Summary
In mammals, cytosine methylation (5mC) is widely distributed throughout the genome, but is notably depleted from active promoters and enhancers. While the role of DNA methylation in promoter silencing has been well documented, the function of this epigenetic mark at enhancers remains unclear. Recent experiments have demonstrated that enhancers are enriched for 5-hydroxymethylcytosine (5hmC), an oxidization product of the Tet family of 5mC dioxygenases and an intermediate of DNA demethylation. These results support the involvement of Tet proteins in regulation of dynamic DNA methylation at enhancers. By mapping DNA methylation and hydroxymethylation at base resolution, we find that deletion of Tet2 causes extensive loss of 5hmC at enhancers, accompanied by enhancer hypermethylation, reduction of enhancer activity, and delayed gene induction in the early steps of differentiation. Our results reveal that DNA demethylation modulates enhancer activity, and its disruption influences the timing of transcriptome reprogramming during cellular differentiation.
Introduction
Cytosine methylation is a well-established epigenetic mechanism essential for genomic imprinting, X chromosome inactivation, silencing of retrotransposons, and lineage-specific expression of developmental regulatory genes (Smith and Meissner, 2013). This epigenetic mark is extensively remodeled during mammalian development and in different tissue lineages (Hemberger et al., 2009; Reik et al., 2001). The establishment, maintenance, and erasure of 5mC depend on several DNA methyltransfeases (DNMTs) and the (Ten-Eleven-Translocation) TET family of protein dioxygenases (Fu and He, 2012; Pfeifer et al., 2013). TET proteins mediate oxidation of 5mC to 5hmC (Tahiliani et al., 2009), which is then further oxidized in a stepwise manner to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (He et al., 2011; Ito et al., 2011; Pfaffeneder et al., 2011). It is now thought that 5hmC, along with 5fC and 5caC, are intermediates of DNA demethylation (Pastor et al., 2013).
While loss of Tet or Dnmt proteins causes global changes in DNA methylation status in mouse embryonic stem cells (mESCs) (Dawlaty et al., 2013; Meissner et al., 2005), the cells nevertheless retain the ability to self-renew (Tsumura et al., 2006), suggesting that the mESC state is quite robust to alterations in DNA methylation. Still, Tet and Dnmt proteins play key roles in development (Dawlaty et al., 2014; Okano et al., 1999). Notably, loss of Dnmt activity causes abnormal mESC differentiation (Sakaue et al., 2010), and loss of Tet1 or Tet2 causes differentiation skewing (Ficz et al., 2011; Koh et al., 2011).
5mC and 5hmC are dynamically regulated both within and across cell types. Notably, 5mC is depleted at distal regulatory elements such as enhancers, where reduction of 5mC is correlated with the activity of these sequences (Hon et al., 2013; Lister et al., 2009; Stadler et al., 2011; Ziller et al., 2013). 5hmC is also significantly enriched at distal cis-regulatory sequences, suggesting that dynamic DNA methylation at these regions is likely mediated by interplays between DNMT-mediated methylation and TET-mediated demethylation processes (Stroud et al., 2011; Szulwach et al., 2011; Yu et al., 2012).
Despite these discoveries, several open questions remain. First, while all TET proteins can oxidize 5mC, how functionally redundant are they? Do different TETs target different regions of the genome, and if so, which ones target enhancers? Second, does enhancer oxidation play a gene regulatory role, perhaps by modulating enhancer activity? Third, previous findings indicate that perturbed oxidation causes skewed differentiation in embryonic stem cells. Does reduced oxidation affect differentiation through epigenetic dysregulation of enhancer activity?
Here, we examined the effects of loss of Tet1 and Tet2 genes on DNA methylation, chromatin modification, and gene expression. By generating base resolution DNA methylation and hydroxymethylation maps, we elucidate a role of Tet2 in enhancer oxidization. Loss of Tet2 leads to dramatic reduction of DNA hydroxymethylation genome-wide and elevated levels of DNA methylation at enhancers. These enhancers exhibit reduced activity, supporting an active role for oxidation at enhancers. Lastly, we provide evidence that disrupted enhancer oxidation during early differentiation causes delayed induction of differentiation genes. Together, our results clarify the functions of Tet1 and Tet2 in mammalian cells, highlight an active role of 5mC oxidation at enhancers, and reveal a role for enhancer DNA methylation in regulating the timing of transcriptome changes during differentiation.
Results
Generation of base resolution maps of 5mC and 5hmC in Tet1−/− andTet2−/− mESCs
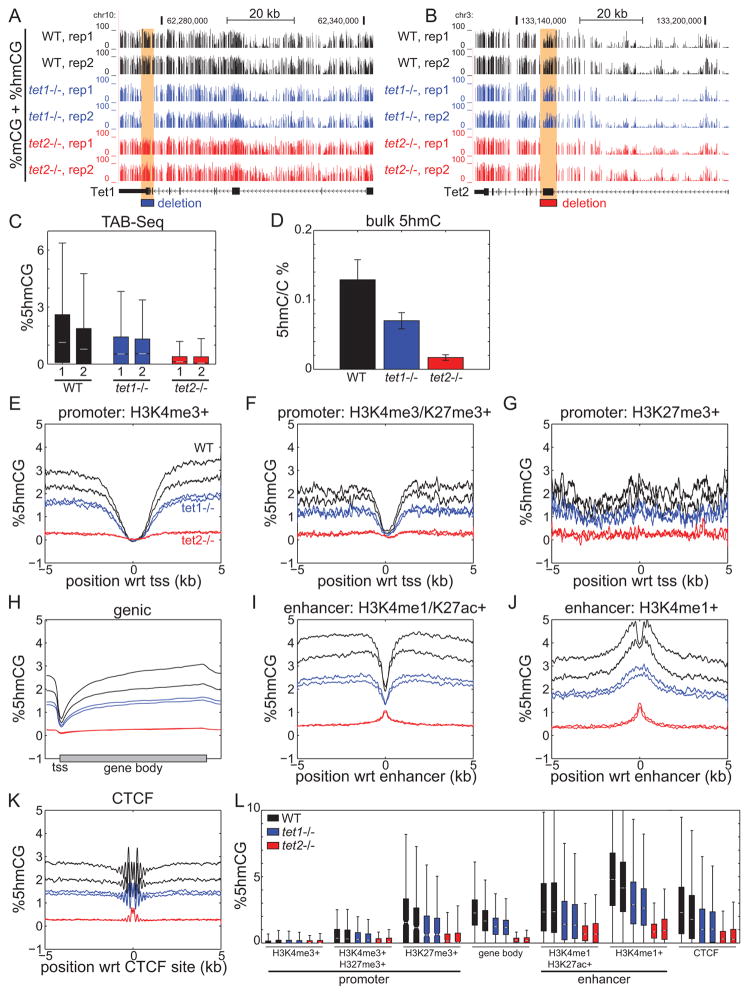
Tet1 and Tet2 can entirely account for 5hmC abundance in mouse ES cells (Dawlaty et al., 2013). To investigate the distinct roles of Tet1 and Tet2 in establishing 5mC and 5hmC patterns, we performed both whole genome bisulfite sequencing and TAB-seq (Lister et al., 2009; Yu et al., 2012) to generate base-resolution 5mC and 5hmC maps in wild-type (WT), Tet1−/−, and Tet2−/− mESCs (Figure 1A–B). Loss of Tet1 results in a 44.0% loss of global 5hmC compared to WT, while Tet2−/− mESCs exhibited more extensive loss of 5hmC (90.7%) (Figure 1C). Bulk quantification of 5hmC by mass spectrometry confirmed these observations (Figure 1D). Consistent with global quantification, we find that loss of Tet2 results in global depletion of 5hmC at promoters, gene bodies, CTCF-bound insulators, and enhancers (Figure 1E–L). In contrast, the pattern of 5hmC in Tet1−/− mESCs parallels that of WT cells, though at a lower abundance. Together, these results suggest that Tet2 is a major hydroxymethylase in mESCs. While these observations are supported by recent findings of Tet2 loss in mESCs and bone marrow (Huang et al., 2014; Li et al., 2011), others have observed less dramatic loss of 5hmC in mESCs (Dawlaty et al., 2013), perhaps highlighting biological variability of TET activity.
Figure 1. Global loss of 5hmC in Tet2−/− mouse ES cells.
(A–B) DNA methylation profiles of the (A) Tet1 and (B) Tet2 genes in Tet1−/− and Tet2−/− ESCs, illustrating the loci targeted for TET deletion. Two biological replicates of bisulfite sequencing are shown. (C) Boxplots of hydroxymethylation abundance for non-overlapping 10-kb bins spanning the mouse genome in WT, Tet1−/−, and Tet2−/− mESCs. (D) Bulk quantification of 5hmC in triplicates. Isolated genomic DNA was digested to single nucleosides and quantified using LC-MS/MS. Average profiles of absolute 5hmC abundance at (E) H3K4me3-only promoters, (F) bivalent H3K4me3/H3K27me3 promoters, (G) H3K27me3-only promoters, (H) gene bodies, (I) active H3K4me1/H3K27ac enhancers, (J) poised H3K4me1-only enhancers, and (K) CTCF-bound insulators. (L) Boxplot quantification of 5hmC abundance in E–J. For boxplots, notches indicate median, boxes extend to the 25th and 75th percentiles, and whiskers extend to non-outliers. In bar charts, error bars indicate standard deviation. wrt, with respect to.
Loss of 5mC oxidation and gain of 5mC at enhancers in Tet2−/− cells
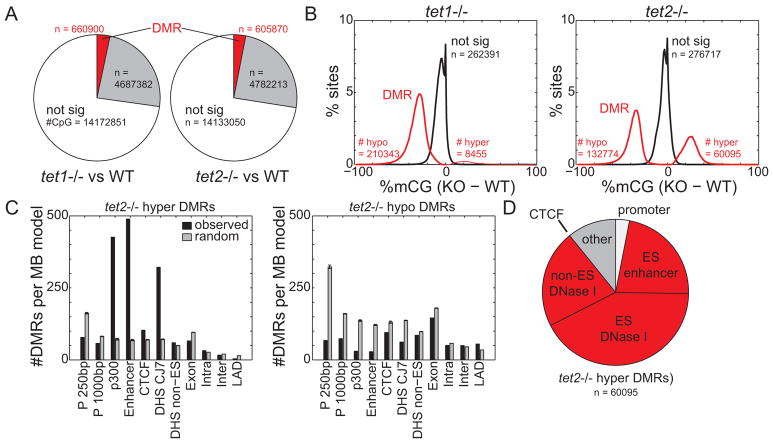
Next, we examined the effect of Tet1 and Tet2 deletion on DNA methylation levels genome-wide. We used a χ2-based statistic to capture cell-specific DNA methylation, and employed a hidden Markov model on this statistic to segment the genome into regions that have either no change or substantially altered levels of DNA methylation in the Tet1−/− and Tet2−/− mESCs (Figure S1A, Table S1). Only 4.4% and 3.8% of the Tet1−/− and Tet2−/− cell genomes, respectively, exhibited significant deviation of DNA methylation from WT cells (Figure 2A, S1A). These differentially methylated regions (DMRs) are short, with median length under 500bp (Figure S1C). In contrast to Tet1−/− cells where the vast majority of DMRs (96.1%) are hypomethylation events (Figure S1B), DMRs in Tet2−/− ESCs are split more evenly among hypermethylation (31.1%) and hypomethylation (68.9%) events. There are more than 7 times as many hypermethylated DMRs in Tet2−/− (60,095) than in Tet1−/− cells (8,455) (Figure 2B, S1B). Despite their abundance, hypomethylated DMRs overlap poorly with known and predicted regulatory elements in mESCs (Figure 2C).
Figure 2. Hypermethylation of enhancers in Tet2−/− cells.
Genome-wide DNA methylation profiles were segmented by a hidden Markov model to identify cell-specific DNA methylation. Shown are (A) the relative abundance of DMRs in Tet1−/− and Tet2−/− cells and (B) the distribution of 5mC change at DMRs of knockout compared to wild-type cells. (C) The relative enrichment of Tet2−/− hyper(left)/hypo(right) DMRs (black) and random sites (gray) at genomic elements, normalized to the total coverage of the element type. “P 250bp” indicates promoter defined as transcription start site +/− 250bp, DHS denotes DNase I hypersensitive sites, and LAD denotes lamina-associated domains (Peric-Hupkes et al., 2010). Random consists of 5 random samplings of genomic loci. (D) The relative abundance of Tet2−/− hyper-DMRs at regulatory elements. Red indicates distal regulatory elements. In bar charts, error bars indicate standard deviation. See also Figure S1 and Table S1.
Previous studies have shown that active enhancers in mESCs contain high levels of 5hmC, reflective of abundant oxidation of 5mC, and correspondingly low abundance of 5mC (Stadler et al., 2011). We therefore hypothesized that loss of Tet2 could lead to reduced oxidation and increased 5mC at enhancers. Indeed, hypermethylated DMRs (hyper-DMRs) in Tet2−/− cells exhibit several hallmarks of enhancers including: evolutionary sequence conservation (Siepel et al., 2005) (Figure S1E), enrichment for enhancer chromatin marks H3K4me1 and H3K27ac (Heintzman et al., 2009) (Figure S1F), and significant overlap with co-activator p300 binding sites (o/e = 5.9, p < 1E-200), DNase I hypersensitive sites (Consortium, 2011) (o/e = 4.5, p < 1E-200), and predicted enhancers (Shen et al., 2012) (o/e = 7.1, p < 1E-200) (Figure 2C). The vast majority (86.0%) of Tet2−/− hyper-DMRs overlap with previously identified enhancers and distal-regulatory elements (Figure 2D). While 70.0% of Tet1−/− hyper-DMRs are also distal regulatory elements (Figure S1D), there are 8.7 times as many in Tet2−/− cells. These results indicate that Tet2 contributes to enhancer hypomethylation by oxidizing 5mC.
Hypermethylation of enhancers with low TF occupancy
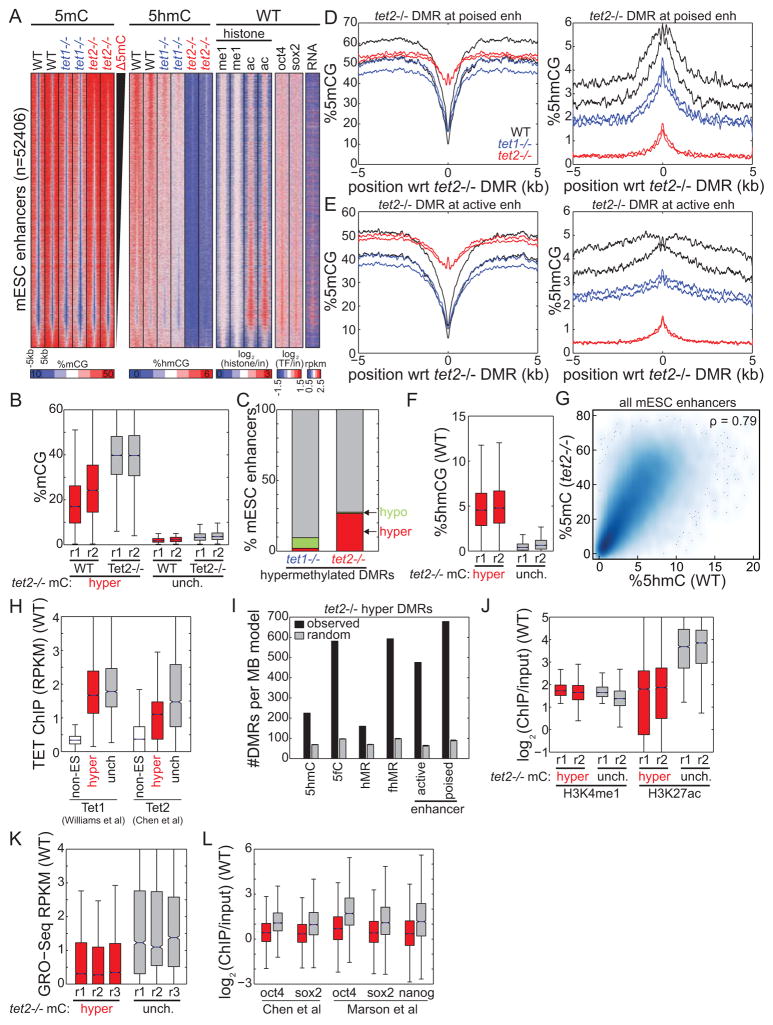
We next wondered which enhancers undergo hypermethylation in Tet2−/− mESCs, and whether this event is dependent on enhancer activity. Focusing on 52,406 enhancers identified based on chromatin modification patterns in mESCs (Figure 3A) (Shen et al., 2012), we found that 26.6% undergo significant hypermethylation in Tet2−/− mESCs, compared to only 2.0% for Tet1−/− cells (Figure 3B–C). Both active and poised enhancers (Creyghton et al., 2010; Hawkins et al., 2011; Rada-Iglesias et al., 2010) in Tet2−/− mESCs exhibit extensive hypermethylation to nearly background genomic levels, with almost complete loss of 5hmC (Figure 3D–E). Active enhancers are >4.5 times more methylated in Tet2−/− (40.6% mCG) than WT (9.0%) and Tet1−/− (10.5%) cells. Similarly, poised enhancers are more than 3.3 times more methylated in Tet2−/− mESCs (%mCGTet2−/− = 44.5% mCG, %mCGTet1−/− = 16.5%, %mCGWT = 13.4%). Expectedly, the enhancers exhibiting the greatest increase of DNA methylation in Tet2−/− cells correspond to those with the highest levels of 5hmC in WT cells (Figure 3F) (prep1 < 1E-15, prep2 < 1E-15), and there is a significant correlation between these two features (Figure 3G) (p < 1E-15). That hypermethylation in TET2 knockout mESC coincides with regions of 5hmC in WT implies that our observations are a direct rather than indirect consequence of Tet2 loss and its roles in 5mC oxidation and DNA de-methylation. Supporting this observation, upon examination of a previously published TET2 ChIP-seq dataset in mESC (Chen et al., 2013), we found that hypermethylated enhancers exhibit significantly more TET2 occupancy than non-ES cell enhancers in WT cells (Figure 3H) (p < 1E-15, Wilcoxon).
Figure 3. 5hmC-containing enhancers are hypermethylated in Tet2−/− cells.
(A) (left) Abundance of 5mC and 5hmC at mESC enhancers in WT, Tet1−/−, and Tet2−/− cells. (right) Enhancer chromatin state (me1: H3K4me1; ac: H3K27ac) and expression state (RNA: Global Run-On) in WT cells is also indicated. Enhancers are ranked by change in methylation state between Tet2−/− and WT cells. (B) For enhancers that are hypermethylated in Tet2−/− cells (red) and those that remain hypomethylated (gray), shown is the enrichment of 5mC in WT and Tet2−/− cells. (C) Quantification of mESC enhancers that are hypermethylated, hypomethylated, or unchanged in knockout cells compared to wild-type. (D–E) Profiles of average (left) 5mC and (right) 5hmC centered at Tet2−/− hyper-DMRs at (D) poised enhancers and (E) active enhancers. (F) For enhancers that are hypermethylated in Tet2−/− cells (red, left) and those that remain hypomethylated (gray, right), shown is the enrichment of 5hmC in wild-type cells. (G) Density plot illustrating the relationship between 5hmC abundance in WT cell and 5mC abundance in Tet2−/− cells. ρ indicates Spearman rank correlation. (H) Quantification of TET enrichment, using data previously mapped by ChIP-Seq (Chen et al., 2013; Williams et al., 2011), at enhancers active in non-ES cells (white), enhancers hypermethylated in Tet2−/− cells (red), and enhancers that remain hypomethylated (grey) in Tet2−/− cells. (I) The relative enrichment of Tet2−/− hyper-DMRs (black) and random sites (gray) at peaks of 5hmC and 5fC enrichment, at domains of 5fC/5hmC (fhMR) or 5hmC alone (hMR), and at active/poised enhancers. Error bars indicate standard deviation. (J–L) For enhancers that are hypermethylated in Tet2−/− cells (red, left) and those that remain hypomethylated (gray, right), shown is the enrichment of (J) active chromatin (H3K4me1 and H3K27ac), (K) nascent RNA transcription (by GRO-Seq) in wild-type cells, and (L) transcription factor binding (OCT4, SOX2, NANOG) (Chen et al., 2008; Marson et al., 2008). Boxplot edges indicate the 25th and 75th percentiles, and whiskers indicate non-outlier extremes. See also Figure S2.
TET proteins can further oxidize 5hmC to 5fC (He et al., 2011; Ito et al., 2011), and loci bearing both 5hmC and 5fC (fhMRs) are thought to mark regions of active de-methylation. Hyper-DMRs are significantly enriched at fhMRs (Figure 3I) (p < 1E-15, normal distribution). Furthermore, consistent with the observation that fhMRs correspond to poised enhancers (Song et al., 2013), we observe that poised enhancers are more likely to exhibit hypermethylation than active enhancers (Figure 3I). In agreement with these observations, Tet2−/−-hypermethylated enhancers exhibit weaker enrichment of histone acetylation in WT cells (Figure 3J). Furthermore, these enhancers show decreased nascent RNA transcription from previously published global run-on sequencing (Melgar et al., 2010) (Figure 3K). Consistent with these observations, hypermethylated enhancers also exhibit significantly weaker occupancy of the ES-cell core transcription factors (TFs) OCT4, SOX2, and NANOG than those that do not change DNA methylation status (Figure 3L) (Boyer et al., 2005). Taken together, these results suggest that weak enhancers with low TF occupancy undergoing active de-methylation by Tet2 in wild-type cells are preferentially hypermethylated to nearly background levels upon loss of Tet2.
Hypermethylation reduces enhancer activity
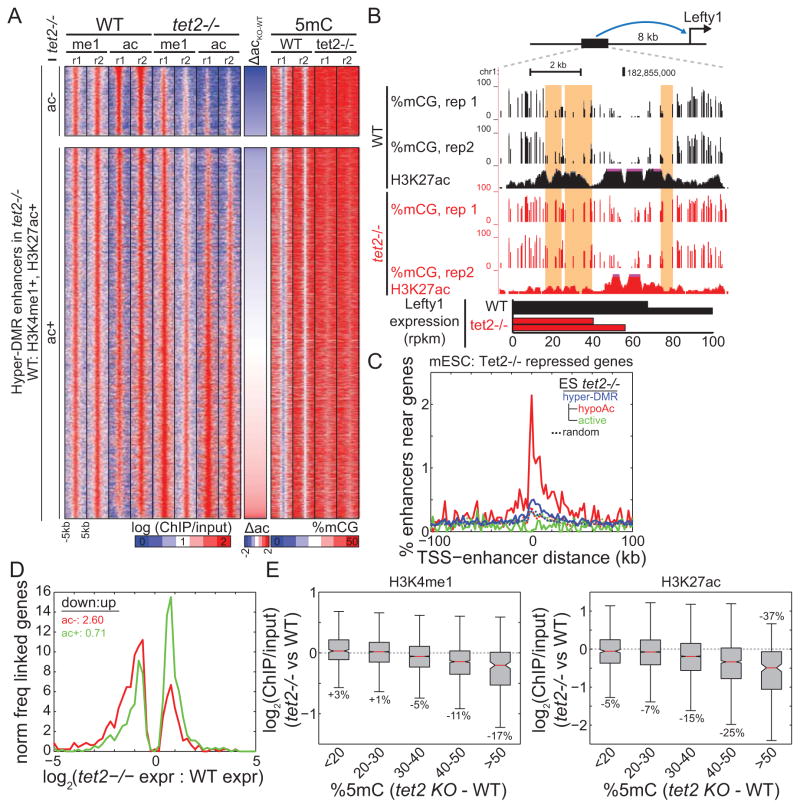
The hypermethylation of a subset of enhancers in Tet2−/− cells offers a unique opportunity to examine the roles of DNA methylation in modulating enhancer activity and gene expression. Since DNA methylation and active chromatin are antagonistic epigenetic marks (Hawkins et al., 2010; Okitsu and Hsieh, 2007), we wondered if increased enhancer DNA methylation may be accompanied by loss of active chromatin. Surprisingly, we find that global levels of both active and repressive chromatin marks at enhancers generally remain unchanged upon hypermethylation (Figure S3A), suggesting that the transcriptional effects of enhancer hypermethylation may be limited. Indeed, we only observe a small fraction (2,710; 15.7%) of Tet2−/− hyper-DMRs that lose H3K27ac compared to WT cells (hypo-Ac) (Figure 4A) (Table S2). For this subset of enhancers, we hypothesized that gain of DNA methylation and loss of active chromatin has a repressive effect on enhancer function. Indeed, an enhancer that physically interacts (Kagey et al., 2010) with the developmental gene Left-Right Determination Factor 1 (Lefty1) is hypermethylated and hypoacetylated in Tet2−/− cells, potentially explaining the significantly decreased expression of this gene (Figure 4B). Several transcription factors including lymphoid enhancer-binding factor 1 (Lef1) and Signal Transducer And Activator Of Transcription 5A (Stat5a) also exhibit hypermethylation/hypo-acetylation of nearby enhancers, which may contribute to the decreased expression of these genes (Figure S2).
Figure 4. Enhancer loss of acetylation and reduced gene expression.
(A) Partitioning of active enhancers hypermethylated in Tet2−/− ESCs into those that (top) lose H3K27ac and (bottom) retain H3K27ac. The middle column indicates difference in H3K27ac, and the right columns indicate DNA methylation abundance. (B) UCSC Genome Browser snapshots of Tet2−/− hyper-DMRs that lose H3K27ac (highlighted yellow) near an enhancer of the differentially expressed (bottom) Lefty1 gene. (C) The enrichment of different enhancer groups near genes differentially repressed in Tet2−/− cells. (D) The distribution of expression change for genes physically interacting with enhancers that (red) lose H3K27ac and (green) retain H3K27ac. The ratio of down-regulated to up-regulated genes is indicated. (E) Difference in enrichment of active chromatin (H3K4me1, left; H3K27ac, right) between Tet2−/− and WT cells, as a function of hypermethylation. Average percentage change in ChIP enrichment is indicated. Boxplot edges indicate the 25th and 75th percentiles, and whiskers indicate non-outlier extremes. See also Figure S3 and Table S2.
Extending our analysis genome-wide, we find that genes down-regulated in Tet2−/− cells are significantly enriched near enhancers with reduced histone acetylation (Figure 4C, S3B) (prand=all = 4.3E-82, prand=expr = 1.5E-17, normal). In contrast, up-regulated genes are significantly depleted (prand=all = 9.3E-4, prand=expr = 7.7E-4, normal). Furthermore, no significant change in gene expression was observed near enhancers with unchanged H3K27ac (prand=all = 0.03, prand=expr = 0.31), despite the presence of hypermethylation at these enhancers. If reduced gene expression is due to a loss of enhancer activity, we would expect repressed genes in Tet2−/− mESC to be physically linked to enhancers exhibiting reduced histone acetylation. To test this hypothesis, we employed a method to predict enhancer-promoter interactions (Jin et al., 2013) using published Hi-C datasets of WT mESCs (Dixon et al., 2012; Selvaraj et al., 2013). We find that enhancers with reduced histone acetylation are more likely to be physically linked to repressed genes than expected by chance (p = 9.3703e-14, binomial), and more frequently than hypermethylated enhancers with no loss of H3K27ac (p = 1.7166e-21, Wilcoxon) (Figure 4D). Further supporting these observations, we find that the hypo-Ac enhancers identified in this system are significantly enriched near genes down-regulated in Tet2 knockdown mESCs (p = 0.0073, normal) (Koh et al., 2011) and in independently derived Tet2−/− mESCs (p = 0.0093, normal) (Dawlaty et al., 2013) (Figure S3C–D). Together, these results implicate the altered methylation state of enhancers as an effector of enhancer activity and the expression of target genes.
Why does hypermethylation of only a subset enhancers correlate with reduced activity? One possible explanation is that at a cell population level, the degree of hypermethylation determines changes of enhancer activity. Consistent with this idea, we find that hypermethylation of most enhancers, which gain on average 30.5% 5mC in Tet2−/− cells, does not contribute to dramatic alterations in enhancer activity as reflected through chromatin structure or the expression of nearby genes (Figure 4C–E). However, when 5mC levels increase beyond a threshold of ~40% mCG, we observe a significant loss of active chromatin modifications, particularly of histone acetylation (Figure 4E). Indeed, enhancers that have ≥50% more methylation in Tet2−/− compared to WT cells also exhibit on average a 37.2% decrease in H3K27ac abundance. However, enhancers with small changes in methylation only exhibit minor loss of H3K27ac. A possible explanation for this observation is that small changes in methylation may reflect only a subset of cells having increased methylation. Since ChIP-Seq is enrichment-based, it is less sensitive to changes in a small population of cells, unlike bisulfite sequencing which uniformly samples the entire population. Thus, while we detect reduced activity at only a subset of hypermethylated enhancers, we cannot exclude the possibility that a larger subset also exhibit reduced activity in the subset of affected cells.
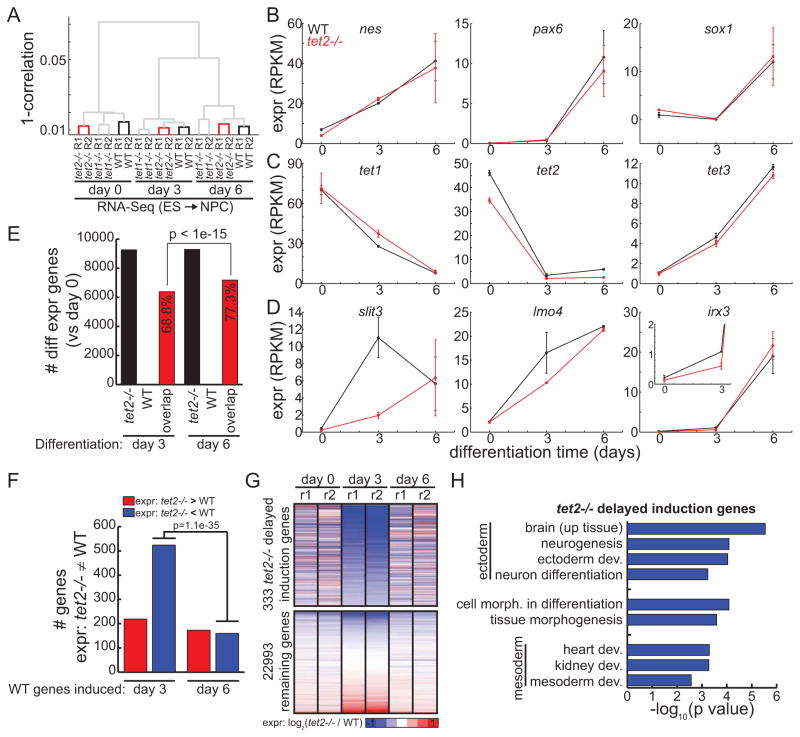
Delayed gene induction in Tet2−/− cells during differentiation
Previous studies have observed skewed differentiation of mESCs deficient for TET1 or TET2 (Ficz et al., 2011; Koh et al., 2011). As enhancers are involved in the early steps of cell fate commitment during ES cell differentiation (Hawkins et al., 2011), we wondered if the altered epigenetic status of enhancers in Tet2−/− cells affects gene expression during differentiation. We performed RNA-seq to measure expression profiles in a time-course of mESC differentiation to neural progenitor cells (NPCs) for 3 and 6 days. Consistent with previous reports, cluster analyses of gene expression profiles in WT and mutant cells showed that NPC differentiation is not globally disrupted by the loss of either protein (Figure 5A). Indeed, markers of neuronal cell fate including nes, pax6, and sox1 are consistently induced in WT and Tet2−/− cells (Figure 5B). Both genotypes exhibited consistent alterations in Tet expression, with increased expression of Tet3 contrasting decreases of Tet1/Tet2 expression (Figure 5C). Interestingly, we also observed discordant expression of genes normally induced in WT cells, notably at day 3 (Figure 5D). These results suggest that expression between WT and Tet2−/− cells is more discordant in early, rather than late, steps of differentiation. Supporting this observation, only 68.6% of genes are commonly differentially expressed in WT and Tet2−/− cells at day 3, noticeably less than the 77.3% observed at day 6 (p < 1e-15) (Figure 5E). To verify that delayed induction is not unique to our system, we also differentiated a previously reported Tet2 knockdown mESC line (Huang et al., 2014) and performed quantitative real-time PCR analysis. Consistent with our observations in TET2−/− mESCs, we also find delayed induction of the three marker genes slit3, lmo4, and irx3 (Figure S4A–B). Locus-specific bisulfite sequencing also indicates that Tet2 knockdown cells exhibit enhancer hypermethylation of DMRs identified in Tet2−/− cells (Figure S4C–E).
Figure 5. Delayed gene induction of Tet2−/− cells during NPC differentiation.
(A) Dendrogram summarizing RNA-Seq experiments during differentiation of mES cells to NPCs. Red indicates Tet2−/− branches; bold black indicates WT branches. (B–C) Expression of (B) neuronal markers and (C) Tet genes during NPC differentiation. (D) Examples of genes exhibiting delayed gene induction in Tet2−/− specifically at differentiation day 3. Error bars indicate standard deviation. (E) The total number of differentially expressed genes in Tet2−/− (black) and WT (white) cells, as compared to undifferentiated ES cells. Shown in red are those genes commonly differentially expressed in these two cells. (F) Of the genes induced in WT cells at day 3 (left) or day 6 (right) during differentiation towards NPCs, shown are the number of genes repressed (blue) or induced (red) in Tet2−/− cells compared to WT. (G) Expression of genes in Tet2−/− relative to WT differentiated cells for delayed induction genes (top) and all other genes (bottom). Genes in (F) that exhibited differential expression in d0 or d6 were removed. (H) Ontology terms enriched for delayed induction genes. See also Figure S4.
To further investigate genes discordantly regulated in Tet2−/− cells during early NPC differentiation, we identified genes induced in WT cells and examined their expression in Tet2−/− cells. Of the genes differentially expressed between WT and Tet2−/− cells at day 3, 70.5% were induced at a smaller degree in Tet2−/− cells than WT cells (p < 1e-15, binomial). This is significantly greater than the 48.0% observed at day 6 (p = 1.1e-35), suggesting that Tet2−/− cells exhibit delayed gene induction during early, but not late, differentiation. Next, we identified 333 genes discordantly regulated in Tet2−/− cells during early NPC differentiation as those that: 1) are induced in WT cells at day 3 compared to day 0, 2) are repressed in Tet2−/− cells at day 3 compared to WT cells, and 3) are not repressed in Tet2−/− cells at days 0 and 6 compared to WT cells (Figure 5G). Functional enrichment analysis of delayed induction genes revealed significant enrichment for neural development and tissue morphogenesis (Figure 5H).
Differential enhancer methylation contributes to delayed gene induction
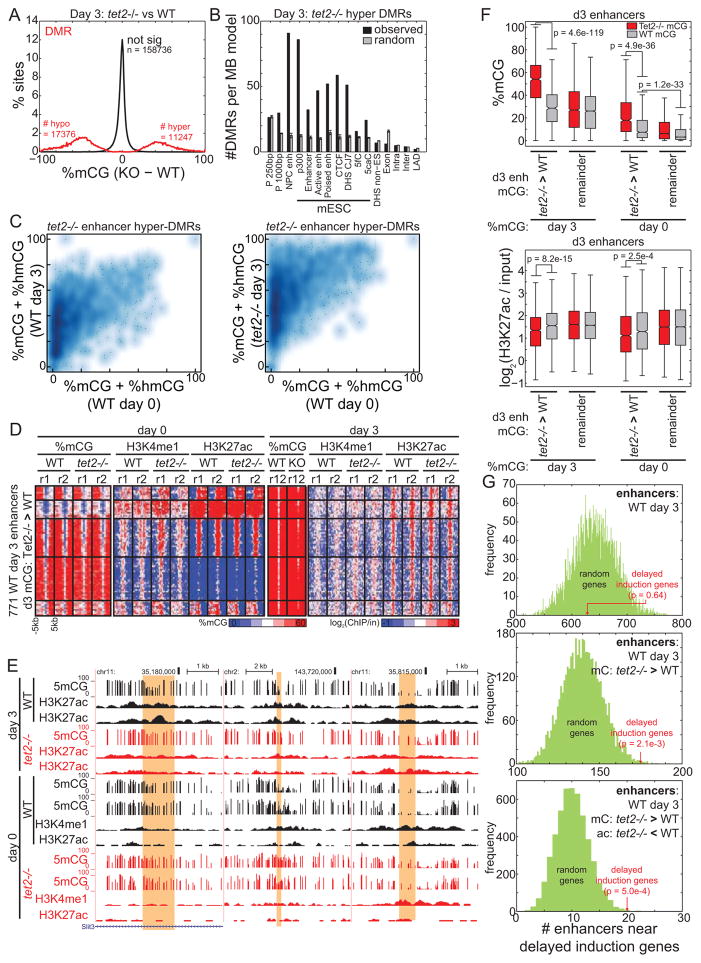
One possible explanation of this delayed gene induction is an altered epigenetic state of enhancers in Tet2−/− cells. To test this possibility, we performed bisulfite sequencing to map DNA methylation at day 3 of differentiation in WT and Tet2−/− cells. Compared to WT cells, the number of Tet2−/− hyper-DMRs at this stage of differentiation (denoted “D3 hyper-DMRs”) had dramatically decreased by 81.3% to only 11,246 regions (Figure 6A). Notably, only 2,192 of these regions (19.5%) were DMRs at day 0, suggesting that Tet2 deficiency has additional effects on the epigenome during differentiation.
Figure 6. Differential enhancer methylation contributes to delayed gene induction.
(A) Distribution of change in 5mC for DMRs identified in Tet2−/− cells compared to WT cells 3 days after mESC differentiation towards NPCs. (B) Relative enrichment of Tet2−/− hyper DMRs from (A) (black) and random sites (gray) at genomic elements, normalized to the total coverage of the element type. Error bars indicate standard deviation. (C) Density heatmap representing the methylation state of hypermethylated enhancers before (x axis) and after (y axis) differentiation in WT (left) and Tet2−/− (right) cells. (D) Heatmap representing the epigenetic state of d3 Tet2−/− hypermethylated enhancers. (E) Genome browser snapshots of DNA methylation and chromatin state at d3 Tet2−/− hypermethylated enhancers (yellow). (F) Boxplots quantifying mCG (top) and quantile-normalized H3K27ac (bottom) at enhancers before (right) and after (left) differentiation. Day 3 Tet2−/− specific hypermethylated enhancers are labeled as “Tet2−/− > WT”, and other active enhancers as “remainder”. Boxplot edges indicate the 25th and 75th percentiles, and whiskers indicate non-outlier extremes. (G) The number of enhancers within TADs containing delayed induction genes is indicated in red, compared to the distribution of 5000 random gene sets. Enhancers are defined as: (top) the set of all WT day 3 active enhancers, (middle) the subset that is hypermethylated in Tet2−/− cells at day 3, and (bottom) the subset with WT specific H3K27ac at day 3.
D3 hyper-DMRs remain significantly enriched at distal regulatory elements, including enhancers (Figure 6B). While many of these enhancers are normally hypermethylated in WT cells during differentiation, they exhibit a greater degree of hypermethylation in Tet2−/− cells (Figure 6C–E). D3 hyper-DMRs are almost twice as methylated in Tet2−/− (median, 54.1%) than WT cells (median, 28.6%) (p = 4.6e-119, Wilcoxon) (Figure 6F). To assess if an epigenetic status predisposes these enhancers to be hypermethylated during differentiation, we examined methylation in day 0 mESCs. Interestingly, these enhancers are weakly hypermethylated in Tet2−/− compared to WT mESCs. Furthermore, in WT mESCs this group of enhancers is significantly hypermethylated compared to other enhancers (p = 1.2e-33, Wilcoxon) (Figure 6F). These observations suggest that D3 hyper-DMRs are pre-disposed to hypermethylation.
Next, we asked if enhancer hypermethylation during differentiation affects enhancer activity. Using histone acetylation as a marker for enhancer activity, we find that enhancers hypermethylated in day 3 exhibit significantly reduced levels of H3K27ac (p = 8.2e-15, Wilcoxon) (Figure 6F). If hypermethylation decreases enhancer activity, one expectation is that target genes will exhibit reduced expression. To test this possibility, we examined enhancer enrichment near genes exhibiting delayed induction in Tet2−/− mESC. Considering that the mammalian genome is partitioned into developmentally stable, mega-base-sized topological domains (TADs) (Dixon et al., 2012), we asked whether hypermethylated enhancers are specifically enriched in the TADs containing genes with delayed induction. While the control set of all active NPC enhancers is not enriched in these TADs (p = 0.64, normal) (Figure 6G), the subset of hypermethylated enhancers is significantly enriched (p = 2.1e-3, normal), as well as subset of hypermethylated enhancers that loses H3K27ac (p = 5.0e-4, normal). Together, these observations provide additional evidence that Tet2 modulates enhancer methylation status and activity during differentiation, and that its loss contributes to delayed induction of differentiation genes and therefore a disrupted timing of early transcriptome reprogramming during differentiation.
Discussion
While DNA hypomethylation is a universal feature of active enhancers (Stadler et al., 2011), the requirement of hypomethylation on enhancer activity is inadequately understood. By examining the epigenome in the context of Tet deletion, we have identified Tet2 as an important mediator of 5mC oxidation and hypomethylation at enhancers. We document a cascade of epigenetic events at enhancers initiating from loss of 5mC oxidation, to gain of methylation, loss of active chromatin modification, and culminating in reduced enhancer activity. This observation has several implications. The first is that DNA methylation changes can be upstream of chromatin changes (or mutually affect each other) in this system. The second is that changes in chromatin structure are sufficient to perturb enhancer activity, in agreement with previous studies (Lee et al., 2013). The third is that Tet2 is a positive regulator of enhancer activity.
Since both Tet1 and Tet2 can catalyze the oxidation of 5mC, their functional distinctions are either through unique temporal activities or genomic targets. Indeed, consistent with our observations, a recent study found that knockdown of Tet1 causes loss of promoter oxidation, while depletion of Tet2 results in loss of 5hmC at some enhancers (Huang et al., 2014). The biased activities of TET proteins could be accomplished by context-dependent genomic binding that may be attributed to distinct domain structures. For example, TET1 contains a CpG-island targeting CXXC domain (Pastor et al., 2011), while the lack of this domain in TET2 potentially opens it to a wider set of genomic loci including enhancers, perhaps through DNA-targeting partners (Ko et al., 2013). Nevertheless, while loss of Tet2 causes extensive epigenetic effects at enhancers, our observations that 1) enhancer 5hmC decreases marginally in Tet1−/− cells, 2) Tet1 is enriched at enhancers, and 3) hyper-DMRs in Tet1−/− mESCs are enhancer-rich suggest that Tet1 plays at least some role in the oxidation of some enhancers.
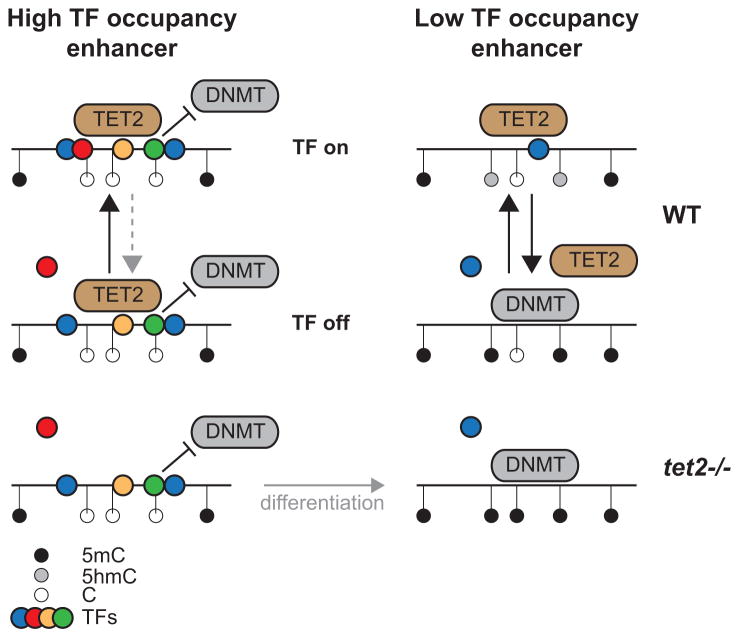
Why are weaker enhancers preferentially targeted for hypermethylation upon Tet2 loss? One possibility relates to a model whereby TF occupancy inhibits de novo DNA methylation (Gebhard et al., 2010), and unmethylated loci not bound by TFs are passively methylated (Figure 7) (Thurman et al., 2012). In contrast, we observe that Tet2 occupancy increases with increasing TF occupancy (Figure 3H). Thus, while TF occupancy is antagonistic to methylation by DNMTs, it may be permissive for Tet2 activity. At one extreme, weak enhancers with lower TF occupancy are subject to increased TF turnover. TF-off states promote passive filling in of DNA methylation, while TF-on states promote Tet2-mediated oxidation of 5mC to 5hmC to balance passive DNA methylation. This equilibrium is broken upon loss of Tet2, in favor of a fully methylated state. In contrast, strong enhancers contain a higher density of TFs. The lower probability of complete TF-off states potentially results in a fully de-methylated state in wild-type cells due to lack of passive DNA methylation, which remains unaltered upon loss of Tet2. Over time, this model would suggest a passive methylation of low occupancy enhancers, while high occupancy enhancers remain unmethylated and active. Similarly, as promoters are more DNase I hypersensitive and therefore more occupied by TFs than enhancers (Boyle et al., 2008), dynamic methylation and de-methylation occurs more frequently at enhancers than promoters. Thus, this model suggests that Tet2 fine-tunes enhancer activity.
Figure 7. Model of enhancer hypermethylation in Tet2−/− cells.
Transcription factors bind to DNA and therefore occlude hypermethylation by DNMTs. Enhancers with high TF occupancy (left) are more resistant to DNMTs, and therefore remain hypomethylated in Tet2−/− cells. However, enhancers with low TF occupancy (right) are prone to hypermethylation, which is balanced by the action of Tet2. Loss of Tet2 causes re-methylation.
The lack of gross phenotypic differences in Tet-deficient mESCs is in agreement with previous studies showing the viability of mESCs lacking DNA methylation (Jackson et al., 2004). Furthermore, the ability of Tet1/2-deficient mESCs to differentiate (Ficz et al., 2011; Koh et al., 2011) and contribute to live animals (Dawlaty et al., 2013) suggests that Tet1 and Tet2 are not required for development or viability. However, we observe that Tet2−/− cells are more transcriptionally distinct from wild-type cells in early rather than late differentiation. This result suggests that enhancer oxidation may play a role in the timing of early regulatory events during lineage commitment to NPCs. Our data links loss of 5mC oxidation to increased methylation of NPC enhancers, which contributes to their decreased activity. However, as all Tet proteins share the same enzymatic activity and may compensate for each other, rapid induction of Tet3 by day 6 of differentiation may offset the early regulatory consequences of losing Tet2. The mechanism for how a subset of enhancers is targeted, how changes in chromatin structure occur, and how these results apply to other systems such as hematopoietic malignancies driven by loss of Tet2 will be subjects of future research.
Experimental Procedures
Cell lines
Tet1−/− ES cell lines were derived from the knockout mouse described previously (Zhang et al., 2013). The derivation of Tet2−/− ESCs is described elsewhere (Hu et al., 2013, submitted). These knockout ESCS have been shown to be pluripotent by chimera formation assay.
Mouse ESC Culture and Differentiation
Mouse ESCs were cultured in feeder-free gelatin-coated plates in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen Cat. No. 11995) supplemented with 15% FBS (GIBCO), 2 mM L-glutamine (GIBCO), 0.1 mM 2-mercaptoethanol (Sigma), 1 x nonessential amino acids (GIBCO), 1,000 units/ml LIF (Millipore Cat. No. ESG1107), 1 x pen/strep (GIBCO), 3 mM CHIR99021 (Stemgent), and 1 mM PD0325901 (Stemgent). Differentiation of ESC towards NPC was performed out as previously described (Bibel et al., 2007). Briefly, differentiation was induced by growing cells in suspension in non-adhesive bacterial dishes in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 0.1 mM 2-mercaptoethanol, 1 x nonessential amino acids, and 1 x pen/strep. After four days, 5 μM of retinoic acid was added in the medium. Medium was changed every two days. Cells are harvested after three days and six days.
methylC-Seq, TAB-Seq, ChIP-Seq, and RNA-Seq
MethylC-Seq and TAB-Seq was performed as previously described (Lister et al., 2009; Yu et al., 2012). Briefly, extracted DNA (DNeasy Kit, Qiagen) was spiked with control DNA (Promega) at 0.5%. For methylC-Seq, the control consisted of unmethylated lambda DNA. For TAB-Seq, two different controls here used: 1) lambda DNA with distinct PCR-amplified regions containing either 5mC, C, or 5hmC; or 2) M.sssI-methylated lambda DNA with 5hmC-amplified pUC19 DNA. Spiked DNA was sonicated (Bioruptor, Diagenode), end-repaired, adenylated, and ligated to Illumina TruSeq sequencing adapters. After 2% agarose gel purification to select fragments of size 200–650 bp, samples were subjected to bisulfite conversion (MethylCode, Invitrogen) and PCR amplification with PfuTurbo Cx Hotstart DNA Polymerase (Agilent). After gel purification, libraries were sequenced on an Illumina Hi-Seq 2000. Reads were mapped to the computationally bisulfite-converted mouse genome (mm9) using bowtie (Langmead et al., 2009) and PCR duplicates were removed with Picard. Only basecalls with Phred score ≥ 20 were considered for analysis. See Supplemental Experimental Procedures.
Chromatin immunoprecipitation was performed out as previously described (Hawkins et al., 2010) with 20μg chromatin and 5μg antibody. The following antibodies were used H3K4me1 (Abcam, Ab8895), H3K4me3 (Active Motif 39159), H3K9me3 (Abcam Ab8898), H3K27ac (Active Motif 39133), H3K27me3 (Active Motif 39155), and H3K36me3 (Abcam Ab9050). Library preparation and sequencing procedures were carried out as described previously (Hawkins et al., 2010) according to Illumina protocols with minor modifications (Illumina, San Diego, CA). Reads were mapped with bowtie (Langmead et al., 2009) (parameters: -v 3-m 1--best strata) and PCR duplicates were removed.
Poly-A tail selected, strand-specific mRNA sequencing was performed as previously described (Parkhomchuk et al., 2009). Reads were mapped with tophat (Trapnell et al., 2009) (parameters:-g 1--library-type=fr-firststrand). Differential gene expression of exonic sense reads was assessed by the edgeR tool (Robinson et al., 2010).
External datasets
Previously published ChIP-Seq data for CTCF and p300 (Shen et al., 2012), as well as DNase I hypersensitivity data (Consortium, 2011), were either previously published or acquired from the mouse ENCODE Project. PhastCons (Siepel et al., 2005) conservation scores from alignments of 29 vertebrate genomes with mouse, as well as definitions of repetitive elements, were acquired from the UCSC Genome Browser. Topological domains in mouse ES cells were previously defined (Dixon et al., 2012).
Enhancers
Wild-type mouse ES cell enhancers were defined as promoter-distal (>5000bp from transcription start sites) DNase I hypersensitive sites ES cells (Consortium, 2011) exhibiting enrichment of both H3K4me1 and H3K27ac (active enhancers) or only H3K4me1 but not H3K27ac. Enrichment was defined as log2(ChIP/input) ≥ 1 and lack of enrichment aslog2(ChIP/input) ≤ 0. To define putative enhancers in day 3 differentiation cells, we merged promoter-distal H3K27ac peaks from replicate experiments, as identified from the MACS peak-finder (Zhang et al., 2008).
Identifying cell type-specific DNA methylation
DMRs were defined using a hidden Markov model approach as described previously (Hon et al., 2013). DMRs with fewer than 10 base-calls spanning CpGs, or with less than 20% absolute methylation difference between knockout and wild-type, were removed. The final set of DMRs are those with p-value <= 0.05 (χ2 distribution, 1-tailed).
Assessing abundance of epigenetic modifications
The abundance of DNA methylation was measured as %mCG, the percentage of methylated cytosines in CpG context. The abundance of ChIP-Seq reads in a given genomic region was measured as a log-ratio of ChIP RPKM to input RPKM, each with a pseudocount of 0.05. We estimated 5mC and 5hmC from methylC-Seq and TAB-Seq experiments. See Supplemental Experimental Procedures.
Supplementary Material
Figure S1, related to Figure 2: Characterization of DMRs.
(A) UCSC Genome Browser snapshots of DNA methylation at Tet2−/− hyper-DMRs. (B) Genomic coverage of DMRs in Tet2−/− and Tet2−/− cells. (C) Length distribution of DMRs. Boxplot edges indicate the 25th and 75th percentiles, and whiskers indicate non-outlier extremes. (D) Relative enrichment of Tet1−/− hyper-DMRs at various genomic features. Error bars indicate standard deviation. (E) Average PhastCons conservation score centered at Tet2−/− DMRs. (F) Enrichment of histone modifications centered at Tet2−/− DMRs.
Figure S2, related to Figure 3: Snapshots of hypermethylated enhancers.
UCSC Genome Browser snapshots illustrating the enrichment of DNA methylation and histone modifications at WT enhancers that lose H3K27ac in Tet2−/− cells. The expression of genes near these enhancers is indicated in the bar chart below.
Figure S3, related to Figure 4: Enhancer enrichment at differentially expressed genes.
(A) For enhancers bearing (left) poised and (right) active chromatin signatures in WT cells that overlap with Tet2−/− hyper-DMRs, shown is the enrichment of various histone modifications. Boxplot edges indicate the 25th and 75th percentiles, and whiskers indicate non-outlier extremes. (B) (left) Schematic illustrating the assignment of differentially expressed genes to enhancers by using local chromatin interactions from Hi-C experiments (Dixon et al., 2012). (right) Enrichment of domains containing Tet2−/− differentially expressed genes with enhancers that lose or retain H3K27ac. Comparisons are made to randomly selected genes (light gray) and randomly selected genes expressed in ESCs (dark gray). Error bars indicate standard deviation. (C–D) Enrichment of hypermethylated hypo-Ac enhancers identified in this study with genes repressed in (C) Tet2 knockdown mESCs (Koh et al., 2011) and (D) independently derived Tet2 knock-out mESCs (Dawlaty et al., 2013).
Figure S4, relatedp to Figure 5: Comparison to Tet2 knockdown cells.
(A) Wild-type and Tet2 knockdown mESCs (Huang et al., 2014) were differentiated towards NPCs, and the expression of three delayed induction genes were monitored by qRT-PCR at day 0 and day 3 after differentiation. Error bars indicate standard deviation. (B) Shown is the relative induction ratio (day 3 / day 0) for delayed induction genes Irx3, Lmo4, and Slit3 in wild-type and Tet2 knockdown cells. (C–E) Locus-specific bisulfite sequencing for DMRs near (C) Irx3, (D) Lmo4, and (E) Slit3. Shown are data for wild-type (left), Tet2 knockdown (middle), and Tet2 knockout cells (right) at DMRs identified in Tet2−/− cells before (top) and after (middle/bottom) differentiation. White circles indicate unmethylated cytosine and black circles indicate methylated cytosine for individual clones. P-values indicated are from applying Fisher’s Exact Test.
Table S1, related to Figure 2: DMRs identified in Tet1−/− and Tet2−/− compared to wild-type cells.
Table S2, related to Figure 4: Enhancers hypermethylated and hypo-acetylated in Tet2−/− compared to wild-type cells.
Highlights.
Base resolution maps of 5hmC and 5mC in WT, Tet1−/− and Tet2−/− mESCs.
Reduced 5hmC and increased 5mC at enhancers in Tet2−/− mESCs.
Hypermethylated enhancers exhibit reduced activity.
Hypermethylation and delayed gene induction during Tet2−/− mESC differentiation.
Acknowledgments
We would like to thank Anjana Rao for donating Tet2 knockdown cells for this study. The research is supported by the Ludwig Institute for Cancer Research and funds from NIH to B.R. and C.H (R01 HG006827). C. He is an investigator of Howard Hughes Medical Institute. The TAB-seq technology has been licensed to Wisegene LLC by the University of Chicago Technology Transfer Office.
Footnotes
Accession Numbers
The data generated in this study has been deposited to the Gene Expression Omnibus, accession GSE48519.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bibel M, Richter J, Lacroix E, Barde YA. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Consortium E. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F, et al. Loss of tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Fu Y, He C. Nucleic acid modifications with epigenetic significance. Curr Opin Chem Biol. 2012;16:516–524. doi: 10.1016/j.cbpa.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard C, Benner C, Ehrich M, Schwarzfischer L, Schilling E, Klug M, Dietmaier W, Thiede C, Holler E, Andreesen R, et al. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res. 2010;70:1398–1407. doi: 10.1158/0008-5472.CAN-09-3406. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Yang C, Antosiewicz-Bourget JE, Lee LK, Ngo QM, Klugman S, Ching KA, Edsall LE, Ye Z, et al. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 2011;21:1393–1409. doi: 10.1038/cr.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013 doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martinez JA, Pape UJ, Jacobsen SE, Peters B, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon J, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza C, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013 doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, Lahdesmaki H, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melgar MF, Collins FS, Sethupathy P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biol. 2010;12:R113. doi: 10.1186/gb-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007;27:2746–2757. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin. 2013;6:10. doi: 10.1186/1756-8935-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue M, Ohta H, Kumaki Y, Oda M, Sakaide Y, Matsuoka C, Yamagiwa A, Niwa H, Wakayama T, Okano M. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr Biol. 2010;20:1452–1457. doi: 10.1016/j.cub.2010.06.050. [DOI] [PubMed] [Google Scholar]
- Selvaraj S, dixon J, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol. 2013 doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, et al. Genome-wide Profiling of 5-Formylcytosine Reveals Its Roles in Epigenetic Priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, Wirbelauer C, Oakeley E, Gaidatzis D, Tiwari V, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, Ye P, Rosa L, Lee YK, Wu HP, et al. Tet1 Regulates Adult Hippocampal Neurogenesis and Cognition. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 2: Characterization of DMRs.
(A) UCSC Genome Browser snapshots of DNA methylation at Tet2−/− hyper-DMRs. (B) Genomic coverage of DMRs in Tet2−/− and Tet2−/− cells. (C) Length distribution of DMRs. Boxplot edges indicate the 25th and 75th percentiles, and whiskers indicate non-outlier extremes. (D) Relative enrichment of Tet1−/− hyper-DMRs at various genomic features. Error bars indicate standard deviation. (E) Average PhastCons conservation score centered at Tet2−/− DMRs. (F) Enrichment of histone modifications centered at Tet2−/− DMRs.
Figure S2, related to Figure 3: Snapshots of hypermethylated enhancers.
UCSC Genome Browser snapshots illustrating the enrichment of DNA methylation and histone modifications at WT enhancers that lose H3K27ac in Tet2−/− cells. The expression of genes near these enhancers is indicated in the bar chart below.
Figure S3, related to Figure 4: Enhancer enrichment at differentially expressed genes.
(A) For enhancers bearing (left) poised and (right) active chromatin signatures in WT cells that overlap with Tet2−/− hyper-DMRs, shown is the enrichment of various histone modifications. Boxplot edges indicate the 25th and 75th percentiles, and whiskers indicate non-outlier extremes. (B) (left) Schematic illustrating the assignment of differentially expressed genes to enhancers by using local chromatin interactions from Hi-C experiments (Dixon et al., 2012). (right) Enrichment of domains containing Tet2−/− differentially expressed genes with enhancers that lose or retain H3K27ac. Comparisons are made to randomly selected genes (light gray) and randomly selected genes expressed in ESCs (dark gray). Error bars indicate standard deviation. (C–D) Enrichment of hypermethylated hypo-Ac enhancers identified in this study with genes repressed in (C) Tet2 knockdown mESCs (Koh et al., 2011) and (D) independently derived Tet2 knock-out mESCs (Dawlaty et al., 2013).
Figure S4, relatedp to Figure 5: Comparison to Tet2 knockdown cells.
(A) Wild-type and Tet2 knockdown mESCs (Huang et al., 2014) were differentiated towards NPCs, and the expression of three delayed induction genes were monitored by qRT-PCR at day 0 and day 3 after differentiation. Error bars indicate standard deviation. (B) Shown is the relative induction ratio (day 3 / day 0) for delayed induction genes Irx3, Lmo4, and Slit3 in wild-type and Tet2 knockdown cells. (C–E) Locus-specific bisulfite sequencing for DMRs near (C) Irx3, (D) Lmo4, and (E) Slit3. Shown are data for wild-type (left), Tet2 knockdown (middle), and Tet2 knockout cells (right) at DMRs identified in Tet2−/− cells before (top) and after (middle/bottom) differentiation. White circles indicate unmethylated cytosine and black circles indicate methylated cytosine for individual clones. P-values indicated are from applying Fisher’s Exact Test.
Table S1, related to Figure 2: DMRs identified in Tet1−/− and Tet2−/− compared to wild-type cells.
Table S2, related to Figure 4: Enhancers hypermethylated and hypo-acetylated in Tet2−/− compared to wild-type cells.