.
5.3 Monte Carlo Cross-Validation
MCCV randomly splits the data that have class labels m times. For every split, one subset (e.g., 75 % of the data) is used as the training set to build a classification model, and the other subset (e.g., 25 % of the data) is used as the test set. The m test data results can be combined to generate an overall confusion matrix and an overall estimate of the model error rate and the confidence interval. Meanwhile, the sensitivity and specificity are computed for every split and can be plotted in an ROC space, which displays the variation of the model performance (e.g., ref. 33). Another approach is to compute the classification error for every test set, and the average is reported as an estimate of the model error rate [34]. The number of splits, m, can be as small as 50–100. As the number of splits increases, MCCV becomes increasingly computationally demanding. Compared with leave-one-out or k-fold cross-validation, the error rate estimate obtained by MCCV has a smaller mean square error [34].
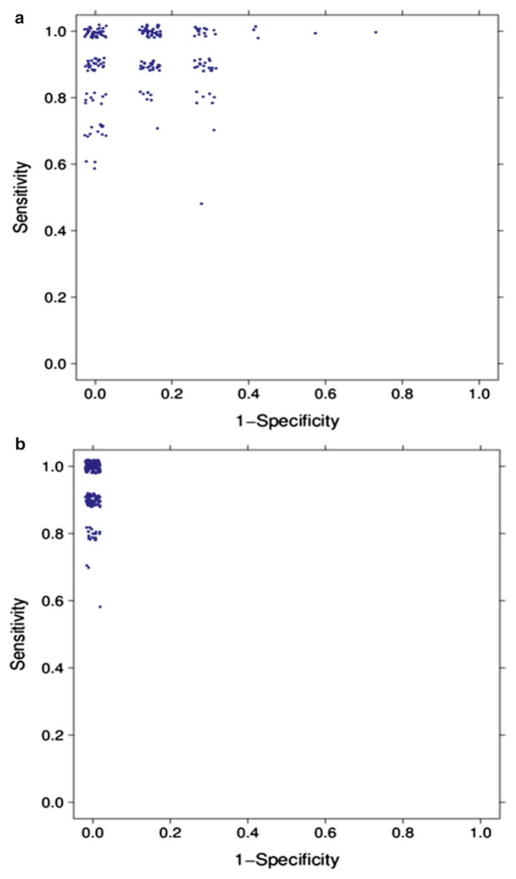
We applied MCCV to evaluate the performance of both the logistic regression model and the SVM model using the four selected metabolites in the liver cancer study. In every split, 20 HCC samples and 15 HCV samples were randomly selected as the training set, and the remaining 10 HCC samples and 7 HCV samples were used as the test set. For each model, we ran 200 iterations of MCCV. Figure 5 shows the values of sensitivity and 1−specificity in the ROC space. Because multiple splits share the same sensitivity and specificity values, the data points show some jitter in the two figures. MCCV results show that SVM has a more robust performance than the logistic regression model.
Fig. 5.
(a) MCCV with 200 iterations for logistic regression model constructed using the four selected metabolites in the liver cancer study; (b) MCCV with 200 iterations for SVM constructed using the four selected metabolites in the liver cancer study
6 Conclusions
This article focuses on the various multivariate statistical techniques for analyzing and modeling MS-based metabolomics data. The advantages and limitations of the various statistical techniques are discussed, as well as possible ways of combining several statistical techniques in one study. Because of the complexity of the metabolomics data and the typically limited number of samples available, we need to pay special attention to avoid over-fitting the data. Furthermore, to ensure that the discoveries are valid, prior knowledge such as results from previous studies and metabolic pathways must be considered along with the statistical analysis results of validation studies.
Acknowledgments
This article was written while one of the authors, Bowei Xi, was on sabbatical leave at the Statistical and Applied Mathematical Sciences Institute (SAMSI, Research Triangle Park, NC). This work is partially funded by NSF DMS-1228348, ARO W911NF-12-1-0558, DoD MURI W911NF-08-1-0238 (BX) and NIH R01GM085291 (DR).
Footnotes
It is interesting to note that different variable selection procedures may select different sets of biomarkers. The reason is that they conduct variable selection from different perspectives. Model based variable selection procedures take into account the joint effect of all metabolites within a specific model framework. PCA utilizes the correlation structure of metabolites. The t-test examines the mean values of the two groups. Hence, it is not surprising that they return different results. Different sets of selected variables are all potential candidates for an effective classification model. A variable selection step may employ one or several statistical techniques to select potential biomarkers and combine the results if multiple techniques are used. Prior knowledge such as previous studies about a disease and metabolic pathways must also be considered along with statistical analysis in variable selection.
References
- 1.The NIST MS database. http://www.hmdb.ca/
- 2.The Metlin Database. http://metlin.scripps.edu/index.php.
- 3.Gu H, Pan Z, Xi B, Hainline B, Shanaiah N, Asiago V, Gowda G, Raftery D. 1H NMR metabolomics study of age profiling in children. NMR Biomed. 2009;22:826–833. doi: 10.1002/nbm.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson R, Wichern DW. Applied multivariate statistical analysis. 5. Prentice-Hall; Englewood Cliffs, NJ: 2002. [Google Scholar]
- 5.Nyamundanda G, Brennan L, Gormley IC. Probabilistic principal component analysis for metabolomic data. BMC Bioinformatics. 2010;11:571. doi: 10.1186/1471-2105-11-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Z, Gu H, Talaty N, Chen H, Shanaiah N, Hainline BE, Cooks G, Raftery D. Principal component analysis of urine metabolites detected by NMR and DESI–MS in patients with inborn errors of metabolism. Anal Bioanal Chem. 2007;387:539–549. doi: 10.1007/s00216-006-0546-7. [DOI] [PubMed] [Google Scholar]
- 7.Wiklund S, Johansson E, Sjstrm L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80:115–122. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 8.Wikoffa WR, Anforab AT, Liub J, Schultzb PG, Lesleyb SA, Petersb EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu H, Pan Z, Xi B, Asiago V, Musselman B, Raftery D. Principal component directed partial least squares analysis for combining nuclear magnetic resonance and mass spectrometry data in metabolomics: application to the detection of breast cancer. Anal Chim Acta. 2011;686:57–63. doi: 10.1016/j.aca.2010.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bretz F, Hothorn T, Westfall P. Multiple comparisons using R. Chapman & Hall; New York: 2011. [Google Scholar]
- 11.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 12.Storey JD. A direct approach to false discovery rates. J Royal Stat Soc Ser B. 2002;64:479–498. [Google Scholar]
- 13.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 14.Bender R, Lange S. Adjusting for multiple testing—when and how? J Clin Epidemiol. 2001;54:343–349. doi: 10.1016/s0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- 15.Baniasadi H, Nagana Gowda GA, Gu H, Zeng A, Zhuang S, Skill N, Maluccio M, Raftery D. Targeted metabolic profiling of hepatocellular carcinoma and hepatitis C using LC-MS/MS. Electrophoresis. 2013;34:2910–2917. doi: 10.1002/elps.201300029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wold S, Antti H, Lindgren F, Öhman J. Orthogonal signal correction of near-infrared spectra. Chemom Intell Lab Sys. 1998;44:175–185. [Google Scholar]
- 17.Liao JG, Chin KV. Logistic regression for disease classification using microarray data: model selection in a large p and small n case. Bioinformatics. 2007;23:1945–1951. doi: 10.1093/bioinformatics/btm287. [DOI] [PubMed] [Google Scholar]
- 18.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park MY, Hastie T. Penalized logistic regression for detecting gene interactions. Biostatistics. 2008;9:30–50. doi: 10.1093/biostatistics/kxm010. [DOI] [PubMed] [Google Scholar]
- 20.R package stepPlr. http://cran.r-project.org/web/packages/stepPlr/
- 21.Cortes C, Vapnik V. Support-vector networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- 22.Steinwart I, Christmann C. Support vector machine. Springer; New York: 2008. [Google Scholar]
- 23.Mahadevan S, Shah SL, Marrie TJ, Slupsky CM. Analysis of metabolomic data using support vector machines. Anal Chem. 2008;80:7562–7570. doi: 10.1021/ac800954c. [DOI] [PubMed] [Google Scholar]
- 24.Zhu J, Rosset S, Hastie T, Tibshirani R. 1-Norm support vector machines. Adv Neural Inf Process Syst. 2004;16:49–56. [Google Scholar]
- 25.Chang CC, Lin CJ. libsvm: a library for support vector machines. 2001 http://www.csie.ntu.edu.tw/~cjlin/libsvm/
- 26.Weka: data mining software in Java. http://www.cs.waikato.ac.nz/ml/weka/
- 27.R package e1071. http://cran.r-project.org/web/packages/e1071/
- 28.Platt J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Adv Large Margin Classifiers. 1999;10:61–74. [Google Scholar]
- 29.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 30.West PR, Weir AM, Smith AM, Donley EL, Cezar GG. Predicting human developmental toxicity of pharmaceuticals using human embryonic stem cells and metabolomics. Toxicol Appl Pharmacol. 2010;247:18–27. doi: 10.1016/j.taap.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning. 2. Springer; New York: 2009. [Google Scholar]
- 32.R package randomForest. http://cran.r-project.org/web/packages/randomForest/
- 33.Carrola J, Rocha CM, Barros AS, Gil AM, Goodfellow BJ, Carreira IM, Bernardo J, Gomes A, Sousa V, Carvalho L, Duarte IF. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J Proteome Res. 2011;10:221–230. doi: 10.1021/pr100899x. [DOI] [PubMed] [Google Scholar]
- 34.Molinaro AM, Simon R, Pfeiffer PM. Prediction error estimation: a comparison of resampling methods. Bioinformatics. 2005;21:3301–3307. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]