Summary
Myocardial infarction (MI), a leading cause of death around the world, displays a complex pattern of inheritance1,2. When MI occurs early in life, the role of inheritance is substantially greater1. Previously, rare mutations in low-density lipoprotein (LDL) genes have been shown to contribute to MI risk in individual families3–8 whereas common variants at more than 45 loci have been associated with MI risk in the population9–15. Here, we evaluate the contribution of rare mutations to MI risk in the population. We sequenced the protein-coding regions of 9,793 genomes from patients with MI at an early age (≤50 years in males and ≤60 years in females) along with MI-free controls. We identified two genes where rare coding-sequence mutations were more frequent in cases versus controls at exome-wide significance. At low-density lipoprotein receptor (LDLR), carriers of rare, damaging mutations (3.1% of cases versus 1.3% of controls) were at 2.4-fold increased risk for MI; carriers of null alleles at LDLR were at even higher risk (13-fold difference). This sequence-based estimate of the proportion of early MI cases due to LDLR mutations is remarkably similar to an estimate made more than 40 years ago using total cholesterol16. At apolipoprotein A-V (APOA5), carriers of rare nonsynonymous mutations (1.4% of cases versus 0.6% of controls) were at 2.2-fold increased risk for MI. When compared with non-carriers, LDLR mutation carriers had higher plasma LDL cholesterol whereas APOA5 mutation carriers had higher plasma triglycerides. Recent evidence has connected MI risk with coding sequence mutations at two genes functionally related to APOA5, namely lipoprotein lipase15,17 and apolipoprotein C318,19. When combined, these observations suggest that, beyond LDL cholesterol, disordered metabolism of triglyceride-rich lipoproteins contributes to MI risk.
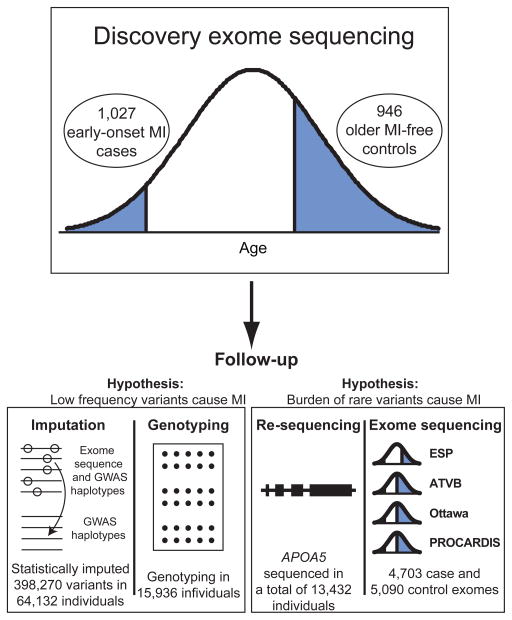
The U.S. National Heart, Lung, and Blood Institute’s Exome Sequencing Project sought to use exome sequencing as a tool to discover novel genes and mechanisms contributing to heart, lung, and blood disorders. Within this program, we designed a discovery study for the extreme phenotype of early-onset MI (Figure 1) since heritability is substantially greater when MI occurs early in life1,2. From eleven studies, we ascertained 1,088 cases with MI at an early age (MI in males ≤50 years old and in females ≤60 years old). As a comparison group, we selected 978 participants from prospective cohort studies who were of advanced age (males ≥60 years old or females ≥70 years old) and free of MI.
Figure 1. Overall design for the Early-Onset Myocardial Infarction Study within the U.S. National Heart, Lung, and Blood Institute’s Exome Sequencing Project.
Whole exome sequencing was performed in 1,973 individuals from the phenotypic extremes. To test the hypothesis that low-frequency variants confer risk for myocardial infarction (MI), we performed follow-up statistical imputation and array-based genotyping of single nucleotide variants. To test the hypothesis that a burden of rare mutations in a gene confers risk for MI, we performed targeted re-sequencing and additional exome sequencing.
We sequenced cases and controls to high coverage by performing solution-based hybrid selection of exons followed by massively parallel sequencing (see Methods)20. We performed several quality control steps to identify and remove outlier samples and variants (see Methods and Supplementary Figures 1–13). Characteristics of the discovery set of 1,027 cases and 946 controls are provided in Supplementary Tables 1–3. Across the autosomes, each participant had an average of 43 nonsense, 7,828 missense, 92 splice-site, 189 indel frameshift, 366 indel non-frameshift, and 103 non-synonymous singleton variants.
We first tested whether low-frequency coding variants (defined here as a single nucleotide variant (SNV) or indel with minor allele frequency (MAF) between 1% and 5%) are associated with risk for MI in the discovery sequencing study. We observed no significant association of MI status with any individual variant (Supplementary Figure 14). We next evaluated the hypothesis that rare alleles (defined here as a SNV or indel with MAF< 1% frequency) collectively within a gene contribute to risk for MI (see Methods). We tested for an excess (or deficit) in cases versus controls of rare, non-synonymous mutations by aggregating together SNVs and indels with MAF < 1% (‘T1’ test) in each gene and comparing the counts in cases and controls21. Empirical P-values were obtained using permutation.
The need to aggregate rare variants poses a challenge: which variants should be aggregated? Ideally, one would aggregate only harmful alleles and ignore benign alleles. To enrich for harmful alleles, we considered three sets of variants: 1) non-synonymous only; 2) a “Deleterious (PolyPhen)” set consisting of non-synonymous after excluding missense alleles annotated as benign by PolyPhen-2 HumDiv software; and 3) “Disruptive” mutations only (nonsense, indel frameshift, splice-site; also referred to as “null” mutations). To account for multiple testing, we set exome-wide significance for this study at P=8 × 10−7, a Bonferroni correction for the testing of ~20,000 genes and three variant sets. When the T1 test was applied across these three sets of alleles in the discovery sequencing study, no gene-based association signal deviated from what we expected by chance (Supplementary Figure 15–22).
We followed up on discovery sequencing results in four ways: (1) statistical imputation; (2) array-based genotyping using the Illumina HumanExome Beadchip (‘Exome’ Chip); (3) targeted re-sequencing; and (4) additional exome sequencing (Figure 1). Imputation and array-based genotyping were utilized to largely evaluate low-frequency variants whereas targeted re-sequencing and exome sequencing were utilized to test the role of rare mutations.
With the first and second follow-up approaches - imputation (n=64,132) and array-based genotyping (n=15,936), we did not identify novel low-frequency variants associated with MI or coronary artery disease (CAD) (see Methods, Supplementary Tables 4–7 and Supplementary Figures 23–27). The top association results for SNVs from array-based genotyping are shown in Supplementary Table 8.
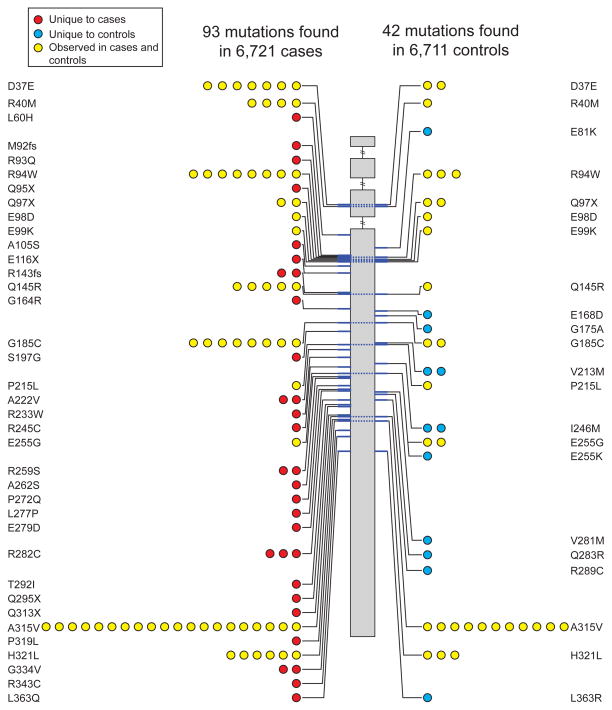
In the third follow-up approach, we re-sequenced several genes in additional cases and controls (see Methods, Supplementary Table 9). After sequencing the exons of APOA5 in 6,721 cases and 6,711 controls, we identified 46 unique non-synonymous or splice-site SNVs or indel frameshifts with allele frequency <1% (Supplementary Table 10). Based on these variants, we observed 93 alternate allele counts in cases and 42 alternate allele counts in controls (P=5 × 10−7; Table 1, Figure 2 and Supplementary Table 10). This burden of rare mutation signal was primarily driven by mutations seen in one or two study participants (Figure 2 and Supplementary Table 10). Carriers of a rare APOA5 mutation had a 2.2-fold higher risk for MI/CAD than non-carriers (Table 1).
Table 1.
Association of a burden of rare mutations in the apolipoprotein A–V (APOA5) gene with risk for early-onset myocardial infarction or coronary artery disease
| Mutation set | N cases/controls | T1 cases | T1 controls | % freq cases | % freq controls | OR | P |
|---|---|---|---|---|---|---|---|
| Non-synonymous | 6,721/6,711 | 93 | 42 | 1.4 | 0.63 | 2.2 | 5 × 10−7 |
| Deleterious (PolyPhen) | 6,721/6,711 | 63 | 31 | 0.94 | 0.46 | 2.0 | 6 × 10−5 |
| Deleterious (Broad) | 6,721/6,711 | 68 | 31 | 1.0 | 0.46 | 2.2 | 2 × 10−5 |
| Deleterious (Strict) | 6,721/6,711 | 10 | 3 | 0.15 | 0.045 | 3.3 | 0.008 |
| Disruptive | 6,721/6,711 | 9 | 2 | 0.13 | 0.03 | 4.5 | 0.007 |
Summary allele counts and carrier frequencies are shown. Only SNVs and indels with minor allele frequency less than 1% were considered in burden analysis. “Deleterious (PolyPhen)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as “possibly damaging” or “probably damaging” by PolyPhen-2 HumDiv software; “Deleterious (Broad)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as deleterious by at least one of the five protein prediction algorithms of LRT score, MutationTaster, PolyPhen-2 HumDiv, PolyPhen-2 HumVar and SIFT; “Deleterious (Strict)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as deleterious by all five protein prediction algorithms; Disruptive defined as nonsense, splice-site or indel frameshift; T1: Alleles with SNVs or indels with minor allele frequency less than 1%; % freq: percentage of cases or controls carrying a T1 allele; OR: odds ratio.
Figure 2. Apolipoprotein A-V (APOA5) mutations discovered after sequencing of 13,432 individuals.
Individual mutations (non-synonymous, indel frameshift and splice-site variants with minor allele frequency less than 1%) are depicted according to genomic position along the length of the APOA5 gene starting at the 5′ end (top). The number of circles on the left and right represents the number of times that mutation is observed in cases or controls, respectively. Dashed lines across the gene connect the same mutation seen in cases and controls. Mutations are shaded in red, blue, or yellow if observed in cases only, controls only, or both cases and controls, respectively.
According to a recent report, consideration of variant sets based on multiple protein prediction algorithms might yield stronger association signals 22. Therefore, in exploratory analyses, we investigated two additional variant sets: 1) “Deleterious (Broad)” as defined by nonsense, splice-site, frameshift, and missense annotated as damaging by at least one of five protein prediction algorithms and 2) “Deleterious (Strict)” as defined by nonsense, splice-site, frameshift, and missense annotated as damaging by all five protein prediction algorithms (see Methods). Carriers of a rare APOA5 “Deleterious (Strict)” mutation had an even higher risk for MI/CAD (3.3-fold, P=0.008).
A burden of rare mutations in APOA5 explains about 0.14% of the total variance for MI and roughly 0.28% of the heritability (assuming that additive genetic factors explain ~50% of the overall variance) (see Methods and Supplementary Table 11). When compared with non-carriers, carriers of rare non-synonymous APOA5 alleles had higher plasma triglycerides (median in carriers was 167 mg/dl versus 104 mg/dl for non-carriers, P=0.007) and lower high-density lipoprotein cholesterol (mean in carriers was 43 mg/dl versus 57 mg/dl for non-carriers, P=0.007) but similar LDL cholesterol (median in carriers was 110 mg/dl versus 108 mg/dl for non-carriers, P=0.66) (Supplementary Table 12).
In the fourth follow-up approach, we performed exome sequencing in additional early-onset MI/CAD cases and controls, bringing the total number of exomes analyzed to 9,793 (Supplementary Tables 13–14). We tested for an excess (or deficit) in cases versus controls of rare mutations in any gene (Supplementary Figure 28 and Supplementary Tables 15–17). At this sample size, rare alleles collectively conferred risk for MI at exome-wide significance in only one gene, namely LDLR (Figure 3).
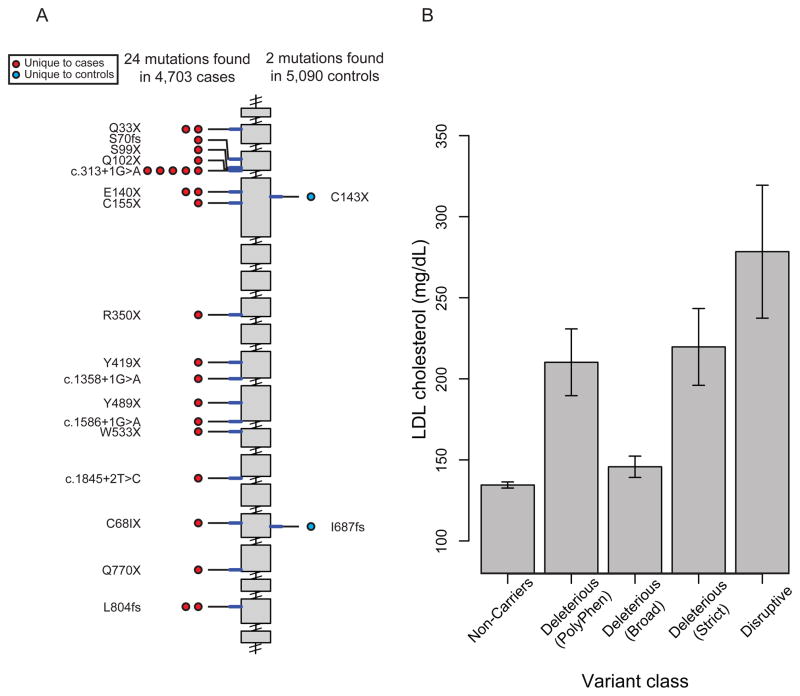
Figure 3. Low-density lipoprotein receptor (LDLR) mutations discovered after sequencing 9,793 individuals.
A. Individual disruptive mutations (nonsense, indel frameshift, and splice-site variants with minor allele frequency less than 1%) are depicted according to genomic position along the length of the LDLR gene starting at the 5′ end (top). The number of circles on the left and right represents the number of times that mutation is observed in cases or controls, respectively. Mutations are shaded in red or blue, if observed in cases only or controls only, respectively. B. Low-density lipoprotein cholesterol level as a function of LDLR gene mutation annotation. Mean (height of bar) and 95 % confidence intervals (error bars) are shown. Each individual is categorized based on mutation annotation as follows. Non-Carriers: carriers without a missense or disruptive mutation; “Deleterious (PolyPhen)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as “possibly damaging” or “probably damaging” by PolyPhen-2 HumDiv software; “Deleterious (Broad)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as deleterious by at least one of five protein prediction algorithms (LRT score, MutationTaster, PolyPhen-2 HumDiv, PolyPhen-2 HumVar and SIFT); “Deleterious (Strict)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as deleterious by all five of the above protein prediction algorithms; Disruptive: carriers of mutations that are nonsense, indel frameshift, or splice-site.
After sequencing the exons of LDLR in 4,703 cases and 5,090 controls, we identified 156 unique non-synonymous, splice-site SNVs and indel frameshifts with allele frequency <1% (Table 2 and Supplementary Table 18). Of these variants, we observed 285 alleles in cases (6.1% of cases) and 208 alleles in controls (4.1% of controls) (1.5-fold effect size, P=4 × 10−6) (Table 2). When restricting analysis to the “Deleterious (PolyPhen)” set, 3.1% of cases and 1.3% of controls carried at least one such rare mutation, for a 2.4-fold effect size (P=1 × 10−11). A higher effect size of 4.2-fold (P=3 × 10−11) was observed when restricting to the “Deleterious (Strict)” set. When restricting to disruptive alleles, 0.51% of cases and 0.04% of controls carried at least one such rare “disruptive” mutation, for a 13-fold effect size (P=9 × 10−5) (Table 2 and Figure 3).
Table 2.
Association of a burden of rare mutations in the low-density lipoprotein receptor (LDLR) gene with risk for early-onset myocardial infarction
| Mutation set | N cases/controls | T1 cases | T1 controls | % freq cases | % freq controls | OR | P |
|---|---|---|---|---|---|---|---|
| Non-synonymous | 4,703/5,090 | 285 | 208 | 6.1 | 4.1 | 1.5 | 4 × 10−6 |
| Deleterious (PolyPhen) | 4,703/5,090 | 148 | 67 | 3.1 | 1.3 | 2.4 | 1 × 10−11 |
| Deleterious (Broad) | 4,703/5,090 | 243 | 158 | 5.2 | 3.1 | 1.7 | 9 × 10−8 |
| Deleterious (Strict) | 4,703/5,090 | 90 | 23 | 1.9 | 0.45 | 4.2 | 3 × 10−11 |
| Disruptive | 4,703/5,090 | 24 | 2 | 0.51 | 0.039 | 13.0 | 9 × 10−5 |
Summary allele counts and carrier frequencies are shown. Only SNVs and indels with minor allele frequency less than 1% were considered in burden analysis. “Deleterious (PolyPhen)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as “possibly damaging” or “probably damaging” by PolyPhen-2 HumDiv software; “Deleterious (Broad)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as deleterious by at least one of the five protein prediction algorithms of LRT score, MutationTaster, PolyPhen-2 HumDiv, PolyPhen-2 HumVar and SIFT; “Deleterious (Strict)” as defined by nonsense, splice-site, indel frameshift, and missense annotated as deleterious by all five protein prediction algorithms; Disruptive defined as nonsense, splice-site or indel frameshift; T1: Alleles with SNVs or indels with minor allele frequency less than 1%; % freq: percentage of cases or controls carrying a T1 allele; OR: odds ratio.
A burden of rare mutations in LDLR explains about 0.24% of the total variance for MI and roughly 0.48% of the heritability (see Methods and Supplementary Table 19). LDL cholesterol level differed based on functional class annotation with the greatest difference seen between carriers of “disruptive” mutations and those who did not carry any non-synonymous mutations (279 mg/dl versus 135 mg/dl, Figure 3 and Supplementary Table 20). Approximately 49% of the LDLR alleles discovered in this study (77 of 156) have been previously associated with familial hypercholesterolemia in LDLR FH databases23 (Supplementary Table 21).
Using these rare variant signals as a guide, we estimated sample sizes that will be required to make similar discoveries. A very large number of samples – at least 10,000 exomes – are required to achieve 80% statistical power at an exome-wide level of statistical significance (Supplementary Figure 29–31).
Here, we show that a burden of multiple rare alleles in two genes - LDLR and APOA5 - contributes to risk for MI. These results suggest several conclusions regarding the inherited basis for MI and rare variant association studies. First, after a DNA sequence-based search across nearly all protein-coding genes in >9,700 early-onset MI cases and controls, LDLR is the strongest association signal, where mutations in the gene account for about 3% of cases. In 1973, Goldstein and colleagues studied survivors of early MI and noted two common lipid abnormalities - hypercholesterolemia and hypertriglyceridemia16. Based on a total cholesterol value exceeding ~285 mg/dl, it was estimated that 4.1% of cases with MI prior the age of 60 had familial hypercholesterolemia; this original estimate is similar to ours based on direct sequencing. In contrast, the prevalence of harmful LDLR mutations in the general population is several orders of magnitude higher than the original estimate (~0.5 – 1.3% in the present study versus 0.1 – 0.2% by Goldstein). Second, the rare variant association signal presented here establishes APOA5 as a bona fide MI gene. Initially discovered through comparative genomics analysis of a region harboring several lipid regulators (i.e., APOA1 and APOC3), the APOA5 locus harbors common variants associated with plasma triglycerides24. Candidate gene studies have associated a common variant at this locus also with MI risk (−1131T>C, APOA5 promoter region, rs662799, MAF of 8%)25,26. However, due to extensive linkage disequilibrium in this region, it had been previously uncertain which gene is responsible for the association with MI. The identification of multiple coding sequence variants within APOA5 clarifies that this gene contributes to MI risk in the population. Third, these data point to a route to MI beyond LDL cholesterol, namely triglyceride-rich lipoproteins27 and the lipoprotein lipase pathway. Genetic variation at two other proteins related to APOA5 function, apolipoprotein C-III18,19,28 and lipoprotein lipase15, have been associated with triglycerides and MI risk. Finally, the present study makes clear that rare variant discovery for complex disease will require the sequencing of thousands of cases and careful statistical analysis. Two reasons for the large sample size requirement are an inability to readily distinguish harmful from benign alleles and the extreme rarity of harmful alleles.
Methods
General Overview of the Exome Sequencing Project (ESP)
Details of the study design of the National Heart, Lung and Blood Institute’s GO Exome Sequencing Project (NHLBI ESP) have been published previously 29. Briefly, the goal of the NHLBI ESP was to discover rare coding variation in genes contributing to heart, lung and blood disorders using next generation sequencing of the protein coding regions of the genome (“exome sequencing”). The study includes five primary groups including: Seattle GO – University of Washington, Seattle, WA; Broad GO – Broad Institute, Cambridge, MA; WHISP – Ohio State University Medical Center, Columbus, OH; Lung GO – University of Washington, Seattle, WA; Heart GO – University of Virginia Health System, Charlottesville, VA and two collaborating groups, WashU GO – Washington University, St. Louis and CHARGE-S GO – University of Texas Health Sciences Center, Houston, TX.
We included samples from several studies: Women’s Health Initiative (WHI); Framingham Heart Study (FHS); Jackson Heart Study (JHS); Multi-Ethnic Study of Atherosclerosis (MESA); Atherosclerosis Risk in Communities (ARIC); Coronary Artery Risk Development in Adults (CARDIA); Cardiovascular Health Study (CHS); Lung Health Study (LHS); COPD Genetic Epidemiology (COPD Gene); Severe Asthma Research Project (SARP); Pulmonary Arterial Hypertension (PAH); Acute Lung Injury (ALI); Cystic Fibrosis (CF); Cleveland Clinic Genebank (CCGB); Massachusetts General Hospital Premature Coronary Artery Disease Study (MGH PCAD); Heart Attack Risk in Puget Sound (HARPS); Translational Research Underlying Disparities in Myocardial Infarction Patients’ Health Status (TRIUMPH) and the PennCath study.
General Overview of the ESP Early-Onset Myocardial Infarction Study
Within the NHLBI ESP, we designed an exome sequencing experiment specifically to study early-onset myocardial infarction (EOMI). We selected EOMI cases and controls from eleven studies, including: ARIC, MESA, CCGB, FHS, HARPS, MGH-PCAD, PennCath, TRIUMPH, WHI, CHS, and JHS (Supplementary Tables 1–3). Samples were selected based on the extreme tails of the phenotypic distribution, in order to enrich for a genetic contribution to disease. EOMI cases were defined as individuals who had a MI at an age of ≤50 for men and ≤60 for women. Controls were selected as individuals with no history of MI at baseline or during follow-up to at least age 60 for men and 70 for women. The study samples, along with case and control definitions, are briefly described below and shown in Supplementary Tables 1–3.
Study and Phenotype Descriptions for ESP EOMI
a. The HeartGO Consortium
HeartGO is a multiethnic consortium consisting of six NHLBI population-based cohorts of men and women: ARIC, CHS, FHS, CARDIA, JHS, and MESA. The age range of participants in these six cohorts spans the spectrum from early adulthood to old age, providing a broad age representation. Each participating cohort in HeartGO has completed ascertainment of multiple phenotypes, including all of the major cardiovascular risk factors (blood pressure, lipids, diabetes status), biomarkers including measures of blood cell counts, subclinical disease imaging, and cardiovascular and lung outcomes including MI and stroke. Participants in all six cohorts provided written informed consent. The NIH database of Genotypes and Phenotypes (dbGaP) site contains further details regarding the phenotypes accessible for each individual HeartGO cohort.
b. Cleveland Clinic GeneBank (CCGB)
The CCGB study is a single-center prospective cohort-based study that enrolled patients undergoing elective diagnostic coronary angiography between 2001 and 2006. Coronary artery disease (CAD) was defined as adjudicated diagnoses of stable or unstable angina, MI (adjudicated definition based on defined electrocardiographic changes or elevated cardiac enzymes), angiographic evidence of ≥ 50% stenosis of one or more major epicardial vessel, and/or a history of known CAD (documented MI, CAD, or history of revascularization).
c. Heart Attack Risk in Puget Sound (HARPS)
The HARPS study is a population-based case-control study that enrolled cases with incident MI presenting to a network of hospitals in the metropolitan Seattle-Puget Sound region of Washington State between 1998 and 2002.
d. Massachusetts General Hospital - Premature Coronary Artery Disease (MGH-PCAD) Study
The MGH-PCAD study is a hospital-based case-control study that enrolled cases hospitalized with early MI at MGH between 1999 and 2004.
e. PennCath
The PennCath study is a catheterization-lab based cohort study from the University of Pennsylvania Medical Center and enrolled subjects at the time of cardiac catheterization and coronary angiography between 1998 and 2003. Persons undergoing cardiac catheterization at either the Hospital of the University of Pennsylvania or Penn Presbyterian Medical Center consented for the PennCath study to identify genetic and biochemical factors related to coronary disease.
f. Translational Research Investigating Underlying Disparities in Acute Myocardial Infarction Patients’ Health Status (TRIUMPH)
The TRIUMPH study is a large, prospective, observational cohort study of consecutive patients with acute MI presenting to 24 US hospitals from April 2005 to December 2008. MI was diagnosed using contemporary definitions 30 and all patients had an elevated troponin blood test.
g. Women’s Health Initiative
The Women’s Health Initiative (WHI) is a major research program that has been ongoing for over 20 years to address the most common causes of death, disability and poor quality of life in postmenopausal women -- cardiovascular disease, cancer, and osteoporosis. Cases were defined based on the occurrence of MI, coronary revascularization, hospitalized angina or death due to coronary disease.
Studies Involved in Follow-up Statistical Imputation, Array-based Genotyping, Targeted Resequencing and Additional Exome Sequencing
a. Statistical Imputation
We performed imputation of single nucleotide variants (SNVs) discovered in the exomes of 786 samples. We imputed exomic SNVs into 64,132 independent samples in 16 studies to test for association of coding SNVs with MI or CAD. The studies are described in Supplementary Table 5.
b. Array-based Genotyping
We performed follow-up genotyping using the Illumina HumanExome Beadchip (“Exome Chip”) array in 15,936 independent samples from seven studies. The studies are described in Supplementary Table 7.
c. Targeted Sequencing
We performed targeted sequencing of the APOA5 gene in an additional 11,414 individuals from five cohorts. The studies are described in Supplementary Table 9.
d. Exome Sequencing-based Follow-up
We leveraged additional exomes from the ESP study and performed exome sequencing in additional individuals from three cohorts. The studies are described in Supplementary Table 13.
Detailed methods for the processing and analysis of samples for the various stages of the project are described below. We describe methods for the different stages of the project, including discovery exome sequencing and follow-up imputation, array-based genotyping, targeted re-sequencing and additional exome sequencing.
Laboratory Methods for Discovery Exome Sequencing in the ESP EOMI Project
a. Exome Sequencing
Exome sequencing was performed at the Broad Institute. Sequencing and exome capture methods have been previously described 29. A brief description of the methods is provided below.
b. Receipt/Quality Control of Sample DNA
Samples were shipped to the Biological Samples Platform laboratory at the Broad Institute of MIT and Harvard. DNA concentration was determined by the Picogreen assay (Invitrogen) before storage in 2D-barcoded 0.75 mL Matrix tubes at −20°C in the SmaRTStore™ (RTS, Manchester, UK) automated sample handling system. We performed initial quality control (QC) on all samples involving sample quantification (PicoGreen), confirmation of high-molecular weight DNA and fingerprint genotyping and gender determination (Illumina iSelect). Samples were excluded if the total mass, concentration, integrity of DNA or quality of preliminary genotyping data was too low.
c. Library Construction and In-Solution Hybrid Selection
Starting with 3μg of genomic DNA, library construction and in-solution hybrid selection were performed as described previously 31. A subset of samples, however, was prepared using the Fisher et al. protocol with some slight modifications. Initial genomic DNA input into shearing was reduced from 3μg to 100ng in 50μL of solution. In addition, for adapter ligation, Illumina paired end adapters were replaced with palindromic forked adapters with unique 8 base index sequences embedded within the adapter.
d. Preparation of Libraries for Cluster Amplification and Sequencing
After in-solution hybrid selection, libraries were quantified using qPCR (KAPA Biosystems) with probes specific to the ends of the adapters. This assay was automated using Agilent’s Bravo liquid handling platform. Based on qPCR quantification, libraries were normalized to 2nM and then denatured using 0.1 N NaOH using Perkin-Elmer’s MultiProbe liquid handling platform. A subset of the samples prepared using forked, indexed adapters was quantified using qPCR, normalized to 2nM using Perkin-Elmer’s Mini-Janus liquid handling platform, and pooled by equal volume using the Agilent Bravo. Pools were then denatured using 0.1 N NaOH. Denatured samples were diluted into strip tubes using the Perkin-Elmer MultiProbe.
e. Cluster Amplification and Sequencing
Cluster amplification of denatured templates was performed according to the manufacturer’s protocol (Illumina) using either Genome Analyzer v3, Genome Analyzer v4, or HiSeq 2,000 v2 cluster chemistry and flowcells. After cluster amplification, SYBR Green dye was added to all flowcell lanes, and a portion of each lane visualized using a light microscope, in order to confirm target cluster density. Flowcells were sequenced either on Genome Analyzer II using v3 and v4 Sequencing-by-Synthesis Kits, then analyzed using RTA v1.7.48, or on HiSeq 2,000 using HiSeq 2,000 v2 Sequencing-by-Synthesis Kits, then analyzed using RTA v1.10.15. All samples were run on 76 cycle, paired end runs. For samples prepared using forked, indexed adapters, Illumina’s Multiplexing Sequencing Primer Kit was also used.
f. Read Mapping and Variant analysis
Samples were processed from real-time base-calls (RTA 1.7 software [Bustard], converted to qseq.txt files, and aligned to a human reference (hg19) using Burrows-Wheeler Aligner (BWA, see URL 32). Aligned reads duplicating the start position of another read were flagged as duplicates and not analyzed (“duplicate removal”). Data was processed using the Genome Analysis ToolKit (GATK v1.1.3 33). Reads were locally realigned (GATK IndelRealigner) and their base qualities were recalibrated (GATK TableRecalibration). Variant detection and genotyping were performed on both exomes and flanking 50 base pairs of intronic sequence using the UnifiedGenotyper (UG) tool from the GATK. Variant data for each sample was formatted (variant call format (VCF)) as “raw” calls for all samples. SNVs and indel sites were flagged using the Variant Filtration walker (GATK) to mark sites of low quality that were likely false positives. SNVs were marked as potential errors if they exhibited strong strand bias (SB >= 0.10), low average quality (quality per depth of coverage (QD) <5.0), or fell in a homopolymer run (HRun > 4). Indels were marked as potential errors for low quality (quality score (QUAL) < 30.0), low average quality (QD < 2.0), or if the site exhibited strong strand bias (SB > -1.0). Samples were considered complete when exome targeted read coverage was ≥20X over ≥80% of the exome target.
g. Data Analysis QC
Fingerprint concordance between sequence data and fingerprint genotypes was evaluated. Variant calls were evaluated on both bulk and per-sample properties: novel and known variant counts, transition-transversion (TS-TV) ratio, heterozygous-homozygous non-reference ratio, and deletion/insertion ratio. Both bulk and sample metrics were compared to historical values for exome sequencing projects at the Broad Institute. No significant deviation of the ESP variants or ESP samples from historical values was noted.
Data Processing, Quality Control and Association Analysis of Discovery Exome Sequencing
a. Variant Calling
Variants (SNVs and indels) were identified and genotyped from recalibrated BAM files 34 using the multi-sample processing mode of the Unified Genotyper tool from the GATK. Variants were first identified and genotyped in random batches of 100 samples. The batches were then merged into a single VCF file using the GATK CombineVariants tool.
b. Variant Annotation
Variants (SNVs and indels) were annotated using the GRCh37.64 database using the SNP effect predictor tool (SnpEff, see URL) 35 and the GATK VariantAnnotator. The primary SnpEff genomic effects that were annotated include: splice-site acceptor, splice-site donor, indel frameshift, indel non-frameshift, nonsense, non-synonymous and synonymous variants. For variants that have different annotations due to multiple transcripts of the gene, the highest impact effect for each variant was taken.
c. Sample Level Quality Control
We performed several quality control steps to identify and remove outlier samples (Supplementary Figures 1–8). First, we required that each sample had a minimum of 20-fold coverage for at least 80% of the targeted bases. Second, we compared self-reported ancestry with that inferred from the sequence data and removed discordant samples. Third, we removed samples with high degree of heterozygosity and low number of singleton counts as this pattern suggests DNA contamination across samples. Fourth, we removed samples with an extremely high number of variants or singletons as this can suggest low quality DNA. Finally, we removed samples exhibiting a mismatch between the reported gender and that inferred from sequence data. Of 2,066 cases and controls sequenced across the exome, we removed 93 samples due to these exclusion criteria.
d. Variant Level Quality Control
QC measures were also performed to remove low quality variants. We assessed population genetics metrics including the TS-TV ratio, the ratio of the number of heterozygous changes to the number of homozygous non-reference changes, and the number of non-synonymous to the number of synonymous changes. This analysis can help filter false positive calls since we expect the true TS-TV to be around ~3.2 in European populations 33, while a set of random SNVs (or false positive variants) should give a random expectation of 0.5. Variants with low depth of coverage (DP) and high percent missingness generally had low TS-TV and heterozygous-homozygous non-reference ratios. Variants were removed if there was DP < 8 average per sample and > 2% missingness (Supplementary Figures 9–12). Distribution of allele frequencies of the SNVs is shown in Supplementary Figure 13.
e. Common Variant Association Analysis
We performed single variant association analysis in our exome sequencing dataset. For SNVs with minor allele frequency (MAF) greater than 5%, we ran logistic regression, after adjusting for 10 principal components while for SNVs with MAF less than 5%, we ran Fisher’s Exact test. We performed association analysis in European Americans and African Americans separately and then performed sample size weighted meta-analysis using METAL 36. The association results are shown in Supplementary Figure 14.
f. Rare Variant Association Analysis
To test whether rare mutations contribute to MI, we performed burden of rare variant analysis on the ~2,000 ESP EOMI exome samples. We performed a variant of the Combined Multivariate Collapsing test 21, that groups the count of alleles of SNVs in cases and controls. Phenotype labels were permuted 100,000 times to assign a statistical significance. We accounted for ethnicity by permuting phenotype labels within each ethnicity. Association analysis was performed using PLINK/SEQ (see URL).
We collapsed variants based on computational predictions from PolyPhen-2 HumDiv 37. Minor allele frequencies were calculated from all available samples sequenced in each study in order to obtain the most accurate MAF estimates. Therefore, calculation of MAF for ESP EOMI 1 and 2 was performed on a larger set of exome samples that were sequenced at the Broad Institute as part of ESP (n=970 exomes for ESP EOMI 1 and n=3014 for ESP EOMI 2). For our burden of rare variant association analysis, we use a MAF threshold of 1% (T1). Furthermore, we use three different types of variant groupings when collapsing by gene. These variant groups are: 1) non-synonymous only; 2) a deleterious set consisting of non-synonymous after excluding missense alleles annotated as benign by PolyPhen-2 HumDiv software; and 3) disruptive (nonsense, indel frameshift, splice-site) mutations only. We also performed the T1 test after collapsing all non-synonymous mutations by KEGG pathways (Supplementary Figure 21 and 22).
Methods for Follow-up Statistical Imputation
a. Construction of Reference Panels and Targeted Imputation Panels
Exome imputations were performed using two reference panels and 16 targeted imputation panels. A total of 697 ESP samples (436 African Americans and 261 European Americans) were used for the first reference panel while 89 samples from the 1000 Genomes Project 38 were drawn for the second reference panel. For the ESP reference panel, all samples from ARIC (n=212), JHS (n=119), MIGen (n=151) and WHI studies (n=41) were genotyped using commercially available Affymetrix 6.0 arrays. Samples from the FHS (n=174) were genotyped using the Affymetrix 5.0 array. The second reference panel was comprised of samples from the 1000 Genomes Project that had genotype data for both low coverage sequencing and high coverage exome sequencing data 38. A total of 89 samples were selected from 6 diverse populations (23 African Ancestry in Southwest US (ASW), 9 Utah residents with Northern and Western European ancestry (CEU), 12 Colombian in Medellin, Colombia (CLM), 25 Mexican Ancestry in Los Angeles, CA (MXL), 17 Toscani in Italia (TSI) and 3 Yoruba in Ibadan, Nigeria (YRI) samples). Low coverage whole genome sequencing, high coverage exome sequencing and targeted exome capture were performed based on standard protocols at the Broad Institute. Details of the sequencing methods and samples have been described previously 38. Imputation was performed into 16 independent study samples with genome-wide genotype data. Study samples were genotyped using commercially available Affymetrix or Illumina genotyping arrays. Further details are described in Supplementary Table 5.
Reference panels were created by merging genotypes from SNVs that span the entire genome (hence, providing a haplotype “scaffold”), with genotypes from SNVs from ESP exome sequencing data. The first reference panel was generated using genotypes from both genome-wide SNV arrays obtained from dbGAP and exome sequencing data. The second reference panel was generated using genotype data for both low coverage sequencing and high coverage exome sequencing data. Both the reference panel and targeted genome-wide panel were phased using the “best guess haplotypes” option in IMPUTE2 39. Haplotype phasing were performed in 5 megabase chunks as recommended by the software tutorial 39.
b. Data Processing, Quality Control and Association Analysis
Imputation of the exome was performed using IMPUTE2. We imputed approximately 400,000 coding SNVs from the reference panels into 28,068 cases and 36,064 controls from 16 different study samples with genome-wide data. Descriptions for the study samples have been reported elsewhere (Supplementary Table 5 for references). We filtered SNVs with MAF < 1% and imputation quality (INFO) < 0.5 from further analysis. The distribution of imputation qualities of the SNVs is shown in Supplementary Figure 23–24. Association testing for CAD/MI was performed using the score method and assuming an additive model in SNPTEST (see URL) 40. Age, sex and the first two principal components were used as covariates when appropriate. We did not observe any indication of excess inflation of test statistics in any of the study samples (Supplementary Table 22). Meta-analysis of study-specific P-values for imputed SNVs was performed using the Z-score method weighted by sample size in METAL (see URL). Beta and standard errors were estimated based on an inverse-weighted meta-analysis. The distribution of association results for the imputation results is shown in Supplementary Figure 25 and top association results in Supplementary Table 6.
Methods for Follow-up Array-Based Genotyping
a. Laboratory Methods
DNA samples were sent to the Broad Institute Genetic Analysis Platform for genotyping and were placed on 96-well plates for processing using the Illumina HumanExome v1.0 SNP array. Genotypes were assigned using GenomeStudio v2010.3 using the calling algorithm/genotyping module version 1.8.4 along with the custom cluster file StanCtrExChp_CEPH.egt. Only samples passing an overall call rate of 98% criteria and standard identity check were released from the Genetic Analysis Platform.
b. Data Processing, Quality Control and Association Analysis
To identify single low frequency SNVs associated with MI or CAD, we performed array-based genotyping using the Illumina Human Exome Beadchip. We genotyped 83,680 sites identified from exome sequencing in 1,027 early-onset MI cases and 946 controls. The samples for genotyping were drawn from the cohorts listed in Supplementary Table 7 and have been previously described. The functional effect of each variant was predicted using the SeattleSeq Annotation server (see URL). For variants having more than one functional class, the most deleterious class was retained.
Several quality control processes were employed to ensure high quality genotypes and samples were used in the association analysis. Samples were excluded for the following criteria: greater than 5% missing genotypes; discordance between inferred gender based on genotype and self-reported gender; inbreeding coefficient less than −0.2 or greater than 0.2; duplicated samples; or proportion of genotypes identical by descent > 0.2. In addition, principal components were calculated using Eigenstrat 4.2 41 and samples were removed if they were found to be statistical population outliers. Variants were removed for the following criteria: MAF = 0%; significant difference between missingness in cases compared with controls; extreme deviation from Hardy-Weinberg equilibrium (P < 1×10−6); or significant association with genotyping plate assignment. All quality control filtering were performed using PLINK (see URL) 42 and R (see URL, The R Project for Statistical Computing, Vienna, Austria).
Association testing for CAD/MI was performed within each study separately using logistic regression with ten principal components of ancestry as covariates. An inverse standard-error weighted meta-analysis was performed to combine results across studies. The association testing was performed using PLINK 42 and the meta-analysis was performed using METAL. There was no indication of an inflation of test statistics across studies (Supplementary Table 23). The stability of logistic regression was assessed by examining the standard error of the beta estimate as a function of minor allele frequency (see Supplementary Figure 32). As shown, logistic regression is unstable for a MAF < 0.05%. Fisher’s Exact test was used for variants with MAF < 0.05%. The top association results are shown in Supplementary Table 8.
Methods for Follow-up Resequencing
a. Selection of Genes
We first selected six associated genes (based on biologic and/or statistical evidence with T1 P < 0.005; APOA5, CHRM5, SMG7, LYRM1, APOC3, NBEAL1) for replication sequencing in the ATVB study (Supplementary Table 24) where all cases had suffered an MI prior to age of 46. We also pursued the same six genes in the Ottawa Heart Study with 552 cases and 586 controls (Supplementary Table 25). One of the genes (APOA5) continued to show significant results and was sequenced in three additional studies (Table 1 and Supplementary Table 26). In total, we performed follow-up sequencing of APOA5 in six study samples, including the Verona Heart Study (VHS), Ottawa Heart Study (OHS), additional exomes from Atherosclerosis, Thrombosis, and Vascular Biology Italian Study Group (ATVB), additional exomes from the ESP EOMI study (ESP EOMI 2), Precocious Coronary Artery Disease study (PROCARDIS), and the Copenhagen City Heart Study and Copenhagen Ischemic Heart Disease Study (CCHS/CIHDS).
b. Laboratory Methods
For the VHS study, genomic DNA was extracted from white blood cells using the salting-out method. The protein-coding regions corresponding to the RefGene transcripts NM_052968 for APOA5 and NM_012125 for CHRM5 were sequenced using in-house designed primers (available on request) and the BigDye Terminator Cycle Sequencing Kit v1.1 on an ABI-3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA). SNVs were called using the Variant Reporter software v1.1 (Applied Biosystems).
For the OHS study, PCR primers were designed, tested and optimized to target the exons and flanking non-coding sequences for each gene. Sequencing reactions were performed using big dye terminator chemistry and chromatograms obtained with an Applied Biosystems ABI 3730XL capillary sequencer. Chromatograms were base-called by using Phred, assembled into contigs by using Phrap, and scanned for SNVs with PolyPhred 43 to identify polymorphic sites. Each read was trimmed to remove low-quality sequence (Phred score <25), resulting in analyzed reads with an average Phred quality of 40. After assembly and variant calling, each polymorphic sites were reviewed by a data analyst using Consed 44 to ensure the quality and accuracy of the variant calls. This process generates sequence-based SNV genotypes with accuracy > 99.9%.
For the PROCARDIS study, a single long range PCR product (LRPCR) was amplified to provide coverage of the APOA5 exonic, intronic and flanking sequences (human reference sequence NCBI build 37 chromosome 11:116,659,905–116,664,331). The LRPCR products were tagged with unique sequence (barcode) adaptors, and processed into 56 short amplicons (Reflex ™ reactions - www.populationgenetics.com) and pooled for multiplex next-generation sequencing (NGS). NGS was performed on a MiSeq™ personal sequencer to >20x coverage across 95% of the APOA5 target region on 1,385 MI cases and 1,499 controls. Paired-end reads were mapped to NCBI build 37 using the BWA and SMALT aligners; variants were identified by the GATK unified genotyper (v1.6.13) and annotated using SnpEff v2.0.5 and the GRCh37.64 database.
For the CCHS/CIHDS study, lightscanner screening and resequencing were performed. Genomic DNA was isolated from frozen whole blood (QiaAmp4 DNA blood mini kit; QIAGEN GmbH, Hilden, Germany). Six PCR fragments were amplified covering the three coding exons and adjacent splice-sites (approximately 20 base-pairs upstream and downstream each exon) of APOA5. Mutational analysis of the PCR products was performed by high resolution melting curve (HRM) analysis using the Lightscanner™ system (Idaho Technology Inc. Salt Lake City, Utah). PCR fragments showing heteroduplex formation by HRM analysis were subsequently sequenced on an ABI 3730 DNA analyzer (Applied Biosystems Inc., Foster City, CA).
c. Data Processing, Quality Control and Association Analysis
After sequencing, variants were annotated using SnpEff or Annovar 45. For each study, only non-synonymous SNVs with MAF < 1% were analyzed. Rare variant burden testing was performed using the T1 test. Meta-analysis was performed to combine evidence across study specific P-values using the sample size weighted Z-score method, implemented in METAL. Association results and a listing of APOA5 mutations discovered from sequencing are described in Table 1 and Supplementary Table 10. P-values for association between APOA5 mutation carrier status and lipid traits were performed using the Mann-Whitney ranksum test. Results are shown in Supplementary Table 12.
Methods for Follow-up Exome Sequencing
a. Laboratory Methods
We performed follow-up exome sequencing in additional samples from three other studies. Sequencing was performed at the Broad Institute, using the same protocols described above for the NHLBI ESP Project.
b. Data Processing, Quality Control and Association Analysis
Variant calling and annotations were performed as described above for the NHBLI ESP EOMI. Quality control of samples was performed using the following steps. To detect mismatched samples, we calculated discordance rates between genotypes from exome sequencing with genotypes from array-based genotyping. We removed samples with discordance rate > 0.02. We tested for sample contamination using verifyBamID 46, which examines the proportion of non-reference bases at reference sites. We removed samples with FREEMIX or CHIPMIX scores > 0.2. Furthermore, we removed outlier samples with too many or too few SNVs (>700 or <5 singletons, >400 or <5 doubletons), and those with too high or low TS-TV (>4 or <3) and heterozygosity (heterozygote to homozygote ratio >6 or <2). Finally, we removed samples with high missingness (>0.1). In total, 202 samples were removed. For quality control of variants, we removed SNPs and indels that had low recalibration scores after running GATK VariantRecalibrator. We also removed SNVs with low coverage (DP < 140000 and quality over depth (QD) < 2) and high missingness (frequency of missing genotypes > 0.02). For quality control of indels, we removed indels that had excessive strand bias (Fisher Strand > 200), high proportion of alternate alleles seen near the ends of reads (ReadPosRankSum < −20), deviation from Hardy-Weinberg equilibrium (InbreedingCoeff < −0.8) and low coverage (QD < 3). Rare variant association analysis was performed using EPACTS (see URL). We performed burden of rare variant analysis using the Efficient Mixed-Model Association eXpedited (EMMAX) Combined Multivariate and Collapsing (CMC) test 47. This approach uses a kinship matrix to take into account population structure. We restricted analyses to SNVs and indels with minor allele frequency < 0.01. Furthermore, we restricted analyses to three different sets of variants: 1) non-synonymous only; 2) a deleterious set consisting of non-synonymous after excluding missense alleles annotated as benign by PolyPhen-2 HumDiv software; and 3) disruptive (nonsense, indel frameshift, splice-site) mutations only.
Estimation of Heritability Explained by a Burden of Rare Mutations in the APOA5 and LDLR Genes
We calculated the heritability explained by a burden of rare mutations in the APOA5 and LDLR genes using the following assumptions. We assumed that the alleles come from a mixture of two distributions: harmless alleles, with no effect on the trait, and null alleles, which destroy the function of the gene, and have an (constant) effect on the trait. We assumed different values for the fraction of null alleles, α (our current expectation for most genes for α is around 1/3–1/2 for missense alleles. Here, we clump missense alleles together with nonsense alleles, which should slightly increase α). The variance explained is sensitive to this parameter. We assumed a liability-threshold model for disease, with an underlying (un-observed) continuous trait representing risk for MI, and MI occurring if risk is above a certain threshold. We assume all null alleles have effect β (in units of standard deviations) on the liability scale. We assumed different values for the prevalence (denoted κ) for early MI (3% to 5%). Results are somewhat sensitive to prevalence; higher prevalence will slightly increase heritability estimates. Given the prevalence, the number of carriers in cases and controls gives us the allele frequency in the population (which is very close to the allele frequency in controls).
We fitted the effect size (β on liability scale) and alleles for different values of α and κ. Results for APOA5 are shown in Supplementary Table 11 and results for LDLR are shown in Supplementary Table 19. For APOA5, β is moderate (up to roughly one standard deviation), with variance explained between 0.08% and 0.17% of the total phenotypic variance (on the liability scale). If we assume the heritability of MI is 50%, a burden of rare mutations in the APOA5 gene may explain 0.16–0.34% of the heritability. For LDLR, for all values, variance explained is between 0.13% and 0.32% of the total phenotypic variance (on the liability scale) and 0.26–0.64% of the heritability.
Sample Size Extrapolations and Power Calculations for Burden of Rare Variants
We evaluated the sample size that is needed to reach genome-wide significance levels (P=2.5 × 10−6) for the T1 test. Our calculations relied on the following assumptions. We assumed that all allelic variants with population frequency less than 1% are causal and have identical effect sizes. We also assumed that all alleles with frequency greater than 1% were benign.
Our calculations differentiate between the allele frequency of a SNV in our exome samples with its true allele frequency in a population. The T1 test compares the number of carriers of an allele for a SNV with sample (rather than population) allele frequency less than 1% among cases and controls. We considered three factors when extrapolating to larger sample sizes. First, we assumed our sample is comprised of 50% cases and 50% controls. Since the prevalence of EOMI is estimated to be 5%, the sample frequency of causal alleles is likely to be higher than the population frequency. Second, some alleles with population frequency below 1% may, by chance, have sampling frequency greater than 1% and therefore be excluded from the test. Third, the true allele frequency of the SNVs in the population is unknown. In contrast to earlier work that relied on population genetics modeling 48, we provide an update on the power needed to detect rare variant signal after considering the three factors above. We calculated liberal and conservative estimates for our sample size extrapolations and power calculations. The conservative estimate was based on the estimate of the total population frequency of all causal alleles (below 1%) that would be unlikely to be excluded from the T1 test due to the sampling frequencies exceeding 1%. Because allele frequency distribution is dominated by rare alleles, for an allele with population frequency x̂, expected population allele frequency is smaller than x.
| (1) |
Therefore, the expected total population frequency of all alleles below frequency x is smaller than the total sampling frequency of alleles below sampling frequency x̂. However, setting x̂ at 1% would result in a liberal rather than conservative estimate because alleles with population frequency below 1% may be excluded from the T1 test as having sampling frequency above 1%. This occurs due to oversampling cases (our sample has 50% of cases at disease prevalence of 5%) and sampling variance. For example, assuming only one causal allele per gene, the power of the T1 test is maximal for the population allele frequency close to 0.5% for a sample of 1,000 cases and 1,000 controls. For a sample of 10,000 individuals, the chance that a risk allele with population frequency of 0.5% would be excluded from the T1 test is below 10−3, making this threshold even more conservative. Therefore, for a conservative estimate, we have assumed that the total population frequency of all causal alleles per gene would equal the total sampling frequency of alleles below 0.5% in the ESP sample. Our liberal estimate assumed that all causal alleles will be included in the T1 test. We assumed that the total population frequency of all causal alleles per gene would equal the total sampling frequency of alleles below 1% in the ESP sample.
Once we extrapolated the number of mutation carriers to 20,000 samples, we then performed power calculations to see how many samples would be needed to reach a genome-wide significance level for the T1 test (P=2.5 × 10−6 after correcting for 18,500 genes). Power calculations were performed by first sampling a genotype at random from the pool of 20,000 simulated samples. Based on the T1 carrier status of the drawn sample, we simulated the phenotype based on a calculated probability. The phenotype was simulated based on a prevalence rate of 5% for disease, carrier status of the random sample and assumed relative risk of 2.0 of the mutation. For T1 carriers, the probability of being a case was calculated as relative risk (RR) of T1 carrier multiplied by prevalence rate of disease (RR * prevalence rate). For non-carriers, the probability of being a case was simply the prevalence rate. The case-control ratio was 1:1. We performed sample size extrapolations for genes with varying number of T1 mutations (25th percentile, median and 75th percentile of carriers with a T1 mutation for all genes discovered in the exome, Supplementary Figures 29–31).
Supplementary Material
Acknowledgments
The authors wish to acknowledge the support of the National Heart, Lung, and Blood Institute (NHLBI) and the National Human Genome Research Institute (NHGRI) of the U.S. National Institutes of Health (NIH) and the contributions of the research institutions, study investigators, field staff and study participants in creating this resource for biomedical research. Funding for Exome Sequencing Project (ESP) was provided by NHLBI grants RC2 HL-103010 (HeartGO), RC2 HL-102923 (LungGO) and RC2 HL-102924 (WHISP). Exome sequencing was performed through NHLBI grants RC2 HL-102925 (BroadGO) and RC2 HL-102926 (SeattleGO). Exome sequencing in the ATVB, PROCARDIS, and Ottawa studies was supported by NHGRI 5U54HG003067-11 to ESL and SG. Cleveland Clinic GeneBank was supported by NIH grants P01 HL076491 and P01 HL098055. SK is supported by a Research Scholar award from the Massachusetts General Hospital (MGH), the Howard Goodman Fellowship from MGH, the Donovan Family Foundation, R01HL107816, and a grant from Fondation Leducq. RD is supported by a Banting Fellowship from the Canadian Institutes of Health Research. NOS is supported, in part, by a career development award from the NIH/NHLBI K08HL114642 and by The Foundation for Barnes-Jewish Hospital. GP is supported by award number T32HL007208 from the NHLBI. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHLBI, NHGRI, or NIH. The Italian ATVB Study was supported by a grant from RFPS-2007-3-644382. A full listing of acknowledgements is provided in Supporting Online Material.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature
URLs. database of Genotypes and Phenotypes, dbGAP, http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap; Genome Analysis Toolkit, http://www.broadinstitute.org/gatk; Burrows-Wheeler Aligner, BWA, http://bio-bwa.sourceforge.net; SnpEff, SNP Effect Predictor, http://snpeff.sourceforge.net/; IMPUTE2, http://mathgen.stats.ox.ac.uk/impute/impute_v2.html; PLINK, http://pngu.mgh.harvard.edu/~purcell/plink/; The R Project for Statistical Computing, R, http://www.r-project.org; PLINK/SEQ, http://atgu.mgh.harvard.edu/plinkseq; Meta-Analysis Helper, METAL, http://www.sph.umich.edu/csg/abecasis/metal; SeattleSeq Annotation Server, http://snp.gs.washington.edu/SeattleSeqAnnotation134/; EPACTS, http://genome.sph.umich.edu/wiki/EPACTS
DNA sequences have been deposited with the NIH dbGAP repository under accessions phs000279 and phs000806.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Author Contributions R.D., N.O.S., H.-H.W., A.B.J., and A.K. carried out the primary data analyses. R.D., N.O.S., L.A.L., G.M.P., P.L.A., J.E.R., B.M.P., D.M.H., J.G.W., S.S.R., M.J.B., R.P.T., L.A.C., S.L.H., H.A., J.A.S., S.C., C.S.C., C.K., R.D.J., E.B., G.R.A., D.A.-B., S.M.S., D.S.S, D.A.N, S.R.S., C.J.O, D.Altshuler, S.G., and S.K. contributed to the design and conduct of the discovery exome sequencing study. S.G., D.N.F., and M.A.D. enabled the exome sequencing, variant calling, and annotation. R.D., N.O.S., H.-H.W., A.B.J., S.D., P.A.M., M.F., A.G., S.D., I.G., R.A., D.G., N.M., O.O., R.R., A.F.S., D.S., J.D., S.E.S., S.S., G.K.H., J.J.K., N.J.S., H.S., J.E., S.H.S., W.E.K., C.T.J., R.A.H., O.R., E.H., W.M., M.E.K., J.H., A.D.J., M.L., G.L.B., M.G., Y.L., T.L.A., G.H., E.M.L., A.R.F., H.A.T., M.A.R., P.D., D.J.R., M.P.R., J.H., W.WH.T, A.P.R., D.Ardissino, D.Altshuler, R.M., A.T.-H., H.W., and S.K. contributed to the design and conduct of the imputation-based validation, genotyping-based validation, and/or the re-sequencing based validation study. S.S. supervised the analysis of exome sequencing data and power analysis. R.D., N.O.S., H.-H.W., S.D., P.M., M.F., A.G., R.A., E.S.L., R.M., H.W., D.Ardissino, S.G., and S.K. contributed to the design and conduct of the replication exome sequencing study. S.G., E.S.L., S.K., D.N., and D.Altshuler obtained funding. D.Altshuler, D.N., S.R., R.J., and M.B. comprised the executive committee of the NHLBI Exome Sequencing Project. C.J.O. and S.K. led the Early-Onset Myocardial Infarction study team within the NHLBI Exome Sequencing Project. R.D., N.O.S., H.-H.W. and S.K. wrote the manuscript.
References
- 1.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones DM, et al. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. J Am Med Assoc. 2004;291:2204–2211. doi: 10.1001/jama.291.18.2204. [DOI] [PubMed] [Google Scholar]
- 3.Lehrman MA, et al. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 5.Soria LF, et al. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci USA. 1989;86:587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia CK, et al. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292:1394–1398. doi: 10.1126/science.1060458. [DOI] [PubMed] [Google Scholar]
- 7.Berge KE, et al. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–1775. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- 8.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 9.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 12.Kathiresan S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nature Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nature Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 15.Deloukas P, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nature Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein JL, Schrott HG, Hazzard WR, Bierman EL, Motulsky AG. Hyperlipidemia in coronary heart disease. II. Genetic analysis of lipid levels in 176 families and delineation of a new inherited disorder, combined hyperlipidemia. J Clin Invest. 1973;52:1544–1568. doi: 10.1172/JCI107332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varbo A, et al. Remnant cholesterol as a causal risk factor for ischemic heart disease. J Am Coll Cardiol. 2013;61:427–436. doi: 10.1016/j.jacc.2012.08.1026. [DOI] [PubMed] [Google Scholar]
- 18.Crosby J, et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jorgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 20.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nature Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leigh SE, Foster AH, Whittall RA, Hubbart CS, Humphries SE. Update and analysis of the University College London low density lipoprotein receptor familial hypercholesterolemia database. Ann Hum Genet. 2008;72:485–498. doi: 10.1111/j.1469-1809.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 24.Pennacchio LA, et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 25.De Caterina R, et al. Strong association of the APOA5-1131T>C gene variant and early-onset acute myocardial infarction. Atherosclerosis. 2011;214:397–403. doi: 10.1016/j.atherosclerosis.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Do R, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nature Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollin TI, et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alpert JS, Thygesen K, Antman E, Bassand JP. Myocardial infarction redefined--a consensus document of The Joint European Society of Cardiology/American College of Cardiology Committee for the redefinition of myocardial infarction. J Am Coll Cardiol. 2000;36:959–969. doi: 10.1016/s0735-1097(00)00804-4. [DOI] [PubMed] [Google Scholar]
- 31.Fisher S, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w ( 1118) ; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sunyaev S, et al. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 38.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 41.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 42.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephens M, Sloan JS, Robertson PD, Scheet P, Nickerson DA. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nature Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- 44.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 45.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91:839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nature Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci USA. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.