Abstract
Salmon lice Lepeophtheirus salmonis are an ecologically and economically important parasite of wild and farmed salmon. In Scotland, Norway, and Eastern Canada, L. salmonis have developed resistance to emamectin benzoate (EMB), one of the few parasiticides available for salmon lice. Drug resistance mechanisms can be complex, potentially differing among populations and involving multiple genes with additive effects (i.e., polygenic resistance). Indicators of resistance development may enable early detection and countermeasures to avoid the spread of resistance. Here, we collect sensitive Pacific L. salmonis and sensitive and resistant Atlantic L. salmonis from salmon farms, propagate in laboratory (F1), expose to EMB in bioassays, and evaluate either baseline (Atlantic only) or induced transcriptomic differences between populations. In all populations, induced responses were minor and a cellular stress response was not identified. Pacific lice did not upregulate any genes in response to EMB, but downregulated degradative enzymes and transport proteins at 50 ppb EMB. Baseline differences between sensitive and now resistant Atlantic lice were much greater than responses to exposures. All resistant lice overexpressed degradative enzymes, and resistant males, the most resistant group, overexpressed collagenases to the greatest extent. These results indicate an accumulation of baseline expression differences related to resistance.
Keywords: drug resistance, emamectin benzoate, polygenic resistance, salmon aquaculture, sea lice, transcriptomics
Introduction
Development of parasiticide resistance in endo- and ectoparasites of importance to veterinary or human health is a major issue globally. Chemical control is ubiquitous across many parasite taxa, with administration ease and initial efficacy leading to over-reliance, and in some cases resistance (Sangster 2001). For example, trichostrongylid nematode parasites of sheep are commonly resistant to the major classes of anthelmintics, which can limit sheep production (Gilleard 2006). Although currently much less severe than that which occurs in nematodes of sheep, resistance to anthelmintics in nematodes of horses and cattle suggests a similar future concern (Kaplan and Vidyashankar 2012). Control of the cattle tick Rhipicephalus microplus is limited by the development of resistance to the acaricides used to treat this and other tick species (Abbas et al. 2014). Alongside agriculturally important parasites, mosquito vectors of malaria are becoming resistant to chemical treatments throughout Africa (Ranson et al. 2011), and the head louse Pediculus humanus capitis is also becoming resistant to commonly used parasiticides in North America, leading to increased infestations (Yoon et al. 2014). New drugs are slow and expensive to develop (Kaplan and Vidyashankar 2012), and thus, the preservation of efficacy of existing treatments is critical (Sangster 2001; Kaplan 2004) through various management and husbandry strategies and reduction of reliance on chemical control (Kaplan 2004; Kaplan and Vidyashankar 2012).
A major challenge to salmon aquaculture is the recent development of resistance of salmon lice to several important chemical control methods. Salmon lice are ectoparasitic copepods that feed on skin, mucus, and blood of wild and farmed salmon causing host damage and potentially facilitating secondary infections (Fast 2014). Parasiticide treatments are used to remove lice from farmed salmon. As a result, the aquaculture industry is largely reliant on chemical control (Johnson et al. 2004; Igboeli et al. 2014) with limited diversity of mechanisms of action, increasing the potential for resistance development (Denholm et al. 2002). A commonly used in-feed treatment is emamectin benzoate (EMB; trade name SLICE™, Merck), a macrocyclic lactone avermectin derivative that is thought to bind parasite glutamate-gated chloride channels causing hyperpolarization in neuromuscular cells, paralysis, and death (Arena et al. 1995; Stone et al. 1999; Glendinning et al. 2011). Medicated feed is given over a 7-day period and EMB accumulates in flesh and mucus (at 70–100 ppb when administered at 50 μg/kg) (Sevatdal et al. 2005). Resistance to EMB has emerged in Lepeophtheirus salmonis in Norway (Espedal et al. 2013), Scotland, and Atlantic Canada (Lees et al. 2008; Jones et al. 2012a, 2013; Carmichael et al. 2013) as well as in Caligus rogercresseyi in Chile (Bravo et al. 2008), whereas lice in Western Canada remain sensitive (Saksida et al. 2013). This is an important issue for the health and welfare of wild and domestic salmon.
Pesticide resistance can occur by direct disruption of target site binding, metabolism, and detoxification of the active product, increased compound efflux, or reduced uptake through the cuticle or digestive lining (i.e., penetration resistance) (Clark et al. 1995; Bonizzoni et al. 2012; Igboeli et al. 2012). Metabolic resistance is typically polygenic, with many small and additive effects, whereas target site disruption can be monogenic, with a large effect (i.e., knockdown resistance) (ffrench-Constant et al. 2004). Resistance mechanisms can also occur together. For example, complementary mechanisms may explain geographic variation in dichlorodiphenyltrichloroethane (DDT) resistance in knockdown-resistant (kdr) mosquitoes (Brooke 2008; Donnelly et al. 2009). Importantly, selective pressures leading to monogenic or polygenic resistance typically differ. Polygenic resistance is favoured when drug exposures occur within a range of tolerance for a portion of the population, whereas monogenic resistance is favoured when exposure occurs outside the range of tolerance (see ffrench-Constant et al. 2004). Genomics and transcriptomics enable exploration of complementary resistance mechanisms, such as metabolism or sequestration (ffrench-Constant et al. 2004; David et al. 2005), and can also be useful in monitoring and managing resistance (Pedra et al. 2004; Vontas et al. 2005; Zhao et al. 2006).
In Atlantic and Pacific L. salmonis subspecies (Yazawa et al. 2008; Skern-Mauritzen et al. 2014), the dynamics, mechanisms, or potential of EMB resistance are active areas of study. Resistant L. salmonis populations were collected from salmon farms in Norway and propagated for four generations, or crossed with a sensitive laboratory strain (Espedal et al. 2013). Pure strains remained resistant through the generations, hybrids were of intermediate sensitivity between resistant and sensitive strains, and no reproductive or survival costs were associated with resistance. Also indicating a lack of costs, Atlantic Canada L. salmonis remained highly resistant over three generations without continued selection (Igboeli et al. 2013). Intermediate hybrid sensitivity suggests polygenic resistance (Espedal et al. 2013) as identified in other species to ivermectin (for review see Clark et al. 1995). In some cases, resistance may be due to increased efflux of EMB by P-glycoprotein (pgp), for example, through upregulation of p-glycoprotein transcription in response to EMB presence (Heumann et al. 2012) or increased baseline levels of p-glycoprotein mRNA (Igboeli et al. 2012). Male-specific p-glycoprotein upregulation in response to EMB has also been identified, with resistant lice increasing expression to a greater extent than sensitive (Igboeli et al. 2013). Baseline and induced (at 200 ppb EMB) transcriptome differences were recently explored in male L. salmonis from Scottish populations with differing EMB sensitivity; the largest effect identified was between populations regardless of EMB presence, and the authors highlighted reduced expression of a GABA-gated chloride channel and a neuronal acetylcholine receptor in the resistant population in response to EMB, among others (Carmichael et al. 2013). Whether EMB resistance in L. salmonis involves multiple factors and the specifics of these factors are yet to be determined.
Here, we apply a 38K oligonucleotide microarray (Sutherland et al. 2012; Yasuike et al. 2012) and reverse-transcription quantitative PCR (RT-qPCR) to profile gene expression responses of sensitive Pacific lice to EMB (0, 10, 25, 50 ppb) and to compare baseline and induced (0, 0.1, 25, 300, 1000 ppb) expression differences between sensitive and resistant Atlantic lice (Igboeli et al. 2013). Sex-specific differences in EMB resistance have been identified (Westcott et al. 2008; Igboeli et al. 2013; Jones et al. 2013), and accordingly, we profile transcriptomic responses in both male and female pre-adult Atlantic L. salmonis. Our study provides the first baseline dataset for EMB transcriptome responses of the Pacific subspecies of L. salmonis and provides new insight into resistance mechanisms in the Atlantic subspecies.
Materials and methods
Pacific salmon lice collection, EMB exposure, and RNA extraction
Pacific L. salmonis were obtained from Atlantic salmon Salmo salar farms near Campbell River, British Columbia (BC), in March 2009. From these individuals, nauplius larvae were hatched and grown to copepodids, which were allowed to attach to laboratory-reared Atlantic salmon as previously described (Sutherland et al. 2014). After 40 days, salmon were sedated with metomidate hydrochloride (Aquacalm, Syndel Laboratories Ltd.) and pre-adult lice were collected (777 individuals). Stock emamectin benzoate (PESTANAL®, Sigma–Aldrich, St. Louis, MO, USA) was prepared to 100 mg/L in methanol and then diluted to working concentrations in seawater. After removal from fish, lice were held briefly in 10°C seawater for <1 h. Approximately 25 pre-adult stage I and II male and female individuals were haphazardly distributed into each of 24 beakers containing 500 ml filtered and aerated seawater (10°C; 30 parts per thousand (ppt) salinity). Groups of six beakers were assigned to four EMB concentrations (0, 10, 25, or 50 ppb; Table1), and temperature was maintained at 10°C by incubation in a water bath. After 24 h, lice from each beaker were collected on a mesh filter and flash-frozen (n = 24 pools). Total RNA was extracted from frozen tissue using TRIzol® (Life Technologies, Carlsbad, CA, USA) followed by RNeasy column purification (Qiagen, Venlo, Netherlands), as per manufacturers' instructions. Purified total RNA was tested by agarose gel electrophoresis for quality and by spectrophotometry (NanoDrop-1000) for purity and quantity. Use of research animals complied with Fisheries and Oceans Canada Pacific Region Animal Care Committee protocol number 09-001.
Table 1.
Experimental design for RNA profiling. Layout of experiment displaying the number of biological replicates for each condition. Biological replicates are individuals for Atlantic and pools of ∽25 lice for Pacific lice
| Subspecies | Stage | Sex | Emamectin benzoate (EMB) resistance | Sample size for each [EMB concentration (ppb)] | ||||
|---|---|---|---|---|---|---|---|---|
| [0] | [10] | [25] | [50] | |||||
| Pacific | Pre-adult | Mixed | Sensitive | 6 | 6* | 6 | 6 | |
| [0] | [0.1] | [25] | [300] | [1000] | ||||
| Atlantic | Pre-adult | Female | Low | 4 | 4 | 4 | 4 | 4 |
| High | 4 | 4 | 4 | 2 | 4 | |||
| Male | Low | 4 | 4 | 3 | 4 | 4 | ||
| High | 4 | 4 | 4 | 4 | 4 | |||
Condition used for RT-qPCR only.
Atlantic salmon lice collection, EMB exposure, and RNA extraction
Atlantic L. salmonis were collected in 2012 from Atlantic salmon farms in Back Bay (resistant) and near Grand Manan, New Brunswick (sensitive) (Jones et al. 2012a). Live lice were moved to the Atlantic Veterinary College (University of Prince Edward Island) and grown on Atlantic salmon for approximately 80–90 days until the extrusion of the third set of egg strings. Larvae from these egg strings were grown to copepodids and then allowed to rear to the pre-adult stage on Atlantic salmon (Covello et al. 2012; Igboeli et al. 2013). Subsequently, salmon were sedated with tricaine methanesulfonate (MS-222), and lice were collected in Petri dishes with seawater (10°C, 33 ppt salinity). Petri dishes were swirled and only adherent lice were used in subsequent studies. EMB exposures were performed as per standardized bioassay protocols (Westcott et al. 2008). There have been no observable differences in gene expression, phenotype, or survival from the concentration of methanol (EMB solvent) in the EMB dilutions (Hoplite laboratory personal communication), and standardized bioassay protocols do not call for a solvent control (Westcott et al. 2008). Therefore, here, we did not include both a seawater and solvent control. Lice from each population were separated by sex, and four individuals were distributed to each of 40 flasks containing seawater (10°C, 33 ppt salinity). The flasks were assigned in duplicate to 20 treatment groups: EMB concentration (0, 0.1, 25, 300, 1000 ppb), sex, and population (resistant, sensitive; Table1). EMB concentrations were selected to include those used in Pacific lice exposures (e.g., 25 ppb) and those known to cause phenotypic effects in resistant Atlantic lice (e.g., 1000 ppb; M. Fast, personal observation). After 24 h, lice from each beaker were flash-frozen individually and kept at −80°C until RNA extraction (Table1). Total RNA was extracted as described above, with the exception of using TURBO DNase treatment (Life Technologies) prior to RNeasy column purification (Qiagen). Purified total RNA was tested by agarose gel and automated electrophoresis (Experion; Bio-Rad, Hercules, CA, USA). Use of research animals for this study was approved by Canadian Animal Care Committee protocol #12-016.
To confirm EMB sensitivity differences between the resistant and sensitive populations, 24-h bioassays were performed on a subset of the parental collection (F0) and on lice propagated in laboratory (F1). For F0 lice, these bioassays were performed in duplicate flasks with 10 lice per flask per condition (i.e., for each sex, population, and concentration combination), and for F1 lice, with five and four lice per condition for resistant and sensitive populations, respectively (F1 EC50 bioassay sample sizes were constrained to reserve lice for RNA profiling bioassays). After 24 h, lice were evaluated as per standards (Westcott et al. 2008) and EC50 values were calculated in GraphPad (v6; Prism, La Jolla, CA, USA).
cDNA preparation and microarray hybridization
For each sample, 825 ng of total RNA was amplified to Cy5-labeled cRNA using the Low-Input Quick Amp system (Agilent; v6.5, Santa Clara, CA, USA). A Cy3-cRNA reference pool used to hybridize alongside samples (Churchill 2002) was generated for the Pacific and Atlantic lice experiments, ensuring the inclusion of at least one individual from all experimental conditions. The sample and reference pool combination was hybridized to oligonucleotide microarrays designed using previously annotated ESTs from both Pacific and Atlantic L. salmonis (Yasuike et al. 2012; eArray design ID 024389; Agilent), scanned on a ScanArray Express (Perkin Elmer, Waltham, MA, USA) and quantified on Imagene (v8.1; BioDiscovery, Hawthorne, CA, USA) as previously reported (Sutherland et al. 2012). For each probe, the background median was subtracted from the foreground, samples were imported into GeneSpring GX11 (Agilent), any negative raw values were changed to 1.0, and each array was normalized by intensity-dependent Lowess normalization (Yang et al. 2002) (Agilent).
Transcriptome analyses
Pacific and Atlantic experiments were analyzed separately. For each experiment, quality control filters retained probes passing the following criteria in at least 65% of the samples in any one condition: no poor quality flags and raw signal ≥ 500 in both channels.
Pacific lice exposures to 0, 25, and 50 ppb (n = 6 pools per condition; Table1) were profiled by microarray (the 10 ppb condition was only included in subsequent RT-qPCR analysis). Differential expression was tested by one-way anova without equal variance assumption (Benjamini–Hochberg multiple test corrected P ≤ 0.01), followed by post hoc Tukey HSD (P ≤ 0.01) and fold change filter (FC ≥ 1.5) between conditions.
Atlantic male and female pre-adult lice from sensitive and resistant populations exposed to 0, 0.1, 25, 300, and 1000 ppb were profiled by microarray (n = 20 conditions; Table1). Differential expression was tested by three-way anova (Benjamini–Hochberg multiple test corrected P ≤ 0.01) using population, sex, and EMB concentration as factors and including interaction effects. Each condition contained four biological replicates except for two conditions that had insufficient high-quality RNA (i.e., male sensitive 25 ppb (n = 3) and female resistant 300 ppb (n = 2); Table1). Probes with a significant three-way interaction effect were removed from all other significant effect lists and clustered by k-means clustering based on similar expression (Euclidean distance metric; 4 clusters; Agilent). Fold changes were calculated based on marginal means for each comparison described below (i.e., controlling for other factors in the model due to unequal sample size in some conditions; Table1). Probes with a significant sex by population interaction were removed from sex or population main effect lists and fold-change-filtered (FC ≥ 1.5) between populations to identify genes specific to one sex (unchanging or discordant in other sex) or regulated to a larger extent in one sex than the other. Probes with a significant main effect of population were fold-change-filtered between resistant and sensitive populations (FC ≥ 1.5). Probes with a significant main effect of EMB concentration were fold-change-filtered against the 0 ppb control (FC ≥ 1.5).
Gene Ontology and pathway enrichment was performed in DAVID Bioinformatics using a modified Fisher's exact test (Huang et al. 2009) (P ≤ 0.05; number of genes in category ≥4), and result redundancy was reduced using GO Trimming (80% soft trim threshold; Jantzen et al. 2011). For this analysis, Entrez IDs assigned to contigs used for probe design (Yasuike et al. 2012) were used. Background lists for enrichment comparisons (i.e., all probes passing quality control) were specific to each experiment (Pacific = 12 085 probes; Atlantic = 15 578). Principal component analysis was performed in GeneSpring using normalized expression values for all probes passing quality control filters in Atlantic lice.
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
Pacific L. salmonis samples described above (including the 10 ppb condition) were used for RT-qPCR analysis. Each SuperScript® III (Life Technologies) reverse-transcription reaction included 2.5 μg total RNA and a 50:50 mix of random hexamers and oligo(dT)20 primers (total per reaction 50 ng), as per manufacturer's instructions. Resultant cDNA samples were diluted 20-fold. A standard curve for primer testing was generated from a sevenfold dilution of a sample from each condition and used in a fivefold, six-point serial dilution. All primers were tested for efficiency between 80% and 110% and for a single product by melt curve analysis and amplicon purification and sequencing as previously reported (Sutherland et al. 2011). qPCR amplification was performed using SsoFast EvaGreen with Low Rox (Bio-Rad) as per manufacturers' instructions for the MX3000P thermocycler (Agilent) using the following thermal regime: 95°C for 5 min (1 cycle); 95°C for 20 s and then 55°C for 30 s (40 cycles), followed by a melt curve (55–95°C reading fluorescence at 0.5°C increments). Samples were run in duplicate and accepted when technical replicates were within 0.5 cycles. A no template control (NTC) and -RT control was run for each gene. Although some contaminating gDNA remained after column purifications, -RT controls for each gene were more than six cycles greater than the most dilute sample for that gene and thus would have a minimal effect on quantification (Laurell et al. 2012). Genes of interest were normalized using structural ribosomal protein s20, a validated L. salmonis normalizer candidate (Frost and Nilsen 2003) in qBASEplus (Biogazelle) using primer-specific efficiencies.
Pacific salmon lice can carry a microsporidian parasite Facilispora margolisi at approximately 50% prevalence (Jones et al. 2012b). As the impact of the microsporidia on L. salmonis or on the L. salmonis response to EMB has not yet been characterized, to ensure that there was no confounded variation due to differences in microsporidian infection among groups, Pacific lice pools used here were tested for F. margolisi using diagnostic primers (Jones et al. 2012b). All pools tested positive. Additionally, a subset of samples from Atlantic L. salmonis was also tested, but no positives were detected, which is consistent with the observation that F. margolisi has not been found in Atlantic L. salmonis (S. Jones, personal observation).
Atlantic lice used in the microarray experiment were also used for RT-qPCR analysis, with the exception of one sensitive female 1000 ppb sample that did not amplify (total n = 76). Total RNA was reverse-transcribed using 400 ng input in iScript™ Reverse Transcription Supermix (Bio-Rad) with a mix of random hexamers and oligo(dT) primers, as per manufacturer's instructions. Subsequently, cDNA samples were diluted 10-fold. Additionally, primer efficiency was performed as described above, except that the standard curve was prepared with an initial 2-fold dilution, and a single product was confirmed for the amplicons by melt curve analysis. qPCR amplification was performed using SsoAdvanced™ SYBR® Green Supermix (Bio-Rad) as per manufacturers' instructions for the Mastercycler ep realplex thermal cycler (Eppendorf) using the following thermal regime: 95°C for 10 min (1 cycle); 95°C for 15 s, primer-specific annealing temperature (see Table S1) for 15 s, and then 72°C for 15 s (40 cycles), followed by a melt curve (55–95 °C reading fluorescence at 0.5°C increments). Samples were run in duplicate (99.63% of technical replicates within 0.5 cycles). As above, NTC did not amplify, and -RT controls were more than 6 Ct above the lowest expressed samples. Candidate normalizer genes included structural ribosomal protein s20 (rps20), elongation factor 1-alpha (ef1a), and vinculin (vcl). The most stable as calculated by geNORM (Vandesompele et al. 2002) were rps20 and ef1a (geNORM M-value and coefficient of variation of 0.339 and 0.118, respectively), and therefore, the geometric mean of these two genes was used to normalize genes of interest in qBASEplus (Biogazelle, Zwijnaarde, Belgium) using primer-specific efficiencies (Table S1).
For both the Pacific and Atlantic samples, the log2 RT-qPCR normalized value and the log2 microarray value for each sample were correlated in R (R Core Team 2014) using linear models with microarray values as the independent variable and RT-qPCR values as the dependent variable to obtain an adjusted R-squared value and slope of the relationship for each gene. Pacific RT-qPCR (which included the 10 ppb condition) was tested for significance by one-way anova and post hoc Tukey HSD in R.
Results
Transcriptomic effects of EMB exposure in Pacific lice
The transcriptional response of Pacific lice to EMB was minimal until 50 ppb (only three differentially expressed probes at 25 ppb). Although no probes were upregulated at 50 ppb, 148 probes were downregulated (relative to either 0 or 25 ppb; Table2). All differentially expressed genes can be found in Table S2. The downregulated genes at 50 ppb (43 unique annotations) was enriched for proteolysis (11 genes; P = 6.7E-06) and included probes annotated as degradative enzymes including trypsin-1, carboxypeptidase B, chymotrypsin, collagenase, acidic mammalian chitinase, hypodermin-B, and others (Table3). Additional enriched functions included lipid metabolism (five genes; P = 0.025) and cation binding (10 genes; P = 0.04), among others (see Table S3); several other enzymes and transporters were downregulated. The suppression at 50 ppb EMB was confirmed by RT-qPCR, and the lack of differential expression until 50 ppb was confirmed by assessing the additional dataset at 10 ppb by RT-qPCR (Fig.1).
Table 2.
Differential expression in Atlantic and Pacific lice responding to emamectin benzoate (EMB). Probes responding to EMB exposure in Atlantic and Pacific lice. Atlantic responses increased until 300 ppb, and Pacific lice response was minimal until 50 ppb EMB
| Subspecies | Condition versus control, ppb | Probes ≥ 1.5-fold | Probes ≥ 2-fold |
|---|---|---|---|
| Atlantic | 0.1 | 22 | 4 |
| 25 | 236 | 44 | |
| 300 | 513 | 118 | |
| 1000 | 474 | 86 | |
| Pacific | 25 | 3 | 3 |
| 50 | 148 | 144 |
Table 3.
Pacific lice degradative enzyme suppression from emamectin benzoate (EMB). Descriptions, corrected P-value, and linear fold change for genes present in Gene Ontology category proteolysis (bold font), or with known degradative function
| Probe ID | Probe description | Corr. P-value | FC EMB 25 vs 0 | FC EMB 50 vs 0 |
|---|---|---|---|---|
| C068R042 | Acidic mammalian chitinase | 0.0023 | – | −3.8 |
| C036R034 | Anionic trypsin-1 | 0.0035 | – | −7.3 |
| C005R080 | Anionic trypsin-2 | 0.0018 | – | −6.4 |
| C102R046 | Aspartic proteinase oryzasin-1 | 0.0025 | – | −5.8 |
| C020R062 | Carboxypeptidase B | 0.0018 | – | −4.5 |
| C009R120 | Cathepsin D | 0.0021 | – | −5.4 |
| C099R025 | Cathepsin K | 0.0045 | – | −6.6 |
| C022R035 | Chymotrypsin BI | 0.0021 | – | −4.6 |
| C123R134 | Collagenase | 0.0018 | – | −3.8 |
| C053R101 | Gamma-glutamyl hydrolase | 0.0018 | – | −6.5 |
| C133R012 | Neprilysin-2 | 0.0026 | – | −3.1 |
| C100R117 | Ovochymase-1 | 0.0018 | – | −9.7 |
| C158R157 | Placental protein 11 | 0.0032 | – | −5.0 |
| C171R022 | Probable cysteine proteinase At3g43960 | 0.0035 | – | −4.0 |
| C080R027 | Putative serine protease K12H4.7 | 0.0021 | – | −3.9 |
| C028R010 | Transmembrane serine protease 8 | 0.0018 | – | −9.5 |
| C042R088 | Trypsin-1 | 0.0018 | – | −2.4 |
| C171R148 | Zinc carboxypeptidase A 1 | 0.0018 | – | −6.7 |
Figure 1.
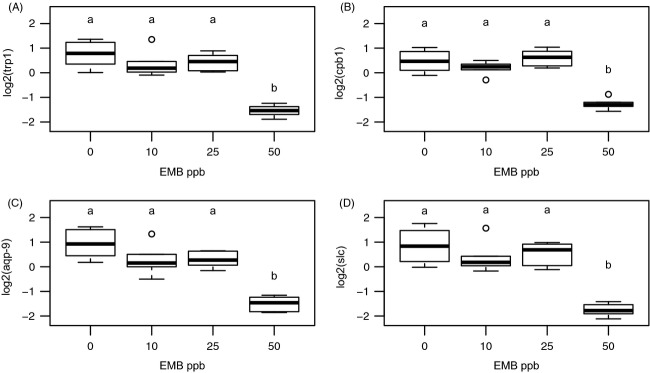
Pacific lice degradative enzyme and transporter suppression. RT-qPCR confirmed the downregulation at 50 ppb of degradative enzymes trypsin-1 (trp-1) and carboxypeptidase-1 (cpb-1), as well as transporters aquaporin-9 (aqp-9) and high-affinity copper transport protein (slc). Conditions that do not share a letter above the boxplot are significantly different from each other (Tukey HSD P ≤ 0.05). Boxplot displays median and interquartile range.
EMB sensitivity differences between Atlantic lice populations
Bioassays confirmed increased resistance in the Back Bay lice relative to those from Grand Manan (Table4) and indicated the highest resistance in males. Furthermore, bioassays of the F1 generation propagated in the laboratory followed similar trends. All F1 lice were healthy-vigorous at 0, 0.1, and 25 ppb EMB. At 300 ppb, four sensitive males were healthy-vigorous, three weak (swimming but not attaching to the beaker), and one immobile (no twitching), whereas nine resistant males were healthy-vigorous and one moribund (immobile and twitching). At 300 ppb, one sensitive female was healthy-vigorous, four weak, and three immobile, whereas six resistant females were healthy-vigorous and four weak. At 1000 ppb, all lice in both populations were immobile.
Table 4.
Survival differences between Atlantic populations. EC50 (ppb) for male and female lice from the two populations with differing emamectin benzoate sensitivity
| Population | EC50 ppb (95% CI) | |
|---|---|---|
| Males | Females | |
| Back Bay (Resistant) | 840 (614, 1047) | 254 (218, 296) |
| Grand Manan (Sensitive) | 63 (11, 352) | 75 (13, 432) |
Comparative influence of sex, population, and EMB on Atlantic lice transcriptomes
The most influential factor affecting gene expression was sex (Fig.2; Table5; PC1: 40.9% of total variation). Population also had a large effect: Males or females clustered by population in the PCA, and many genes differed between populations (Table5). The only sex–population combination to show a large effect from EMB dose in the PCA was the resistant females (Fig.2).
Figure 2.
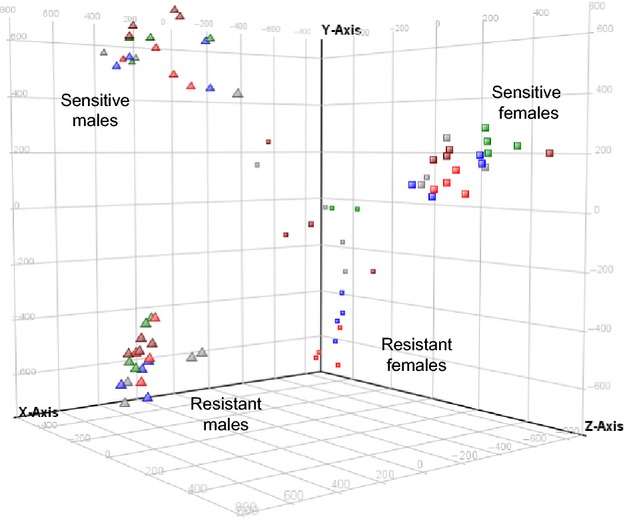
Principal component analysis of samples based on gene expression. Male and female lice samples separated along the x-axis (PC1: 40.9% of variation), and populations separated by y- and z-axes (PC2: 28.3% and PC3: 20.7%, respectively). A separation of emamectin benzoate (EMB) doses is identifiable in the resistant females only. Colors display EMB concentration (red = 0; blue = 0.1; gray = 25; green = 300; brown = 1000), and shape displays the sex (triangle = male; square = female); population is labeled beside each cluster.
Table 5.
Overview of factors influencing Atlantic lice transcriptomes. Numbers of significant probes shown for each effect (sex, population (Pop), and emamectin benzoate concentration (EMB conc)) and interaction prior to fold change filters. Probes with a three-way interaction are not included in two-way interaction or main effect lists, and probes with a significant two-way interaction are not included in the main effect lists involved in the interaction
| Comparison | Number of probes |
|---|---|
| Sex * Pop * EMB conc | 151 |
| Sex * Pop | 8242 |
| Sex * EMB conc | 26 |
| Pop * EMB conc | 19 |
| Sex | 4683 |
| Pop | 3699 |
| EMB conc | 1413 |
The main objective of this study is to identify genes related to EMB resistance, and therefore, the most relevant genes are those (i) differing in baseline expression between populations consistently in both sexes (main effect population); (ii) differing in baseline expression between populations inconsistently in sexes (sex by population interaction); (iii) responding to EMB exposure in both sexes and populations (main effect EMB); and (iv) responding to EMB exposure specifically in one sex–population combination (three-way interaction of sex, population, and EMB). Genes in these four categories will be presented in the next four sections.
Genes differing in baseline expression between populations in both sexes (Atlantic)
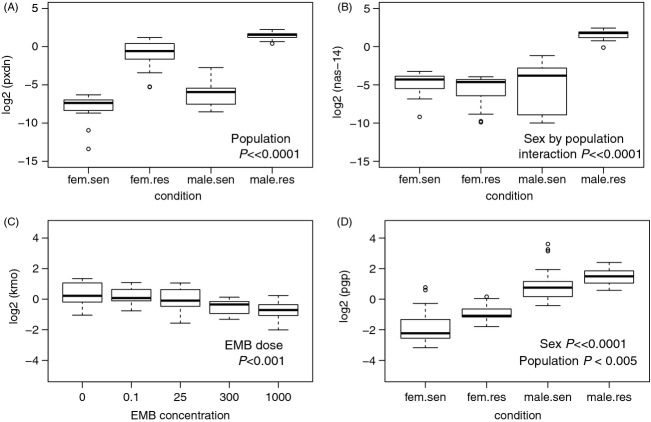
A large number of genes were differentially expressed between populations consistently in both sexes, regardless of EMB presence (main effect population; Table5). Resistant lice overexpressed 446 probes relative to sensitive (141 of which were greater than 2.5-fold). The highest overexpressed genes in resistant lice were peroxidasin homolog (two probes; >140-fold; Fig.3A), collagenase, ovochymase-1, trypsin-1, phospholipid hydroperoxide glutathione peroxidase (mitochondrial), cathepsin D, and carboxypeptidase B (>5-fold; Table S2). Overexpressed genes in resistant lice were enriched for lipid metabolic process (16 genes; P = 1.1E-4), response to chemical stimulus (13 genes; P = 0.005), catalytic activity (58 genes; P = 0.003), and serine-type peptidase activity (seven genes; P = 0.002), among others (Table S3).
Figure 3.
Expression of genes potentially related to resistance in Atlantic lice. (A) The expression of peroxidasin homolog (pxdn) was highly overexpressed in the resistant population for both sexes, and (B) zinc metalloproteinase nas-14 (nas-14) was specifically overexpressed in resistant males. (C) In contrast to the strong differences between populations, the effect of emamectin benzoate dose was minor, although several genes were differentially expressed, including kynurenine-3 monooxygenase (kmo). (D) As identified in previous candidate gene approaches, here, p-glycoprotein (pgp) was overexpressed in the resistant population and had highest expression in males. Data in (A–C) are from the microarray and (D) RT-qPCR.
Sensitive lice overexpressed 549 probes relative to resistant (134 of which were greater than 2.5-fold). The highest overexpressed probes did not have annotation (six probes > 100-fold), and the highest annotated probe was serine protease inhibitor dipetalogastin (10-fold). Overexpressed genes in sensitive lice were enriched for calcium ion binding (17 genes P = 2E-6), system development (21 genes; P < 0.001), cell differentiation (19 genes; P < 0.001), extracellular region (10 genes; P = 0.02), microtubule binding (five genes; P < 0.005), and endopeptidase activity (nine genes; P < 0.02), among others (Table S3). The endopeptidase activity genes included papilin, stubble, calpain 11, proteasome subunit alpha type 4, and others; this category was composed of different genes than those in the serine-type peptidase activity category that was enriched in the resistant lice overexpression list.
Genes differing in baseline expression between populations in only one sex (Atlantic)
Resistance to EMB may be sex dependent (Igboeli et al. 2013; Jones et al. 2013; Whyte et al. 2013), and therefore, it is worthwhile to consider genes that differ between populations in only one sex (i.e., not differentially expressed or discordant in the other).
Specifically overexpressed in resistant males (but not in resistant females) were 1217 probes. Highly overexpressed were metalloproteinases zinc metalloproteinase nas-6 and nas-4 (FC > 50) as well as 72 kDa type IV collagenase, trypsin-like serine protease, and matrix metalloproteinase-9 (FC > 10; Table S2; Fig.3B). In comparison, 1048 probes were overexpressed in sensitive males, including many unannotated probes, or those annotated as chitin_bind_4 and cuticle protein cp14 (FC > 45).
Specifically overexpressed in resistant females (but not in resistant males) were 936 probes. Highly overexpressed were tristetraproline, trypsin-1, hemicentin-1, trypsin-4, and von Willebrand factor D and EGF domain-containing protein (FC > 50). In comparison, 913 probes were overexpressed in sensitive females. Many of these were unannotated but several with high fold change included tyrosine aminotransferase, 72 kda type IV collagenase, histone-lysine N-methyltransferase setd7, vitellogenin-2, and heparan sulfate 2-O-sulfotransferase pipe (FC > 5).
Additionally, there was a subset of genes differentially expressed between populations that were concordantly regulated in both sexes, but to a larger extent in one sex than the other. Male resistant lice overexpressed 146 probes to a greater extent than the female resistant lice; this list was enriched for proteolysis (Table S3) and included chorion peroxidase heavy chain (380-fold in males, 120-fold in females), anionic trypsin-1, matrix metalloproteinase-9 (12-fold in males, 1.8-fold in females), among others (Table S2). In comparison, female resistant lice overexpressed 41 probes to a greater extent than the male lice, but this list had no functional enrichment. Male sensitive lice overexpressed 105 probes to a greater extent than female sensitive lice; this list was enriched for transition metal ion binding (Table S3) and included a disintegrin and metalloproteinase with thrombospondin motifs 12, 18, and 20 (>20-fold in males, >6-fold in females), as well as venom allergen 3 (>60-fold in males, >12-fold in females), among others (Table S2). Female sensitive lice overexpressed 73 probes to a greater extent than male sensitive lice, including histone-lysine N-methyltransferase setd7 (25-fold in females, 3-fold in males), nuclear pore membrane glycoprotein 210 (54-fold in females, 3-fold in males), protein disulfide-isomerase A4 (6-fold in females, 2-fold in males), and gamma-aminobutyric acid receptor subunit beta-like (2.5-fold in females, 1.7-fold in males).
Genes responding to EMB in both sexes and populations (Atlantic)
Relatively few genes responded to EMB dose. Some genes responded in both sexes and populations consistently, although most had low fold change (e.g., only 38 probes ≥ 2.5-fold at 300 ppb). Of these genes, the number of differentially expressed genes increased with EMB dose until 300 ppb; at 1000 ppb, the number and identity of differentially expressed genes were similar to that at 300 ppb (Table2; Table S2).
Among the most significantly upregulated genes from EMB exposure were ring finger protein nhl-1, and alpha- and beta-taxilin (FC ≥ 1.5 at 25, 300, and 1000 ppb; P ≤ 5E-9). However, even these genes did not have high fold change. Upregulation was also identified for genes involved in ion binding or transport, including voltage-gated potassium channel subunit beta-2, sarcoplasmic calcium-binding protein (beta chain), calcium-activated potassium channel slowpoke, caldesmon, and calmodulin.
Genes downregulated in response to EMB exposure in Atlantic lice also only had moderate fold change, including several metalloproteinases (e.g., zinc metalloproteinase nas-4 and nas-15, matrix metalloproteinase-9, astacin, 72 kDa type IV collagenase) and transporters (e.g., solute carrier family 25 member 38, ABC transporter G family member 20, and low-affinity cationic amino acid transporter; Table S2). Additionally, a probe annotated as kynurenine 3-monooxygenase was downregulated at 300 and 1000 ppb (FC > 1.5; P < 0.001; Fig.3C).
Genes responding to EMB specifically in one sex–population combination (Atlantic)
A strong expression change over EMB doses was identified specifically in resistant females. These genes are likely to have contributed to the separation of resistant female samples by dose in the PCA (Fig.2). To further characterize these 151 probes, they were clustered into four clusters with similar expression (Fig.4A). Clusters (i) and (iii) contain genes with elevated expression in resistant female controls (0 ppb) and decrease either to the level of the sensitive lice or to the level of all other conditions, respectively. Cluster (iv) contains genes with low expression in resistant female controls that increases to the level of the sensitive lice. This cluster contains probes annotated as glutenin high molecular weight subunits, plasmodium histidine-rich protein, a disintegrin and metalloproteinase with thrombospondin motifs 20, adhesive plaque matrix protein, and several unknowns. Probes present in each cluster are shown in Table S2. The expression of a disintegrin and metalloproteinase with thrombospondin motifs 20 was confirmed by RT-qPCR (Fig.4B).
Figure 4.
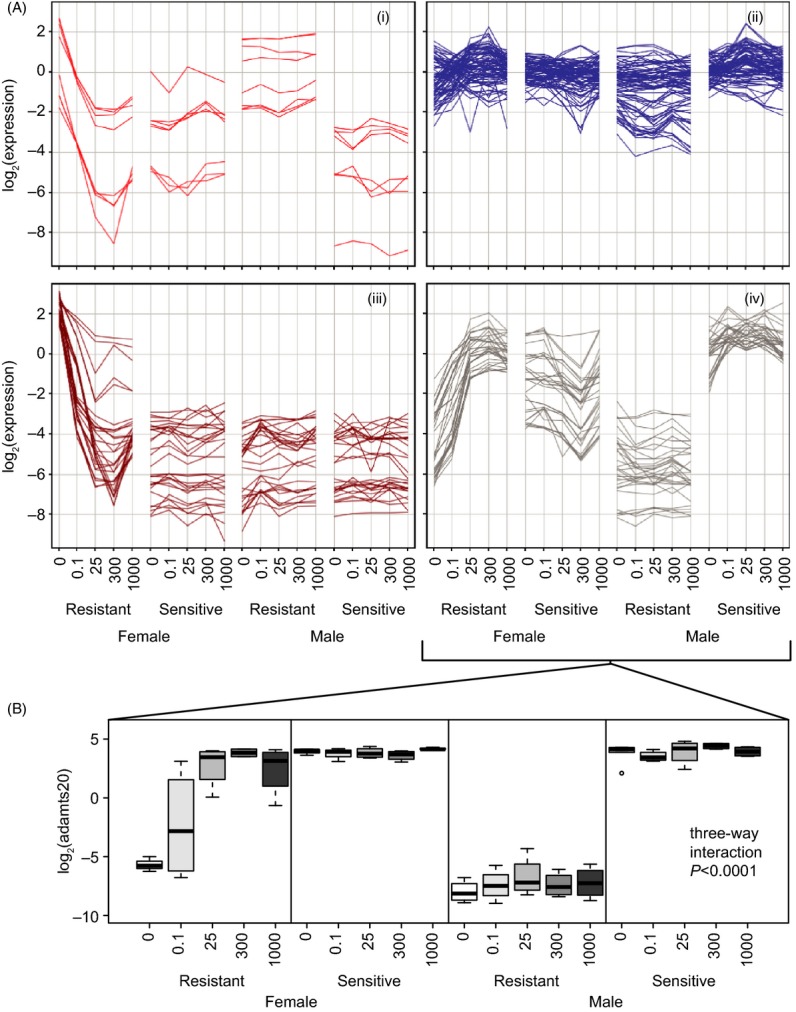
Genes responding to emamectin benzoate (EMB) specifically in resistant females. (A) Genes changing with EMB dose differently depending on sex and population were clustered based on expression into four clusters (i–iv). These genes were mainly comprised of genes responding to EMB exposure in the female resistant population. (B) The expression of a gene within cluster (iv), a disintegrin and metalloproteinase with thrombospondin motifs 20 (adamts20) was confirmed by RT-qPCR.
Additional RT-qPCR exploration
The expression data from the microarray were validated with RT-qPCR for the Pacific (as discussed above; Fig.1) and Atlantic lice, and most genes correlated well between the methods (Fig. S1). This included the degradative enzymes adamts20, pxdn, nas14, and cpb and transporters slc and aqp-9, as well as genes with low fold change (e.g., hspb1, kcnab2, txlnb, kmo; see Table S1 for full names). From these comparisons, it appeared that the RT-qPCR had a larger range than the array for the Atlantic lice (as per the slope of the relationship), whereas the opposite was true for the Pacific lice. The reason for this is not clear, but the correlation between methods was good in both subspecies. Additionally, to further test for evidence of the cellular stress response in Pacific lice, we evaluated the expression of heat-shock protein 90 using previously characterized primers (Sutherland et al. 2012). However, consistent with the microarray analysis, no differential expression was identified at 10, 25, or 50 ppb EMB (data not shown).
Several interesting candidates from previous work on EMB resistance in L. salmonis were evaluated by RT-qPCR in the Atlantic lice, including p-glycoprotein (Igboeli et al. 2013), as well as neuronal acetylcholine receptor and GABA-gated chloride channel (Carmichael et al. 2013). p-glycoprotein was not significantly affected by EMB dose (main effect or interaction; P > 0.1), but had higher expression in males than females (P < 0.0001), and higher expression in the resistant population in both sexes (main effect population P < 0.005; Fig.3D), but no significant sex by population interaction effect. The expression of GABA-gated chloride channel was dependent on the population, sex, and dose of EMB and slightly increased over the doses of EMB in most population/sex combinations (except in male resistant lice; Fig. S2A). Over the doses of EMB neuronal acetylcholine receptor did not change substantially and did not have a large baseline difference between populations (Fig. S2B).
Discussion
The regional differences in the sensitivity to EMB among populations of Atlantic L. salmonis (Igboeli et al. 2013) provide a system to investigate transcriptomic differences contributing to resistance mechanisms (e.g., polygenic resistance; ffrench-Constant et al. 2004). Polygenic resistance is likely in EMB-resistant L. salmonis (Espedal et al. 2013). The inclusion of a sensitive Pacific population in this study provides a first dataset of the induced transcriptomic response of Pacific lice to 10–50 ppb EMB, which can be indirectly compared to the Atlantic responses. However, differences between the exposures methods (e.g., pooling of multiple males and females in Pacific samples, and individual profiling in Atlantic samples) and the lice biology (e.g., presence of microsporidia infections in the Pacific lice) between the two analyses are important to consider while comparing these responses.
Lack of induced transcriptional responses from EMB
Atlantic lice baseline expression differences were much greater in number and fold change than induced responses, in agreement with the relative contributions of baseline and induced (200 ppb EMB) transcriptomic differences in Scottish EMB-resistant and EMB-sensitive male L. salmonis (Carmichael et al. 2013). Interestingly, Pacific lice did not upregulate any genes in response to EMB; however, a suite of genes were downregulated in only the highest Pacific EMB dose (50 ppb). Some induced responses in resistant lice may occur; upregulation of p-glycoprotein occurred in response to EMB in resistant populations of L. salmonis (Igboeli et al. 2013). However, EMB-induced fold change may be moderate; Chilean EMB-resistant Caligus rogercresseyi only had moderate differences in expression of detoxification (e.g., cytochrome P450s) or transport genes (multidrug resistance protein 1 (mrp1); Pgp1) as detected by semi-quantitative PCR (Cárcamo et al. 2011).
There was little evidence of an induced cellular stress response in the transcriptomic response to EMB in either subspecies (Atlantic or Pacific) even though many lice were exhibiting phenotypic signs of stress in bioassays at 300 and especially 1000 ppb. The lack of transcriptomic signatures of stress from the EMB exposure is a notable difference from the cellular stress response observed during salinity perturbations of copepodid L. salmonis (Sutherland et al. 2012). EMB-resistant Chilean C. rogercresseyi did not upregulate heat-shock protein from EMB (Cárcamo et al. 2011), and no stress response was identified in the spot prawn Pandalus platyceros exposed to EMB, although other biological functions were affected (e.g., transcription/translation control; Veldhoen et al. 2012). It is probable that other parasiticides with other mechanisms of action would induce different responses than those viewed here; for example, the antioxidant response of Caligus rogercresseyi was induced by the pyrethroid deltamethrin, which was attributed to increases in free radicals during the biodegradation of deltamethrin within the louse (Chavez-Mardones and Gallardo-Escárate 2014). Stress response genes were induced in response to the pyrethroid permethrin in pyrethroid-resistant A. gambiae (Vontas et al. 2005). Given the phenotypic variation that occurs even in end point detection of bioassays (Westcott et al. 2008) and in treatment response analysis (Jones et al. 2013), it will be important to continue to profile louse responses to EMB and other parasiticides, building on markers identified here, in previous work (Carmichael et al. 2013; Igboeli et al. 2013; Chavez-Mardones and Gallardo-Escárate 2014), or on markers associated with louse stress (Sutherland et al. 2012).
Degradative enzymes and other potential resistance candidates
A main finding of the present work was the overexpression of degradative enzymes in resistant relative to sensitive lice (in both sexes), as well as the highest overexpression of many of these genes specifically in the resistant males. The highest resistance in EC50 bioassays was viewed in resistant males in the current study and in previous work (Igboeli et al. 2013), and therefore, elevated expression of degradative enzymes is associated with the most resistant condition. Interestingly, at higher doses of EMB, many of these enzymes were downregulated in both Pacific and Atlantic salmon. The resistant population may therefore be less impacted by decreases in these genes following high doses of EMB exposure. In salmon lice, secretory degradative enzymes are likely important during feeding on optimal hosts (Fast et al. 2003) and possibly during other functions.
The association of degradative enzymes with resistant phenotypes has been viewed in other species. For example, elevated baseline expression of peptidases was identified in pyrethroid-resistant Anopheles gambiae (Vontas et al. 2005) and in both field- and laboratory-selected DDT-resistant Drosophila melanogaster (Pedra et al. 2004). Functional enzymatic studies have also indicated this result, for example, with increased proteolytic activity in an insecticide-resistant house fly Musca domestica (Ahmed et al. 1998). Both trypsin and chymotrypsin were overexpressed in a deltamethrin-resistant population of the mosquito Culex pipiens pallens, and cotransfection and stable expression of these two proteins in cell culture increased cell viability in response to deltamethrin relative to controls (Gong et al. 2005). It is not clear what the function of the degradative enzymes would be in the resistance phenotype. It has been proposed that increased peptidase activity may generate energy to alleviate costs of metabolic detoxification of drugs (Ahmed et al. 1998). However, energetic costs have yet to be identified in EMB-resistant L. salmonis populations (Espedal et al. 2013), and costs are not always associated with resistance. For example, EMB-resistant green lacewing Chrysoperla carnea were reported to have more rapid development, increased fecundity, and other indicators of increased fitness relative to the sensitive control (Mansoor et al. 2013). Therefore, the reason for increased degradative enzyme expression in resistant populations requires further investigation.
Genes responding to EMB exposure in both populations may also be involved in EMB protection; it is probable that the two Atlantic lice populations used in this study are more EMB resistant than a completely sensitive strain (Igboeli et al. 2013), such as the Pacific population profiled here. Preresistant Atlantic L. salmonis incurred 74–100% mortality from bioassay concentrations of 30–100 ppb EMB (Tribble et al. 2007), a higher level of sensitivity than that observed in the sensitive population used here (see Table4 and Igboeli et al. 2013). Among the few genes that increased as a result of EMB exposure, two of the most significant were alpha- and beta-taxilin, which are involved in calcium-dependent exocytosis of neuroendocrine cells (Nogami et al. 2003) and potentially in promoting motor nerve regeneration (Itoh et al. 2004). As EMB acts to disrupt neurotransmission through hyperpolarization, the induction of these transcripts is noteworthy. The downregulation by EMB of kynurenine 3-monooxygenase is of interest because inhibition of this protein leads to an increase in the concentration of kynurenic acid and a decrease in the concentration of glutamate (Zwilling et al. 2011), the ligand for the target site of EMB (Arena et al. 1995). However, the fold changes of these transcripts were moderate, so this possible effect would have to be explored further. The identification of these genes previously unexplored in relation to EMB resistance exemplifies the potential for transcriptomics in identifying unexpected genes that may be associated with resistance (Pedra et al. 2004; Vontas et al. 2005).
In Pacific lice, the only identified response to EMB was a downregulation of genes at 50 ppb (including an enrichment for peptidase functions). Atlantic lice also decreased degradative enzymes over the concentrations of EMB, although not to the same extent. Downregulation of degradative enzymes in Pacific lice may be due to the interruption of signals for continued production of these enzymes by EMB, for example, through disruption of calcium signaling. For example, experimental calcium (Ca2+) influx into a rat exocrine pancreatic cell line decreased chymotrypsin, amylase, and carboxypeptidase-a1 expression (but increased trypsin) (Stratowa and Rutter 1986). It is also possible that downregulation is related to EMB-induced molting (Waddy et al. 2002), as during molting suppression, it can occur for digestive enzymes trypsin and chymotrypsin (Vanwormhoudt et al. 1995; Klein et al. 1996; Sanchez-Paz et al. 2003). However, the presence of many other genes such as transporters in this list suggests this is not the case. Alternately, EMB may be acting as a antinutritional factor, which can reduce peptidase activity (e.g., in the cotton pest Heliothis zea; Klocke and Chan 1982), or may be reducing available energy stores leading to suppression of degradative enzymes similar to that which can occur during starvation (e.g., in white shrimp Penaeus vannamei; Muhlia-Almazan and Garcia-Carreno 2002). The suppression of a diverse range of transcripts further indicates interrupted signaling, but more work would need to be performed to confirm this.
Elevated p-glycoprotein expression in emamectin benzoate-resistant organisms has been identified in multiple species, including L. salmonis (Igboeli et al. 2013). Here, we also identified the highest baseline expression of p-glycoprotein in the most resistant condition (resistant males). However, here, we did not find evidence of pgp increasing over EMB doses. Our results also confirm the large influence from strain and low influence of EMB dose in Scottish resistant and sensitive L. salmonis (Carmichael et al. 2013). Additionally, in the Scottish populations, downregulation of degradative functions were also identified in response to EMB (e.g., hydrolase activity). In the present study, the highest fold change downregulation of degradative enzymes from EMB exposure occurred in the most sensitive population (Pacific L. salmonis), and in the Scottish L. salmonis, this was also identified in the sensitive strain (e.g., matrix metalloproteinase-9 and metalloproteinase; Carmichael et al. 2013). Here, we did not find the same downregulation trends for neuronal acetylcholine receptor and GABA-gated chloride channel expression in resistant populations using the primers from the original study. However, we did identify lower baseline expression of a probe annotated as gamma-aminobutyric acid receptor subunit beta-like in the resistant population than the sensitive in both sexes (largest fold change in females), albeit with lower fold change than many other genes with population differences in the study. These findings may indicate differences in resistance mechanisms between Atlantic Canada and Scottish L. salmonis populations.
The large number of unknown genes in L. salmonis produces a challenge for the interpretation of L. salmonis transcriptomics. A potential approach to improving L. salmonis transcriptome interpretation may be through further characterization of co-expressed gene clusters noted here and in previous studies (Eichner et al. 2008; Carmichael et al. 2013) and using the relation to clusters and responses to environmental variables for the annotation of unknowns (Pavey et al. 2012). Continued effort in this regard may improve the interpretation of existing and future salmon lice transcriptome analysis.
Relevance for aquaculture
The present work provides insights into the evolution of emamectin benzoate resistance in salmon lice with implications for aquaculture. Specifically, this study provides further support for the polygenic nature of emamectin benzoate resistance. This may occur incrementally among those lice exposed to a sublethal dose (ffrench-Constant et al. 2004). There are several possibilities how this may occur; for example, feeding differences may result in differential ingestion of the parasiticide by the hosts and different exposure levels to lice (Igboeli et al. 2014). The identification of markers of resistance development is an important contribution of this work. In addition to the candidates observed here, the evaluation of single nucleotide polymorphisms (SNPs) and other genomic changes associated with resistance will be an important next step.
The correlation of the resistance phenotype with elevated expression of degradative enzymes may have implications on the biology and pathology of lice. Currently, we understand that in lice, degradative enzymes are likely to function in feeding efficiency, immune evasion, and potentially in pathogenicity (Fast et al. 2003); however, it will be useful to continue to characterize the specific roles of these enzymes. The possibility that elevated expression of degradative enzymes in the resistant lice has an impact on the host–parasite interaction is an important avenue for future work.
Conclusions
Polygenic resistance mechanisms may provide EMB protection in L. salmonis, and the present study identifies some potential mechanisms, most notably the association between high expression of degradative enzymes and the resistant phenotype. Induced transcriptional responses to EMB were minor and had low fold changes in comparison with baseline differences between populations differing in EMB sensitivity. Sensitive Pacific lice responded only with downregulation of enzyme and transporter gene expression. Higher doses of EMB also resulted in downregulation of degradative enzymes in Atlantic lice. Neither subspecies responded to EMB exposure with a cellular stress response. Future work on single nucleotide polymorphism (SNP) differences among populations will continue to improve our understanding of EMB resistance in L. salmonis, in particular the potential role for target site mutation. The interpretation of L. salmonis transcriptome responses (in the present study and others) may be improved with further annotation and characterization of genes and co-expressed gene clusters through meta-analysis of existing and forthcoming transcriptome studies.
Acknowledgments
This work was funded by Genome British Columbia, the Natural Sciences and Engineering Research Council of Canada, the Province of British Columbia, the Department of Fisheries and Oceans Canada (DFO), the University of Victoria, Grieg Seafood, Mainstream Canada, Marine Harvest, Atlantic Canada Opportunities Agency (ACOA), Novartis Animal Health, and Innovation Prince Edward Island. BJGS was supported by an NSERC CGS fellowship. Thanks to S Jantzen and D Sanderson for analysis and sample preparation assistance, E Rondeau for amplicon sequencing, and E Kim for Pacific lice culturing and sample preparation. Thanks to the members of the Hoplite laboratory at Atlantic Veterinary College, Dr. M. Llewellyn, Prof. Louis Bernatchez, and two anonymous reviewers for comments on an earlier version of the manuscript.
Data archiving statement
Gene expression data has been uploaded to GEO under the accession GSE56024.
Supporting Information
Figure S1. RT-qPCR confirmation of microarray expression.
Figure S2. Exploratory RT-qPCR on previously identified targets.
Table S1. Primer table.
Table S2. Differentially expressed genes.
Table S3. Gene Ontology (GO) enrichment.
Literature Cited
- Abbas RZ, Zaman MA, Colwell DD, Gilleard J. Iqbal Z. Acaricide resistance in cattle ticks and approaches to its management: the state of play. Veterinary Parasitology. 2014;203:6–20. doi: 10.1016/j.vetpar.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Ahmed S, Wilkins RM. Mantle D. Comparison of proteolytic enzyme activities in adults of insecticide resistant and susceptible strains of the housefly M. domestica L. Insect Biochemistry and Molecular Biology. 1998;28:629–639. doi: 10.1016/s0965-1748(98)00061-7. [DOI] [PubMed] [Google Scholar]
- Arena JP, Liu KK, Paress PS, Frazier EG, Cully DF, Mrozik H. Schaeffer JM. The mechanism of action of avermectins in Caenorhabditis elegans – correlation between activation of glutamate-sensitive chloride current, membrane binding, and biological activity. Journal of Parasitology. 1995;81:286–294. [PubMed] [Google Scholar]
- Bonizzoni M, Afrane Y, Dunn WA, Atieli FK, Zhou GF, Zhong DB, Li J, et al. Comparative transcriptome analyses of deltamethrin-resistant and -susceptible Anopheles gambiae mosquitoes from Kenya by RNA-seq. PLoS ONE. 2012;7:e44607. doi: 10.1371/journal.pone.0044607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo S, Sevatdal S. Horsberg TE. Sensitivity assessment of Caligus rogercresseyi to emamectin benzoate in Chile. Aquaculture. 2008;282:7–12. [Google Scholar]
- Brooke BD. kdr: can a single mutation produce an entire insecticide resistance phenotype? Transactions of the Royal Society of Tropical Medicine and Hygiene. 2008;102:524–525. doi: 10.1016/j.trstmh.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Cárcamo JG, Aguilar MN, Barrientos CA, Carreño CF, Quezada CA, Bustos C, Manríquez RA, et al. Effect of emamectin benzoate on transcriptional expression of cytochromes P450 and the multidrug transporters (Pgp and MRP1) in rainbow trout (Oncorhynchus mykiss) and the sea lice Caligus rogercresseyi. Aquaculture. 2011;321:207–215. [Google Scholar]
- Carmichael SN, Bron JE, Taggart JB, Ireland JH, Bekaert M, Burgess STG, Skuce PJ, et al. Salmon lice (Lepeophtheirus salmonis) showing varying emamectin benzoate susceptibilities differ in neuronal acetylcholine receptor and GABA-gated chloride channel mRNA expression. BMC Genomics. 2013;14:408. doi: 10.1186/1471-2164-14-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Mardones J. Gallardo-Escárate C. Deltamethrin (AlphaMax™) reveals modulation of genes related to oxidative stress in the ectoparasite Caligus rogercresseyi: implications on delousing drug effectiveness. Aquaculture. 2014;433:421–429. [Google Scholar]
- Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nature Genetics. 2002;32:490–495. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- Clark JM, Scott JG, Campos F. Bloomquist JR. Resistance to avermectins – extent, mechanisms, and management implications. Annual Review of Entomology. 1995;40:1–30. doi: 10.1146/annurev.en.40.010195.000245. [DOI] [PubMed] [Google Scholar]
- Covello JM, Friend SE, Purcell SL, Burka JF, Markham RJF, Donkin AW, Groman DB, et al. Effects of orally administered immunostimulants on inflammatory gene expression and sea lice (Lepeophtheirus salmonis) burdens on Atlantic salmon (Salmo salar. Aquaculture. 2012;366:9–16. [Google Scholar]
- David JP, Strode C, Vontas J, Nikou D, Vaughan A, Pignatelli PM, Louis C, et al. The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4080–4084. doi: 10.1073/pnas.0409348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholm I, Devine GJ, Horsberg TE, Sevatdal S, Fallang A, Nolan DV. Powell R. Analysis and management of resistance to chemotherapeutants in salmon lice, Lepeophtheirus salmonis (Copepoda: Caligidae) Pest Management Science. 2002;58:528–536. doi: 10.1002/ps.482. [DOI] [PubMed] [Google Scholar]
- Donnelly MJ, Corbel V, Weetman D, Wilding CS, Williamson MS. Black WC. Does kdr genotype predict insecticide-resistance phenotype in mosquitoes? Trends in Parasitology. 2009;25:213–219. doi: 10.1016/j.pt.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Eichner C, Frost P, Dysvik B, Jonassen I, Kristiansen B. Nilsen F. Salmon louse (Lepeophtheirus salmonis) transcriptomes during post molting maturation and egg production, revealed using EST-sequencing and microarray analysis. BMC Genomics. 2008;9:126. doi: 10.1186/1471-2164-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espedal PG, Glover KA, Horsberg TE. Nilsen F. Emamectin benzoate resistance and fitness in laboratory reared salmon lice (Lepeophtheirus salmonis. Aquaculture. 2013;416:111–118. [Google Scholar]
- Fast MD. Fish immune responses to parasitic copepod (namely sea lice) infection. Developmental and Comparative Immunology. 2014;43:300–312. doi: 10.1016/j.dci.2013.08.019. [DOI] [PubMed] [Google Scholar]
- Fast MD, Burka JF, Johnson SC. Ross NW. Enzymes released from Lepeophtheirus salmonis in response to mucus from different salmonids. Journal of Parasitology. 2003;89:7–13. doi: 10.1645/0022-3395(2003)089[0007:ERFLSI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- ffrench-Constant RH, Daborn PJ. Le Goff G. The genetics and genomics of insecticide resistance. Trends in Genetics. 2004;20:163–170. doi: 10.1016/j.tig.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Frost P. Nilsen F. Validation of reference genes for transcription profiling in the salmon louse, Lepeophtheirus salmonis, by quantitative real-time PCR. Veterinary Parasitology. 2003;118:169–174. doi: 10.1016/j.vetpar.2003.09.020. [DOI] [PubMed] [Google Scholar]
- Gilleard JS. Understanding anthelmintic resistance: the need for genomics and genetics. International Journal for Parasitology. 2006;36:1227–1239. doi: 10.1016/j.ijpara.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Glendinning SK, Buckingham SD, Sattelle DB, Wonnacott S. Wolstenholme AJ. Glutamate-gated chloride channels of Haemonchus contortus restore drug sensitivity to ivermectin resistant Caenorhabditis elegans. PLoS ONE. 2011;6:e22390. doi: 10.1371/journal.pone.0022390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong MQ, Shen B, Gu Y, Tian HS, Ma L, Li XL, Yang MX, et al. Serine proteinase over-expression in relation to deltamethrin resistance in Culex pipiens pallens. Archives of Biochemistry and Biophysics. 2005;438:53–62. doi: 10.1016/j.abb.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Heumann J, Carmichael S, Bron JE, Tildesley A. Sturm A. Molecular cloning and characterisation of a novel P-glycoprotein in the salmon louse Lepeophtheirus salmonis. Comparative Biochemistry and Physiology. Toxicology & Pharmacology. 2012;155:198–205. doi: 10.1016/j.cbpc.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT. Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Igboeli OO, Fast MD, Heumann J. Burka JF. Role of P-glycoprotein in emamectin benzoate (SLICE®) resistance in sea lice, Lepeophtheirus salmonis. Aquaculture. 2012;344:40–47. [Google Scholar]
- Igboeli O, Burka JF. Fast MD. Sea lice population and sex differences in P-glycoprotein expression and emamectin benzoate resistance on salmon farms in the Bay of Fundy, New Brunswick, Canada. Pest Management Science. 2013;70:905–914. doi: 10.1002/ps.3620. [DOI] [PubMed] [Google Scholar]
- Igboeli O, Burka JF. Fast MD. Lepeophtheirus salmonis: a persisting challenge for salmon aquaculture. Animal Frontiers. 2014;4:22–32. [Google Scholar]
- Itoh S, Uyeda A, Hukuoka Y, Fujimori KE, Matsuda A, Ichinose S, Kobayashi H, et al. Muscle-specific protein MDP77 specifically promotes motor nerve regeneration in rats. Neuroscience Letters. 2004;360:175–177. doi: 10.1016/j.neulet.2004.02.058. [DOI] [PubMed] [Google Scholar]
- Jantzen SG, Sutherland BJG, Minkley DR. Koop BF. GO trimming: systematically reducing redundancy in large Gene Ontology datasets. BMC Research Notes. 2011;4:267. doi: 10.1186/1756-0500-4-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Treasurer JW, Bravo S, Nagasawa K. Kabata Z. A review of the impact of parasitic copepods on marine aquaculture. Zoological Studies. 2004;43:229–243. [Google Scholar]
- Jones PG, Hammell KL, Dohoo IR. Revie CW. Effectiveness of emamectin benzoate for treatment of Lepeophtheirus salmonis on farmed Atlantic salmon Salmo salar in the Bay of Fundy, Canada. Diseases of Aquatic Organisms. 2012a;102:53–64. doi: 10.3354/dao02517. [DOI] [PubMed] [Google Scholar]
- Jones SRM, Prosperi-Porta G. Kim E. The diversity of microsporidia in parasitic copepods (Caligidae: Siphonostomatoida) in the Northeast Pacific Ocean with description of Facilispora margolisi n. g., n. sp. and a new Family Facilisporidae n. fam. Journal of Eukaryotic Microbiology. 2012b;59:206–217. doi: 10.1111/j.1550-7408.2011.00614.x. [DOI] [PubMed] [Google Scholar]
- Jones PG, Hammell KL, Gettinby G. Revie CW. Detection of emamectin benzoate tolerance emergence in different life stages of sea lice, Lepeophtheirus salmonis, on farmed Atlantic salmon, Salmo salar L. Journal of Fish Diseases. 2013;36:209–220. doi: 10.1111/jfd.12022. [DOI] [PubMed] [Google Scholar]
- Kaplan RM. Drug resistance in nematodes of veterinary importance: a status report. Trends in Parasitology. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kaplan RM. Vidyashankar AN. An inconvenient truth: global worming and anthelmintic resistance. Veterinary Parasitology. 2012;186:70–78. doi: 10.1016/j.vetpar.2011.11.048. [DOI] [PubMed] [Google Scholar]
- Klein B, LeMoullac G, Sellos D. VanWormhoudt A. Molecular cloning and sequencing of trypsin cDNAs from Penaeus vannamei (Crustacea, Decapoda): use in assessing gene expression during the moult cycle. International Journal of Biochemistry & Cell Biology. 1996;28:551–563. doi: 10.1016/1357-2725(95)00169-7. [DOI] [PubMed] [Google Scholar]
- Klocke JA. Chan BG. Effects of cotton condensed tannin on feeding and digestion in the cotton pest, Heliothis zea. Journal of Insect Physiology. 1982;28:911–915. [Google Scholar]
- Laurell H, Iacovoni JS, Abot A, Svec D, Maoret J-J, Arnal J-F. Kubista M. Correction of RT-qPCR data for genomic DNA-derived signals with ValidPrime. Nucleic Acids Research. 2012;40:e51. doi: 10.1093/nar/gkr1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees F, Baillie M, Gettinby G. Revie CW. The efficacy of emamectin benzoate against infestations of Lepeophtheirus salmonis on farmed Atlantic salmon (Salmo salar L.) in Scotland, 2002–2006. PLoS ONE. 2008;3:e1549. doi: 10.1371/journal.pone.0001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoor MM, Abbas N, Shad SA, Pathan AK. Razaq M. Increased fitness and realized heritability in emamectin benzoate-resistant Chrysoperla carnea (Neuroptera: Chrysopidae) Ecotoxicology. 2013;22:1232–1240. doi: 10.1007/s10646-013-1111-8. [DOI] [PubMed] [Google Scholar]
- Muhlia-Almazan A. Garcia-Carreno FL. Influence of molting and starvation on the synthesis of proteolytic enzymes in the midgut gland of the white shrimp Penaeus vannamei. Comparative Biochemistry and Physiology. Part B, Biochemistry & Molecular Biology. 2002;133:383–394. doi: 10.1016/s1096-4959(02)00163-x. [DOI] [PubMed] [Google Scholar]
- Nogami S, Satoh S, Nakano M, Shimizu H, Fukushima H, Maruyama A, Terano A, et al. Taxilin; a novel syntaxin-binding protein that is involved in Ca2 + -dependent exocytosis in neuroendocrine cells. Genes to Cells. 2003;8:17–28. doi: 10.1046/j.1365-2443.2003.00612.x. [DOI] [PubMed] [Google Scholar]
- Pavey SA, Bernatchez L, Aubin-Horth N. Landry CR. What is needed for next-generation ecological and evolutionary genomics? Trends in Ecology & Evolution. 2012;27:673–678. doi: 10.1016/j.tree.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Pedra JHF, McIntyre LM, Scharf ME. Pittendrigh BR. Genome-wide transcription profile of field- and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7034–7039. doi: 10.1073/pnas.0400580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. Vienna, Austria: R: A Language and Environment for Statistical Computing; 2014. http://www.R-project.org/ (accessed on 10 December 2014) [Google Scholar]
- Ranson H, N'Guessan R, Lines J, Moiroux N, Nkuni Z. Corbel V. Pyrethroid resistance in African anopheline mosquitoes: what are the implications for malaria control? Trends in Parasitology. 2011;27:91–98. doi: 10.1016/j.pt.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Saksida SM, Morrison D, McKenzie P, Milligan B, Downey E, Boyce B. Eaves A. Use of Atlantic salmon, Salmo salar L., farm treatment data and bioassays to assess for resistance of sea lice, Lepeophtheirus salmonis, to emamectin benzoate (SLICE®) in British Columbia, Canada. Journal of Fish Diseases. 2013;36:515–520. doi: 10.1111/jfd.12018. [DOI] [PubMed] [Google Scholar]
- Sanchez-Paz A, Garcia-Carreno F, Muhlia-Almazan A, Hernandez-Saavedra NY. Yepiz-Plascencia G. Differential expression of trypsin mRNA in the white shrimp (Penaeus vannamei) midgut gland under starvation conditions. Journal of Experimental Marine Biology and Ecology. 2003;292:1–17. [Google Scholar]
- Sangster NC. Managing parasiticide resistance. Veterinary Parasitology. 2001;98:89–109. doi: 10.1016/s0304-4017(01)00425-3. [DOI] [PubMed] [Google Scholar]
- Sevatdal S, Magnusson A, Ingebrigtsen K, Haldorsen R. Horsberg TE. Distribution of emamectin benzoate in Atlantic salmon (Salmo salar L.) Journal of Veterinary Pharmacology and Therapeutics. 2005;28:101–107. doi: 10.1111/j.1365-2885.2004.00629.x. [DOI] [PubMed] [Google Scholar]
- Skern-Mauritzen R, Torrissen O. Glover KA. Pacific and Atlantic Lepeophtheirus salmonis (Kroyer, 1838) are allopatric subspecies: Lepeophtheirus salmonis salmonis and L. salmonis oncorhynchi subspecies novo. BMC Genetics. 2014;15:32. doi: 10.1186/1471-2156-15-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J, Sutherland IH, Sommerville CS, Richards RH. Varma KJ. The efficacy of emamectin benzoate as an oral treatment of sea lice, Lepeophtheirus salmonis (Kroyer), infestations in Atlantic salmon, Salmo salar L. Journal of Fish Diseases. 1999;22:261–270. [Google Scholar]
- Stratowa C. Rutter WJ. Selective regulation of trypsin gene expression by calcium and by glucose starvation in a rat exocrine pancreas cell line. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:4292–4296. doi: 10.1073/pnas.83.12.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland BJG, Jantzen SG, Sanderson DS, Koop BF. Jones SRM. Differentiating size-dependent responses of juvenile pink salmon (Oncorhynchus gorbuscha) to sea lice (Lepeophtheirus salmonis) infections. Comparative Biochemistry and Physiology. Part D, Genomics & Proteomics. 2011;6:213–223. doi: 10.1016/j.cbd.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Sutherland BJG, Jantzen SG, Yasuike M, Sanderson DS, Koop BF. Jones SRM. Transcriptomics of coping strategies in free-swimming Lepeophtheirus salmonis (Copepoda) larvae responding to abiotic stress. Molecular Ecology. 2012;21:6000–6014. doi: 10.1111/mec.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland BJG, Koczka KW, Yasuike M, Jantzen SG, Yazawa R, Koop BF. Jones SRM. Comparative transcriptomics of Atlantic Salmo salar, chum Oncorhynchus keta and pink salmon O. gorbuscha during infections with salmon lice Lepeophtheirus salmonis. BMC Genomics. 2014;200:15. doi: 10.1186/1471-2164-15-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribble ND, Burka JF. Kibenge FSB. Evidence for changes in the transcription levels of two putative P-glycoprotein genes in sea lice (Lepeophtheirus salmonis) in response to emamectin benzoate exposure. Molecular and Biochemical Parasitology. 2007;153:59–65. doi: 10.1016/j.molbiopara.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A. Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwormhoudt A, Sellos D, Donval A, Plairegoux S. Lemoullac G. Chymotrypsin gene expression during the intermolt cycle in the shrimp Penaeus vannamei (Crustacea – Decapoda) Experientia. 1995;51:159–163. [Google Scholar]
- Veldhoen N, Ikonomou MG, Buday C, Jordan J, Rehaume V, Cabecinha M, Dubetz C, et al. Biological effects of the anti-parasitic chemotherapeutant emamectin benzoate on a non-target crustacean, the spot prawn (Pandalus platyceros Brandt, 1851) under laboratory conditions. Aquatic Toxicology. 2012;108:94–105. doi: 10.1016/j.aquatox.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Vontas J, Blass C, Koutsos AC, David JP, Kafatos FC, Louis C, Hemingway J, et al. Gene expression in insecticide resistant and susceptible Anopheles gambiae strains constitutively or after insecticide exposure. Insect Molecular Biology. 2005;14:509–521. doi: 10.1111/j.1365-2583.2005.00582.x. [DOI] [PubMed] [Google Scholar]
- Waddy SL, Burridge LE, Hamilton MN, Mercer SM, Aiken DE. Haya K. Emamectin benzoate induces molting in American lobster, Homarus americanus. Canadian Journal of Fisheries and Aquatic Sciences. 2002;59:1096–1099. [Google Scholar]
- Westcott JD, Stryhn H, Burka JF. Hammell KL. Optimization and field use of a bioassay to monitor sea lice Lepeophtheirus salmonis sensitivity to emamectin benzoate. Diseases of Aquatic Organisms. 2008;79:119–131. doi: 10.3354/dao01887. [DOI] [PubMed] [Google Scholar]
- Whyte SK, Westcott JD, Elmoslemany A, Hammell KL. Revie CW. A fixed-dose approach to conducting emamectin benzoate tolerance assessments on field-collected sea lice, Lepeophtheirus salmonis. Journal of Fish Diseases. 2013;36:283–292. doi: 10.1111/jfd.12055. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J. Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuike M, Leong J, Jantzen SG, von Schalburg KR, Nilsen F, Jones SR. Koop BF. Genomic resources for sea lice: analysis of ESTs and mitochondrial genomes. Marine Biotechnology. 2012;14:155–166. doi: 10.1007/s10126-011-9398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa R, Yasuike M, Leong J, von Schalburg KR, Cooper GA, Beetz-Sargent M, Robb A, et al. EST and mitochondrial DNA sequences support a distinct pacific form of salmon louse, Lepeophtheirus salmonis. Marine Biotechnology. 2008;10:741–749. doi: 10.1007/s10126-008-9112-y. [DOI] [PubMed] [Google Scholar]
- Yoon KS, Previte DJ, Hodgdon HE, Poole BC, Kwon DH, El-Ghar GEA, Lee SH, et al. Knockdown resistance allele frequencies in North American head louse (Anoplura: Pediculidae) populations. Journal of Medical Entomology. 2014;51:450–457. doi: 10.1603/me13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JZ, Collins HL, Li YX, Mau RFL, Thompson GD, Hertlein M, Andaloro JT, et al. Monitoring of diamondback moth (Lepidoptera: Plutellidae) resistance to spinosad, indoxacarb, and emamectin benzoate. Journal of Economic Entomology. 2006;99:176–181. doi: 10.1603/0022-0493(2006)099[0176:MODMLP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Zwilling D, Huang SY, Sathyasaikumar KV, Notarangelo FM, Guidetti P, Wu HQ, Lee J, et al. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell. 2011;145:863–874. doi: 10.1016/j.cell.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. RT-qPCR confirmation of microarray expression.
Figure S2. Exploratory RT-qPCR on previously identified targets.
Table S1. Primer table.
Table S2. Differentially expressed genes.
Table S3. Gene Ontology (GO) enrichment.