Abstract
Saccharomyces cerevisiae chronological life span (CLS) is determined by a wide variety of environmental and genetic factors. Nutrient limitation without malnutrition, i.e. dietary restriction, expands CLS through the control of nutrient signaling pathways, of which TOR/Sch9 has proven to be the most relevant, particularly under nitrogen deprivation. The use of prototrophic wine yeast allows a better understanding of the role of nitrogen in longevity in natural and more demanding environments, such as grape juice fermentation. We previously showed that acetyltransferase Gcn5, a member of the SAGA complex, has opposite effects on CLS under laboratory and winemaking conditions, and is detrimental under the latter. Here we demonstrate that integrity of the SAGA complex is necessary for prolonged longevity, as its dismantling by SPT20 deletion causes a drop in CLS under both laboratory and winemaking conditions. The sch9Δ mutant is long-lived in synthetic SC medium, as expected, and the combined deletion of GCN5 partially suppresses this phenotype. However it is short-lived in grape juice, likely due to its low nitrogen/carbon ratio. Therefore, unbalance of nutrients can be more relevant for life span than total amounts of them. Deletion of RTG2, which codes for a protein associated with Gcn5 and is a component of the mitochondrial retrograde signal, and which communicates mitochondrial dysfunction to the nucleus, is detrimental under laboratory, but not under winemaking conditions, where respiration seems not so relevant for longevity. Transcription factor Rgm1 was found to be a novel CLS regulator Sch9-dependently.
Introduction
The yeast Saccharomyces cerevisiae is a very useful biotechnological tool thanks to its ability to perform alcoholic fermentation, the metabolic process underlying baking, brewing, winemaking and bioethanol production. However, industrial strains show some genetic differences to the strains used in laboratories [1]. Commercial wine yeasts are prototrophs and can produce all their amino acids from a single nitrogen source [2], while laboratory strains are generally mutants in the genes involved in amino acid or nitrogen-base biosynthesis. Overall, industrial yeast strains are more robust and more stress-tolerant to the environmental challenges they face during winemaking, particularly initial high sugar concentration (around 20%), low nitrogen and oxygen levels, and high final ethanol content [3].
S. cerevisiae has been widely used as a eukaryotic model for studying the molecular mechanisms that modulate life span given their high conservation from yeast to mammals [4,5]. In yeast two models of aging, replicative life span (RLS) and chronological life span (CLS), occur. RLS is defined as the number of daughter cells produced by a mother cell, whereas CLS is defined as the capacity of stationary cells to maintain viability in a nondividing state. CLS is the longevity model of postmitotic cells that constitute bulk of tissue in mammals. From the industrial point of view, studying chronological longevity is relevant when a yeast culture no longer divides, as occurs at the end of alcoholic fermentation. Accumulation of damaged proteins and mitochondria with time can cause cell death in both aging types [4], and metabolites, such as ethanol and acetic acid, have been shown to be pro-aging factors in chronological aging
Various regulatory mechanisms are important for determining longevity, including nutrient signaling pathways, acetylation/deacetylation machinery (mainly sirtuins), stress responses and autophagy. Nutrient signaling pathways regulate cell growth and proliferation, metabolism and stress responses. They allow cells to not only stimulate metabolism and growth when nutrients are present, but to also enter the stationary phase during nutrient starvation periods, thus improving long-term survival. The main environmental alteration that extends longevity is decreased nutrient supply without inducing malnutrition, which is called dietary restriction. In yeast, this can occur by reducing the intake of nitrogen or carbon sources, and involves the Ras/cAMP/PKA and TOR/Sch9 pathways [6]. TOR (Target Of Rapamycin), and its related kinase Sch9, control cell growth and metabolism in response to nutrients, which highlights the response to nitrogen availability. The use of chemical inhibitors of TOR, such as rapamycin, or mutations in TOR/Sch9 pathway proteins causes diminished pathway activity that promotes an extension of yeast longevity [7,8]. We previously found that the chemical inhibition of TOR extends CLS under winemaking conditions [9]. In starvation, S. cerevisiae induces autophagy, a highly conserved catabolic process in eukaryotes, which allows the recycling of intracellular components by degradation in the lytic compartment (vacuoles in yeast) [10]. Thus, the cell may obtain nutrients to allow survival in nutritional shortage. However, excessive autophagy can lead to cell death, so it is necessary to keep the process within a physiological range [11]. For instance, in grape juice fermentation autophagy promotes chronological aging [9].
Environmental conditions are also a key factor for onset of CLS. Winemaking fermentation conditions by industrial S. cerevisiae strains vastly differ from standard laboratory environments. Grape juice is very rich in sugars (up to 20–25%), but poor in nitrogen sources [12]. Therefore, cell division arrest occurs when the carbon source is plentiful. In fact most sugar consumption and ethanol production happens when cells are nondividing or dying, so this is a very interesting process from the biotechnological viewpoint. Cell death also happens under high ethanol and low oxygen conditions, so different rules may apply to molecular aging mechanisms. In previous works, we have shown that the mutation of acetyltransferase Gcn5 inhibits autophagy under laboratory and industrial conditions [9]. It plays a positive role in lifespan under standard laboratory conditions of growth in synthetic complete (SC) medium, but its deletion extends CLS under winemaking conditions, suggesting that physiological conditions affect the way that some mechanisms, such autophagy, control aging. Ethanol production in the stationary phase is a key factor for CLS under both laboratory and industrial conditions [13,14]. Gcn5 forms part of the SAGA (Spt-Ada-Gcn5 acetyltransferase) complex, involved in gene expression, from the start of transcription to mRNA transport. The SAGA complex is composed of four modules, two of them with enzymatic activity, the acetyltransferase module (HAT) and the deubiquitinylase (DUB) module (the latter interacts with RNA polymerase II), the TAF module, and a structural module, SPT [15]. Strains lacking genes encoding the DUB module (SGF73, SGF11 and UBP8) have an extended replicative life span [16]. That is not the case for other SAGA components, like GCN5. Acetyltransferase Gcn5 is also associated with another protein complex, SLIK (SAGA-like) [17], which includes Rtg2, a component not found in SAGA. Rtg2 is a central component of the yeast retrograde response pathway [18], which allows communication between mitochondria and the nucleus for the response to mitochondrial stress [19]. The retrograde response lies at the nexus of metabolic regulation, stress resistance, chromatin-dependent gene regulation and aging. Rtg2 is required for the replicative life span extension caused by mitochondrial malfunction [20]. Gcn5 modulates the retrograde response as deletion of GCN5 prevents increased replicative longevity caused by induction of the retrograde response [21].
We herein analyzed the role of the proteins that may interact with acetyltransferase Gcn5 on lifespan regulation under standard laboratory conditions and during winemaking, and both physical and genetic interactions were identified. We found that the integrity of the SAGA complex is relevant for longevity and autophagy. We further investigated the relationship between both Gcn5 and the Sch9 kinase and the retrograde response in chronological aging. Gcn5 is relevant for the CLS extension caused by Sch9 deletion, so it may orchestrate the gene expression pattern set by the kinase. The role of Sch9 under winemaking conditions is to promote life span, probably due to nitrogen starvation conditions.
Materials and Methods
Yeast strains and growth media
S1 Table lists the industrial wine yeasts used in this work. Haploid strain C9 (Mat a, ho::loxP) was a gift from Michelle Walker [22]. Industrial wine yeast L2056 was kindly provided by Lallemand Inc. (Montreal, Canada). Gene disruptions were performed by using recyclable selection marker loxP-kanMX-loxP from plasmid pUG6 [23]. The marker was eliminated by transforming with cre recombinase-containing plasmid YEp351-cre-cyh [24]. S2 Table lists the oligonucleotides employed to amplify deletion cassettes and to check transformants. The petite strains were obtained by growth in SD medium with ethidium bromide (10 μg/mL), where yeast lost the functional mitochondria and became a strictly aerobic petite strain, uncapable of growth in nonfermentable carbon sources (e.g., glycerol)[25].
For yeast growth, YPD medium (1% yeast extract, 2% bactopeptone, 2% glucose) was used. SC medium contained 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% glucose and 0.2% drop-out mix with all the amino acids [26]. SD-N is as SC with no ammonium sulfate and amino acids. SC N 1/25 is like SC, with 25-fold less ammonium sulfate and amino acids. Solid plates contained 2% agar and 20 μg mL-1 geneticin or 0.1 μg mL-1 cycloheximide. Red grape juice (Tempranillo variety) was a gift from Bodegas J. Belda (Fontanars dels Alforins, Spain). It was sterilized overnight with 500 μg/L of dimethyl dicarbonate.
Yeast growth conditions and chronological life span measurements
For the CLS experiments done under laboratory conditions, precultures of selected strains were grown overnight on YPD and were then inoculated in SC media at an OD600 of 0.1. After 3 days of growth at 30°C, aliquots were taken, diluted and plated. Colonies were counted and the percentage of survival was calculated by taking day 3 of growth as 100% survival.
For the microvinification experiments, the cells from 2-day cultures in YPD were inoculated at a final concentration of 106 cells/mL in filled-in conical centrifuge tubes with 30 mL of grape juice. Incubation was done with very low shaking at 24°C. Vinification progress was followed by determining cell viability and sugar consumption, as previously described [27]. Survival plots were drawn by taking the highest cell viability point (around 2–5 days) as 100% survival.
Metabolite determinations and Western blotting
Reducing sugars during fermentation were measured by the reaction to DNS (dinitro-3,5-salycilic acid)[28]. Ethanol was measured with the kits provided by r-Biopharm following the manufacturer’s instructions.
For the autophagy measurements, Ald6 levels were detected by Western blot [29]. At different growth times in SD-N medium, cells were taken and broken with one volume of glass beads in a buffer containing Tris-HCl 0.1 M pH 7.5, NaCl 0.5 M, MgCl2 0.1 M, NP40 1% (v/v), PMSF 10 mM and protease inhibitors (complete Mini, EDTA-free from Roche). Protein concentration was measured by the Bradford method using the Bio-Rad Protein assay following the manufacturer’s instructions. Extracts were diluted in loading buffer for SDS-PAGE (Tris-HCl 240 mM pH 6.8, SDS 8% (p/v), glycerol 40%, β-mercaptoethanol 10%). After electrophoresis, the gel was blotted onto PVDF membranes for the Western blot analysis with an Invitrogen mini-gel device. The anti-ALDH antibody was obtained from Rockland (Gilberstville, USA) and the anti-ADH antibody was obtained from Acris (Hiddenhausen, Germany). The ECL Western blotting detection system (Amersham) was used following the manufacturer´s instructions.
Results
SAGA complex components Ubp8 and Spt20 regulate longevity and autophagy
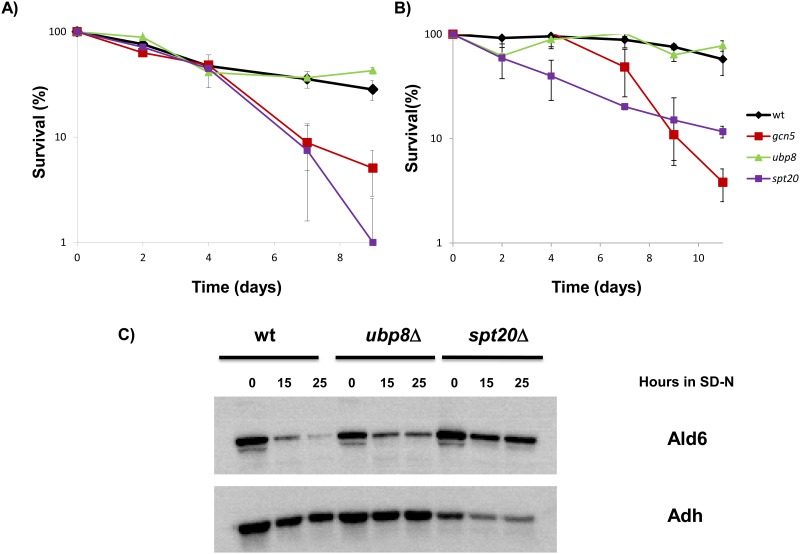
In a previous work, we demonstrated the role of acetyltransferase Gcn5 in autophagy and in CLS control [9]. In order to study the role of other SAGA complex members in those processes, we proceeded to generate deletion mutants for the genes coding for SAGA proteins in the C9 wine strain to analyze the chronological aging profile in their absence. The C9 strain is a haploid derivative of the diploid L2056 commercial wine yeast strain [22], where the construction of single and double mutants is easier, thus it was used throughout this work. We deleted the UBP8 gene that codes for the deubiquitinylation activity of the complex and the SPT20 gene, which codes for the structural Spt20 component, whose absence dismantles the SAGA complex [30]. First, the chronological aging of these strains was studied in standard laboratory minimal complete medium (SC). The wild-type strain and the ubp8Δ mutant have a similar chronological aging profile (Fig. 1A), suggesting that the deubiquitinylase activity of SAGA is not relevant for longevity. However, the spt20Δ mutant presents shorter maximum CLS than the parental strain, which is a similar result to that observed for the gcn5Δ mutant. Therefore, the enzymatic activity of Gcn5 that is involved in longevity seems to occur in the context of SAGA. In order to study the relevance of these genes in a dietary restriction context caused by nitrogen depletion, a similar medium with 25-fold less nitrogen was tested (SC 1/25N), which mimics the low nitrogen conditions of grape musts for winemaking (Fig. 1B). In this case, growth was slower, so the time point corresponding to day 7 was taken as 100% survival instead of day 3. Life span was extended with a low nitrogen concentration for the wild-type strain when compared to the rich medium, as expected under the dietary restriction condition, and the mean life span (50% viability) was extended by 2.5-fold (S3 Table). Once again, the UBP8 deletion mutant had a similar profile to the parental strain (Fig. 1B), which reinforces the conclusion that this activity of the SAGA complex does not seem to play a relevant role in longevity under high or low nitrogen conditions. The mean life span was extended in the gcn5Δ mutant when compared to SC medium, but this extension was not observed in the spt20Δ mutant, which even reduced it (from 3.5 days to 3 days; S3 Table). Therefore, the integrity of the SAGA complex may play a role in the extension of longevity caused by the nitrogen dietary restriction. Thus Gcn5 and Spt20 are relevant for longevity, independently of the nitrogen content of the medium as the strains deleted for these proteins have shorter maximum life spans in any media when compared to the parental strain.
Fig 1. Spt20 plays a role in life span and autophagy control.
A) Survival curves in minimal complete medium SC for the SAGA complex mutants in wine strain C9. Cell number at day 3 after inoculation was considered to be 100% viability. All the experiments were carried out in triplicate and the mean and standard deviation are shown. B) Survival curves in low nitrogen (SC N 1/25) medium for the same mutants. Cell numbers at day 7 were considered to be 100% viability. C) Western blot detection of Ald6 in the wild type and the ubp8Δ and spt20Δ mutants in minimal medium with no nitrogen source (SD-N). Alcohol dehydrogenase (Adh) was used as the loading control.
As mentioned in the Introduction, acetyltransferase Gcn5 controls autophagy in wine yeast [9], as demonstrated by the stabilization of the levels of cytosolic aldehyde dehydrogenase Ald6, a selective marker of autophagy in response to nitrogen starvation [31]. Experiments to analyze autophagy in mutants ubp8Δ and spt20Δ (Fig. 1C) were conducted by the Western Blot detection of Ald6 and by using a C9 strain, where the gene coding for the mitochondrial ALD4 was deleted to prevent cross-detection. Alcohol dehydrogenase (Adh) was used as loading control. This protein is not degraded by autophagy [31], but interstingly its basal levels are decreased in the spt20Δ mutant. The ubp8Δ mutant presents a small defect in autophagy, with slightly higher Ald6 levels than the wild-type strain. The results for the spt20Δ mutant indicate a more marked defect in autophagy, with high Ald6 levels after nitrogen depletion. This indicates that the integrity of the SAGA complex is important for autophagy, and that its loss might participate in the shortening of CLS in this mutant under laboratory conditions.
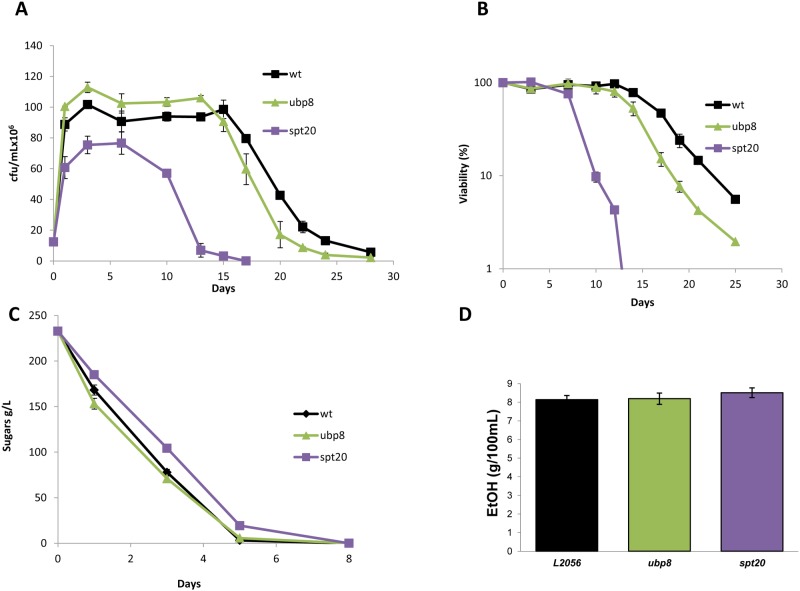
Next, in order to analyze the effect of these mutations under winemaking conditions, the two copies of UBP8 and SPT20 genes were deleted from commercial wine strain L2056 and the mutants were grown in a natural medium, red grape juice. Cell growth during fermentation is shown in Fig. 2A. The ubp8Δ mutant growth profile is similar to the wild type and no significant differences in maximum cell growth were found, although viability dropped faster in the last stages. Mutation of the SPT20 gene led to a dramatic change in the growth profile, with significantly reduced total cell growth and accelerated loss of viability. Slower growth in YPD rich medium was also observed (data not shown), which indicates that poor growth of the mutant is not dependent on the media. Cell viability at day 3, where cell density of the spt20Δ mutant peaked, was taken as 100% survival to plot the CLS curves of these strains (Fig. 2B). The CLS of ubp8Δ strain was shorter if compared to the parental strain. Therefore, Ubp8 has an impact on life span in natural media, which contrasts with the absence of effects observed in the laboratory media. Once again, the spt20Δ mutant displayed a more markedly reduced CLS as compared to the wild-type strains, which also happened in SC medium. These results demonstrate that SAGA integrity is relevant to achieve full CLS under different environmental conditions, whereas the effects of the deubiquitynilation activity of Ubp8 are growth medium-dependent.
Fig 2. Spt20 plays a role in growth and aging under winemaking conditions.
A) Growth curves for wine strain L2056 and its derivatives spt20Δ and ubp8Δ showing the number of viable cells (cfu/mL) determined by plate counting at different times during winemaking in natural grape juice. Experiments were performed at least in triplicate, and errors bars show the standard deviation (SD). B) Survival curves for the same strains. The cell numbers at day 3 in panel A were taken as 100% viability. C) Sugar consumption profiles during fermentation. D) Ethanol production at the end of grape juice fermentation. Ethanol was measured when sugars were completely consumed (below 2 g/l).
To follow the evolution of vinification and to determine the impact of these mutations on the ability to complete wine fermentation, samples were taken at different times during fermentative growth and the level of reducing sugars (glucose and fructose) were determined (Fig. 2C). The sugar consumption of the spt20Δ mutant was lower than consumption of the parental strain, probably due to the low maximum cell number and the viability lost in this mutant. The ubp8Δ mutant, however, showed a similar consumption rate to the parental strain, which is consistent with the similar maximum cell number and viability during the fermentation period, and indicates that there the deletion of this gene had no major impact on global metabolism. The impact that the lack of these proteins in ethanol production (a well-known pro-aging metabolite [13]) was also determined. Fig. 2D shows that no mutant led to significant differences in the final amount of ethanol, so overall fermentative metabolism was not challenged and differences in life spans were not due to differences in ethanol concentration.
The experiments performed with the mutants constructed in commercial wine strain L2056 were repeated for their equivalent in the haploid C9 strain, and both ubp8Δ and spt20Δ gave the same results, with deletion of SPT20 causing lower maximum cell density (S1A Fig.). Both deletions also caused a shorter maximum CLS (S1B Fig.) and no differences in ethanol production were observed (S1D Fig.). Once more, spt20Δ showed slower sugar assimilation (S1C Fig.). Therefore as similar results were obtained in both genetic backgrounds, the C9 strain was selected to perform the remaining experiments for simplicity.
Role of the retrograde response in chronological aging
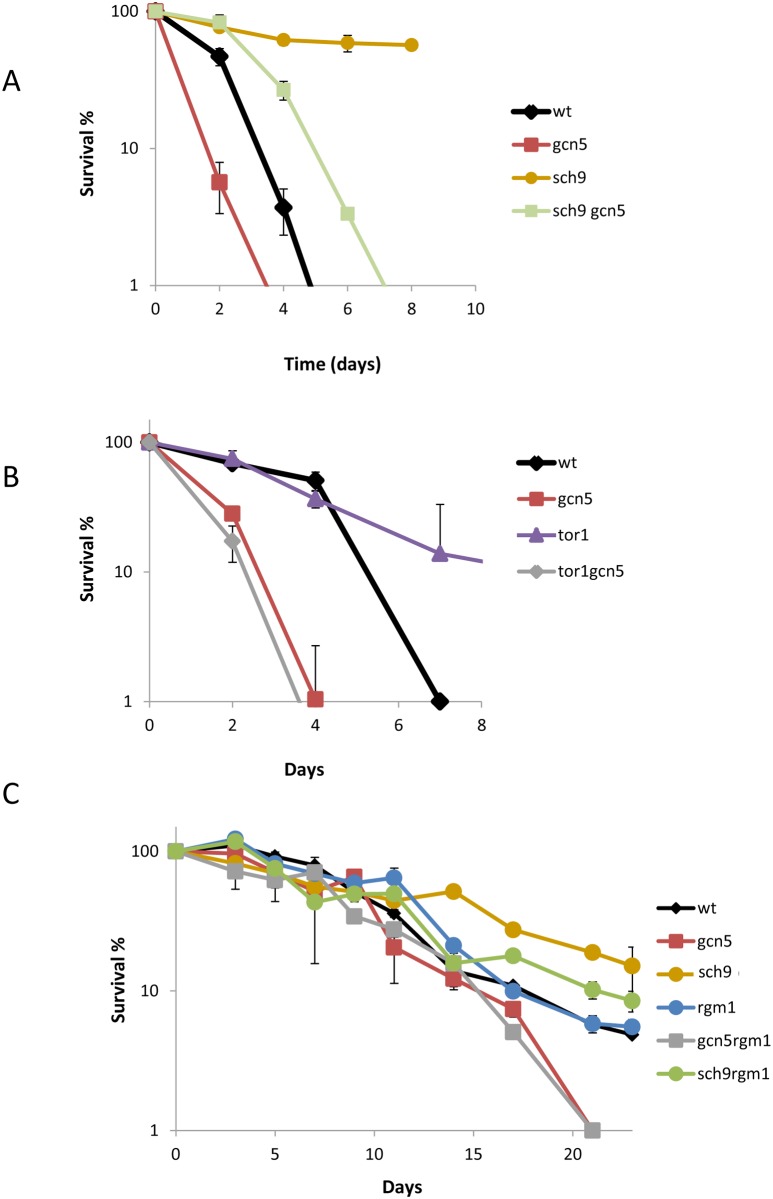
SLIK is a complex similar to SAGA and contains Gcn5, Ubp8 and Spt20, but also includes the Rtg2 protein. Rtg2 belongs to the retrograde response pathway, which informs the nucleus of mitochondrial status. For this reason, we decided to study the relationship between both the RTG2 and GCN5 genes in the chronological life span context in wine yeast under different environmental conditions (Fig. 3). First, the simple rtg2Δ and gcn5Δ mutations, and the double rtg2Δ gcn5Δ mutant, were tested for CLS in laboratory medium SC (Fig. 3A). Deletion of RTG2 causes sharp reduction in CLS if compared to the wild-type strain, and the effect was much stronger than for the gcn5Δ mutant. Therefore, the retrograde response is necessary to achieve full life span during long-term growth in SC, when respiratory metabolism occurs. Lifespan reduced even more when both mutations were combined in the double mutant, indicating that deleterious effects of both mutations take place, at least partially, through independent pathways. Next the behavior of these mutants was tested during grape juice fermentation and, in this case (Fig. 3B), deletion of RTG2 slightly extended life span and, as previously described, also GCN5 deletion. The double rtg2Δ gcn5Δ mutant further extended CLS. Therefore, the effect of these two mutations, be it contrary under the two different growth conditions, is also additive and suggests two independent pathways, but both seemed to similarly react to changes in the environment by increasing or decreasing life span, depending on the medium. The RTG2 and GCN5 mutations did not cause relevant defects in fermentative metabolism, as reflected by the similar sugar consumption rate and ethanol production (S2 Fig.). Therefore, signaling from the mitochondria is detrimental for CLS when metabolism is purely fermentative, as occurs in wine fermentation.
Fig 3. Deletions of RTG2 and GCN5 have additive effects.
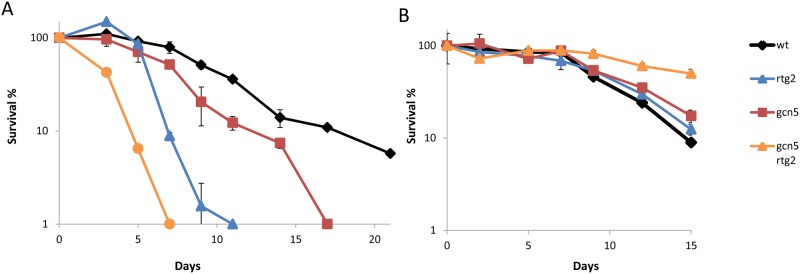
A) Survival curves of mutants rtg2Δ and gcn5Δ in SC medium. Conditions as in Fig. 1A. B) Survival plots during natural grape juice fermentation. Conditions as in Fig. 2B. Experiments were performed in triplicate. Error bars show the standard deviation (SD).
Interaction between Gcn5 and nutrient sensing kinase Sch9
GCN5 deletion blocks the CLS extension caused by partial TOR/Sch9 inhibition during winemaking [9]. Sch9 is a key kinase in which several pathways, such as TOR, Snf1 and stress by sphingolipids [32,33] signaling, converge to control lifespan. Hence the relationship between Gcn5 and Sch9 was studied. The CLS phenotypes of the single mutants analyzed in standard SC medium were opposite; whereas the gcn5Δ mutant had a shortened CLS if compared to the wild type, as mentioned above (Fig. 1A), the sch9Δ mutant had a significantly prolonged lifespan, as expected (Fig. 4A). When both deletions took place in the double mutant, the CLS extension caused by SCH9 deletion was partially blocked. Therefore, it seems that at least part of the mechanisms promoting longevity in the sch9Δ mutant requires Gcn5 activity. A similar experiment was performed with the tor1Δ mutant (Fig. 4B), which also displayed extended maximum life span, but behaved as the parental strain in mean CLS terms. Once again, the combined deletion of GCN5 blocked this CLS extension, and completely so in this case, which suggests a closer functional relationship between these two proteins. Therefore, it seems clear that the expression changes and mechanisms triggered by TOR/Sch9 inhibition and producing CLS extension require the function of acetyltransferase Gcn5, at least partially.
Fig 4. Effect of the TOR/Sch9 pathway on chronological life span in the gcn5Δ mutant in SC medium.
A) Survival curves of the wild type and the gcn5Δ, sch9Δ and double mutants. The cell numbers at day 3 after inoculation were taken as 100% viability. B) Survival curves of the wild type and the gcn5Δ, tor1Δ and double mutants. C) Survival curves of the single and double mutants in genes RGM1, GCN5 and SCH9. Experiments were performed in triplicate. Error bars show the standard deviation (SD).
Given the important role of Sch9 in CLS, we searched the Saccharomyces Genome Database (SGD) to find the physical and genetic connections between Gcn5 and Sch9. Sch9 interacts physically and genetically to transcription factor Rgm1 [34], which has been linked to subtellomeric binding [35]. RGM1 also interacts genetically to GCN5 [36], so we further investigated the role and interaction of this gene in aging. Deletion of RGM1 did not significantly alter CLS (Fig. 4C). When combined with GCN5 deletion, the phenotype of the double mutant was similar to the single gcn5Δ mutation, thus suggesting that Rgm1 plays no role in CLS regulation. Nevertheless, the CLS extension displayed by the sch9Δ mutant was partially blocked by the absence of Rgm1p, as observed in the double sch9Δ rgm1Δ. Therefore, Rgm1 may not play a relevant role during cell growth, but might be important for longevity in starvation, which are mimicked by the sch9Δ mutation.
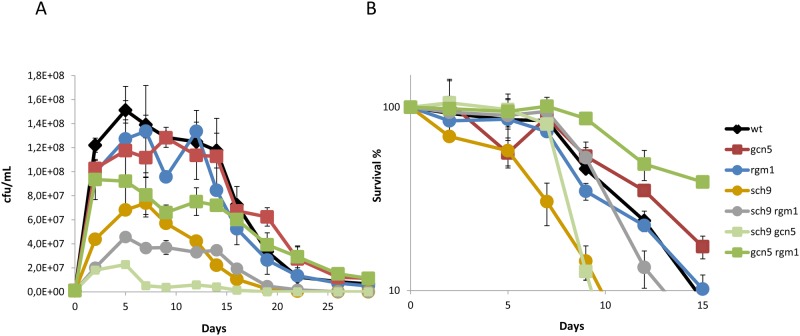
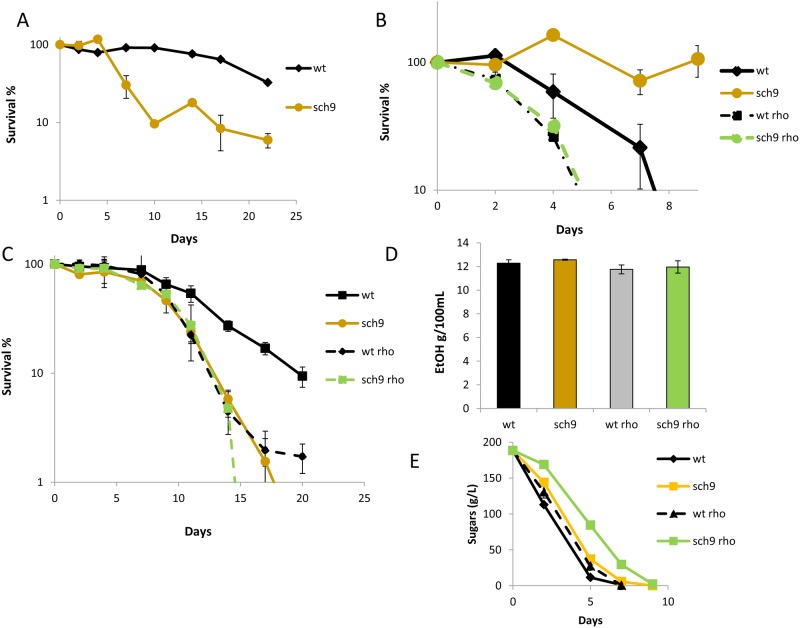
The CLS phenotype of these mutants was analyzed during wine fermentation in natural grape juice (Fig. 5), where major differences in growth profiles were observed (Fig. 5A). The sch9Δ mutation caused a strong growth defect if compared to the wild type, and the remaining single mutants obtained very low cell densities. This fact indicates that Sch9 is relevant for growth in wine fermentations, as expected for a pathway that promotes protein synthesis. This phenotype became more striking when combining the sch9Δ mutation with rgm1Δ and particularly to gcn5Δ, where cell numbers were very small. The rgm1Δ and gcn5Δ single mutants had similar growth profiles, and were also similar to the parental strain, although their combination gave small cell numbers. For this experiment, 100% viability to obtain the CLS plot profile was fixed at day 7, the day when the viability of sch9Δ peaked (Fig. 5B). Deletion of RGM1 led to no significant change in lifespan, as happened in SC medium, while GCN5 deletion extended CLS under the grape juice fermentation conditions, as previously described [9]. Surprisingly, SCH9 deletion brought about shortened CLS (both mean and maximum, see S3 Table), and the opposite effect was observed in SC (Fig. 4A). This result indicates that Sch9 inhibition does not extend life span independently of the environmental conditions, and that its function is influenced by growth conditions, as previously observed for other mutants in longevity genes under winemaking conditions [9,37]. When the SCH9 and GCN5 deletions were combined, maximum life span sharply reduced, which confirms the complex functional interaction between these two proteins. It is noteworthy that RGM1 deletion was able to extend life span in the short-lived sch9Δ mutant and also in long-lived gcn5Δ, thus reinforcing the hypothesis that acetyltranferase Gcn5 plays a negative role in longevity under some environmental conditions, such as winemaking. For metabolite production (S3 Fig.), no variation in the final ethanol concentration was noted, despite the very different growth and death profiles of theses strains. In sugar consumption terms, mutants sch9Δ and gcn5Δ, and particularly the combination of both, showed a slower sugar metabolism profile, probably due to their lower cell densities, although they were all able to complete fermentation.
Fig 5. Sch9 is required for full life span under winemaking conditions.
A) Growth curves for mutant wine yeast sch9 Δ, gcn5Δ and sch9Δ and their combinations, during natural grape juice fermentation. The number of viable cells (cfu/mL) by plate counting at different times during winemaking evolution was determined. B) Survival curves considering day 7 in panel A) to be 100% viability. Experiments were performed in triplicate. Error bars show the standard deviation (SD).
Physiological cell status modulates the role of Sch9 in aging
Given the significant and unexpected reduction of CLS during grape juice fermentation caused by SCH9 deletion, the phenotype of the sch9∆ mutant was further investigated. Winemaking conditions differ for growth in laboratory medium in several aspects, particularly in the low nitrogen and high sugars concentrations present in the medium, and also in the low oxygen environment that favors fermentative over-respiratory metabolism. To mimic low nitrogen, we tested the effect of SCH9 deletion on SC medium containing 25-fold fewer amino acids and ammonium [9] (Fig. 6A). Under these conditions, the sch9Δ mutant was more short-lived than the parental strain, which contrasts with the CLS extension observed in standard SC medium (Fig. 4A). Therefore under the unbalanced low nitrogen and high sugar conditions present in natural grape juice and in synthetic SC 1/25 N, Sch9 is required to achieve full life span. It has been pointed out that repression of TOR/Sch9 produces life span extension by promoting respiration [38,39]. Such behavior would have little effect under the low respiration conditions of grape juice fermentation, and would explain the opposite effect of SCH9 deletion. To gain a better understanding of the role that respiration plays under such conditions, petite mutants (rho o), which are unable to grow in a nonfermentable carbon source such as glycerol, were obtained for wild-type wine strain C9 and its sch9Δ derivative. These strains were grown in SC to obtain the CLS profile (Fig. 6B). The C9 petite rho o mutant has a reduced CLS, indicating that under these conditions with plenty of aeration, mitochondria and respiration are necessary to retain full longevity. Interestingly, lack of active mitochondria has an impact on CLS during fermentation, which is the opposite result to that observed in RLS [20]: CLS is shorter in the petite strain compared to the grande normal strain. The grande sch9Δ mutant has an extended life span, as expected, but the petite version of the sch9Δ mutant has the same CLS as the wild-type petite strain. This finding therefore confirms that respiration is essential for the role of Sch9 in controlling CLS under standard laboratory conditions.
Fig 6. CLS extension by mitochondrial function during winemaking requires Sch9.
A) Survival curves of the wild type and the sch9Δ mutant in SC containing 25-fold less nitrogen. 100% viability was taken at day 7. B) Survival plot of the rho o strains derived from the same strains in SC medium. 100% viability was taken at day 3. C) Survival plots in grape juice fermentation of the strains tested in panel B). D) Ethanol production during grape juice fermentation is shown in panel C). E) The sugar consumption profile for the aforementioned fermentation. Experiments were performed in triplicate. Error bars show the standard deviation (SD).
Next these petite strains were tested during grape juice fermentation (Fig. 6C). Under these conditions, the wild-type rho o mutant also had a reduced CLS. Therefore even during fermentative metabolism with a high sugar concentration, mitochondria play a positive role in longevity, which may not be related to its function in respiration. The combination of deleting the SCH9 and petite mutations did not further alter CLS, which suggests a functional relationship between both factors (Fig. 6C). Regarding the metabolic status of the petite cells, they produced similar amounts of ethanol at the end of fermentation (Fig. 6D), and the rate of sugar consumption was slightly lower than that of the wild-type strain, although both completed fermentation at the same time (Fig. 6E). As indicated above, deletion of SCH9 slowed down sugar assimilation, and this effect increased when the mutant was also petite, although fermentation was finally completed. Therefore, mitochondria have no major impact on carbon metabolism during grape juice fermentation, but it impacts CLS.
To further investigate the relationship between the Sch9 kinase and mitochondria, we focussed on the involvement of the retrograde response as it is known that the TOR pathway is a regulator of the retrograde pathway [18]. Mutations in SCH9 and RTG2 genes were combined, and the CLS profiles were studied in SC laboratory medium (Fig. 7A) and in grape juice (Fig. 7B). In SC, SCH9 deletion caused CLS extension, whereas RTG2 led to CLS shortening, as previously described. The opposite behaviors were observed when CLS was determined in grape juice, where RTG2 deletion caused CLS extension and SCH9 deletion led to CLS shortening. In both cases, however, the double sch9Δ rtg2Δ mutant had an intermediate life span, with higher longevity than the parental strain in SC medium (Fig. 7A) and a shorter one in grape juice (Fig. 7B). These results suggest that the effect of the retrograde response on life span requires Sch9 to be fully channeled. Under winemaking conditions, the situation was reversed. RTG2 deletion caused CLS extension, which was blocked by the mutation of the SCH9 gene, that was short-lived compared to the wild-type strain. This confirms the idea that Sch9 is required to transmit information from the mitochondria that carry the retrograde response and to bring about a change in CLS.
Fig 7. Sch9 is required for CLS regulation through the retrograde response.
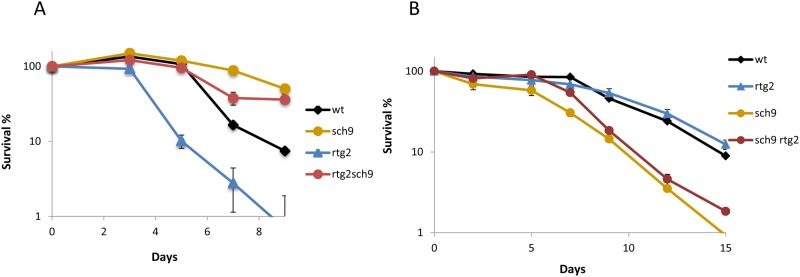
A) Survival curves of the wild type and the single and double sch9Δ and rtg2Δ mutants in SC medium. B) Survival plot of the same strains during grape juice fermentation. Experiments were performed in triplicate. Error bars show the standard deviation (SD).
Discussion
The environmental conditions for S. cerevisiae growth determine the fate of the yeast population in the death phase. It is well-known that growth in a medium with reduced glucose content, i.e., calorie restriction, extends both CLS and RLS [4,5]. When a nonfermentable carbon source is used, longevity is also extended. The effect of nitrogen source depletion on life span is more complex, probably due to the variety of nitrogen sources that yeasts can use (from ammonia to any of the 20 amino acids). The fact that most life span experiments have been performed with laboratory auxotrophic strains spells complexity due to potential intracellular metabolic unbalances. When we used prototrophic industrial wine strains, which are capable of growing with ammonia or with any of the 20 amino acids as a single nitrogen source, we observed that global nitrogen reduction (by reducing both ammonia and amino acids) extended CLS during winemaking fermentation [9,40]. Starvation of selected amino acids has a complex impact on life span, with some acting as pro-aging factors and others have an anti-aging effect [41–43]. In any case, nutrient sensing pathways are the master regulators of life span extension caused by dietary restriction, and TOR/Sch9 is particularly devoted to the cellular response to the nitrogen composition of growth medium. Wine strains respond to carbon source restriction by showing typical CLS extension [44].
Some cellular mechanisms have an invariable effect on life span, regardless of the growth conditions. Examples of such consistent effects are PKA activity, which has negative effect on life span under both laboratory and winemaking conditions; and the stress response, which is positive for longevity [40]. However, we have already described other processes, accepted as being necessary to achieve full chronological life span under laboratory conditions, and are detrimental to CLS during grape juice fermentation. For instance, deletion of the ATG7 gene abolishes autophagy and then causes a reduced CLS in laboratory medium, as expected, but extends life span under winemaking conditions, which indicates that autophagy is detrimental for longevity during grape juice fermentation [29]. However, deletion of the SPT20 gene, and then dismantling the SAGA complex, block autophagy (Fig. 1C), but CLS shortening is observed in the spt20Δ mutant in both SC medium (Fig. 1A) and grape juice (Fig. 2B). Given the relevant role of the SAGA complex in other cellular processes, such as stress response [45], it is believed that any effect of autophagy on life span under winemaking conditions can be overcome when SAGA integrity is lost. Given this variety of cellular outcomes, a picture of aging as a very complex trait where many genetic and environmental factors can act together to achieve full life span, as has emerged in recent years.
This work describes some other mutations in the genes that code for life span-controlling proteins, which play opposite roles in different growth conditions and environments, proteins that are involved in distinct cellular regulatory mechanisms. One of these mechanisms is retrograde pathways, which respond to failure in mitochondrial electrochemical potential and send information to the nucleus to activate the transcription of nuclear genes with a mitochondrial function [18,46]. Rtg2 is an essential component of this pathway and its deletion in wine yeasts produces a drop in CLS in SC medium under aeration (Fig. 3A), which is consistent with the need for respiration when sugars in the medium are consumed. However, the same rtg2Δ mutant displays an even slightly extended CLS in winemaking, where respiration is not required (Fig. 3B), and the energy waste of expressing respiratory proteins under such conditions may be detrimental for cell survival. However, this simplified view of mitochondrial function does not offer a complete explanation of its interplay with life span as petite mutants are incapable of respiration and have a shorter CLS in both SC and also in grape juice (Fig. 6). The behavior of petite mutants if compared to the inhibition of the retrograde response by RTG2 deletion suggests that mitochondrial integrity and/or metabolic roles, other than respiration itself, might be relevant to achieve full life span during grape juice fermentation because it was not affected significantly by loss of mitochondria-nucleus signaling.
The effect of the SCH9 deletion on CLS in winemaking is a challenge for the accepted role of nutrient sensing pathways in aging (Fig. 5). The Sch9 kinase integrates various pathways to promote growth and protein synthesis, and its deletion extends both CLS and RLS in laboratory media [7,8]. This is also the case of wine yeast when CLS is assayed during growth in SC medium (Fig. 4A), but not during grape juice fermentation, where the sch9Δ mutant shows shortened life span (Fig. 5). The specific media composition components causing this difference can be argued, such as the particular high carbon/low nitrogen ratio of natural grape juice, which determines a clearly different dietary restriction. To support this interpretation, SCH9 deletion also shortened CLS (Fig. 6A) in the reproducing CLS experiments performed in a modified SC with very low total nitrogen. Thus it can be concluded that nutritional unbalances are key factors in the impact of SCH9 deletion on CLS as a sch9Δ mutant extends longevity during synthetic grape juice fermentation with a high nitrogen and low lipid composition, and this situation promotes cell death [47]. It has been suggested that the life span extension caused by Sch9 inhibition may be due to an increased respiratory rate [39]. The effect of SCH9 deletion on life span during winemaking also seems to be dependent on mitochondrial integrity since a petite version of the sch9Δ mutant displayed no further decrease in CLS, but reduced the CLS of the wild-type strain (Fig. 6B). In grape juice or SC with low nitrogen, poor amino abundance acids may complicate protein translation, promoted by Sch9. Such shortage in translation may be detrimental when sugars are plenty and respiration is irrelevant. This may explain why double mutant sch9Δ rtg2Δ more resembles the sch9Δ mutant (short-lived) than the rtg2Δ mutant (long-lived) under winemaking conditions (Fig. 7B). Finally, a relationship exists between TOR/Sch9 and the SAGA complex as gcn5Δ mutation prevents, at least partially, CLS extension in SC medium caused by deletions SCH9 and TOR1. That suggests that the gene expression changes caused by the TOR/Sch9 pathway are channeled, at least partially, through the SAGA complex. During grape juice fermentation, the gcn5Δ sch9Δ double mutant displayed a shortened CLS like the sch9Δ simple mutant, which suggests that the role of SAGA in gene expression is also dependent on growth conditions. SCH9 and GCN5 interact with a new life span regulator, transcription factor RGM1. Although Rgm1 seems to have no impact on longevity during normal cellular growth, it appears to be important for CLS in starvation, like those mimicked by the sch9Δ mutation (Fig. 5). It has been recently established a link between SAGA and TOR pathway. The ribosomal transcription factor Ifh1 is acetylated by Gcn5 and its phosphorylation is mediated by TORC1 to modulate replicative life span [48].
Supporting Information
A) Growth curves for wine strain C9 and its derivatives spt20Δ and ubp8Δ showing the number of viable cells (cfu/mL) determined by plate counting at different times during winemaking in synthetic grape juice. Experiments were performed at least in triplicate, and errors bars show the standard deviation (SD). B) Survival curves for the same strains. The cell numbers at day 3 in panel A were taken as 100% viability. C) Sugar consumption profiles during fermentation. D) Ethanol production at the end of grape juice fermentation. Ethanol was measured when sugars were completely consumed (below 2 g/l).
(PDF)
Sugar consumption profiles during fermentation (A) and ethanol production at the end of grape juice fermentation described in Fig. 3B.
(PDF)
Sugar consumption profiles during fermentation (A) and ethanol production at the end of grape juice fermentation described in Fig. 5.
(PDF)
(DOCX)
Oligonucleotide pair a/b were used to amplify the kanMX-containing disruption cassette for each gene. Oligonucleotide c hybridizes to the promoter, oligonucleotide d to the coding sequence and oligonucleotide e to the terminator of each gene. Oligonucleotide K2 matches the selection marker kanMX. Pair c/K2 would give a PCR product is the selected gene is disrupted. Pair c/d will give a PCR product if a copy of the gene is still present in the cell. Pair c/e would give a small PCR product if the selection marker has been eliminated after recombinase cre induction.
(DOCX)
Data was obtained with the Prism GraphPad software package.
(DOCX)
Acknowledgments
This work has been funded by a grant from the Spanish Ministry of Science (AGL2011–24353). CP was supported by an F.P.I. fellowship.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work has been funded by a grant from the Spanish Ministry of Science (AGL2011-24353). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bisson LF, Karpel JE, Ramakrishnan V, Joseph L (2007) Functional genomics of wine yeast Saccharomyces cerevisiae. Adv Food Nutr Res 53: 65–121. [DOI] [PubMed] [Google Scholar]
- 2. Fleet GH (1993) Wine microbiology and biotechnology Chur; Philadelphia, Pa.: Harwood Academic Publishers; x, 510 p. p. [Google Scholar]
- 3. Rachidi N, Barre P, Blondin B (2000) Examination of the transcriptional specificity of an enological yeast. A pilot experiment on the chromosome-III right arm. Curr Genet 37: 1–11. [DOI] [PubMed] [Google Scholar]
- 4. Kaeberlein M (2010) Lessons on longevity from budding yeast. Nature 464: 513–519. 10.1038/nature08981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fontana L, Partridge L, Longo VD (2010) Extending healthy life span—from yeast to humans. Science 328: 321–326. 10.1126/science.1172539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaeberlein M, Burtner CR, Kennedy BK (2007) Recent developments in yeast aging. PLoS Genet 3: e84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD (2001) Regulation of longevity and stress resistance by Sch9 in yeast. Science 292: 288–290. [DOI] [PubMed] [Google Scholar]
- 8. Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, et al. (2005) Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196. [DOI] [PubMed] [Google Scholar]
- 9. Orozco H, Matallana E, Aranda A (2012) Wine yeast sirtuins and Gcn5p control aging and metabolism in a natural growth medium. Mech Ageing Dev 133: 348–358. 10.1016/j.mad.2012.03.013 [DOI] [PubMed] [Google Scholar]
- 10. Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467. 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- 11. Sampaio-Marques B, Felgueiras C, Silva A, Rodrigues F, Ludovico P (2011) Yeast chronological lifespan and proteotoxic stress: is autophagy good or bad? Biochem Soc Trans 39: 1466–1470. 10.1042/BST0391466 [DOI] [PubMed] [Google Scholar]
- 12. Ribéreau-Gayon P, Dubourdieu D, Donèche B (2006) Handbook of enology. Chichester, West Sussex, England; Hoboken, NJ: John Wiley; [Google Scholar]
- 13. Orozco H, Matallana E, Aranda A (2012) Two-carbon metabolites, polyphenols and vitamins influence yeast chronological life span in winemaking conditions. Microb Cell Fact 11: 104 10.1186/1475-2859-11-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, et al. (2005) Sir2 blocks extreme life-span extension. Cell 123: 655–667. [DOI] [PubMed] [Google Scholar]
- 15. Rodriguez-Navarro S (2009) Insights into SAGA function during gene expression. EMBO Rep 10: 843–850. 10.1038/embor.2009.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCormick MA, Mason AG, Guyenet SJ, Dang W, Garza RM, et al. (2014) The SAGA histone deubiquitinase module controls yeast replicative lifespan via Sir2 interaction. Cell Rep 8: 477–486. 10.1016/j.celrep.2014.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, et al. (2002) The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol 22: 8774–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jazwinski SM (2013) The retrograde response: when mitochondrial quality control is not enough. Biochim Biophys Acta 1833: 400–409. 10.1016/j.bbamcr.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Z, Butow RA (2006) Mitochondrial retrograde signaling. Annu Rev Genet 40: 159–185. [DOI] [PubMed] [Google Scholar]
- 20. Kirchman PA, Kim S, Lai CY, Jazwinski SM (1999) Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics 152: 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim S, Ohkuni K, Couplan E, Jazwinski SM (2004) The histone acetyltransferase GCN5 modulates the retrograde response and genome stability determining yeast longevity. Biogerontology 5: 305–316. [DOI] [PubMed] [Google Scholar]
- 22. Walker ME, Gardner JM, Vystavelova A, McBryde C, de Barros Lopes M, et al. (2003) Application of the reuseable, KanMX selectable marker to industrial yeast: construction and evaluation of heterothallic wine strains of Saccharomyces cerevisiae, possessing minimal foreign DNA sequences. FEMS Yeast Res 4: 339–347. [DOI] [PubMed] [Google Scholar]
- 23. Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Delneri D, Tomlin GC, Wixon JL, Hutter A, Sefton M, et al. (2000) Exploring redundancy in the yeast genome: an improved strategy for use of the cre-loxP system. Gene 252: 127–135. [DOI] [PubMed] [Google Scholar]
- 25. Curran BP, Carter BL (1986) Alpha-factor enhancement of hybrid formation by protoplast fusion in Saccharomyces cerevisiae II. Curr Genet 10: 943–945. [DOI] [PubMed] [Google Scholar]
- 26. Adams A, Kaiser C, Cold Spring Harbor Laboratory (1998) Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Plainview, N.Y.: Cold Spring Harbor Laboratory Press; xiv, 177 p. p. [Google Scholar]
- 27. Zuzuarregui A, del Olmo ML (2004) Expression of stress response genes in wine strains with different fermentative behavior. FEMS Yeast Res 4: 699–710. [DOI] [PubMed] [Google Scholar]
- 28. Robyt JF, Whelan WJ (1972) Reducing value methods for maltodextrins. I. Chain-length dependence of alkaline 3,5-dinitrosalicylate and chain-length independence of alkaline copper. Anal Biochem 45: 510–516. [DOI] [PubMed] [Google Scholar]
- 29. Orozco H, Matallana E, Aranda A (2012) Wine yeast sirtuins and Gcn5p control aging and metabolism in a natural growth medium. Mech Ageing Dev 133: 348–358. 10.1016/j.mad.2012.03.013 [DOI] [PubMed] [Google Scholar]
- 30. Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, et al. (1999) Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol 19: 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Onodera J, Ohsumi Y (2004) Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J Biol Chem 279: 16071–16076. [DOI] [PubMed] [Google Scholar]
- 32. Huang X, Liu J, Dickson RC (2012) Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet 8: e1002493 10.1371/journal.pgen.1002493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu JY, Lin YY, Sheu JC, Wu JT, Lee FJ, et al. (2011) Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell 146: 969–979. 10.1016/j.cell.2011.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, et al. (2004) A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 18: 2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mak HC, Pillus L, Ideker T (2009) Dynamic reprogramming of transcription factors to and from the subtelomere. Genome Res 19: 1014–1025. 10.1101/gr.084178.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, et al. (2010) The genetic landscape of a cell. Science 327: 425–431. 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Orozco H, Matallana E, Aranda A (2013) Genetic manipulation of longevity-related genes as a tool to regulate yeast life span and metabolite production during winemaking. Microb Cell Fact 12: 1 10.1186/1475-2859-12-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS (2011) Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab 13: 668–678. 10.1016/j.cmet.2011.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lavoie H, Whiteway M (2008) Increased respiration in the sch9Delta mutant is required for increasing chronological life span but not replicative life span. Eukaryot Cell 7: 1127–1135. 10.1128/EC.00330-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Orozco H, Matallana E, Aranda A (2012) Oxidative stress tolerance, adenylate cyclase, and autophagy are key players in the chronological life span of Saccharomyces cerevisiae during winemaking. Appl Environ Microbiol 78: 2748–2757. 10.1128/AEM.07261-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, et al. (2009) Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell 8: 353–369. 10.1111/j.1474-9726.2009.00469.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mirzaei H, Suarez JA, Longo VD (2014) Protein and amino acid restriction, aging and disease: from yeast to humans. Trends Endocrinol Metab. 10.1016/j.tem.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu Z, Song L, Liu SQ, Huang D (2013) Independent and additive effects of glutamic acid and methionine on yeast longevity. PLoS One 8: e79319 10.1371/journal.pone.0079319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murakami CJ, Wall V, Basisty N, Kaeberlein M (2011) Composition and acidification of the culture medium influences chronological aging similarly in vineyard and laboratory yeast. PLoS One 6: e24530 10.1371/journal.pone.0024530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Daniel JA, Grant PA (2007) Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res 618: 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jazwinski SM (2005) The retrograde response links metabolism with stress responses, chromatin-dependent gene activation, and genome stability in yeast aging. Gene 354: 22–27. [DOI] [PubMed] [Google Scholar]
- 47. Tesniere C, Delobel P, Pradal M, Blondin B (2013) Impact of nutrient imbalance on wine alcoholic fermentations: nitrogen excess enhances yeast cell death in lipid-limited must. PLoS One 8: e61645 10.1371/journal.pone.0061645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cai L, McCormick MA, Kennedy BK, Tu BP (2013) Integration of multiple nutrient cues and regulation of lifespan by ribosomal transcription factor Ifh1. Cell Rep 4: 1063–1071. 10.1016/j.celrep.2013.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Growth curves for wine strain C9 and its derivatives spt20Δ and ubp8Δ showing the number of viable cells (cfu/mL) determined by plate counting at different times during winemaking in synthetic grape juice. Experiments were performed at least in triplicate, and errors bars show the standard deviation (SD). B) Survival curves for the same strains. The cell numbers at day 3 in panel A were taken as 100% viability. C) Sugar consumption profiles during fermentation. D) Ethanol production at the end of grape juice fermentation. Ethanol was measured when sugars were completely consumed (below 2 g/l).
(PDF)
Sugar consumption profiles during fermentation (A) and ethanol production at the end of grape juice fermentation described in Fig. 3B.
(PDF)
Sugar consumption profiles during fermentation (A) and ethanol production at the end of grape juice fermentation described in Fig. 5.
(PDF)
(DOCX)
Oligonucleotide pair a/b were used to amplify the kanMX-containing disruption cassette for each gene. Oligonucleotide c hybridizes to the promoter, oligonucleotide d to the coding sequence and oligonucleotide e to the terminator of each gene. Oligonucleotide K2 matches the selection marker kanMX. Pair c/K2 would give a PCR product is the selected gene is disrupted. Pair c/d will give a PCR product if a copy of the gene is still present in the cell. Pair c/e would give a small PCR product if the selection marker has been eliminated after recombinase cre induction.
(DOCX)
Data was obtained with the Prism GraphPad software package.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.