Abstract
Anderson Disease (ANDD) or Chylomicron Retention Disease (CMRD) is a rare, hereditary lipid malabsorption syndrome associated with mutations in the SAR1B gene that is characterized by failure to thrive and hypocholesterolemia. Although the SAR1B structure has been resolved and its role in formation of coat protein II (COPII) coated carriers is well established, little is known about the requirement for SAR1B during embryogenesis. To address this question, we have developed a zebrafish model of Sar1b deficiency based on antisense oligonucleotide knockdown. We show that zebrafish sar1b is highly conserved among vertebrates, broadly expressed during development, and enriched in the digestive tract organs, brain and craniofacial skeleton. Consistent with ANDD symptoms of chylomicron retention, we found that dietary lipids in Sar1b deficient embryos accumulate in enterocytes. Transgenic expression analysis revealed that Sar1b is required for growth of exocrine pancreas and liver. Furthermore, we found abnormal differentiation and maturation of craniofacial cartilage associated with defects in procollagen II secretion, and absence of select, neuroD-positive neurons of the midbrain and hindbrain. The model presented here will help to systematically dissect developmental roles of Sar1b and to discover molecular and cellular mechanisms leading to organ-specific ANDD pathology.
Keywords: SAR1B, chylomicrons, lipid absorption, COPII, Anderson Disease, Chylomicron Retention Disease
INTRODUCTION
Role of SAR1B in chylomicron and lipid transport
Dietary lipids are absorbed primarily by enterocytes, where they are processed in the endoplasmic reticulum (ER) to triacylglycerols (TAG) [1]. Newly synthesized TAGs are packaged into prechylomicrons and exit ER to the Golgi complex for further processing [2]. ER-to-Golgi protein transport is mediated primarily by the COPII complex, consisting of five core proteins: Sar1 GTPase and Sec23–Sec24 of the inner coat, and Sec13–Sec31 of the outer coat [3]. Subsequently, prechylomicron transport vesicles fuse to Golgi membranes [4]. Lipoprotein cargo is further transported to numerous target tissues, including liver, muscle, adipose tissue and brain [5]. Although protein secretion has been extensively studied in vitro, lipoprotein cargo secretion mechanisms are largely unknown primarily because of lack of animal models to address these complex questions [6].
Anderson Disease
Chylomicron Retention Disease, also known as Anderson Disease (CMRD/ANDD, OMIM 246700), is a rare autosomal recessive disorder characterized by failure to absorb dietary lipids and lipid soluble vitamins, shown to be caused by mutations in the SAR1B gene [7, 8]. Patients are mostly diagnosed early in life based on collective symptoms of steatorrhea, failure to thrive, and hypocholesterolemia with normal TAG, along with small intestine biopsy and histological analysis showing lipid droplets in the cytoplasm [7, 9]. However, some patients are diagnosed later in life [10] and go undetected until their children are diagnosed with ANDD. Recent case reports besides the gut and liver involvement have implicated the heart, skeletal muscle, bone, adipose tissue and pancreas, although the full spectrum of affected organs is unknown [9–11]. The combination of multiple organ involvement, age of onset/diagnosis and lack of phenotype-genotype relationship in patients complicates diagnosis and treatment of the disease, which can be effectively managed by prompt implementation of low-fat diets and supplements of lipid-soluble vitamins [9].
The pleiotropic nature of the syndrome and lack of published mutations affecting the Sar1b gene in vertebrate animal models prompted us to establish a zebrafish model using morpholino-based global knockdown strategies. Zebrafish has been previously used for studying lipid metabolism, benefiting from accessibility of the gastrointestinal organs to live imaging [12–16].
The purpose of this study is to develop a vertebrate animal model system that could be used to (1) characterize the pathophysiology of Sar1b deficiency, (2) study the mechanisms of lipid malabsorption in Sar1 deficient animals, and (3) establish an in vivo model for future study of human SAR1B variants. We show that Sar1b deficiency leads to developmental deficits in multiple organs including gut, pancreas and liver of the gastrointestinal (GI) tract, but also skeletal dysmorphology due to failure of cartilage differentiation and maturation, and CNS nuclei specification. Our findings stress the possibility that ANDD patients’ failure to thrive might not only stem from diarrhea and malnutrition but also multi-organ developmental deficits.
MATERIALS AND METHODS
Gene nomenclature
Gene names in the text are according to nomenclature guidelines, with human genes in capital italic (e.g. SAR1B) and zebrafish in lowercase italic (sar1b). Protein names are in roman font in the sentence case.
Zebrafish lines
AB strain zebrafish were raised and maintained under standard laboratory conditions at 28.5°C [17]. For whole mount in situ hybridization experiments, embryos were raised in 0.2 mM 1-phenyl-2-thiourea (Sigma) to block pigmentation.
The transgenic fish Tg:[(fabp10a:dsRed;ela3A:GFP)gz15] line was generated by the Gong laboratory [18] and shared with us by the Leach laboratory. The transgenic fish Tg:[(P0-pax6b:GFP)ulg515] (abbreviated pax6b:GFP) was generated by the Martial laboratory [19] and shared with us by the Chen laboratory. For live imaging WT and sar1b-MO larvae were anesthetized in Tricaine (Sigma) and mounted in low-melt agarose. All confocal imaging in this manuscript was taken with a Zeiss LSM510 inverted confocal microscope (Vanderbilt Cell Imaging Shared Resource), while all other imaging was taken with a Zeiss AxioImager Z1. All experiments were conducted in accordance with the guidelines established by the IACUC at Vanderbilt University.
Cloning and sequencing
Zebrafish sar1b cDNA (NCBI Reference Sequence: NM_001024377.1) was cloned using the primers 5′-GGCGGATCCTGAGAGCGGAGTTTGTCCAC-3′ and 5′-GCCTCTAGATGTGTTTAGTCGATGTACTGA-3′ (MWG Operon). The cDNA without the morpholino targeting site was subcloned from this plasmid for use with the rescue experiments. A 110 bp region of the sar1b 5′UTR and coding region was cloned using the primers 5′-CCCCTCGAGTTCTCCGGTGTTTCCTCATTG-3′ and 5′-CCCCCGCGGACTATAAAT CCAATCAAATATG-3′. Zebrafish sar1a cDNA (NM_001017882) was cloned using the primers 5′-GCAGTGTTTCGCCTGCTTAC-3′ AND 5′-GCTTACCTGTCACTAAACTGG-3′. Human SAR1B cDNA (NM_001033503) was cloned using the primers 5′-GGATATGTCCTTCATATTTGATTG-3′ and 5′-TGTGTTAATCAATGTACTGTGC-3′. Zebrafish sar1b and human SAR1B are located in syntenic regions of chromosome 21 and 5, respectively.
Cartilage proteoglycan staining
Alcian Blue staining was performed as previously described [20]. 5 dpf larvae were fixed in 4% PFA, bleached in H2O2/KOH solution, stained overnight in 0.1% Alcian Blue in 70% EtOH/1% HCL, and then de-stained in acidic ethanol (70% ethanol, 5% HCl).
Whole mount in situ hybridization
Whole-mount in situ hybridization with probes recognizing col2a1 and sox9a [21, 22], neuroD, dlx2, foxa3, trypsin, pax6b, nkx2.2, prox1, and ceruloplasmin (http://zfin.org/) [23] were performed as previously described [24, 25]. The sar1b probe was made by cloning 862 nucleotides from the 3′ UTR of the sar1b cDNA into the pGEM-T Easy Vector with the primers 5′-TTGACAAACCCGAGGCCATC-3′ and 5′-CATTCCACCGCTGCTCCATG-3′.
Immunofluorescence and lectin staining
Type II collagen and wheat germ agglutinin (WGA) staining was performed as previously described [20] using 1:200 diluted primary antibody against collagen type II (Polysciences) and 1:200 WGA–Alexa-Fluor-555 conjugate (Molecular Probes), followed by 1:300 Alexa Fluor 488-conjugated secondary antibody (Molecular Probes) and TO-PRO-3 (Molecular Probes). For acetylated tubulin staining, 5 dpf larvae were fixed overnight in Prefer Fixative (Anatech), rinsed, post-fixed for 10 minutes in 4% PFA, and then permeabilized in 0.5% triton X-100 for 10 minutes. Larvae were then bleached in H2O2/KOH solution for 20 minutes and then washed in blocking solution. 1:250 primary antibody against acetylated tubulin (Sigma) [26] was applied overnight at 4°C followed by 1:300 Alexa Fluor 488-conjugated secondary antibody (Molecular Probes) for 3 hours at RT. After washing, larvae were post-fixed in 4% PFA for 20 minutes and then cleared in glycerol overnight. Images are presented as maximum intensity z-projections.
Morpholino Oligonucleotides
Antisense morpholino oligonucleotides (MOs) (Gene Tools, Corvallis, OR) were designed to target the sar1b 5′UTR sequence (sar1b-MO): 5′-CAGTCCAACGGGGATCAATGAGGAA-3′. The sar1b-MO targets bp 41–65 of the sar1b 5′UTR, which is 45 bp upstream of the ATG site. This target site is unique to sar1b and is not present in sar1a. 1 nl was injected into 1–2 cell stage embryos at increasing doses (0.4 ng–3 ng) to determine optimal concentrations. To confirm morpholino-targeting efficiency, 60 pg of sar1b-eGFP mRNA (see Fig. 1J, K) was co-injected with 0.4 ng and 1 ng of sar1b-MO, and loss of eGFP expression was analyzed. All data presented in figures are at 0.4 ng, determined to be the optimal concentration. Control morpholino provided by manufacturer did not generate morphological changes.
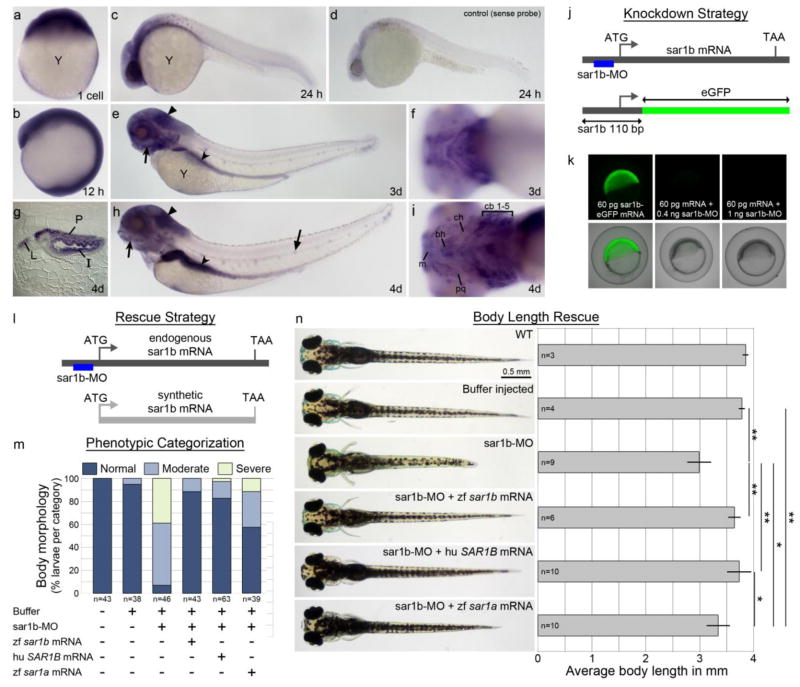
Figure 1. Sar1b is expressed in select neural tissues, pharyngeal arches and gut.
Sar1b mRNA expression by whole mount in situ hybridization at 1cell stage (a), 12 hpf (b), 24 hpf (c), 3 dpf (e, f) and 4 dpf (g–i). Embryos are oriented with head towards the left in lateral views (c, d, e, h), ventral views (f, i) and sagittal section of the mid-gut region at 4 dpf (g). At 24 hpf, expression is concentrated within the ventral brain region (c) and not detected by the sense probe (d). Pigment cells appear as dark spots in C and D. By 3 dpf, transcripts are present throughout the brain, eye, lateral line organ (flat arrow) and enriched in pharyngeal arches (concave arrow) gut (concave arrowhead), and cerebellar plate (flat arrowhead) (e). By 4 dpf the expression persists in the same locations (h), including intestinal epithelium, pancreas, and liver (g), as well as the pharyngeal arches (i). Targeting efficiency of sar1b-MO (J–K). (j) Schematic representation of the knockdown strategy showing sar1b mRNA and position of the translation blocking morpholino (sar1b-MO). The sar1b 5′UTR-eGFP fusion protein mRNA construct contains the sar1b-MO binding site. (k) Double injected embryos with fusion construct and increasing doses of sar1b-MO show almost complete knockdown of GFP expression at 0.4 ng (middle panel) and complete absence of GFP expression at 1 ng of sar1b-MO (right panel). Targeting specificity of sar1b-MO (L–O). (l) Schematic representation of the rescue strategy showing sar1b mRNA and synthetic sar1b mRNA, which lacks the sar1b-MO binding site and which is used for rescue experiments. (m) Double-blinded phenotypic categorization of overall morphology at 4 dpf according to body length, head size, and pectoral fin shape. 54.3% of sar1b-MO injected embryos exhibit a moderate phenotype (representative example in panel N), while 88.4% of embryos co-injected with sar1b-MO and sar1b mRNA are morphologically normal at 4 dpf. Human (hu) SAR1B mRNA rescues morphogenesis similarly to zebrafish (zf) sar1b, while zf sar1a mRNA partially compensates for sar1b knockdown. (n) Images of live embryos with body length quantification for rescue experiments. Sar1b morphants exhibit reduced body length while sar1b-MO + zf sar1b mRNA or hu SAR1B mRNA co-injected embryos do not have significant reduction of body length at 4 dpf. zf sar1a partially rescues body length in sar1b-MO injected embryos. Error bars are ± SD. Abbreviations: y, yolk, m, Meckel’s cartilage, bh, basihyal, pq, palatoquadrate, ch, ceratohyal, cb, ceratobranchials 1–5, I, intestine, L, liver, P, pancreas; zf, zebrafish; hu, human. * p < 0.05, ** p < 0.01; One-way ANOVA standard weighted means analysis with Tukey’s test.
Dietary lipid clearance assay
The lipid clearance assay was modified from Schlegel and Stainier [12]. Live 5 dpf larvae were incubated with 10% chicken egg yolk (CEY) suspension in 0.3x Danieau buffer for 6 hours at 28.5°C and then rinsed in several washes in fresh egg water and then either processed immediately for lipid staining or fasted 16 hours and then processed. To ensure uniform micelle formation before feeding, CEY suspension was vortexed for 5 minutes, force pipetted, and then examined with a 100x objective before feeding. After feeding, larvae were processed using detergent-free conditions. They were fixed in 4% PFA, rinsed in PBS, bleached in H2O2/KOH solution, washed in 60% 2-propanol, and stained in 0.3% Oil Red O (Sigma) in 60% 2-propanol for 3 hours at room temperature, followed by washing in water. For analyzing lipids in enterocytes, larvae were fed as described above, fixed in 4% PFA, and then embedded in OCT. Cryosections were collected using a Leica CM-1900 cryotome and stained with Oil Red O. For TEM analysis of lipid droplets, larvae were processed as previously described [31] and imaged on a Phillips CM-12 Transmission Electron Microscope provided by the VUMC Cell Imaging Shared Resource.
Cholesterol absorption assay
NBD-cholesterol (Invitrogen) stock solution was added to EY suspension to a final concentration of 3 μg/mL and vortexed for 1 minute. Incorporation into EY micelles was confirmed using a 100x objective with fluorescence. Larvae were fed for 4 hours at 28.5°C, rinsed briefly in fresh Danieau buffer, anesthetized and then mounted in 1% low melting point agarose on glass bottom dishes with the intestinal bulb secured to the bottom of the dish. Confocal images of the anterior intestine near the swim bladder were collected.
Quantification of transgenic fish organ size
Live embryos were anesthetized and mounted in 1% low melting point agarose. Confocal z-stacks were collected and maximum intensity projections were used to calculate 2-D area using ImageJ.
RESULTS
Sar1 genes and their expression during embryonic development
Vertebrate genomes from fish to mammals contain two Sar1 paralogs, Sar1a and Sar1b. We cloned the zebrafish sar1a and sar1b genes and found that both are highly evolutionarily conserved. Zebrafish sar1b shares 91% identity and 98% similarity with the human gene and is located in a syntenic region on chromosome 21. The 11.6 kb locus contains 8 exons that code for a predicted 198 amino acid peptide (Supplementary Fig. 1).
Sar1b is selectively expressed throughout development (Fig. 1). Initially, transcripts are maternally deposited into the egg (Fig. 1A) and zygotically expressed throughout gastrulation in all germ layers (Fig. 1B). During organogenesis, highest expression levels are found in the central nervous system and eye (Fig. 1C). At 3 days of development the transcripts are highly enriched in the craniofacial skeleton and the alimentary tract (Fig. 1E, F), a trend that continues throughout day 4 (Fig. 1G–I). On the contrary its close paralog, sar1a, is expressed ubiquitously throughout development (Supplementary Fig. 2). Thus, it appears that sar1 genes are expressed in overlapping domains, with sar1b being enriched in the CNS, craniofacial cartilage, gut and other organs of the digestive system.
In order to develop an in vivo model system to characterize the pathophysiology of the Sar1b deficiency, we designed a morpholino-labeled antisense oligonucleotide targeting the 5′UTR of the sar1b transcript (Fig. 1J). The morpholino (MO) blocked translation of a Sar1b reporter construct (Fig. 1K), and we have used in all of the experiments presented here the lowest effective dose (0.4 ng).
To test specificity of the MO, we conducted rescue experiments in a double-blinded fashion by co-injecting synthetic RNA. We injected either buffer alone as a control, sar1b-MO alone, or co-injected sar1b-MO along with zebrafish sar1b mRNA lacking the MO target sequence (Fig. 1L), human SAR1B mRNA, or zebrafish sar1a mRNA. Injected embryos were analyzed at 4 dpf and grouped into normal, moderate, or severe phenotypic categories, according to body length, head size, and pectoral fin morphology (Fig. 1M), and the identity of the treatment groups was revealed after phenotypic categorization. Sar1b morphant embryos presented shortened body length, a malformed jaw, and kinked pectoral fins (7% normal, 54% moderate (as shown in Fig. 1N), 39% severe). These morphant phenotypes were efficiently rescued by co-injection of synthetic zebrafish sar1b mRNA (87% normal, 13% moderate) or human SAR1B mRNA (83% normal, 14% moderate, 3% severe) (Fig. 1M). Zebrafish sar1a mRNA co-injection partially rescued the morphant phenotypes (57% normal, 31 % moderate, 12% severe) but less effectively than sar1b. To quantitatively assess rescue efficiency, we measured body length (Fig. 1N), which showed a similar trend as observed with phenotypic categorization.
Sar1b-deficient zebrafish fail to absorb dietary lipids
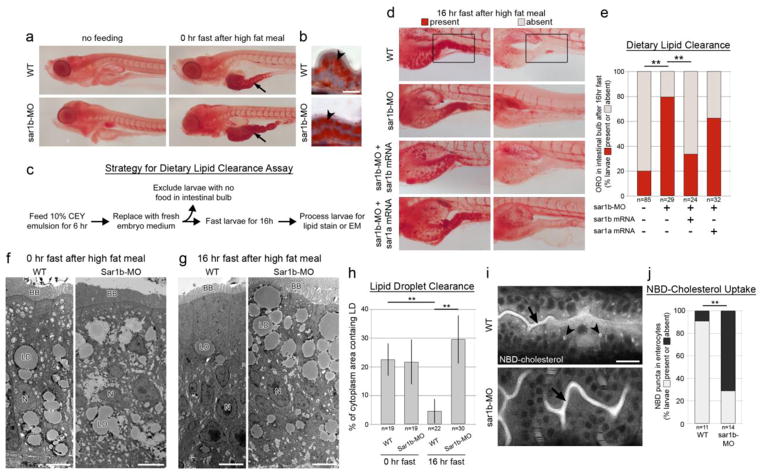
The hallmark of ANDD is failure of dietary lipid absorption due to impaired secretion from enterocytes. To test the efficiency of lipid absorption in Sar1b morphants, we used a dietary lipid clearance assay [12]. First, we fed 5-day old larvae with a high fat meal (chicken egg yolk suspension) and assayed lipid content in enterocytes with Oil Red O (ORO). We found that WT and Sar1b-morphant enterocytes formed abundant lipid droplets in response to feeding (Fig. 2A–B). We then tested the efficiency of clearing ingested lipids from the intestinal bulb using a pulse-chase approach (Fig. 2C). We fed larvae for 6 hours and then removed the feeding solution and fasted the animals for 16 hours before processing them for ORO staining. To exclude any effects of feeding behavior differences, we excluded larvae that did not participate in feeding, which can be visually confirmed because of the transparency of the zebrafish gut [16]. We found that 80% of WT larvae cleared lipids from the intestinal bulb after fasting. After Sar1b depletion, however, only 21% of larvae cleared ingested lipids. Dietary lipid retention in Sar1b morphants could be efficiently rescued with sar1b expression (p < 0.001, Fisher’s exact test) but not sar1a expression (p = 0.17). Transmission electron microscopy analysis showed that WT and sar1b morphant enterocytes both formed lipid droplets in response to feeding, but Sar1b morphants failed to clear lipid droplets after fasting (Fig. 2F–H). These findings confirmed that Sar1b morphants are able to ingest and uptake dietary lipids into enterocytes but have diminished capability to secrete lipid droplets, a phenotype resembling that of ANDD patients [9].
Figure 2. Sar1b knockdown impairs dietary lipid absorption and cholesterol uptake.
(a) Whole-mount detergent-free Oil Red O (ORO) staining of WT and sar1b morphant embryos under non-fed fasting conditions (left) and immediately after feeding a high fat meal (10% chicken egg yolk) [36]. After feeding, the intestinal bulb contains abundant lipids (arrow). (b) ORO staining of gut cryosections after feeding shows that lipids accumulate within enterocytes (concave arrowhead). (c) Schematic of experimental design and workflow for testing dietary lipid clearance in zebrafish larvae. Sar1b knockdown impairs dietary lipid clearance from enterocytes (D–H). (d) ORO staining of WT, sar1b-MO, and sar1b-MO + mRNA co-injected larvae fasted 16 hours after feeding. Left panels show representative examples of larvae with lipids remaining in the gut after fasting (present), while right panels show larvae that have cleared lipids from the intestinal bulb (absent). (e) Quantification of ORO presence in intestinal bulb after lipid clearance assay. Sar1b knockdown significantly impairs dietary lipid clearance, which can be rescued with sar1b mRNA expression. Sar1a mRNA does not effectively compensate for sar1b knockdown. (f) Transmission electron microscopy (TEM) analysis of WT and sar1b morphant larvae immediately after feeding 10% CEY. Lipid droplets (LD) are present in the enterocyte cytoplasm after a high fat meal [37]. Scale bars represent 5 μm. (g) TEM of WT and sar1b morphant larvae after lipid clearance assay. WT enterocytes contain few LDs compared to sar1b morphants [38, 39]. Scale bars represent 5 μm. (h) Quantification of lipid droplet abundance in enterocyte cytoplasm from TEMs. (i) NBD-cholesterol is present in the intestinal lumen of WT and morphants (arrows) when fed a high fat meal spiked with the cholesterol-fluorophore conjugate, but cholesterol positive inclusions are present only in the WT enterocytes (arrowheads) and absent in 71.4% of Sar1b morphants. Scale bars represent 20 μm. (j) Quantification of NBD fluorescence in enterocytes after feeding. ** p<0.01, Fisher’s exact test (E, J) or two way ANOVA with Tukey’s test (H). Error bars are ± SD. Abbreviations: BB, brush border; LD, lipid droplet; N, nucleus.
Another characteristic of ANDD is hypocholesterolemia, which may result directly from cholesterol malabsorption in enterocytes or secondarily from impaired liver function. To test whether Sar1b is required for dietary cholesterol absorption, we fed larvae with NBD-cholesterol, a fluorescent cholesterol analog, in chicken egg yolk suspension (NBD-Ch+EY) for 4 hours and then imaged live enterocytes using confocal microscopy. We found that 91% of WT larvae fed with NBD-Ch+CEY exhibited fluorescent puncta within enterocytes (Fig. 2I–J). In contrast, only 28% of Sar1b morphants had up-taken NBD-cholesterol into enterocytes (Fisher’s exact test, p<0.001). Both WT and Sar1b morphants ingest NBD-cholesterol normally, as indicated by high levels of fluorescence in the intestinal lumen. These findings provide for the first time direct evidence that Sar1b might be required for cholesterol uptake into enterocytes and suggests that hypocholesterolemia in ANDD patients may not be an exclusive result of defects in lipid metabolism.
Collectively, our findings show that Sar1b depletion in zebrafish closely resembles ANDD deficits. Conservation of this major ANDD phenotype in zebrafish could establish a practical animal model to study lipid malabsorption and test effective therapeutic strategies.
Sar1b is essential for digestive tract development
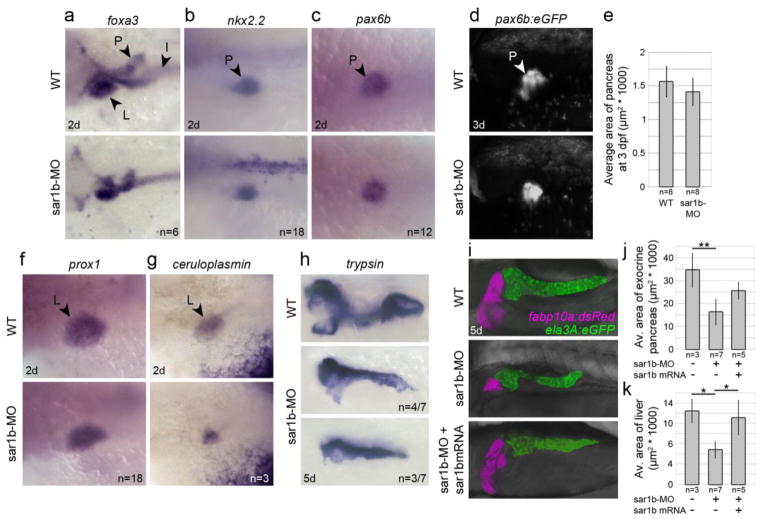
Sar1b is highly expressed in the zebrafish gut, pancreas and liver. Patients with ANDD were described to exhibit exocrine pancreas deficiency and sporadic hepatomegaly [8, 27]. This phenotype was ascribed to chronic diarrhea and altered lipid metabolism postnatally and not to developmental defects. To test whether digestive organs develop normally in Sar1b depleted embryos, we examined expression of patterning and differentiation markers of liver and pancreas [28]. Using whole mount in situ hybridization with foxa3 (a transcription factor directing GI tract patterning), we found that the gut primordium, including liver and pancreas buds, are largely unaffected (Fig. 3A) at 2 dpf. Similarly, pancreas patterning markers nkx2.2 and pax6b (Fig. 3B–C) were largely unaffected at 2 dpf, and at 3 dpf in live pax6b:eGFP transgenic fish (Fig. 3D–E). Liver patterning was also not affected in Sar1b morphants, using prox1 and ceruloplasmin in situ probes at 2 dpf (Fig. 3F–G). However, at 5 dpf the exocrine pancreas, as marked by trypsin probe (Fig. 3H) and in live LiPan transgenic line (Fig. 3I–J), failed to grow and expand caudally [18], and liver growth was also significantly reduced (Fig. 3I, K), which could be partially rescued with injection of synthetic sar1b mRNA (Fig. 3I–K). The intestinal bulb and adjacent anterior intestine was correctly patterned, but slightly distended as marked by ifabp gene expression, and hematoxylin and eosin staining of enterocytes revealed normal columnar epithelia and brush border morphology (our unpublished observations). These data indicate that Sar1b is not essential for patterning of the digestive organs, but is required for their growth.
Figure 3. Sar1b knockdown results in reduced size of digestive organs.
(a–c) Transcripts of foxa3, nkx2.2, and pax6b detected by whole-mount in situ hybridization show no significant differences in spatiotemporal patterning and specification of pancreatic precursors at 2 dpf. (d–e) Expression of the (pax6b:GFP) transgene shows modest but insignificant reduction in pancreas size at 3 dpf. (f–g) Transcripts of prox1 and ceruloplasmin show no significant differences in spatiotemporal patterning and specification of liver precursors at 2 dpf. (h) Transcripts of trypsin show reductions in pancreas size and change in lobular shape at 5 dpf. (i–k) Expression of the liver (fabp10a:dsRed) and pancreas (ela3A:eGFP) transgenes show consistently reduced size in 5 dpf Sar1b morphants, which can be partially rescued with -sar1b mRNA expression. Error bars are ± SD. Abbreviations: L, liver, P, pancreas, I, intestinal bulb. * p < 0.05, ** p < 0.01; two-tailed unpaired t-test (E) or One-way ANOVA standard weighted means analysis with Tukey’s test (J–K).
Sar1b-depletion affects brain patterning
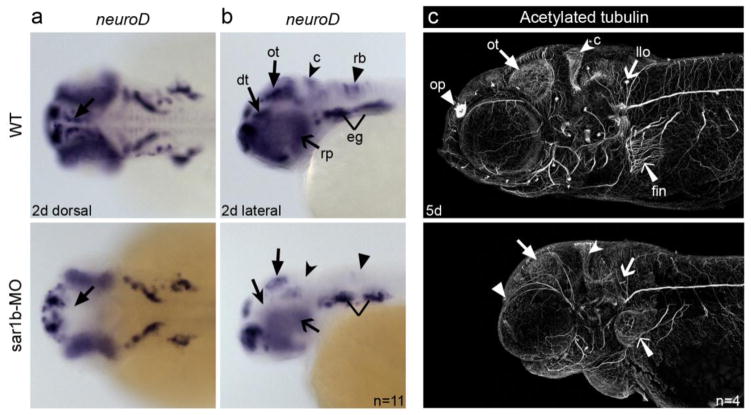
ANDD patients were reported to present with cerebellar ataxia and sensory neuropathy symptoms, which were attributed to Vitamin E deficiency due to dietary malabsorption [11]. To test whether the neuronal deficits are secondary to dietary depravation, or alternatively, are the primary developmental defect occurring before the fish begin food intake, we conducted a whole mount mRNA in situ hybridization to assess expression of neuroD, a bHLH transcription factor specifying progenitor populations of neuronal precursors in retina, epibranchial ganglia, cerebellum, hindbrain rhombomeres, thalamus and optic tectum (Fig. 4). We found normal neuroD expression in epibranchial ganglia, but expression was significantly reduced in retina, cerebellum, hindbrain rhombomeres, thalamus, and optic tectum (Fig. 4A–B).
Figure 4. Depletion of Sar1b disrupts neural development.
(a, b) Whole mount in situ hybridization with riboprobe detecting neuroD transcripts in 2 dpf embryos shows relatively normal spatio-temporal expression in epibranchial ganglia (eg), but reduced or absent expression demarcating neuroprogenitors in the dorsal thalamus (dt), optic tectum (ot), cerebellum (c) and hindbrain rhombomeres (rb), as well as retinal progenitors (rp). Dorsal (a) and (b) lateral views. (c) Maximum intensity z-projections of acetylated tubulin stained axonal tracks reveals reduction in staining in the olfactory pits (op), lateral line (llo) sensory patches of the head and pectoral fin innervation (fin). The forebrain and the optic tectum (ot) are reduced in size and shifted anteriorly in the morphants.
These findings suggest that Sar1b function is required in select neural stem cells of the brain. Because neuronal deficits appear before the larvae become dependent on dietary lipid absorption, we conclude that the observed phenotypes are most likely due to primary developmental defects. Moreover, the position of neurons in epibranchial ganglia matched the corresponding developmental stage, indicating that the deficits do not represent developmental delays in morphants. Acetylated tubulin staining revealed the presence of all major axonal tracks, although the positioning and density of the axonal projections in the optic tectum and rhombic lip appear to be underdeveloped (Fig. 4C). These findings reveal potential for broad developmental deficits in the CNS, but their functional implications need to be further investigated in patients and animal models.
Sar1b knockdown disrupts skeletal morphogenesis
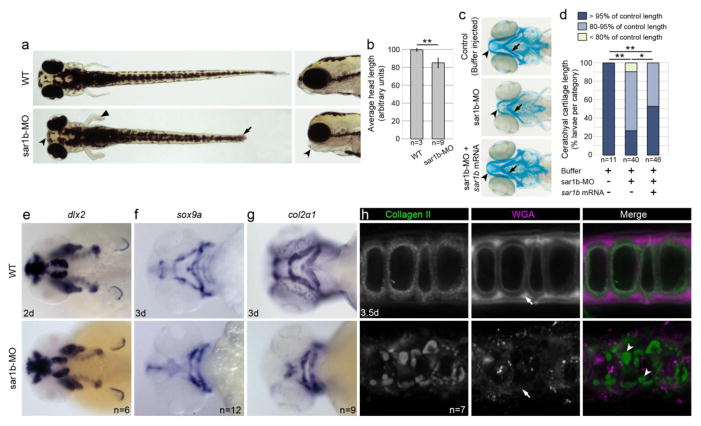
Depletion of Sar1b resulted in shorten body length, kinked pectoral fins and a compact head, particularly along the rostro-caudal axis (Fig. 5A–B). The craniofacial skeleton was significantly smaller and malformed as evident in Alcian blue stained preparations at 5 days of development (Fig. 5C–D). To determine at what developmental stage the first craniofacial deficits can be detected, we used whole mount mRNA in situ hybridization with probes for transcription factors dlx2 and sox9a [29] marking craniofacial primordia at 2 dpf and 3 dpf, respectively (Fig. 5E–F). Although we did not observe any deficits at 2 dpf, the time of neural crest migration, at 3 dpf the craniofacial cartilage primordia of sar1b morphants were already smaller than the corresponding structures in control siblings.
Figure 5. Sar1b knockdown results in skeletal dysmorphology.
(a) Live images of WT and sar1b morphants show shorter head in anteroposterior direction (concave arrowhead), kinked pectoral fins (flat arrowhead) and a shorter overall body length (arrow) in dorsal and lateral head views. (b) Quantification of anteroposterior head length measurements at 4 dpf. (c) Alcian blue stained head skeleton preparations revealed a shortened Meckel’s (concave arrowhead) and malformed ceratohyal (Ch) cartilage elements (arrow) of Sar1b-MO compared to control embryos, while sar1b-MO + sar1b mRNA co-injection rescued cartilage morphology. (d) Quantification of Ch cartilage length of Alcian blue stained embryos. Control length refers to the average length of buffer-injected embryos. (e) Unchanged dlx2 expression in Sar1b-MO shows that neural crest cell migration and patterning are normal in sar1b morphant embryos at 2 dpf. (f) Chondrogenic differentiation marker sox9a, (g) and its target gene col2a1 show reduced size and malformed morphant cartilages compared to WT. (h) Immunofluorescence staining with antibody recognizing Collagen II and of N-glycans using wheat germ agglutinin (WGA) shows intracellular inclusions of procollagen type II (arrowheads) and reduced staining intensity of N-glycans within extracellular matrix (arrows). Error bars are ± SD. * p < 0.05, ** p < 0.01; two-tailed unpaired t-test (b) or Fisher’s exact test (d).
Our prior works on Sec23a and Sec24D genes, two components of the COPII inner coat, revealed similar deficits in sox9a expression in zebrafish mutants [20, 30]. To test whether the phenotype stems from deficits in cartilage ECM secretion, we analyzed the expression of chondrogenic differentiation markers, including col2a1, the primary component of cartilage ECM, and hsp47, the collagen II specific chaperon required for its secretion from the ER. We noted smaller and misshapen cartilage elements expressing comparable levels of col2a1 transcripts (Fig. 5G). However, expression of hsp47 was highly upregulated in cartilage elements, consistent with patterns observed in other COPII mutants (our unpublished observations). The deficits in cartilage differentiation were likely a consequence of collagen II backlog in the ER as confirmed by immunofluorescence antibody labeling and the staining for N-glycosylated proteins by wheat germ agglutinin (WGA) of craniofacial chondrocytes, which revealed deficits in ECM matrix deposition and intracellular accumulation of collagen (Fig. 5H).
DISCUSSION
Sar1b Gene and Genetics of Anderson Disease
Here we present the first vertebrate animal model for Anderson Disease (ANDD), a rare genetic disease associated with mutations in the SAR1B gene. The zebrafish genome, like the human genome, contains two sar1 paralogs, sar1a and sar1b. The two paralogs differ by only 20 residues in the 198 amino acid peptides in humans [27] and by 25 residues in zebrafish. Interestingly, none of the divergent residues were found to be mutated in described patients [8], suggesting that the specialized function of Sar1b may not depend on the divergent sequence differences. The zebrafish and human Sar1b are 91% identical and 98% similar (Supplemental Fig. S1), prompting us to use zebrafish Sar1b depletion to model the pathophysiology of ANDD. Using a gene knockdown strategy, we show that Sar1b loss-of-function presents with phenotypes similar to ANDD, and leads to reduction of dietary lipid absorption as well as other developmental defects that are consistent with ANDD symptoms.
To date no mutations in SAR1A have been associated with ANDD or any other human syndrome, and it is unknown what phenotypes could be associated with SAR1A variants. We found that although sar1a is ubiquitously expressed in zebrafish embryos (Supplementary Fig. S2), its knockdown does not lead to gross morphological deficits (our unpublished observations). However, the combined depletion of Sar1a and Sar1b results in phenotypes similar to Sar1b knockdown, but with more severe phenotypes (our unpublished observations). Furthermore, overexpression of Sar1a is able to partially rescue Sar1b deficiency, suggesting that the two genes play largely similar roles in secretion with possible Sar1a partial compensation for Sar1b. This hypothesis may explain the broadly variable phenotype in spectrum and severity and is further supported by compensatory increase of SAR1A expression in enterocytes of ANDD patients [8]. Additionally, Sar1 paralog- and cargo-specific transport could explain the rather modest and tissue-specific manifestations of the disease. Potential transcriptional control responsible for fine-tuning of SAR1 gene expression has not been investigated and could explain unique and redundant functions of SAR1 genes in lipid absorption and more broadly in COPII-dependent transport of other cargos [31]. Further investigation of paralog-specific functions will be needed to address these interesting questions [32].
Sar1b Function in Lipid and Cholesterol Absorption
ANDD diagnosis is made based on collective clinical symptoms of lipid malabsorption, hypocholesterolemia with normal serum triglycerides, and histology of small intestine biopsy samples showing accumulation of lipid droplets in enterocytes [9]. Although hypocholesterolemia is a hallmark of ANDD, its mechanism is unknown [27], and despite major research interest in cholesterol homeostasis and its role as a risk factor in cardiovascular diseases, numerous questions remain unanswered. For example, the mechanism by which cholesterol in intestinal micelles crosses the brush-border barrier is being investigated [2], and the role of Sar1b in cholesterol transit to the basolateral membrane of the enterocyte is also actively pursued [33].
In our model we fed a high-fat diet to Sar1b-deficient zebrafish larvae and observed that the enterocytes contained abundant lipid droplets (Fig. 2B, F), consistent with symptoms observed in ANDD patients. This shows that Sar1b morphant enterocytes are able to uptake lipids. When we spiked the dietary lipids with a fluorescent cholesterol analog, however, a significantly higher percentage of Sar1b-deficient larvae were negative for intracellular cholesterol analog compared to wild-type siblings (Fig. 2I), suggesting that Sar1b might play a role in cholesterol absorption. Because ANDD is considered a lipid absorption disease, one might intuitively conclude that hypocholesterolemia is caused by a similar mechanism as lipid malabsorption in ANDD patients. If this was true, then one would expect cholesterol to be backlogged in chylomicrons in Sar1b-deficient enterocytes. Our findings, however, indicate that Sar1b is needed for cholesterol uptake into enterocytes, suggesting unexpected and complex function in cholesterol homeostasis. Studies of COPII-dependent transport of proteins that are required for cholesterol uptake, such as cholesterol esterase secretion from the pancreas and NPCL1 translocation to the enterocyte brush border [2], may further clarify the mechanisms of cholesterol malabsorption in ANDD patients. Dietary cholesterol uptake represents less than a third of daily cholesterol supply, with the majority coming from endogenous production by the liver and peripheral tissues. Thus, malabsorption alone may not explain the severe and consistent hypocholesterolemia observed in ANDD patients. Further functional studies will be required to elucidate the mechanisms of Sar1b-dependent hypocholesterolemia.
Pancreas Function in ANDD
Besides lipid malabsorption, ANDD patients often present with exocrine pancreatic insufficiency (EPI) and require exocrine pancreatic enzymes supplemented to their diet to attain normal growth [8, 9]. Prevailing interpretations of EPI conclude that chronic diarrhea and ensuing lipid malabsorption is responsible for reduced pancreatic function [8]. In addition to this possibility, EPI could stem from reduced secretory capacity of the exocrine pancreas (COPII-dependent protein secretion), along with developmental deficits that lead to reduced pancreatic mass. This possibility is supported by our data. In Sar1b-depleted animals, we observed significant reduction in pancreas size and lobular structure ranging from very severe to mild (representative phenotypes shown in Fig. 3 H–I) that could explain the variable penetrance of this phenotype in patients. Our finding that variable reductions in pancreas size occur during development and before feeding may suggest that steatorrhea is not a primary mechanism of EPI in ANDD patients.
Sar1b in Skeletal Morphogenesis
Prior Northern Blot analysis of adult human tissue RNA samples has shown SAR1B expression in the small intestine, liver, skeleton and heart muscle [7]. Here, we have expanded the spectrum of the sar1b expression and deficiency phenotypes to include skeletal cartilage. Although French patients had decreased bone mineral density, this was attributed to poor nutritional regime due to malabsorption and not to an intrinsic role of SAR1B in bone homeostasis [9]. We show that lack of Sar1b leads to deficiencies in chondrocyte capacity to produce and maintain type II collagen, a predominant matrix component of cartilage (Fig. 5). Thus it is possible that osteopenia and osteoporosis observed in some patients might be a consequence of COPII-deficiency that impairs ECM secretion in skeletal tissues [34].
Conclusions
Remarkably little is known about COPII-dependent lipid transport and the molecular mechanisms controlling these processes. Although biochemical studies have helped to clarify the mechanisms of Sar1b-dependent chylomicron transport, lack of genotype-phenotype correlation in ANDD complicates understanding of Sar1b function in vivo. The ANDD model presented here will help to address numerous outstanding questions related to pathophysiology of Sar1b-deficiency. Additionally, this model provides entry points to discover mechanisms of other complex traits. Obesity and other metabolic diseases such as atherosclerosis present formidable challenges to functionally dissect underlying molecular mechanisms because of small contributions of common and rare genetic variants, which do not grant sufficient quantitative power to explain the disease phenotype. Zebrafish models offer a unique experimentally amenable system to advance understanding of rare genetic diseases and metabolic disorders [35]. Zebrafish genetic models can be used to perform chemical biology screens to discover novel compounds that may be used not just to treat ANDD but other disorders of cholesterol synthesis and homeostasis. The study presented in this manuscript sets the stage for future investigations and development of new high-throughput tools to address this problem, and importantly, a better understanding of the pathophysiology of SAR1B deficiency will lead to early differential diagnosis of new cases.
Supplementary Material
Key Messages.
Sar1b depletion phenotype in zebrafish resembles Anderson Disease deficits
Sar1b deficiency results in multi-organ developmental deficits
Sar1b is required for dietary cholesterol uptake into enterocytes
Acknowledgments
We thank C. Guthrie for excellent animal care, K. Zavalin for expert technical assistance, and Lisette Maddison for sharing plasmids and transgenic fish. We are indebted to Kevin Ess and Antonis Hatzopoulos for critical reading of the manuscript. This manuscript is dedicated in memory of our colleague JR Minkel. This work was supported in part by the Zebrafish Initiative of the Vanderbilt University Academic Venture Capital Fund, the NIH NIDCR grant R01 DE018477 (E.W.K.). D.S.L. was supported by NRSA F31DE022226 and T32HD007502 Training Program in Developmental Biology; D.B.M was supported by T32GM008554, the Cellular, Biochemical and Molecular Sciences Training Program.
Footnotes
DISCLOSURE
The authors declare that they do not have any competing or financial interests.
References
- 1.Abumrad NA, Davidson NO. Role of the Gut in Lipid Homeostasis. Physiol Rev. 2012;92:1061–1085. doi: 10.1152/physrev.00019.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iqbal J, Hussain MM. Intestinal lipid absorption. Am J Physiol - Endocrinol Metab. 2009;296:E1183–E1194. doi: 10.1152/ajpendo.90899.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller EA, Barlowe C. Regulation of coat assembly—sorting things out at the ER. Curr Opin Cell Biol. 2010;22:447–453. doi: 10.1016/j.ceb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqi SA, Gorelick FS, Mahan JT, Mansbach CM. COPII proteins are required for Golgi fusion but not for endoplasmic reticulum budding of the pre-chylomicron transport vesicle. J Cell Sci. 2003;116:415–427. doi: 10.1242/jcs.00215. [DOI] [PubMed] [Google Scholar]
- 5.Walther TC, Farese RV. Lipid Droplets and Cellular Lipid Metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansbach CM, Siddiqi SA. The Biogenesis of Chylomicrons. Annu Rev Physiol. 2010;72:315–333. doi: 10.1146/annurev-physiol-021909-135801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones B, Jones EL, Bonney SA, Patel HN, Mensenkamp AR, Eichenbaum-Voline S, Rudling M, Myrdal U, Annesi G, Naik S, et al. Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nat Genet. 2003;34:29–31. doi: 10.1038/ng1145. [DOI] [PubMed] [Google Scholar]
- 8.Georges A, Bonneau J, Bonnefont-Rousselot D, Champigneulle J, Rabès JP, Abifadel M, Aparicio T, Guenedet JC, Bruckert E, Boileau C, et al. Molecular analysis and intestinal expression of SAR1 genes and proteins in Anderson’s disease (Chylomicron retention disease) Orphanet J Rare Dis. 2011;6:1. doi: 10.1186/1750-1172-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peretti N, Roy CC, Sassolas A, Deslandres C, Drouin E, Rasquin A, Seidman E, Brochu P, Vohl M-C, Labarge S, et al. Chylomicron retention disease: A long term study of two cohorts. Mol Genet Metab. 2009;97:136–142. doi: 10.1016/j.ymgme.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Silvain M, Bligny D, Aparicio T, Laforêt P, Grodet A, Peretti N, Ménard D, Djouadi F, Jardel C, Bégué J, et al. Anderson’s disease (chylomicron retention disease): a new mutation in the SARA2 gene associated with muscular and cardiac abnormalities. Clin Genet. 2008;74:546–552. doi: 10.1111/j.1399-0004.2008.01069.x. [DOI] [PubMed] [Google Scholar]
- 11.Bernard G, Panisset M, Sadikot AF, Chouinard S. Chylomicron retention disease: Dystonia as a new clinical feature. Mov Disord. 2010;25:1755–1756. doi: 10.1002/mds.23165. [DOI] [PubMed] [Google Scholar]
- 12.Schlegel A, Stainier DYR. Microsomal Triglyceride Transfer Protein Is Required for Yolk Lipid Utilization and Absorption of Dietary Lipids in Zebrafish Larvae. Biochemistry. 2006;45:15179–15187. doi: 10.1021/bi0619268. [DOI] [PubMed] [Google Scholar]
- 13.Anderson JL, Carten JD, Farber SA. Chapter 5 - Zebrafish Lipid Metabolism: From Mediating Early Patterning to the Metabolism of Dietary Fat and Cholesterol. In: William H, Detrich MW, LIZ, editors. Methods Cell Biol. Academic Press; 2011. pp. 111–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carten JD, Bradford MK, Farber SA. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev Biol. 2011;360:276–285. doi: 10.1016/j.ydbio.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maddison LA, Chen W. Nutrient Excess Stimulates β-Cell Neogenesis in Zebrafish. Diabetes. 2012;61:2517–2524. doi: 10.2337/db11-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walters JW, Anderson JL, Bittman R, Pack M, Farber SA. Visualization of Lipid Metabolism in the Zebrafish Intestine Reveals a Relationship between NPC1L1- Mediated Cholesterol Uptake and Dietary Fatty Acid. Chem Biol. 2012;19:913–925. doi: 10.1016/j.chembiol.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 18.Korzh S, Pan X, Garcia-Lecea M, Winata C, Pan X, Wohland T, Korzh V, Gong Z. Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev Biol. 2008;8:84. doi: 10.1186/1471-213X-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delporte FM, Pasque V, Devos N, Manfroid I, Voz ML, Motte P, Biemar F, Martial JA, Peers B. Expression of zebrafish pax6b in pancreas is regulated by two enhancers containing highly conserved cis-elements bound by PDX1, PBX and PREP factors. BMC Dev Biol. 2008;8:53. doi: 10.1186/1471-213X-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarmah S, Barrallo-Gimeno A, Melville DB, Topczewski J, Solnica-Krezel L, Knapik EW. Sec24D-Dependent Transport of Extracellular Matrix Proteins Is Required for Zebrafish Skeletal Morphogenesis. PLoS One. doi: 10.1371/journal.pone.0010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sachdev SW, Dietz UH, Oshima Y, Lang MR, Knapik EW, Hiraki Y, Shukunami C. Sequence analysis of zebrafish chondromodulin-1 and expression profile in the notochord and chondrogenic regions during cartilage morphogenesis. Mech Dev. 2001;105:157–162. doi: 10.1016/s0925-4773(01)00417-8. [DOI] [PubMed] [Google Scholar]
- 22.Montero-Balaguer M, Lang MR, Sachdev SW, Knappmeyer C, Stewart RA, De La Guardia A, Hatzopoulos AK, Knapik EW. The mother superior mutation ablates foxd3 activity in neural crest progenitor cells and depletes neural crest derivatives in zebrafish. Dev Dyn. 2006;235:3199–3212. doi: 10.1002/dvdy.20959. [DOI] [PubMed] [Google Scholar]
- 23.Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Howe DG, Knight J, Mani P, Martin R, Moxon SAT, et al. ZFIN: enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 2011;39:D822–D829. doi: 10.1093/nar/gkq1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller II, Knapik EW, Hatzopoulos AK. Expression of the protein related to Dan and Cerberus gene—prdc—During eye, pharyngeal arch, somite, and swim bladder development in zebrafish. Dev Dyn. 2006;235:2881–2888. doi: 10.1002/dvdy.20925. [DOI] [PubMed] [Google Scholar]
- 25.Müller II, Melville DB, Tanwar V, Rybski WM, Mukherjee A, Shoemaker MB, Wang W-D, Schoenhard JA, Roden DM, Darbar D, et al. Functional modeling in zebrafish demonstrates that the atrial-fibrillation-associated gene GREM2 regulates cardiac laterality, cardiomyocyte differentiation and atrial rhythm. Dis Model Mech. 2013;6:332–341. doi: 10.1242/dmm.010488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Granero-Moltó F, Sarmah S, O’Rear L, Spagnoli A, Abrahamson D, Saus J, Hudson BG, Knapik EW. Goodpasture Antigen-binding Protein and Its Spliced Variant, Ceramide Transfer Protein, Have Different Functions in the Modulation of Apoptosis during Zebrafish Development. J Biol Chem. 2008;283:20495–20504. doi: 10.1074/jbc.M801806200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charcosset M, Sassolas A, Peretti N, Roy CC, Deslandres C, Sinnett D, Levy E, Lachaux A. Anderson or chylomicron retention disease: Molecular impact of five mutations in the SAR1B gene on the structure and the functionality of Sar1b protein. Mol Genet Metab. 2008;93:74–84. doi: 10.1016/j.ymgme.2007.08.120. [DOI] [PubMed] [Google Scholar]
- 28.Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DYR. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- 29.Barrallo-Gimeno A, Holzschuh J, Driever W, Knapik EW. Neural crest survival and differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development. 2004;131:1463–1477. doi: 10.1242/dev.01033. [DOI] [PubMed] [Google Scholar]
- 30.Lang MR, Lapierre LA, Frotscher M, Goldenring JR, Knapik EW. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat Genet. 2006;38:1198–1203. doi: 10.1038/ng1880. [DOI] [PubMed] [Google Scholar]
- 31.Melville DB, Montero-Balaguer M, Levic DS, Bradley K, Smith JR, Hatzopoulos AK, Knapik EW. The feelgood mutation in zebrafish dysregulates COPII-dependent secretion of select extracellular matrix proteins in skeletal morphogenesis. Dis Model Mech. 2011;4:763–776. doi: 10.1242/dmm.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unlu G, Levic DS, Melville DB, Knapik EW. Trafficking mechanisms of extracellular matrix macromolecules: Insights from vertebrate development and human diseases. Int J Biochem Cell Biol. 2014;47:57–67. doi: 10.1016/j.biocel.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siddiqi S, Siddiqi SA, Mansbach CM. Sec24C is required for docking the prechylomicron transport vesicle with the Golgi. J Lipid Res. 2010;51:1093–1100. doi: 10.1194/jlr.M002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melville DB, Knapik EW. Traffic jams in fish bones: ER-to-Golgi protein transport during zebrafish development. Cell Adhes Migr. 2011;5:114–118. doi: 10.4161/cam.5.2.14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vacaru A, Unlu G, Spitzner M, Mione M, Knapik E, Sadler K. In vivo cell biology using zebrafish – providing insights into vertebrate development and disease. J Cell Sci. 127:1–11. doi: 10.1242/jcs.140194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlombs K, Wagner T, Scheel J. Site-1 protease is required for cartilage development in zebrafish. PNAS. 2003;100:14024–14029. doi: 10.1073/pnas.2331794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, Lee B, Buhman KK, Cheng J-X. A dynamic, cytoplasmic triacylglycerol pool in enterocytes revealed by ex vivo and in vivo coherent anti-Stokes Raman scattering imaging. J Lipid Res. 2009;50:1080–1089. doi: 10.1194/jlr.M800555-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy CC, Levy E, Green PH, Sniderman A, Letarte J, Buts JP, Orquin J, Brochu P, Weber AM, Morin CL, et al. Malabsorption, hypocholesterolemia, and fat-filled enterocytes with increased intestinal apoprotein B. Chylomicron retention disease. Gastroenterology. 1987;92:390–399. doi: 10.1016/0016-5085(87)90133-8. [DOI] [PubMed] [Google Scholar]
- 39.Dannoura AH, Berriot-Varoqueaux N, Amati P, Abadie V, Verthier N, Schmitz J, Wetterau JR, Samson-Bouma M-E, Aggerbeck LP. Anderson’s Disease Exclusion of Apolipoprotein and Intracellular Lipid Transport Genes. Arterioscler Thromb Vasc Biol. 1999;19:2494–2508. doi: 10.1161/01.atv.19.10.2494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.