Summary
Evolutionarily conserved SNARE (Soluble N-ethylmaleimide sensitive factor Attachment protein REceptors) proteins form a complex that drives fusion between membranes in eukaryotes. SNARE complexes are disassembled by the ATPase NSF (N-ethylmaleimide Sensitive Factor), together with SNAP (Soluble NSF Attachment Protein) proteins, making individual SNAREs available for a subsequent round of fusion. Here we report structures of ATP- and ADP-bound NSF, and the NSF/SNAP/SNARE (20S) supercomplex determined by single-particle electron cryomicroscopy at near-atomic to sub-nanometer resolution without imposing symmetry. Large, potentially force-generating, conformational differences exist between ATP- and ADP-bound NSF. The 20S supercomplex exhibits broken symmetry, transitioning from six-fold symmetry of the NSF ATPase domains, to pseudo four-fold symmetry of the SNARE complex. SNAPs are interacting with the SNARE complex with an opposite structural twist, suggesting an unwinding mechanism. The interfaces between NSF, SNAPs, and SNAREs exhibit characteristic electrostatic patterns, suggesting how one NSF/SNAP species can act on many different SNARE complexes.
Introduction
Membrane fusion is essential for many physiological processes in eukaryotic cells, including protein and membrane trafficking, hormone secretion, and neurotransmission1,2. The evolutionarily conserved SNARE (Soluble N-ethylmaleimide sensitive factor Attachment protein REceptors) proteins play a key role in these processes. Specific combinations of SNARE proteins are located on opposite membranes. Upon zippering into a highly stable four-helix bundle—the SNARE complex, they provide the energy for membrane fusion3,4. These combinations of SNARE proteins depend on the source of vesicles and the identity of target membranes, but other factors also contribute to the specificity of the membrane targeting5. To maintain the pool of individual SNARE proteins, the ATPase NSF (N-ethylmaleimide Sensitive Factor), together with SNAP (Soluble NSF Attachment Protein), together with the adaptor protein SNAP (Soluble NSF Attachment Protein), disassembles post-fusion and non-productive SNARE complex into individual protein using the energy from ATP hydrolysis6.
NSF was the first protein found to play a key role in eukaryotic trafficking7,8. NSF is a member of AAA+ (ATPases Associated with diverse cellular Activities) superfamily of ATPases9. NSF forms a homomeric hexamer with a molecular weight of ~500 kDa, with each protomer consisting of an N-terminal domain (termed N) and two ATPase domains (termed D1 and D2) (Fig. 1a). The D1 domains are responsible for the majority of the ATPase activity of NSF whereas the D2 domains are primarily responsible for hexamerization10. The N domains are involved in SNAP and, possibly, SNARE binding11. Prior to ATP hydrolysis, NSF, SNAP, and SNARE complex form the so-called 20S supercomplex6.
Figure 1. 3D maps of ATP- and ADP-bound NSF.
a, Domain diagram of the NSF protomer. b, Different views of the sharpened map (6.5 σ) of ATP-bound NSF filtered to a resolution of 4.2 Å with each domain color coded to match the domain diagram in panel a. A single chain of NSF (protomer) is colored in gold to help with visualization. The density of one D1 domain (subsequently referred to as the D1 domain in Chain F) is not well resolved (see black arrow). c, Different views of the unsharpened map (1.8 σ) of ATP-bound NSF showing the positions of the N domains. d, Different views of the sharpened map (6.8 σ) of ADP-bound NSF filtered to a resolution of 7.6 Å with each domain color coded to match the domain diagram in panel a. A single chain of NSF (protomer) is colored in gold to help with visualization. e, Different views of unsharpened map (1.3 σ) of ADP-bound NSF showing the positions of the N domains. The gap in the D1 ring is indicated by a black arrow.
Individual components of 20S supercomplex have been structurally characterized, including the crystal structures of several SNARE complexes3,12–16, SNAPs17,18, and the D2 and N domains of NSF19–22. Structural studies of full-length NSF and the 20S supercomplex have also been carried out using electron microscopy (EM) including quick-freeze/deep-etch, negative-staining and cryo-EM23–26. However, due to the low resolution limits of these studies, the detailed molecular architecture of the 20S supercomplex is unknown and critical questions remain to be answered: how the adaptor protein SNAP recognizes SNARE complexes, and how many SNAPs are involved; how one NSF/SNAP species disassembles many different SNARE complexes in a promiscuous fashion; and what is the molecular mechanism of disassembly?
Here we present the structures of full-length NSF in two different nucleotide states (ATP- and ADP-bound, at 4.2 Å and 7.6 Å resolution, respectively), and structures of two different 20S supercomplexes involving different SNARE substrates determined by single particle cryo-EM, ranging from 7.6 to 8.4 Å resolution. The EM structures revealed large conformational differences of NSF between ATP- and ADP-bound states, and upon binding to SNAPs and SNAREs. We confirmed by site-directed mutagenesis that the molecular interfaces between SNAPs, SNAREs, and NSF play important roles in disassembly, and propose that recognition at these interfaces is based on characteristic electrostatic patterns. Based on these new insights we speculate about molecular mechanisms of NSF-mediated SNARE complex disassembly.
ATP- and ADP-bound NSF structures
We developed a new purification scheme to address the heterogeneity of NSF samples caused by mixtures of nucleotide states. In essence, hexameric NSF was monomerized by completely removing the bound nucleotides through size exclusion chromatography (Methods and Extended Data Fig. 1a, b). The resulting NSF protomers could be reassembled into hexamers in the presence of desired nucleotide. The reassembled ATP- and ADP-bound NSF hexamers were studied by single particle cryo-EM; EDTA was included to prevent hydrolysis. Our reassembled NSF hexamers are functionally active (Extended Data Fig. 1g and Methods).
The reconstruction of ATP-bound NSF is shown in Figure 1b, c and Extended data Figure 2. The reconstruction has an estimated overall resolution27 of 4.2 Å after masking out flexible N domains (Extended Data Fig. 2e and Extended Data Table 1). All D2 domains, and five out of six D1 domains were well resolved in the final 3D density map (Fig. 1b and Extended Data Figs. 2d and 3a–c). Consistent with the estimated resolution, we observed grooves in α-helices, β-strands within β-sheets, and backbone zigzags corresponding to ~3.8 Å Cα distances, along with densities of some aromatic side chains (Extended Data Fig. 3a, b). The crystal structure of the ATP-bound NSF D2 hexamer19 was readily docked into the corresponding EM density, followed by refinement. The densities of the D1 domains were of sufficient quality to build and refine a de novo atomic model of the D1 domain with bound ATPs. The D1 domain is a typical AAA+ module with two subdomains (α and α/β) and motifs that are generally found in ATPases (Extended Data Fig. 3d, e). The α2 helix of the D1 domain is bent (Extended Data Fig. 3d), a distinctive feature compared to the straight α2 helices found in the D2 domain of NSF as well as in both D1 and D2 domains of the closest related relative, the AAA+ ATPase valosin containing protein (VCP/p97)28–30.
The 3D reconstruction of ADP-bound NSF is shown in Figure 1d, e. The overall estimated resolution27 is 7.6 Å (Extended Data Fig. 4 and Extended Data Table 1), with well-resolved tubular densities for α-helices (Fig. 1d ). To obtain an atomic model of ADP-bound NSF, we docked our EM structure of the D1 domain protomer (obtained from ATP-bound NSF) and the crystal structure of the D2 domain hexamer19 into the corresponding EM densities, followed by refinement.
The cryo-EM datasets of the NSF particles used in this study were of sufficient quality to determine and refine 3D reconstructions to high resolution without imposing any symmetry, which turned out to be critical (Extended Data Fig. 5, see Supplementary Information for a detailed discussion).
Asymmetric features of ATP- and ADP-bound NSF
Both the ATP- and ADP-bound structures of NSF are organized into three layers: two rings consisting of six D2 domains and six D1 domains, respectively, and a layer of six (four) N domains for ATP (ADP)-bound NSF (Figs. 1c, e, and 2a, b). For ADP-bound NSF, the remaining two N domains are flipped along the sides of the ATPase rings with well resolved densities compared to the N domains atop the D1 ring, leaving little doubt as regards the identity of these two densities (Fig. 1e and Extended Data Figs. 4c, e and 7c).
Figure 2. Structures of ATP- and ADP-bound NSF.
a, Side-view of ATP-bound NSF. b, Side-view of ADP-bound NSF. The six protomer chains are rainbow colored counterclockwise based on the relative positions of the D1 domains to the D2 ring in the ATP-bound NSF model; the chain with the closest distance between D1 and D2 domains is named Chain A (red). Nucleotides are shown as grey surfaces. See Methods for generation and refinement of the atomic models. c, A schematic diagram showing the topology of ATPase rings of ATP- and ADP-bound NSF, respectively. D1 rings are colored according to the models shown in panel a and b.
For ATP-bound NSF, the D2 ring is planar and approximately six-fold symmetric. The D1 ring is reminiscent of a right-handed “split washer”, with each chain stepping up about 5 Å as manifested by the relative positions of the α2 helix in the D1 domains (Fig. 2c and Extended Data Fig. 6a). Chain F (purple) is an exception, which does not step up relative to Chain E (blue), but instead slightly steps down towards Chain A (red); there is a large step down from Chain F to Chain A. The D1 domain of Chain F was not as well resolved in the density map as the other D1 domains (Fig. 1b), indicating its potential flexibility (Supplementary Video 1). However, the density for this domain is clear enough to indicate that the α subdomain has a different position relative to the α/β subdomain, compared to the other five D1 domains that can all be well superposed (Extended Data Fig. 6c). Densities for ATP molecules are clearly visible in the nucleotide binding pockets of the D1 domains of Chains A through E, and in all D2 domains (Extended Data Fig. 3f, g); however, there is no clear density in the nucleotide binding pocket of the D1 domain of Chain F.
For ADP-bound NSF, the D2 ring slightly deviates from a near perfect six-fold symmetric conformation, producing a small gap (Extended Data Fig. 7a). The D1 ring is more expanded and planar compared to ATP-bound NSF (Extended Data Figs. 6b, 7b), with a large opening between Chains A and F which coincides with the small gap in the D2 ring, i.e., it forms an open “flat washer” (Fig. 2c). The structures of all six D1 domains can be well superposed, but adopt different orientations relative to the D2 domains (Extended Data Fig. 6b, d).
When superposing the α/β subdomains of all the D1 domains of both ATP- and ADP-bound NSF (except for the flexible Chain F in ATP-bound NSF), the α7 helix in the α subdomain is translated between the ATP and ADP-bound states (Extended Data Fig. 6e). The nucleotides are likely absent in the D1 ring of ADP-bound NSF since the conformations would not favor binding of nucleotides because of possible clashes between the nucleotide and the translated α7 helix (Extended Data Fig. 6e). This conformational change of the α subdomain is correlated with the large difference of the D1 ring between the ATP- and ADP-bound states. The ATP loaded D1 ring is more compact, with a total interface area of 5938 Å2 compared to that of 3746 Å2 in the ADP-bound state, resembling a spring-like transition from a “loaded” split-washer state to a “relaxed” open-flat-washer state (Extended Data Fig. 7b and Supplementary Video 2). This transition is further correlated with outward rotations of the D1 domains of Chains A and B and the changes in their N domains (Extended Data Figs. 6b and 7b, c). For a detailed comparison of our NSF structures with other members of the AAA+ family, see Supplementary Information.
Structures of the 20S supercomplex
We prepared 20S supercomplex consisting of AMPPNP-bound hexameric NSF, αSNAP, and neuronal SNARE complex. The neuronal SNARE complex consists of Syntaxin-1A, Synaptobrevin-2/VAMP-2 (vesicle-associated membrane protein 2), and SNAP-25 (synaptosomal-associated protein 25) (Methods and Extended Data Fig. 1c–f). We used a truncated neuronal SNARE complex (green shaded fragments in Fig. 3a), identical to the one of which the high-resolution structure had been determined12; it was chosen because it remains monomeric in solution even at high concentration, and is sufficient for reversible assembly and disassembly (Extended Data Fig. 1g). 3D classification of the 20S supercomplex produced four different reconstructions that each represents an asymmetric molecular state of 20S, referred to as states I, II, IIIa and IIIb (see Methods). The corresponding refined maps of the 20S supercomplex (without symmetry) have an overall resolution ranging from 7.6 to 8.4 Å after gold-standard refinement by RELION (Extended Data Fig. 8f)27,31.
Figure 3. 3D map and structure of state I of the 20S supercomplex.
a, Domain diagrams of the 20S supercomplex consisting of NSF, αSNAP, Synaptobrevin-2 (Syb-2), Syntaxin-1A, and SNAP-25. The truncated neuronal SNARE complex consists of four SNARE domains (green) from Synaptobrevin-2, Syntaxin-1A and SNAP-25 (two SNARE domains). The boundaries of domains and lengths of proteins are indicated above the domain diagrams. Boundaries of the fragments of the truncated neuronal SNARE complex are indicated below the diagrams. b, Different views of the sharpened EM map (4.8 σ) of state I of the 20S supercomplex filtered to a resolution of 7.6 Å, with each domain color coded to match the domain diagram in a. c, Three-dimensional model of state I of the 20S supercomplex. Each model is in the same orientation as in b.
The structures of the four states of the 20S supercomplex have the same overall architecture, so we discuss state I as a representative of all states in the following; detailed differences between the states are discussed later. The 20S supercomplex resembles a tower with different domains organized into layers (Fig. 3b). At the base of the tower (in the orientation shown in Fig. 3b, middle panel) are the D2 and D1 ATPase rings of NSF, and at the top of the tower is a “spire”, made up of four αSNAP molecules and one SNARE complex, surrounded by the six N domains of NSF. The 20S supercomplex is a striking example of broken symmetry: The approximate six-fold symmetry at the base of the complex is progressively violated in the D1 and N domains, allowing the complex to transit from six-fold symmetry to a pseudo four-fold symmetry at the top (Fig. 3b and Extended Data Fig. 8e).
The four α-helix bundle of the SNARE complex at the center of the spire is clearly visible along with its characteristic twisted left-handed grooves, although the chemical identity of each polypeptide chain cannot be uniquely assigned at the available resolution (Fig. 3b and Extended Data Fig. 3l). Although the densities of the six N domains of NSF are not as well resolved compared to the other components of the supercomplex, they are significantly better defined than those of ATP- and ADP-bound NSF alone. For about half of the six N domains, the density has a characteristic kidney shape as expected from the crystal structure of the N-domain (Extended Data Fig. 3j)21,22. To obtain an atomic model of the entire 20S supercomplex, we docked the crystal structures of the D2 and N domain19, 21, our cryo-EM structure of the D1 domain (Fig. 2a), a homology model of αSNAP derived from the crystal structure of yeast homologue Sec1717, and the crystal structure of the truncated neuronal SNARE complex12 into the EM map. Real-space rigid body minimization was carried out, followed by reciprocal space refinement as described in Methods. The resulting model fit the density well (Fig. 3c, Extended Data Fig. 3h–l, and Supplementary Video 3).
ATPase rings are tightened upon SNAP/SNARE binding
When comparing the ATPase domains of the AMPPNP-bound 20S supercomplex to those of ATP-bound NSF, there are similar overall features, along with some important differences. The overall root-mean-square-deviation (RMSD) of the main chain atoms of D1 and D2 ATPase rings is 4 Å, based on a superposition of the D1 ring. The D2 ring of 20S supercomplex is approximately six-fold symmetric, whereas the D1 ring has a split-washer-like arrangement, similar to that of ATP-bound NSF (Extended Data Fig. 7d). Overall, the ATPase rings of the 20S supercomplex adopt a tighter conformation than NSF alone (Extended Data Fig. 7e). While there is a uniform increase in interface areas for the D2 ring, the changes are more complex for the D1 ring (Extended Data Fig. 7f). These interface area changes are correlated with the conformational differences of the α subdomain of Chain A and the α/β subdomain of Chain F (see Extended Data Fig. 3d for definition of these subdomains), as illustrated by the relative positions of α7 helices in the two structures (Extended Data Fig. 7f inset). Overall, the ATPase rings of NSF are tightened upon binding to αSNAPs and SNARE complex, resembling a spring being loaded (Supplementary Video 4).
Four states of the 20S supercomplex
While the D1/D2 ATPase rings of NSF are very similar among the four states, the αSNAP-SNARE spire and the N domains differ (Fig. 4a). The four states were grouped into three observed “patterns” (I, II, and III) based on the mode of interaction between αSNAP molecules and N domains (Fig. 4b). Considering that each αSNAP can interact with either one or two nearby N domains, one expects a total of nine theoretical patterns taking into account the “split-washer” asymmetry of the D1 ring (see Supplementary Discussion). However, only three patterns consistently emerged in the 3D classification. One explanation for this phenomenon is that the position of the αSNAP-SNARE spire is not random, which might favor certain patterns over the others. Indeed, the center of the spire—the SNARE complex—is always located close to Chains E and F, which are at the raised edge of the D1 split washer (arrows in Fig. 4b), indicating possible interactions between the pore loops (YVG motif)32 of D1 domains and the SNARE complex. For pattern III, two subclasses were refined separately, resulting in states IIIa and IIIb. The main difference between the two states involves the relative position of the spires, not the pattern of αSNAP and N domain interactions (Supplementary Video 5). Thus, the 20S supercomplex exhibits four major states, which are mainly characterized by the patterns of the N domains and the position of the αSNAP-SNARE spire, whereas the conformations of the spire and base themselves do not differ much.
Figure 4. Top views of the four states of 20S supercomplex.
The identified four states were aligned with respect to the D1 rings. a, Sharpened EM maps (state I: 4.7 σ, state II: 4.5 σ, state IIIa: 4.2 σ, state IIIb: 4.0 σ). b, Schematic drawings to help with visualization. The N domains and αSNAP molecules are shown as spheres and squares, respectively. The D1 rings are rainbow colored using the same scheme as in Fig. 2, with black arrows indicating the split between Chain A and Chain F. The D2 rings are omitted for clarity. Each N domain is labeled with its corresponding chain identifier. The numbers of particles that contributed to the reconstruction of each state are listed.
Multi-modal interactions between N domains and αSNAP
The structures of the four states of 20S reveal eight instances of αSNAP molecules that are interacting with two N domains of NSF, and another eight instances of αSNAP molecules that are interacting with one N domain (Fig. 4). When superposing αSNAP molecules separately based on the 1:2 and 1:1 binding scenarios, two distinct N-domain binding sites on the surface of the C-terminal region of αSNAP appear in the case of 1:2 binding, whereas in the case of 1:1 binding, the N domains bind somewhere between the two sites (Fig. 5a). The electrostatic potential surface of the C-terminal region of αSNAP is quite negative, and both distinct binding sites are located in this negatively charged area (Fig. 5b). The N domains of NSF interact with either of the two sites on αSNAP with the same positively charged area (Fig. 5c). The two interfaces involve five positively charged residues of the N domain of NSF and eight negatively charged residues of the C-terminal region of αSNAP (Fig. 5d).
Figure 5. Interactions between αSNAPs and N domains.
a, Superposition of αSNAP molecules and N domains from the structures of the four states of 20S (Fig. 4). In eight cases one αSNAP interacts with two N domains (left). In the other eight cases one αSNAP interacts with one N domain (right). The superposition was performed with respect to the αSNAP molecules. The two distinct binding sites are highlighted by dotted boxes. b, Electrostatic potential surface of αSNAP. The two binding sites of the N domains are highlighted. c, Electrostatic potential surface of the N domain. Only the surface area interacting with αSNAP molecules is shown. d, Ribbon representation of αSNAP and N domain interactions. The SNARE complex is shown to help with visualization. Side chains of charged residues involved in the interactions are shown, with Asp and Glu colored in red and Arg and Lys colored in blue. e, Kinetic curves of the fluorescence dequenching-based SNARE complex disassembly assay using wildtype αSNAP and specified αSNAP mutants. f, Corresponding initial SNARE complex disassembly rates.
Previous mutagenesis studies suggested that certain positively charged residues of the N domains are important for αSNAP and SNARE binding33, and that the C-terminal region of αSNAP is important for 20S formation34, so our structures of the 20S supercomplex now provide the molecular explanation for these previous results. To further validate our observed interfaces with a functional assay, we performed mutagenesis of αSNAP based on our 20S structures, and used a fluorescence dequenching-based assay to monitor the kinetics of SNARE complex disassembly35. Indeed, αSNAP mutants of either Site I (D217A/E249K/E252K/E253K) or Site II (ΔC) showed impaired kinetics of SNARE complex disassembly as well as slower initial reaction rates (Fig. 5e, f). Simultaneous mutations of both sites resulted in completely inactive NSF/SNAP.
Promiscuous interactions between αSNAP and SNARE complex
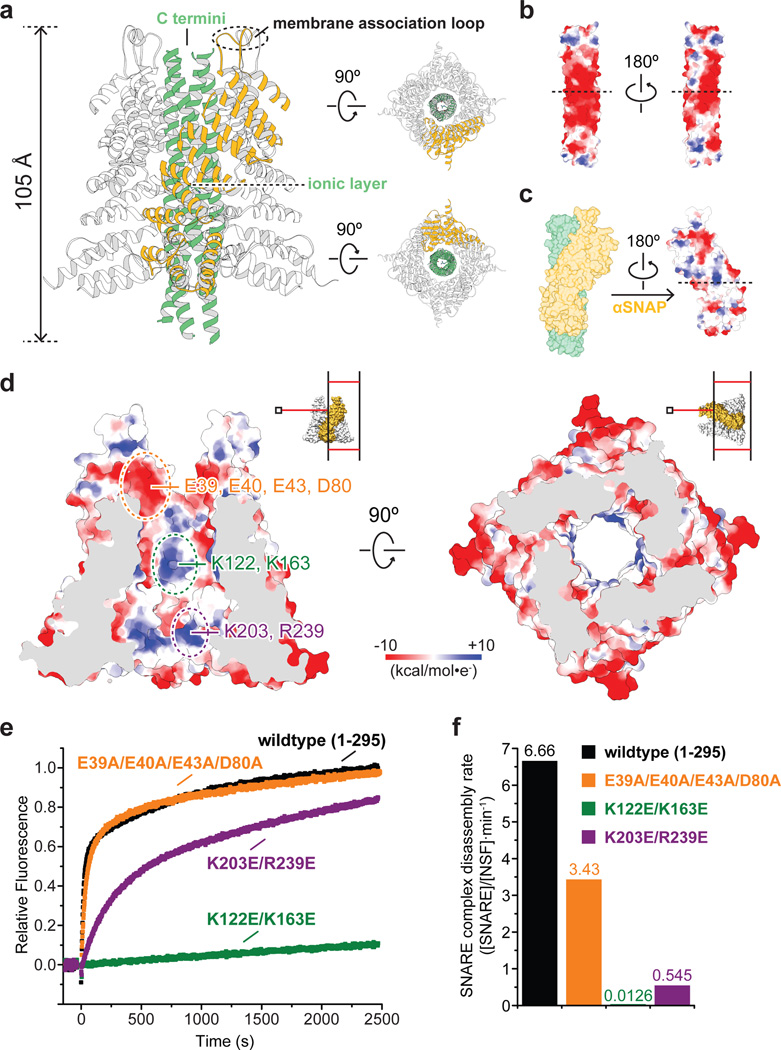
In the structures of the 20S supercomplex, four αSNAP molecules wrap around the SNARE complex in a four-fold rotational symmetric arrangement, despite the fact that the SNARE complex itself is only pseudo-symmetric, i.e. the four α-helices consist of different peptide chains (Fig. 6a). Even more remarkably, the αSNAP “barrel” has a right-handed twist in contrast to the left-handed twist of the four α-helix bundle of the SNARE complex.
Figure 6. Interactions between αSNAP molecules and SNARE complex.
a, A ribbon representation of the αSNAP-SNARE subcomplex for state I of 20S supercomplex. b, Electrostatic potential surface of the SNARE complex. Dotted lines indicate the location of the ionic layer3,40 at the center of the SNARE complex. c, Electrostatic potential surface of αSNAP. Only the surface area interacting with the SNARE complex is shown. The dotted lines indicate the location of the ionic layer of the SNARE complex. d, Cross sections of the electrostatic potential surface of the αSNAP barrel. Three regions that may interact with the SNARE complex are highlighted and labeled. The locations of the cross sections are illustrated in the insets. e, Kinetic curves of the fluorescence dequenching-based SNARE complex disassembly assay using wildtype αSNAP and specified αSNAP mutants that are potentially defect in SNARE interactions. f, Corresponding initial SNARE complex disassembly rates.
We infer that the C termini of the SNARE complex are facing upward since they must accommodate the transmembrane α-helices of the SNAREs Syntaxin-1A and Synaptobrevin-2 (transmembrane domains were not included in the constructs used). This inference is consistent with the orientations of αSNAPs, the hydrophobic loops of which are pointing up for possible membrane association36 (Fig. 6a). Our structures provide a molecular explanation for the previous finding that the trans-SNARE complex before membrane fusion is resistant to NSF disassembly37: the αSNAP/SNARE subcomplex would not be able to form since the trans-SNARE complex is likely in a half-zippered state38.
The electrostatic potential surface of the SNARE complex has a highly conserved pattern with negative charges at the center (Fig. 6b)39. The interacting surface of αSNAP has two extruding positively charged residues (K122, K163) close to the central ionic layer (zero layer)3, 40 of the SNARE complex, and another two close to the C-terminal region of αSNAP (K203, R239) (Fig. 6c, d). Previous mutagenesis studies suggested the importance of K122, K163, and K20341. To further test the functional importance of these possible interactions, we mutated these two groups of residues, as well as a conserved concave negative patch close to the N terminus of αSNAP (E39, E40, E43, D80), and performed the SNARE complex disassembly assay. Remarkably, the K122E/K163E mutant was completely inactive and the K203E/R239E mutant showed impaired kinetics (Fig. 6e, f). The negative patch mutant E38A/E40A/E43A/D80A affected kinetics to a lesser degree, slightly decreasing initial reaction rates using SNARE constructs used in this study; the presence of a membrane may make this interaction between SNAPs and SNAREs more important36.
Variable αSNAP stoichiometry
To investigate how NSF disassembles a different SNARE complex, we prepared a SNARE complex consisting of VAMP-7 (vesicle-associated membrane protein 7), Syntaxin-1A and SNAP-25 (referred to as the V7-SNARE complex); this complex includes the N-terminal Habc domain of Syntaxin-1A and the N-terminal Longin domain of VAMP-7 (Fig. 7a). We assembled the supercomplex with NSF and αSNAP (referred to as the V7-20S supercomplex, Extended Data Fig. 9a–c). This complex is functionally active since the V7-SNARE complex is disassembled upon hydrolysis39. We determined a cryo-EM reconstruction to an estimated resolution of 8 Å without imposing any symmetry (Extended Data Fig. 9d–f). Only two αSNAP molecules bound to the rather inclined SNARE complex bundle (Fig. 7b, c). Note that the two N terminal domains of the V7-SNARE complex and two of the six N domains of NSF were not visible. Although the spire is not as well resolved as in the 20S supercomplex (as indicated by the lack of separation of the four α-helices of the V7-SNARE complex), the EM reconstruction of V7–20S revealed that NSF can use fewer αSNAP molecules and readjust the N domains when binding to different SNARE complexes. While the supercoil axis of the truncated neuronal SNARE complex in the 20S structures is approximately perpendicular to the plane of the ATPase rings, in the V7-20S structure the V7-SNARE complex is angled at 76 degrees relative to the plane of ATPase rings (Fig. 7b, d). Despite these differences, the mode (e.g., right-handed twist) by which αSNAP interacts with SNARE complex and the location of the SNARE complex atop the D1 ATPase ring in V7-20S are similar to those of 20S (compare Fig. 3c and Fig. 7c).
Figure 7. 3D map and structure of V7-20S supercomplex.
a, Domain diagram of V7-SNARE complex. Transmembrane domains were not included in the complex (grey). The two N-terminal domains of VAMP-7 and Syntaxin-1A are highlighted in red boxes. b, Different views of the 3D map (4.5 σ) colored similarly to Fig. 3b. The angle between the long axis of V7-SNARE complex and the plane of ATPase rings is approximately 76° as shown in the inset. c, Corresponding views of the atomic model fit to the EM density map of V7-20S. d, An illustration of the top view similar to Fig. 4b. The location of the N termini of the V7-SNARE α-helix bundle is indicated by a green dotted circle. Note that each of the two aSNAP molecules interacts with two NSF N domains.
To further confirm the stoichiometry of the αSNAP-SNARE subcomplexes in solution, we conducted composition-gradient multi-angle light scattering (CG-MALS) experiments using the two different SNARE complexes mixed with αSNAP in a composition gradient, respectively (Extended Data Fig. 10a, b). The CG-MALS data analysis revealed that αSNAP binds to the truncated neuronal SNARE complex at a maximum ratio of 4:1, whereas it binds to V7-SNARE complex at a maximum ratio of 2:1 (Extended Data Fig. 10c–d). In solution, multiple species of the αSNAP-SNARE subcomplex are in equilibrium, but the EM structures of both 20S and V7-20S suggest that NSF catches the saturated complex in both cases (Extended Data Fig. 10e, f).
NSF mediated SNARE complex disassembly
Using our cryo-EM structures, we propose a working model of NSF mediated SNARE complex disassembly (Fig. 8). Starting with cis-SNARE complex (i.e., with both transmembrane domains in the same membrane), the first step (Fig. 8a, b) is the binding of SNAP molecules. Our EM structures suggest that depending on the SNARE complex, up to four SNAP molecules are present. A stoichiometry higher than 4:1 is unlikely, though, due to packing considerations. Dozens of SNARE proteins exist in an eukaryote, but there are only a few SNAP and very few NSF species5,18,39,42. Based on our 20S structures we propose that one NSF/SNAP species can disassemble all SNARE complexes (including yeast SNAREs14,43) using shape and characteristic electrostatic pattern recognition of SNARE complexes by SNAPs, rather than specific “lock into key” interactions (Fig. 6).
Figure 8. Models of NSF-mediated SNARE complex disassembly.
The model consists of four stages (a–d). The model refers to the neuronal SNARE complex (consisting of Synaptobrevin-2 (Syb2), Syntaxin-1A (Stx1A), and SNAP-25) and αSNAPs, but the model is also applicable to other SNARE complexes, along with a different number of SNAP molecules as observed in V7-20S (see Supplementary Discussion for SNAP species and stoichiometry). The N domains and chain F of the D1 domains in panel b and four of the N domains in panel d are blurred to indicate flexibility as suggested by Cryo-EM density maps.
The second step of our model (Fig. 8b, c) is the binding of NSF, i.e., the formation of 20S supercomplex. Upon binding to the SNAP-SNARE subcomplex, which acts like a fastener, both NSF ATPase rings are tightened, akin to a loaded spring (Extended Data Fig. 7d–f and Supplementary Video 4). The N domains are immobilized due to the interactions with SNAP molecules; characteristic electrostatic patterns may also play a role in these interactions. The opposing twists of SNAP molecules and the SNARE four α-helix bundle in both the 20S and V7-20S supercomplexes (Fig. 6a), along with the existence of four distinct molecular states (Fig. 4), suggests that the 20S supercomplex exerts a torque to unwind or loosen the SNARE complex while switching between the four states. This step requires ATP hydrolysis to initiate the movement of the NSF N domains, and subsequent force transmission via SNAPs. A second possibility is that the four states represent independent binding modes: each would apply a force on its own upon binding to the SNAP-SNARE subcomplex to unwind or loosen the SNARE complex.
The final step of our model (Fig. 8c, d) is the hydrolysis of multiple ATP molecules. We observed large conformational differences of NSF between the ATP- and ADP-bound states (Figs. 1e and Extended Data Fig. 7a–c), suggesting large motions of NSF upon ATP-hydrolysis. The changes of the D1 ring involve a 20 Å vertical movement of the pore loops (Extended Data Fig. 6a, b), which may apply a shear force to the SNARE complex; the opening of the D1 ring from a split washer to a open flat washer, together with a motion of the N domains towards the sides of the ATPase rings could apply a pulling force. Taken together, these forces completely disassemble SNARE complex into individual SNARE proteins. Interestingly, exactly two N domains are flipped in ADP-bound NSF (Chains A and B). These two chains are at the lower edge of the D1 split washer (Fig. 2c). During the transition towards the ADP-bound flat ring, the D1 domains of Chains A and B rotate outwards (Supplementary Video 4 and Extended Data Fig. 6b), so they might be in a position favored for the motion of the N domains. Also in our 20S structure, two N domains bind to one SNAP, and together they may be able to exert a larger force compared to 1:1 binding. We speculate that state II is a likely precursor of ADP-bound NSF. Finally, the opening of the D1 ring in ADP-bound NSF may serve as an exit for individual SNARE proteins since a pore translocation mechanism is unlikely (see Supplementary Discussion). Our model predicts that the release of individual SNARE proteins and SNAPs would initiate nucleotide exchange and restart the cycle.
Methods
Protein expression and purification
Chinese hamster NSF with a tobacco etch virus (TEV) protease cleavable N-terminal His-tag was expressed from pPROEX-1 vector in E. coli. BL21(DE3)-RIL cells (Agilent Technologies) at 25 °C overnight using autoinducing LB medium44. After collecting the cells by centrifugation, they were resuspended in Lysis Buffer (50 mM TrisCl, pH 8.0, 300 mM NaCl, 50 mM imidazole, and 0.5 mM TCEP), and were subjected to sonication and centrifugation. The cleared lysate was loaded onto a HisTrap column (GE Healthcare), and washed with Lysis Buffer. NSF was eluted using Elution Buffer (Lysis Buffer supplemented with 350 mM imidazole). The fresh elution was pooled, concentrated, and supplemented with 1 mM EDTA, 1mM ATP, and 10% glycerol immediately to prevent aggregation and precipitation. The concentrated protein was immediately loaded onto a Superdex 200 16/60 column (GE Healthcare) that was pre-equilibrated with SEC Buffer (50 mM TrisCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM ATP, 1mM DTT, and 10% glycerol). Fractions containing hexameric NSF was separated from aggregated NSF eluted from the void volume. Concentrated hexameric NSF was loaded onto a Superdex 200 16/60 column that was pre-equilibrated with Monomerization Buffer (50 mM sodium phosphate, pH 8.0, 150 mM NaCl, 0.5 mM TCEP). Depending on the amount of proteins, this step needed to be repeated for 3–4 times until all the NSF proteins were eluted as monomers (Extended Data Fig. 1a, b). Monomerized NSF fractions were pooled and supplemented with TEV protease, and incubated overnight at 4 °C. Tag-cleaved NSF was run through a HisTrap column to remove the TEV protease and the cleaved tags. Note that our method differs from that previously reported, which used Apyrase to monomerize NSF24. Monomerized NSF was frozen and stored at −80 °C for future use. To reassemble the hexameric NSF in a specific nucleotide state, e.g., ATP or ADP, monomerized NSF was dialyzed at 4 °C overnight in Reassembly Buffer (50 mM TrisCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM nucleotide, 1mM DTT, and 10% glycerol). The concentrated dialysate was loaded onto a Superdex 200 16/60 column that was pre-equilibrated with Reassembly Buffer for a final clearance. For cryo-EM sample vitrification, the final SEC buffer did not contain glycerol, and the protein samples were concentrated to ~15 mg/mL. The functional activity of purified NSF was tested by a gel-based disassembly assay (Extended Data Fig. 1g) and by a fluorescence-dequenching based SNARE-complex disassembly assay described below. Note that some of the classical disassembly assays in the field were performed with NSF and nucleotide initially in the absence of Mg2+, and then the reaction was triggered by the addition of Mg2+, for example reference 36, establishing that NSF is active when initially prepared in complex with nucleotide in the absence of Mg2+.
Rat αSNAP was expressed and purified as described before35.
Truncated neuronal SNARE complex containing rat SNAP-25 (7–83), SNAP-25 (141–204), Syntaxin-1A (191–256), and His-tagged Synaptobrevin-2 (28–89) was cloned in the Duet expression system (Novagen). The complex was expressed in E. coli. BL21-DE3 cells at 30 °C overnight using autoinducing LB medium44. After collecting the cells by centrifugation, they were resuspended in Lysis Buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 50 mM imidazole, and 0.5 mM TCEP), and were subjected to sonication and centrifugation. The cleared lysate was loaded onto a HisTrap column (GE Healthcare), and washed with Lysis Buffer, Urea Buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 50 mM imidazole, 7.5 M urea, and 0.5 mM TCEP) and Wash Buffer (Lysis Buffer supplemented with additional 10 mM imidazole) sequentially. The SNARE complex was eluted using Elution Buffer (Lysis Buffer supplemented with 350 mM imidazole). The fresh elution was pooled and dialyzed in Dialysis Buffer (50 mM sodium phosphate, pH 8.0, 50 mM NaCl, 3 M urea, and 0.5 mM TCEP) at 4 °C overnight for anion exchange chromatography. The anion exchange chromatography was performed in buffers containing 3 M urea (Buffer A: 50 mM sodium phosphate, pH 8.0, 50 mM NaCl, 3 M urea, and 0.5 mM TCEP; Buffer B: 50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 3 M urea, and 0.5 mM TCEP) using a linear gradient of NaCl. The peak fractions were pooled, concentrated, and loaded onto a Superdex 75 16/60 column (GE Healthcare) that was pre-equilibrated with SEC Buffer (50mM TrisCl, pH 8.0, 150 mM NaCl, 0.5 mM TCEP). The peak fractions were pooled and supplemented with TEV protease, and incubated overnight at 4 °C. The tag-cleaved complex was subjected to a second round of anion exchange chromatography (Buffer A: 50 mM TrisCl, pH 8.0, 50 mM NaCl, and 0.5 mM TCEP; Buffer B: 50 mM TrisCl, pH 8.0, 500 mM NaCl, and 0.5 mM TCEP), and further purified by a final size exclusion chromatography in the SEC Buffer. The purified truncated neuronal SNARE complex was tested for disassembly activity using a SDS-PAGE gel based assay as previously described (Extended Data Fig. 1g)35,45.
V7-SNARE complex containing rat full-length SNAP-25 (1–206), Syntaxin-1A (1–265), and His-tagged VAMP-7 (1–190) was cloned and expressed similarly to the truncated neuronal SNARE complex. However, the first anion exchange chromatography step for the truncated neuronal SNARE complex was omitted since the V7-SNARE complex showed less tendency to precipitate during purification. The other purification steps were the same as for the truncated neuronal SNARE complex.
To assemble the 20S supercomplex, hexameric NSF loaded with AMPPNP was mixed with αSNAP and truncated neuronal SNARE complex in a 1:10:2 mole ratio and incubated on ice for 30 minutes. The mixture was then concentrated and purified by size exclusion chromatography using a Superdex 200 10/300 column (GE Healthcare) pre-equilibrated with 20S Buffer (50mM TrisCl, pH 8.0, 150 mM NaCl, 1 mM AMPPNP, 1 mM EDTA, and 1 mM DTT) (Extended Data Fig. 1c–f). The resulting peak fractions containing the 20S supercomplex were pooled and concentrated to a final concentration of ~15 mg/mL for the cryo-EM studies. The assembly and purification protocol for V7-20S supercomplex were essentially the same, except that hexameric NSF loaded with ATP was used.
Sample vitrification
Initial attempts to image the reassembled NSF and 20S supercomplex by cryo-EM were hindered by preferential orientations. 2D class averages showed that most of the particles were in end-on views, which is insufficient for structure determination (Extended Data Fig. 8a inset). When the samples were deposited to a thin layer of carbon on top of the holey carbon grid, many side views were observed under FEI Tecnai TF20 operated at 200 kV. However, the contrast of the particles was significantly weaker. To ensure a sufficient number of side-view particles suspended in vitreous ice, samples were incubated in a buffer containing a small amount of detergent before plunge freezing. Multiple detergents were screened, and particles in Nonidet P-40 or its substitutes displayed the most side views. The final protein solution contained 0.05% of Nonidet P-40. However, this buffer condition dramatically reduces the particle density in the hole (~100 fold). In order to achieve a reasonable particle distribution, we concentrated the protein sample to approximately 15 mg/mL. This concentration exceeds the usual requirement for cryo-EM. However, even at such high concentration, we observed few particles in areas where ice thickness was considered to be thin and ideal for cryo-EM. Therefore, images were collected from holes with ice that was considered to be thick by general cryo-EM consensus. Quantifoil Cu R1.2/1.3 grids (Quantifoil Micro Tools GmbH, Germany) were washed in chloroform for one hour and air-dried overnight. Aliquots of 2.5 µL samples were loaded onto the grids. Because the buffer contained detergent, the protein solution spread relatively well over the grid surface without glow discharge. Grids were blotted for 3 to 4 s and plunge frozen in liquid ethane cooled by liquid nitrogen using a FEI Vitrobot (FEI Company).
Cryo-EM data collection
Grids were transferred to TF30 Polara equipped with a field emission source and operated at 300kV. Images were recorded on a Gatan K2 Summit direct electron detector operated in super-resolution counting mode following the established dose fractionation data acquisition protocol46. Images were recorded at a nominal magnification of 31000×, corresponding to a calibrated super resolution pixel size of 0.61 Å on the specimen. The dose rate on the detector was set to be ~8.2 counts (corresponding to ~10.9 electrons) per physical pixel per second. At this setting, the coincidence loss is about 11.5% and the total loss, including the loss due to imperfect detector quantum efficiency, is about 25%47. The total exposure time was 6 seconds, leading to a total accumulated dose of 44 e−/Å2 on the specimen. Each image was fractionated into 30 frames, each with an accumulation time of 0.2 s. Dose-fractionated images were recorded using a semi-automated acquisition program UCSFImage4 (written by Xueming Li). Defocus values ranged from −1.8 to −2.8 µm.
Image processing
Super-resolution counting images were 2 × 2 binned by Fourier cropping for motion correction, resulting in a pixel size of 1.22 Å. Motion corrected frames were summed to a single micrograph for subsequent processing46. Defocus values were determined for each micrograph using CTFFIND348. A semi-automated procedure similar to a previous work was used to pick particles49. Briefly, for each dataset, ~2,000 particles were manually picked to calculate 2D class averages. Unique 2D class averages were selected as templates for automated particle picking. Picked particles were visually inspected. Obviously falsely picked particles were removed. 2D classification was performed using RELION and SPIDER50. Initial 3D models were generated using the common lines method51. 3D classification and gold-standard refinement were performed using RELION27. The initial model from the common lines method was low-pass filtered to 60 Å and used as the starting model for the initial auto-refinement, which generated a consensus model. This consensus model was again filtered to 60 Å and used for 3D classification. We used prior knowledge of NSF to distinguish “good” classes from “bad” classes: 3D class averages that showed incorrect features of NSF were considered as bad, including the wrong numbers of apparent D1 and/or D2 domains or a seriously deteriorated D2 ring. All good class averages had the correct number (six) of all the domains and well-defined D2 ring density.
For 20S, good 3D class averages showed the correct number of all the domains, and well-defined densities of the αSNAP-SNARE spire and D2 ATPase ring. Particles in distinct good 3D classes were refined separately, yielding four final reconstructions (Fig. 4a). The four final reconstructions belong to three “patterns” (I, II, and III) based on the mode of interactions between αSNAP and N domains. When we tested different settings of the 3D classification procedure27, other patterns were either rarely populated or showed deteriorated features in some domains, preventing refinement of the pattern to a reasonable resolution. Therefore, these other patterns may represent minor populations if they actually exist. Note that when we sub-classified pattern I or II, we also observed multiple states, but the differences between them were much smaller than those between states IIIa and IIIb. Therefore we did not refine those subclasses individually. The observed conformational heterogeneity may have prevented crystallization of the 20S supercomplex despite extensive efforts by many laboratories.
No symmetry was assumed throughout the entire process for all NSF and 20S reconstructions (except for demonstrating the detrimental effect of including symmetry, Extended Data Fig. 5, see also Supplementary Discussion). The solvent area of raw particles was set at zero for 3D classification (zero_mask=true). The solvent area was filled with random noise for auto-refinement (zero_mask=false). The result from auto-refinement was slightly worse when zero_mask was set to true. For ATP-bound NSF sample, continued refinement using the same particles but summed from frame #2 to #18 (the first frame is #1) improved the resolution from 4.4 Å to 4.2 Å. As recommended by the RELION documentation, “relion_postprocess” was used to generate a soft mask, which was applied to the two half maps before FSC was calculated. The resolution reported in Extended Data Table 1 was estimated from the masking-effect-corrected FSC curve using FSC = 0.143 criterion52. B-factor sharpening was carried out by “relion_postprocess” implementing an automated B-factor fitting algorithm53. The local resolution was estimated using ResMap on unsharpened and unfiltered maps54. Detailed information for each final reconstruction is summarized in Extended Data Table 1.
Model building and refinement
The two half-maps of ATP-bound NSF were summed to a single unsharpened map. The summed map was then sharpened manually using a series of negative B-factors by XMIPP55. The sharpened maps were used for model building in COOT56. The crystal structure of the D2 domain of NSF (PDB accession code: 1nsf)19 in complex with ATP-Mg2+ was first docked as the hexamer observed in the crystal structure into the density map as a single rigid body. Minor adjustments of the backbone and side chains of the D2 domain were manually performed using COOT. The nucleotide conformation observed in the crystal structure matched the EM density (Extended Data Fig. 3f), demonstrating that the absence of Mg2+ in the assembly of ATP-bound NSF did not affect the conformation of the nucleotide. A homology model of the D1 domain protomer generated by SWISS-MODEL57 based on the crystal structure of N-D1 domain of p97 (PDB accession code: 1e32)58 was initially placed into the best of the six copies of D1 densities. However, the detailed fit to the observed densities of the D1 domains was relatively poor and it required complete retracing the main chain and side chains in order to obtain a better fit (starting from the structure of the D2 domain of p97 as the homology model did not produce a better initial fit to the density map and it would have still required complete retracing). The rebuilt D1 domain was then docked into the other five copies of D1 density. Densities for ATP were visible in 11 out of the 12 possible nucleotide binding pockets (no clear density for nucleotide was seen in the D1 domain of Chain F), and models of ATP were fit to these densities. Rigid body minimization was carried out for the α/β and α subdomains of each D1 domain separately using COOT, followed by manual adjustment, residue by residue. To complete the model, the crystal structure of NSF N domain (PDB accession code: 1qcs)21 was docked into the corresponding densities of the unsharpened map without any fitting; the quality of these densities was good enough to determine the approximate positions of the N domains. The partial model containing the D1 and D2 domains was subjected to reciprocal space refinement. Amplitudes of the summed map were corrected by frequency-dependent scaling factor determined by comparing the experimental maps with a reference map calculated from the full model59,60. A soft-edged mask was generated based on the built atomic model (including only the D1 and D2 domains) and applied to the scaled map. Most solvent regions and the density corresponding to N domains were masked out. The masked maps were put into an artificial unit cell with P1 symmetry and converted to MTZ format using CCP4 program sftools61. The resulting reflection files were used to perform maximum likelihood refinement using PHENIX62 with secondary structure restraints, reference model restraints, and automatic optimization of experimental/stereochemistry weights. The reference models were generated from the built models using geometry minimization function in PHENIX. The refined D1 and D2 domains were combined with the docked N domains to produce a complete model.
To model ADP-bound NSF, the crystal structure of the D2 domain and the refined EM structure of the D1 domain (from ATP-bound NSF) were docked into the sharpened map. Rigid body minimization was carried out for the α/β and α subdomains of each ATPase domain (6 D1 and 6 D2) separately using COOT. Densities for ADP were clearly visible for four nucleotide binding sites of the D2 domains (except those in Chains A and F). The resulting model was refined using PHENIX as described above for the ATP-bound structure. The model was completed by docking the N domains into the unsharpened map, without fitting and further refinement; the quality of these densities was good enough to determine the approximate positions of the N domains.
For 20S supercomplex structure, the model of ATP-bound NSF containing the D1 and D2 domains was docked into each of the four sharpened maps corresponding to the four states of 20S (Fig. 4). Rigid body minimization was carried out for the α/β and α subdomains of each ATPase domain (6 D1 and 6 D2) separately using COOT56. The crystal structure of the NSF N-domain (PDB accession code: 1qcs)21 were docked into the peripheral densities. The SNARE complex was easily identified and modeled using the crystal structure of truncated neuronal SNARE complex (PDB accession code: 1n7s)12. A homology model of αSNAP generated by SWISS-MODEL57 based on the crystal structure of yeast Sec17 (PDB accession code: 1qqe)17 was docked into the remaining densities surrounding the SNARE complex. The resulting models were refined in reciprocal space using PHENIX62 as described above.
The fitting of the V7-20S supercomplex was performed similarly, except that only two αSNAPs and four out of six N domains were modeled based on the observed densities in the EM map. The crystal structure of the closely-related truncated neuronal SNARE complex12 was used for model building since there is no crystal structure of the V7-SNARE complex available. The Habc domain of Syntaxin-1A and the Longin domain of VAMP-7 were not visible. The model of V7-20S supercomplex was not further refined in reciprocal space.
For cross-validation of overfitting, we followed the procedures published before63. Briefly, the coordinates of refined ATP-bound NSF model (only D1 and D2 domains) were displaced randomly by 0.1 Å using PHENIX (PDB Tools) to remove potential model bias. The displaced model was then refined against one of the half maps in reciprocal space. FSC curves were calculated between the resulting model and half map 1 (“work”, i.e., used for refinement), the resulting model and half map 2 (“free”, i.e., not used for refinement), and the resulting model and the summed map. The lack of separation between “work” and “free” FSC curves suggested that the model was not overfitted.
MolProbity64 was used for evaluating the geometry of the models. The model statistics of the refined models are summarized in Extended Data Table 2 (note that R values are arbitrary since they depend on the exact definition of the P1 cell, so they are not provided in the table). Molecular graphics and analyses were performed with either PyMOL (The PyMOL Molecular Graphics System, Version 1.7.0.5 Schrödinger, LLC.) or UCSF Chimera package65. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). All maps presented were B-factor sharpened and filtered by RELION using the B-factors listed in Extended Data Table 1, unless otherwise mentioned.
Flourescence dequenching-based SNARE complex disassembly assay
The details of the assay were published previously35. Briefly, soluble rat neuronal SNARE complex containing Syntaxin-1A (1–265, S249C, K253C), full-length SNAP-25 (1–206), and Synaptobrevin-2 (1–96), was labeled with Oregon Green (forming covalent linkages with two cysteine residues close to the C-terminus of Syntaxin-1A, and four native cysteine residues of SNAP-25). The disassembly reactions were carried out using FlexStation II (Molecular Devices) in a 384-well plate with a reaction volume of 60 µL. Each condition (different αSNAP mutants) was divided into four replicas, and the average was plotted. A final concentration of 400 nM Oregon Green-labeled SNARE complex, 2 µM αSNAP, and 85 nM NSF was included in the reaction buffered with 50 mM TrisCl, pH 8.0, 20 mM NaCl, 2 mM ATP, 2 mM MgCl2, and 0.5 mM TCEP. To measure the initial SNARE complex disassembly rate, the first 20 data points after adding the NSF were used for linear regression analysis, except for αSNAP mutants D217A/E249K/E252K/E253K/ΔC, and K122E/K163E, for which the first 1000 data points were used because the slope was close to zero. Note that the disassembly rate of wild type proteins (Fig. 5f) is comparable to that of previous experiments35,39, illustrating that our purification method of NSF produces fully active protein.
Composition-gradient multi-angle light scattering (CG-MALS)
The experiments were performed in Tris buffered saline (TBS; 17 mM Tris, 50 mM NaCl, pH 8.0) with or without 0.1 mM TCEP. After dilution to the appropriate stock concentration, proteins were filtered using a syringe-top filter (0.02 µm, Anotop, Whatman). CG-MALS experiments were performed with a Calypso II composition gradient system (Wyatt Technology Corporation) to prepare different compositions of protein and buffer, and deliver them to an online UV/Vis detector (Waters Corporation) and DAWN HELEOS II multi-angle light scattering detector (Wyatt). Inline filter membranes with 0.03 µm pore size were installed in the Calypso for additional sample and buffer filtration. An automated CG-MALS method was performed, consisting of single component concentration gradients for each species to quantify any self-association and a dual-component “crossover” gradient to assess the interaction between αSNAP and each SNARE complex (Extended Data Fig. 10a). For each composition, The Calypso system prepared an aliquot of protein solution, injected it into the detectors and stopped the flow for 60–240 seconds to allow the reaction to come to equilibrium within the MALS detector flow cell. Equilibrium light scattering and concentration data were fit to an appropriate association model using the CALYPSO software (Wyatt).
Extended Data
Extended Data Figure 1. Purification of recombinant NSF bound to specific nucleotide and 20S supercomplex.
a, Schematic diagram of the purification steps of NSF. Chromatography columns and buffer conditions are provided. b, Size exclusion chromatograms corresponding to the colored steps in panel a. Major peaks are labeled. c, A scheme showing the purification steps of 20S supercomplex. d, Size exclusion chromatogram of the 20S supercomplex. Major peaks are labeled. e, SDS-PAGE gel of fractions collected in panel d. The samples were not boiled. f, SDS-PAGE gel of the same fractions as in e. The samples were boiled. g, SDS-PAGE-based SNARE disassembly assay of the truncated neuronal SNARE complex. This complex is stable in SDS without boiling. Disassembly by NSF/αSNAP is observed as a function of time.
Extended Data Figure 2. 3D reconstruction of ATP-bound NSF by single-particle cryo-EM.
a, A representative electron micrograph (out of 1150 micrographs) of ATP-bound NSF particles in vitreous ice. b, Selected 2D class averages (6 out of 50). c, Plots of the angular distribution of particle projections. The radius of the sphere at each projection direction is proportional to the number of particle images assigned to it. Two alternative views are shown: with the Z-axis pointing out or the Y-axis pointing out. Two corresponding re-projection images of the final map are shown under the plots. d, Selected slice views of the final reconstruction. The slice numbers are indicated. e, FSC curves for the 3D map after RELION post-processing. The resolution is estimated to be 4.2 Å by the gold-standard refinement criterion, as indicated by the red arrow. The FSC curve between the refined atomic model and the 3D map is shown in blue. f, FSC curves for cross-validation. Black, FSC model versus summed map (full dataset); green, FSC model versus half map 1 (used for test refinement); orange, model versus half map 2 (not used for test refinement). See methods for details. g, 3D map colored according to the local resolution as estimated by ResMap.
Extended Data Figure 3. Representative densities from the EM reconstructions of ATP-bound NSF (a–g) and the 20S supercomplex (h–l).
Maps shown in panels a–f were sharpened by XMIPP using a B-factor of −123Å2. a, Representative density (black mesh, 7.8 σ) for an α-helix and a β-strand of the D1 domain with the refined model (colored sticks) superposed. b, Representative density (black mesh, 7.0 σ) of the β-sheet of the α/β subdomain of D1 (Chain C) with the refined model (yellow Cα ribbon) superposed. c, Density (black mesh, 7.0 σ) and model (yellow Cα ribbon) for the D1 and D2 domains of chain B. Note that the linker between the two domains is well resolved. d, De novo model (yellow cartoon) of the D1 domain built from the cryo-EM map (black mesh, 7.0 σ). The arrangement of the subdomains and nucleotide is illustrated in the inset. The pore loop (YVG motif) and two α-helices: α2 from the α/β subdomain, and α7 from the α subdomain are highlighted in the red dotted boxes. e, Density (black mesh, 6.5 σ) and model (yellow Cα ribbon) of the ATP binding pocket of the D1 domain (Chain C). Motifs that are typical for AAA+ ATPase are indicated. f, Superposition of the crystal structure of ATP-bound D2 domain (PDB accession code: 1nsf, colored sticks and balls) with Mg2+,19 and the EM map (black mesh, 7.6 σ) of ATP-bound NSF (density of Chain C). The crystal structure was docked into the density as a rigid body without any refinement. Note that no Mg2+ was present in the EM samples, but the ATP molecule and the protein coordinates from the crystal structure match the EM density well. g, Nucleotide binding sites of the D1 domains from ATP-bound NSF. The density (translucent surface, Chain A–E: 8.2 σ, Chain F: 5.0 σ) of each D1 domain is shown together with the built model in ribbon representation. The nucleotide binding pockets are highlighted by dotted circles. Five out of the six D1 domains show clear density for ATP. h–l, Representative density (translucent surface, 4.7 σ) from reconstruction for state I of 20S supercomplex with the model (cartoon) superposed. All densities are representative except for the N domain in panel j, which represents the better-resolved half of the N domain densities (12 out of 24 cases).
Extended Data Figure 4. 3D reconstruction of ADP-bound NSF by single-particle cryo-EM.
a, A representative electron micrograph (out of 840 micrographs) of ADP-bound NSF particles in vitreous ice. b, Selected 2D class averages (6 out of 30). c, Selected focused 2D class averages (5 out of 10). The first image shows the focused classification mask, which locates the flipped N domains. d, Plots of angular distribution of particle projections. The radius of the sphere at each projection direction is proportional to the number of particle images assigned to it. Two alternative views are shown: with the Z-axis pointing out towards the viewer or the Y-axis pointing out. Two corresponding re-projection images of the final map are shown under the plots. e, Selected slice views of the final reconstruction. Slice numbers are indicated. f, FSC curve for the 3D map after RELION post-processing. The resolution is estimated to be 7.6 Å by the gold-standard refinement criterion. g, 3D map colored according to the local resolution as estimated by ResMap.
Extended Data Figure 5. Detrimental effect of imposing C6 symmetry on the EM reconstruction of NSF and C3 symmetry on the EM reconstruction of the 20S supercomplex.
a, For the ADP-bound NSF maps, in order to visualize densities of the N domains, an unsharpened map is displayed (translucent surface, C1: 1.2 σ, C6: 0.6 σ) together with the sharpened map using no symmetry (C1) or C6 symmetry during reconstruction (colored surface, C1: 5.9 σ, C6: 7.0 σ). For the reconstruction that uses C6 symmetry, symmetric densities for the N domains at top and side positions appear in the unsharpened map, however, these densities cannot be matched to the crystal structure of the N domain. Likewise, the D1 domains appear compressed and cannot be fit well using the structure of the D1 domain that we obtained by asymmetric reconstruction. b, Reconstruction of state I of the 20S supercomplex without symmetry (C1) or with C3 symmetry. Maps are shown in colored surfaces similar to Fig. 3 (C1: 4.7 σ, C3: 4.9 σ). The C3 averaging causes the D1 domains to display alternating up and down positions. The density for the SNARE complex is a featureless rod without the characteristic left-handed twist of the four α-helix bundle. Densities for only three SNAPs emerge, but without any interpretable features (e.g. grooves between helices), preventing a match with the crystal structure of the known homolog of αSNAP, Sec17. The N domain densities are weak and none of them exhibit the expected kidney shape.
Extended Data Figure 6. Comparison of AAA+ ATPase domains from ATP- and ADP-bound NSF structures.
a, Unrolling of the ATPase domains of ATP-bound NSF. Two orthogonal views are shown. Individual chains are aligned based on the D2 domains (white) to show the split-washer arrangement of the D1 domains.b, Unrolling of the ATPase domains of ADP-bound NSF. Individual chains are aligned as in panel a. Dotted boxes in panel a and b highlight the α2 helices of the D1 domains in order to help with visualization of the relative positions. The six protomer chains are rainbow colored as in Fig. 2. c, Overlay of the six D1 domains of ATP-bound NSF, using the α/β subdomains (white) for the superposition. The relative positions of α7 helices from α subdomains are illustrated in the inset. d, Corresponding superposition of the ADP-bound NSF D1 domains. e, Overlay of the five D1 domains (without Chain F) of ATP-bound NSF (grey), and six D1 domains of ADP-bound NSF (white) using α/β subdomains for the superposition. The α7 helices from the α subdomains are highlighted by red dotted boxes. The relative translation of the α7 helices between the ATP-bound state and the ADP-bound state is shown in the inset.
Extended Data Figure 7. Comparison of ATP- and ADP-bound NSF structures (a–c), and ATPase domains of ATP-bound NSF and 20S supercomplex (d–f).
a–c, Surface representations of the D2, D1 and N domains of ATP- and ADP-bound NSF (looking down from the N side of the NSF hexamer). The maximum diameters of the D2 and D1 rings, and the interface areas (calculated by PISA66) between ATPase domains are indicated. Each protomer chain is colored as in Fig. 2. The D1 ring is also shown in panel c and colored white to help with visualization. d–f, The ATPase domains of the structure of the 20S supercomplex (state I) were superposed on the ATP-bound NSF using the D1 ring as the reference for the fit. Six protomer chains from ATP-bound NSF are rainbow colored counterclockwise from the top based on the relative positions of the D1 domains to the D2 ring. The ATPase domains of the 20S supercomplex are colored in white and grey. Note that the density of Chain F in the EM reconstruction of ATP-bound NSF alone is poorly resolved (Fig. 1b), whereas in the 20S reconstruction it is well defined, although the overall resolution of the 20S reconstruction is lower. d, Side views. e, Top view of the D2 rings. Each individual D2 domain is labeled. Percentages of interface area change (from NSF to 20S) between the D2 domains are provided in the figure. The interface areas between the D2 domains are similar in the NSF and 20S structures, except for a significant increase (12%) between Chains D and E for 20S compared to NSF alone. f, Top view of the D1 rings. Each D1 domain is labeled, with the split between Chains A and F indicated by a black arrow. The translation of the α7 helix in α subdomain of Chain A is illustrated in the inset. Percentages of interface area change (from NSF to 20S) between the D1 domains are shown. Three stay the same; the one between Chains A and B decreases, whereas those between Chains E and F, and Chains F and A increase significantly.
Extended Data Figure 8. 3D reconstruction of 20S supercomplex by single-particle cryo-EM.
a, A representative electron micrograph (out of 610 micrographs) of the 20S supercomplex in its original purification buffer recorded using the TF20 microscope and the TVIPS TemCam-F816 CMOS camera. The inset shows selected 2D class averages (5 out of 50). b, A representative electron micrograph (out of 2459 micrographs) of 20S supercomplex in the buffer containing additional 0.05% Nonidet P-40 recorded using the TF30 Polara microscope and the Gatan K2 Summit detector. c–g are results from this imaging condition. c, Selected 2D class averages (6 out of 50). d, Plots of angular distribution of particle projections. The radius of the sphere at each projection direction is proportional to the number of particle images assigned to it. Two alternative views are shown, one has the Z-axis pointing out, the other has the Y-axis pointing out. Two corresponding re-projection images of the final map are shown under the plots. e, Selected slice views of the final reconstruction. Slice numbers are indicated. Slices from different layers are framed in different colors: SNAREs and αSNAPs: yellow, N domains: pink, D1 ring: blue, and D2 ring: purple. f, FSC curves for the 3D maps of the four states after RELION post-processing. The estimated resolution ranges from 7.6 Å to 8.4 Å as estimated by the gold-standard refinement criterion. g, 3D map colored using local resolution estimated by ResMap. The right panel shows a cut-through view of the interior of the map. c–e and g are results from a subclass representing state I.
Extended Data Figure 9. Purification and 3D reconstruction of V7-20S supercomplex.
a, Size exclusion chromatogram of the V7-20S supercomplex. Major peaks are labeled. Only fraction 10 was concentrated and used for single-particle cryo-EM. b, SDS-PAGE gel of fractions collected in panel a. The samples were not boiled. c, SDS-PAGE gel of the same fractions as in panel b. The samples were boiled. d, A representative electron micrograph (out of 993 micrographs) of the 20S supercomplex containing V7-SNARE complex. e, Selected 2D class averages (6 out of 50). f, FSC curves for the 3D maps after RELION post-processing. The estimated resolution is 8.0 Å as estimated by the gold-standard refinement criterion.
Extended Data Figure 10. CG-MALS characterization of αSNAP-SNARE subcomplex.
a, Concentration gradient setup for the experiment that measures the binding between αSNAP and truncated neuronal SNARE complex. b, Measured molar mass for different components. Note that there were two independent runs for αSNAP over the specified concentration ranges. c, Measured molecular mass of αSNAP-SNARE (truncated) subcomplex converted from light scattering over the concentration gradient. The experimental data are represented by blue dots. Simulated curves with different αSNAP:SNARE (truncated) stoichiometry are shown. The best fit is 4:1. d, Measured molecular mass of the αSNAP-V7-SNARE subcomplex calculated from light scattering over the concentration gradient. The experimental data are represented by blue dots. Simulated curves with different αSNAP:V7-SNARE stoichiometry are shown. The best fit is 2:1. e, Calculated mole fractions of different αSNAP-SNARE (truncated) species over the concentration gradient based on 4:1 stoichiometry. f, Calculated mole fractions of different αSNAP-V7-SNARE species over the concentration gradient based on 2:1 stoichiometry.
Extended Data Table 1.
3D reconstructions of NSF and 20S supercomplex by single particle cryo-EM
| NSP (ATP) | NSF (ADP) | 20S | V7-20S | ||||
|---|---|---|---|---|---|---|---|
| Electron microscope | TF30 Polara | TF30 Polara | TF30 Polara | TF30 Polara | |||
| Accelerating Voltage(kV) | 300 | 300 | 300 | 300 | |||
| Defocus range (µm) | −1.8 – −2.8 | −1.8 – −2.8 | −1.8 – −2.8 | −1.8 – −2.8 | |||
| Electron dose (e−/Å2) | 44 (26.4*) | 44 | 44 | 44 | |||
| Pixel size (Å) | 1.2156 | 2.4312 | 2,4312 | 2.4312 | |||
| Particles processed | 89,289 | 30,848 | 116,082 | 65,126 | |||
| Particles refined | 50,781 | 12,830 | State I | State II | State IIIa | State IIIb | 32,100 |
| 29,717 | 21,489 | 15,249 | 14,991 | ||||
| Resolution of unmasked map (Å) | 6.4 | 9.2 | 8.6 | 8.9 | 9.4 | 9.2 | 9.2 |
| Resolution of masked map (Å)† | 4.2 | 7.6 | 7.6 | 7.8 | 8.4 | 8.2 | 8.0 |
| Map sharpening B-factor (Å2)‡ | −129 | −479 | −428 | −601 | −612 | −734 | −395 |
The accumulated dose of the first 18 frames.
Resolution of masked map is estimated from Masking-effect-corrected FSC curves.
B-factor automatically determined by RELION.
Extended Data Table 2.
Statistics of model refinement.
| NSF(ATP) | NSF(ADF) | 20S supercomplex | ||||
|---|---|---|---|---|---|---|
| Model composition | ||||||
| Total atoms | 21,712 | 21,407 | 41,435 | |||
| Peptide chains | 6 | 6 | 14 | |||
| Protein residues | 2,906 | 2,887 | 5,431 | |||
| Refinement | ||||||
| Unit cell(P1) | ||||||
| a=b=c(Å) | 311.2 | 311.2 | 311.2 | |||
| α=β=γ(°) | 90 | 90 | 90 | |||
| Resolution(Å) | 4.2 | 7.6 | state I | state II | state IIIa | state IIIb |
| 7.6 | 7.8 | 8.4 | 8.2 | |||
| R.m.s. deviations | ||||||
| Bond lengths(Å) | 0.008 | 0.011 | 0.009 | 0.009 | 0.009 | 0.009 |
| Bond angels(°) | 1.762 | 1.976 | 1.596 | 1.567 | 1.585 | 1.570 |
| Ramachandran plot | ||||||
| Favored (%) | 93.0 | 92.8 | 89.6 | 88.9 | 89.3 | 89.2 |
| Outliers(%) | 2.0 | 2.2 | 2.3 | 2.5 | 2.3 | 2.3 |
| MolProbity | ||||||
| Overall score | 2.62 | 2.54 | 2.54 | 2.58 | 2.59 | 2.57 |
| Rotamer outliers (%) | 1.8 | 0.8 | 0.9 | 1.0 | 1.1 | 1.1 |
Supplementary Material
Acknowledgements
We thank W.I. Weis for critical discussions and reading of the manuscript; S. Kenrick from Wyatt Technology Corporation for technical help with CG-MALS analysis. This research was supported in part by the National Institutes of Health (R37MH63105 to A.T.B., and a subcontract from Grant #5-U01AI082051-05 to A.T.B.; R01GM082893, R01GM098672 and P50GM082250 to Y.C.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Contributions M.Z., Q.Z., and D.J.C. expressed and purified all protein samples; M.Z. and S.W. collected cryo-EM data; S.W. processed cryo-EM data; M.Z. built and refined the atomic models on the basis of cryo-EM maps; M.Z. performed the disassembly assay; S.V. performed CG-MALS experiment; M.Z., S.W., Y.C., and A.T.B. designed experiments, analyzed data and wrote the manuscript.
Author Information 3D cryo-EM density maps of ATP- and ADP-bound NSF, and the 20S supercomplex have been deposited in the Electron Microscopy Data Bank (EMDB), with accession codes EMD-6204 (NSF-ATP), EMD-6205 (NSF-ADP), EMD-6206 (20S-state I), EMD-6207 (20S-state II), EMD-6208 (20S-state IIIa), EMD-6209 (20S-state IIIb) and EMD-6210 (V7-20S). The coordinates of atomic models of ATP- and ADP-bound NSF, the 20S supercomplex have been deposited in the Protein Data Bank (PDB) under the accession codes , 3j94, 3j95, 3j96, 3j97, 3j98 and 3j99. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Wickner W, Schekman R. Membrane fusion. Nat. Struct. Mol. Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Südhof TC. Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–690. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 4.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 5.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 6.Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 7.Block MR, Glick BS, Wilcox CA, Wieland FT, Rothman JE. Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc. Natl. Acad. Sci. U. S. A. 1988;85:7852–7856. doi: 10.1073/pnas.85.21.7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malhotra V, Orci L, Glick BS, Block MR, Rothman JE. Role of an N-ethylmaleimide-sensitive transport component in promoting fusion of transport vesicles with cisternae of the Golgi stack. Cell. 1988;54:221–227. doi: 10.1016/0092-8674(88)90554-5. [DOI] [PubMed] [Google Scholar]
- 9.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 10.Matveeva E, He P, Whiteheart S. N-Ethylmaleimide-sensitive fusion protein contains high and low affinity ATP-binding sites that are functionally distinct. J. Biol. Chem. 1997;272:26413–26418. doi: 10.1074/jbc.272.42.26413. [DOI] [PubMed] [Google Scholar]
- 11.Whiteheart S, Schraw T, Matveeva E. N-ethylmaleimide sensitive factor (NSF) structure and function. Int. Rev. Cytol. 2001;207:71–112. doi: 10.1016/s0074-7696(01)07003-6. [DOI] [PubMed] [Google Scholar]
- 12.Ernst JA, Brunger AT. High resolution structure, stability, and synaptotagmin binding of a truncated neuronal SNARE complex. J. Biol. Chem. 2003;278:8630–8636. doi: 10.1074/jbc.M211889200. [DOI] [PubMed] [Google Scholar]
- 13.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strop P, Kaiser SE, Vrljic M, Brunger AT. The structure of the yeast plasma membrane SNARE complex reveals destabilizing water-filled cavities. J. Biol. Chem. 2008;283:1113–1119. doi: 10.1074/jbc.M707912200. [DOI] [PubMed] [Google Scholar]
- 15.Antonin W, Fasshauer D, Becker S, Jahn R, Schneider TR. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 2002;9:107–111. doi: 10.1038/nsb746. [DOI] [PubMed] [Google Scholar]
- 16.Zwilling D, et al. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J. 2007;26:9–18. doi: 10.1038/sj.emboj.7601467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice L, Brunger A. Crystal structure of the vesicular transport protein Sec17: implications for SNAP function in SNARE complex disassembly. Mol. Cell. 1999;4:85–95. doi: 10.1016/s1097-2765(00)80190-2. [DOI] [PubMed] [Google Scholar]
- 18.Bitto E, et al. Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family. Proteins. 2008;70:93–104. doi: 10.1002/prot.21468. [DOI] [PubMed] [Google Scholar]
- 19.Yu RC, Hanson PI, Jahn R, Brunger AT. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat. Struct. Biol. 1998;5:803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]
- 20.Lenzen C, Steinmann D, Whiteheart S, Weis W. Crystal Structure of the Hexamerization Domain of N-ethylmaleimide-Sensitive Fusion Protein. Cell. 1998;94:525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- 21.Yu R, Jahn R, Brunger A. NSF N-terminal domain crystal structure: models of NSF function. Mol. Cell. 1999;4:97–107. doi: 10.1016/s1097-2765(00)80191-4. [DOI] [PubMed] [Google Scholar]
- 22.May AP, Misura KM, Whiteheart SW, Weis WI. Crystal structure of the amino-terminal domain of N-ethylmaleimide-sensitive fusion protein. Nat. Cell Biol. 1999;1:175–182. doi: 10.1038/11097. [DOI] [PubMed] [Google Scholar]
- 23.Furst J, Sutton RB, Chen J, Brunger AT, Grigorieff N. Electron cryomicroscopy structure of N-ethyl maleimide sensitive factor at 11 A resolution. EMBO J. 2003;22:4365–4374. doi: 10.1093/emboj/cdg420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang L-F, et al. Structural characterization of full-length NSF and 20S particles. Nat. Struct. Mol. Biol. 2012;19:268–275. doi: 10.1038/nsmb.2237. [DOI] [PubMed] [Google Scholar]
- 25.Moeller A, et al. Nucleotide-dependent conformational changes in the N-Ethylmaleimide Sensitive Factor (NSF) and their potential role in SNARE complex disassembly. J. Struct. Biol. 2012;177:335–343. doi: 10.1016/j.jsb.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanson P, Roth R, Morisaki H, Jahn R, Heuser J. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 27.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 29.Huyton T, et al. The crystal structure of murine p97/VCP at 3.6A. J. Struct. Biol. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 31.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 33.Zhao C, Matveeva EA, Ren Q, Whiteheart SW. Dissecting the N-ethylmaleimide-sensitive factor: required elements of the N and D1 domains. J. Biol. Chem. 2010;285:761–772. doi: 10.1074/jbc.M109.056739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnard RJ, Morgan A, Burgoyne RD. Stimulation of NSF ATPase activity by alpha-SNAP is required for SNARE complex disassembly and exocytosis. J. Cell Biol. 1997;139:875–883. doi: 10.1083/jcb.139.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cipriano DJ, et al. Processive ATP-driven substrate disassembly by the N-ethylmaleimide-sensitive factor (NSF) molecular machine. J. Biol. Chem. 2013;288:23436–2345. doi: 10.1074/jbc.M113.476705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winter U, Chen X, Fasshauer D. A conserved membrane attachment site in alpha-SNAP facilitates N-ethylmaleimide-sensitive factor (NSF)-driven SNARE complex disassembly. J. Biol. Chem. 2009;284:31817–31826. doi: 10.1074/jbc.M109.045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber T. SNAREpins Are Functionally Resistant to Disruption by NSF and alphaSNAP. J. Cell Biol. 2000;149:1063–1072. doi: 10.1083/jcb.149.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zorman S, et al. Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. Elife. 2014:e03348. doi: 10.7554/eLife.03348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vivona S, et al. Disassembly of all SNARE complexes by N-ethylmaleimide-sensitive factor (NSF) is initiated by a conserved 1:1 interaction between α-soluble NSF attachment protein (SNAP) and SNARE complex. J. Biol. Chem. 2013;288:24984–24991. doi: 10.1074/jbc.M113.489807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marz KE, Lauer JM, Hanson PI. Defining the SNARE complex binding surface of alpha-SNAP: implications for SNARE complex disassembly. J. Biol. Chem. 2003;278:27000–27008. doi: 10.1074/jbc.M302003200. [DOI] [PubMed] [Google Scholar]
- 42.Clary DO, Griff IC, Rothman JE. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell. 1990;61:709–721. doi: 10.1016/0092-8674(90)90482-t. [DOI] [PubMed] [Google Scholar]
- 43.Wilson DW, et al. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature. 1989;339:355–359. doi: 10.1038/339355a0. [DOI] [PubMed] [Google Scholar]
References
- 44.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 45.Fasshauer D, Eliason WK, Brunger AT, Jahn R. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 1998;37:10354–10362. doi: 10.1021/bi980542h. [DOI] [PubMed] [Google Scholar]
- 46.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Zheng SQ, Egami K, Agard DA, Cheng Y. Influence of electron dose rate on electron counting images recorded with the K2 camera. J. Struct. Biol. 2013;184:251–260. doi: 10.1016/j.jsb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mindell Ja, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 49.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 51.Shaikh TR, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 54.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheres SHW, Núñez-Ramírez R, Sorzano COS, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–w258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Shaw A, Bates P, Newman R. Structure of the AAA ATPase p97. Mol. Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 59.Penczek P, Ban N, Grassucci RA, Agrawal RK, Frank J. Haloarcula marismortui 50S subunit-complementarity of electron microscopy and X-Ray crystallographic information. J. Struct. Biol. 1999;128:44–50. doi: 10.1006/jsbi.1999.4157. [DOI] [PubMed] [Google Scholar]
- 60.Saad A, et al. Fourier amplitude decay of electron cryomicroscopic images of single particles and effects on structure determination. J. Struct. Biol. 2001;133:32–42. doi: 10.1006/jsbi.2001.4330. [DOI] [PubMed] [Google Scholar]
- 61.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.