Abstract
DNA methyltransferase (DNMT) inhibitor 5-aza-2′-deoxycytidine (5-aza-CdR) is able to cause DNA demethylation in the genome and induce the expression of silenced genes. Whether DNA demethylation can affect the gene expression of stem/progenitor cells has not been understood. Mouse utricle epithelia-derived progenitor cells (MUCs), which possess stem cell features as previously described, exhibit a potential DNA methylation status in the genome. In this study, MUCs were treated with 5-aza-CdR to determine whether DNMT inhibitor is able to induce the differentiation of MUCs. With 5-aza-CdR treatment for 72 hr, MUCs expressed epithelial genes including Cdh1, Krt8, Krt18, and Dsp. Further, hair cell genes Myo7a and Myo6 increased their expressions in response to 5-aza-CdR treatment. The decrease in the global methylated DNA values after 5-aza-CdR treatment indicated a significant DNA demethylation in the genome of MUCs, which may contribute to remarkably increased expression of epithelial genes and hair cell genes. The progenitor MUCs then turned into an epithelial-like hair cell fate with the expression of both epithelial and hair cell genes. This study suggests that stem cell differentiation can be stimulated by DNA demethylation, which may open avenues for studying stem cell fate induction using epigenetic approaches.
Keywords: 5-aza-2′-deoxycytidine, demethylation, epigenetics, epithelial, hair cell, methylation, prosensory cell, stem cell
Introduction
In the mammalian auditory system, inner ear sensory hair cells are the primary receptors for auditory and balance signals. However, mature mammalian hair cells have very limited regeneration ability; therefore, loss of hair cell usually causes permanent hearing loss, balance impairment, and other inner ear disorders in humans, which affects the daily activity of at least 10% of the population. In recent animal studies, hair cell regeneration has obtained promising progresses owing to the advances in stem cell biology [1–6]. Stem cells are considered a likely cell source for hair cell replacement because they possess the ability of self-renewal and pluri/multipltency [2]. In the mammalian inner ear, sphere-forming progenitors have been identified in the cochlea and utricles [7, 8]. In addition, a subpopulation of supporting cells has been reported to transdifferentiate into new hair-like cells [7, 9–11]. We have recently found that adult mouse utricle sensory epithelial cells underwent epithelial-mesenchymal transit to become prosensory-like cells (MUCs) that were able to be maintained in culture for at least 50 passages [12]. We found that the expression of epithelial genes Cdh1 (encoding E-Cadherin) and Krt18 (encoding cytokeratin) was completely shut down in MUCs. In the meantime, MUCs expressed prosensory genes, including Six1, Lfng, Isl1, Bmp4, Dlx5, and Pax2, as well as genes shown in both prosensory cells and supporting cells, including Sox2, P27kip1, Jag1, and Notch1 [9, 10, 13–15], which indicates that MUCs may possess features of prosensory cells [16]. Prosensory cells are considered to be hair cell progenitors because they develop into inner ear epithelial hair cells and supporting cells during normal development [15, 17]. To generate epithelial hair cells from prosensory-like MUCs, one of the important steps in our hair cell regeneration strategy is to induce MUCs to up-regulate the expression of epithelial genes such as Cdh1. However, how to stimulate MUCs to express epithelial genes is still an open question.
In recent studies, epigenetic modification becomes one of the most popular areas to study gene expression, in which the expression and/or phenotype of genes are heritably changed while their DNA sequences remain constant [18–20]. DNA methylation is one of the common approaches that are used to epigenetically silence gene expression. In eukaryotes, the additional methyl group is transferred from S-adenosyl methionine (SAM) to 5′-cytosine catalyzed by DNA methyltransferase (DNMT). There are three functional DNMTs, including DNMT1, DNMT3A, and DNMT3B. DNMT1 takes charge of genome methylation maintenance, and DNMT3A and DNMT3B are responsible for de novo methyltransferase activity [21–25]. Once DNA methylation occurs at the promoter sequence, it interferes the interaction between transcription factors and the promoter, which subsequently inhibits gene transcription [20, 26].
DNA methylation has a reverse reaction called DNA demethylation, which re-activates the expression of genes silenced by DNA methylation [23]. The DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-aza-CdR) is a cytosine analogue that acts as a suicide substrate for DNMTs. When 5-aza-CdR enters the cell, it incorporates into genomic DNA and binds to DNMTs, which irreversibly inhibits the activity of DNMTs and causes passive demethylation [27–29]. The demethylation effect of 5-aza-CdR is genome-wide, and its effect on the promoter allows the newly synthesized DNA being transcribed to mRNA [28, 30]. It has been demonstrated that 5-aza-CdR-induced demethylation is able to stimulate Cdh1 expression in cervical cancer [23], prostate cancer [31], acute leukemia [32], and esophageal cancer [33]. Further, 5-aza-CdR treatment is able to induce global demethylation in normal fibroblast cell line, which stimulates gene expression in both short-term and long-term activation [30]. DNA methylation/demethylation of stem cell has been recent studied [34, 35], but it is still a largely understudied area. For instance, whether DNA demethylation is able to activate epithelial gene expression in stem/progenitor cells has not been determined. In this study, we treated prosensory-like MUCs with 5-aza-CdR to determine whether 5-aza-CdR was able to stimulate MUCs to express epithelial genes such as Cdh1. Success of this study will provide evidence for the generation of epithelial hair cells from prosensory-like MUCs using an epigenetic approach, which may also open avenues for DNA methylation/demethylation research in stem cell biology.
Materials and methods
Cell culture and demethylation treatment
MUCs were cultured in DMEM/F12 GlutaMAX™ with 10% fetal bovine serum (FBS, all from Invitrogen) in an incubator supplied with 5% CO2 at 37°C as previously described [16]. When MUCs reached approximately 80–90% confluence, 5-aza-2′-deoxycytidine (5-aza-CdR, Sigma) was added to culture medium at 1, 2, and 4 μM. In the control group, vehicle (DEME/F12) was applied to MUC cultures. MUCs were observed daily using phase contrast microscopy and digital images were captured using a digital camera. After 48 hr of 5-aza-CdR treatment, half of the culture medium was replaced and MUCs were maintained for another 24 hr. In this study we investigated the effect of 5-aza-CdR on MUC gene expression at 72 hr after treatment, which was determined based on previous publications [30,32, 36].
Viability assay
To determine whether 5-aza-CdR was toxic to MUCs, the viability of MUCs was assessed with calcein and propidium iodide (PI, all from Invitrogen). After 72 hr of 5-aza-CdR treatment, MUCs in the treatment and control groups were incubated in the culture medium containing 2 μM calcein and 0.3 μg/ml PI for 30 min in a CO2 incubator. MUCs were observed using epifluorescence microscopy and photographed using a cooled CCD camera (n=5/group). Analysis of variance (ANOVA) was used for analysis and P<0.05 was determined as the criterion of statistical significance in this study.
Genomic DNA extraction and genomic methylated DNA quantification
After MUCs were treated with 5-aza-CdR or vehicle for 72 hr, genomic DNA (gDNA) was extracted using Flexi DNA Kit (Qiagen) according to the manufacturer’s protocol. Quantification of genomic methylated DNA was examined by MethylFlash Methylated DNA Quantification Kit (Epigentek) using genomic DNAs of both 5-aza-CdR-treated and control MUCs. A methylated DNA positive control was artificially synthesized and provided with the kit by the manufacturer, in which 50% of the DNAs have been methylated at 5-cytosine. A standard curve was generated by diluting this positive control to a series of concentrations at 0, 0.5, 1.0, 2.0, 5.0, and 10.0 μg/ml following the manufacturer’s protocol. The relative fluorescence unite (RFU) was read at 530EX/590EM nm using a Gemini EM fluorescence microplate reader (Molecular Devices). The percentage of 5-methylcytosin (5-mC) was used to quantify the genomic methylation level of each sample. A relative quantification of 5-mC% and the absolute amount of 5-mC were calculated using manufacturers’ methods (n=4). A two-tailed Student’s t-test was used to compare the quantification of 5-mC% in the genome of treatment and control MUCs.
RNA extraction, reverse transcription PCR (RT-PCR), and real time quantitative RT-PCR (quantitative PCR)
After 72 hr of 5-aza-CdR treatment, MUCs from treatment and control groups were harvested and the total RNA was extracted using RNeasy Mini Kit (Qiagen), followed by cDNA synthesis using a QuantiTect Reverse Transcription Kit (Qiagen) according to manufacturers’ protocols. RT-PCR was performed on a thermal cycler (Eppendorf) using GoTaq® Green Master Mix (Promega) with primers listed in Table 1. PCR products were electrophoresed and imaged using a ChemiDoc-It® 2 imaging system (UVP). A Bio-Rad CFX system was applied for quantitative PCR using SsoAdvanced™ SYBR® Green Supermix (Bio-rad) (n=3). The melting temperature (Tm) and efficiency of the primers were evaluated in quantitative PCR analyses. In this study, primers with the efficiency of approximately 90% to 110% and the melting peak of 80–90°C in melting curve analysis were selected for quantitative PCR study. The mean of quantification cycle (Cq) was calculated by Bio-Rad CFX Manager software using a regression mode. The housekeeping gene Gapdh that exhibited approximately the same Cq values (difference ≤ one cycle) in the control and treatment groups were qualified for being used as calibrator references. The relative expression levels of studied genes were delta/delta Cq values normalized with internal control gene Gapdh. In the quantitative analysis, relative gene expression change ≥ 2-fold was considered to be of biomedical importance in this study. A two-tailed Student’s t-test was used to compare the expressions of genes between treatment and control groups. Cq value >40 was considered no gene expression in the sample and was not qualified for delta/delta value or statistical analysis.
Table 1.
Primers used in RT-PCR, quantitative PCR, and Net-MSP
| Gene | Forward: 5′-3′ | Reverse: 5′-3′ | Product length (bp) |
|---|---|---|---|
| Gapdh | GGCCGCATCTTCTTGTGCAGT | TGCAAATGGCAGCCCTGGTGA | 111 |
| Cdh1 | ATTCAAAGTGGCGACAGACGGC | ACCTGGGTACACGCTGGGAAACAT | 223 |
| Krt8 | CAAGGTGGAACTAGAGTCCCG | CTCGTACTGGGCACGAACTTC | 187 |
| Krt18 | ACTCCGCAAGGTGGTAGATGA | TCCACTTCCACAGTCAATCCA | 162 |
| Dsp | AGCCCTTTACAAGGCCATCAGCGT | TGTTCCACTGAACCAGCGTCCACA | 198 |
| Snai1 | ATGCACATCCGAAGCCACACG | TGGAGCAAGGACATGCGGGAGAA | 245 |
| Snai2 | CATCCTTGGGGCGTGTAAGTC | GCCCAGAGAACGTAGAATAGGTC | 186 |
| Zeb1 | ACTGCAAGAAACGGTTTTCCC | GGCGAGGAACACTGAGATGT | 127 |
| Zeb2 | CCACGCAGTGAGCATCGAA | CAGGTGGCAGGTCATTTTCTT | 131 |
| Fn1 | GTGACACTTATGAGCGCCCTA | CCACTTGTCGCCAATCTTGTA | 137 |
| Vim | AAGCCGAAAGCACCCTGCAGTCAT | AGGTCAGGCTTGGAAACGTCCACA | 202 |
| Cdh2 | ATGCCCTGAATGGAATGCTGCGGT | GCTGTGGCTGTGTTTGAAAGGCCA | 211 |
| Hes1 | AGCACAGAAAGTCATCAAAGCC | ATGTCTGCCTTCTCTAGCTTGG | 142 |
| Dlx5 | CACCACCCGTCTCAGGAATC | GCTTTGCCATAAGAAGCAGAGG | 125 |
| Jag1 | CCTCGGGTCAGTTTGAGCTG | CCTTGAGGCACACTTTGAAGTA | 150 |
| P27kip1 | GCGGTGCCTTTAATTGGGTC | TTCGGGGAACCGTCTGAAAC | 197 |
| Isl1 | ATGATGGTGGTTTACAGGCTAAC | TCGATGCTACTTCACTGCCAG | 174 |
| Lfng | GCCGTCAAGACCACCAGAAAG | GGTCATACTCCACAGCCATCTT | 208 |
| Sox2 | GCGGAGTGGAAACTTTTGTCC | CGGGAAGCGTGTACTTATCCTT | 157 |
| Bmp4 | CATGAGGGATCTTTACCGGCTC | TCTCCAGATGTTCTTCGTGATGG | 140 |
| Pax2 | GACGAGCACCACTCTACCTG | GATGGCTGTATGGGTTGCCT | 125 |
| Myo7a | GCACTTCATCATCGGCAACGGCAT | GCTGCTCTTGGATGGGTTGTGTGT | 100 |
| Myo6 | GGCATCGTCCCAAGAGATTTTC | CCACAATGTCAAAGTTCGGTACA | 150 |
| Atoh1 | CCCGTCCTTCAACAACGACAAG | AGGTGATGGTGGTCATTTTTGC | 156 |
| Methylated Cdh1 | GTTTTTAGTTAATTAGCGGCGTC | ACACTAAACTCGAATACGATCGAA | 175 |
| Unmethylated Cdh1 | GTTTTTAGTTAATTAGTGGTGTTGG | CACTAAACTCAAATACAATCAAA | 174 |
Bisulfite conversion reactions and Nested-methylation-specific PCR (Nested-MSP)
Genomic DNA of both 5-aza-CdR-treated and control MUCs were converted using an EpiTect Bisulfite Conversion Kit (Qiagen) according to the manufacturer’s protocol. After conversion reactions, gDNAs were eluted, and the bisulfite-converted gDNA was stored at −20 °C for the following studies. The methylation pattern of epithelial gene Cdh1 was studied using a nested-PCR approach with a MSP Kit (Qiagen) (n=3). The primers for MSP were listed in Table 1. In electrophoresis analysis of the MSP products, the relatively expressions of methylated Cdh1 were normalized by the expressions of unmethylated Cdh1 in the treatment and control groups. The MSP products were imaged using a ChemiDoc-It® 2 imaging system (UVP).
Results
Viability of MUCs following 5-aza-CdR treatment
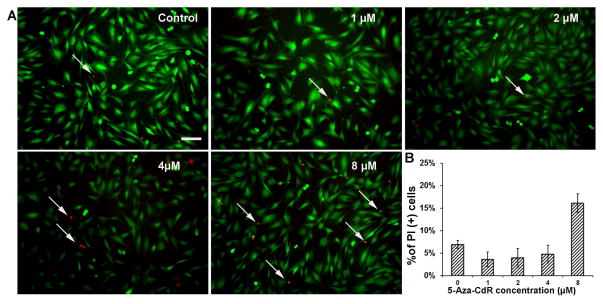
When MUCs were treated with 5-aza-2′-deoxycytidine (5-aza-CdR) for 72 hr, the viable cells were stained with calcein that was shown with green fluorescence (Fig. 1A). Quantitative study revealed that approximately 93.08% ± 0.93%, 96.41% ± 1.62%, 96.06% ± 2.09%, 95.24% ± 1.97%, and 83.89% ± 2.02% of MUCs were labeled by calcein when they were treated with medium containing vehicle (DMEM/F12), 1, 2, 4, and 8 μM 5-aza-CdR respectively (Fig. 1B). ANOVA indicated significant difference in the number of viable cells in these groups (P<0.05). Tukey post hoc test suggested that 8 μM 5-aza-CdR-treated MUCs showed a significantly decreased number of viable MUCs (P<0.05), while there is no significant difference among other groups (P>0.05). The dead cells were indicated by propidium iodide (PI) staining and shown in red fluorescence (Fig. 1A). Approximately 6.92% ± 0.93%, 3.59% ± 1.62%, 3.94% ± 2.09%, 4.76% ± 1.97%, and 16.11% ± 2.02% of MUCs were PI positive when exposed to control medium, 1, 2, 4, and 8 μM 5-aza-CdR respectively (Fig. 1B), which was statistically significant (P<0.05; ANOVA). Tukey post hoc test indicated that a remarkably increased number of dead MUCs were identified in 8 μM 5-aza-CdR group (P<0.05). Overall, we observed that 4 μM 5-aza-CdR did not significantly affect the viability of MUCs; therefore, 4 μM was selected as an optimal 5-aza-CdR concentration in the following studies.
Fig. 1. Viability of MUCs following 5-aza-CdR treatment.
A. Calcein and PI were used to determine the viability of MUCs treated with control medium and medium containing 1–8 μM 5-aza-CdR. The viable and dead MUCs were stained with calcein (green) and PI (red) respectively and imaged using epifluorescence microscopy. Scale bar: 10 μM.
B. The percentage of viable cells in the control and 5-aza-CdR treatment groups. Approximately 93.08% ± 0.93%, 96.41% ± 1.62%, 96.06% ± 2.09%, 95.24% ± 1.97%, 83.89% ± 2.02% of MUCs were labeled by calcein when they were treated with medium containing vehicle, 1, 2, 4, and 8 μM 5-aza-CdR respectively. ANOVA suggested significant difference in the groups (P<0.05). Tukey post hoc test suggested that 8 μM 5-aza-CdR-treated MUCs showed a significantly decreased number of viable MUCs (P<0.05), while there is no significant difference among other groups.
Global methylation level and expression of Dnmt1 of MUCs following 5-aza-CdR treatment
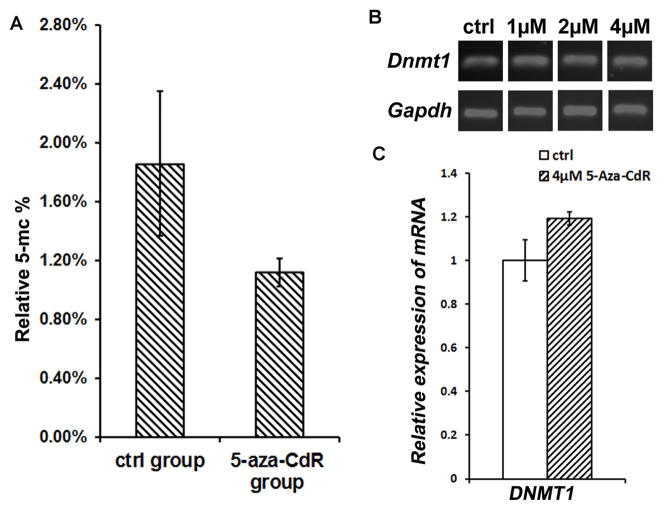
The relative quantification of genome-wide methylated DNA (5-mC%) of control MUCs was 1.31% ± 0.35% (Fig. 2A). After 72 hr of 4 μM 5-aza-CdR treatment, the relative 5-mC% of MUCs decreased to 0.79% ± 0.07% (Fig. 2A), which was statistically significant (P<0.05; Student’s t-test). The absolute amount of 5-mC in control MUCs was 1.84 ± 0.49 ng, while it decreased to 1.13 ± 0.10 ng after 4 μM 5-aza-CdR treatment for 72 hr. Student’s t-test indicated statistically significant difference in the absolute amount of 5-mC between the control and 5-aza-CdR-treated MUCs (P<0.05). These data suggested that 5-aza-CdR treatment caused a significant genome-wide demethylation in prosensory-like MUCs.
Fig. 2. 5-aza-CdR decreased global methylation level of MUCs but did not significantly influence the expression of Dnmt1.
A. 4 μM 5-aza-CdR caused a significant genome-wide demethylation in prosensory-like MUCs. The relative quantification of genome-wide methylated DNA (5-mC%) for control and 4 μM 5-aza-CdR treated MUCs was 1.31% ± 0.35% and 0.79% ± 0.07% respectively. Statistical analysis indicated significant difference in the genome demethylation between the control and 5-aza-CdR-treated MUCs (P<0.05; Student’s t-test).
B. RT-PCR study showed that the expression of DNA methyltransferase DNMT1 did not significantly changed in response to 5-aza-CdR treatment.
C. Quantitative PCR study suggested that the normalized relative gene expression values of Dnmt1 of MUCs in the 5-aza-CdR treatment group was slightly increased (approximately 1.2 fold higher, less than 2 fold; therefore not a significant change).
To determine whether 5-aza-CdR affected the expression of DNA methyltransferase 1 (DNMT1), the maintenance DNMT, we studied the expression of Dnmt1 of MUCs following 5-aza-CdR treatment. Reverse transcription PCR (RT-PCR) suggested that the expression of Dnmt1 in 5-aza-CdR-treated MUCs did not significantly change after 5-aza-CdR treatment (Fig. 2B). In quantitative PCR, the normalized relative expression value of Dnmt1 in 5-aza-CdR treatment group was slightly higher than the control group (approximately 1.2 fold higher, which was less than 2-fold and considered not biomedically significant; Fig. 2C).
Expression of prosensory and hair cell genes following 5-aza-CdR treatment
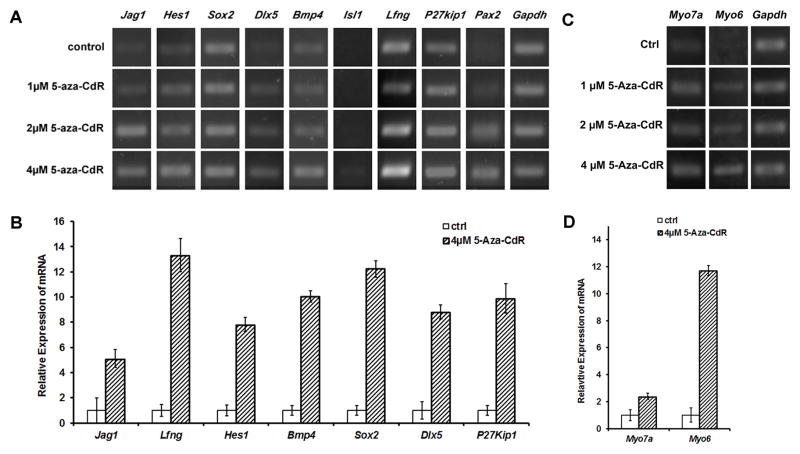
In response to 5-aza-CdR treatment, expressions of prosensory genes, including Jag1, Hes1, Sox2, Dlx5, Bmp4, Lfng, P27kip1, and Pax2, were up-regulated compared to control MUCs in RT-PCR studies (Fig. 3A). Further, 5-aza-CdR exerted concentration-dependent effects on gene up-regulation. For example, 2 and 4 μM 5-aza-CdR treatment seemed to remarkably up-regulate gene expressions than 1 μM 5-aza-CdR on prosensory genes including Jag1, Hes1, Dlx5, Bmp4, and Pax2 (Fig. 3A). Furthermore, quantitative PCR study revealed that normalized relative expression values of Jag1, Hes1, Sox2, Dlx5, Bmp4, Lfng, and P27kip1 of 5-aza-CdR-treated MUCs were 5.12 ± 0.72, 7.81 ± 0.56, 12.25 ± 0.65, 8.81 ± 0.57, 10.04 ± 0.45, 13.32 ± 1.32, 9.89 ± 1.16 fold higher than those of control MUCs respectively (Fig. 3B). Statistical analysis showed significant difference (P<0.05; Student’s t-test).
Fig. 3. MUCs up-regulated the expression of prosensory and hair cell genes following 5-aza-CdR treatment.
A. RT-PCR study showed that the expression of prosensory genes Jag1, Hes1, Sox2, Dlx5, Bmp4, Lfng, P27kip1, and Pax2 was up-regulated following 5-aza-CdR treatment. In addition, 2 and 4 μM 5-aza-CdR seemed to exert remarkable effects on the expression of studied prosensory genes.
B. Quantitative study indicated that normalized relative expression values of Jag1, Hes1, Sox2, Dlx5, Bmp4, Lfng, and P27kip1 of 5-aza-CdR-treated MUCs were 5.12 ± 0.72, 7.81 ± 0.56, 12.25 ± 0.65, 8.81 ± 0.57, 10.04 ± 0.45, 13.32 ± 1.32, and 9.89 ± 1.16 fold higher than those of control MUCs, and statistical analysis showed significant difference (P<0.05; Student’s t-test)
C. RT-PCR revealed that the expressions of hair cell gene Myo7a and Myo6 were up-regulated in 5-aza-CdR-treated MUCs.
D. Quantitative PCR showed that normalized relative gene expression values of Myo7a and Myo6 of MUCs in the 5-aza-CdR group were 2.37 ± 0.24 and 11.71 ± 0.37 fold higher than those of MUCs in the control group. The statistical analysis indicated that the expression level of Myo7a between the treatment and control groups was statistical significant (P<0.05; Student’s t-test).
When MUCs were treated with 5-aza-CdR, expressions of hair cell genes Myo7a and Myo6 were up-regulated in RT-PCR compared to control MUCs. Moreover, 4 μM 5-aza-CdR caused distinctly increased expression of Myo7a (Fig. 3C). In quantitative PCR study, normalized relative expression values of Myo7a and Myo6 in 5-aza-CdR group were 2.37 ± 0.24 and 11.71 ± 0.37 fold higher than those in control group (Fig. 3D). Statistical analysis indicated that the expression level of Myo7a between the treatment and control groups was statistically significant (P<0.05; Student’s t-test).
Up-regulated expressions of epithelial genes and the methylation pattern of Cdh1 following 5-aza-CdR treatment
RT-PCR studies revealed that expressions of epithelial genes, including Cdh1, Krt8, and Krt18 were up-regulated after MUCs were treated with 5-aza-CdR (Fig. 4A). For example, the expression of Cdh1 was not detected in RT-PCR when MUCs were treated with control medium. Following 1 and 2 μM 5-aza-CdR treatment, Cdh1 expression was slightly up-regulated, while the expression of Cdh1 was significantly increased in response to 4 μM 5-aza-CdR treatment (Fig. 4A). Further, quantitative PCR confirmed that the studied epithelial genes Cdh1, Krt8, Krt18, and Dsp showed expression changes along with the concentration of 5-aza-CdR treatment, which suggested that 5-aza-CdR may have a dose-dependent effect on the epithelial gene expression in MUCs. In quantitative PCR, normalized relative expressions of Cdh1 and Krt18 of 4 μM 5-aza-CdR-treated MUCs were 13.64 ± 0.05 and 13.57 ± 3.68 fold higher than those of control MUCs respectively. Statistical analysis revealed significant difference between 5-aza-CdR and control groups (P<0.05; Student’s t-test). Additionally, expression of Krt8 and Dsp was not detected in the control group with 40 cycles of quantitative PCR. However, the expressions of these two genes were up-regulated with their mean Cq values of 31.68 ± 0.65 and 33.90 ± 0.26 respectively. These studies indicate that 5-aza-CdR caused up-regulated expression of epithelial genes in MUCs.
Fig. 4. The expression of epithelial genes following 5-aza-CdR treatment.

A. RT-PCR showed that MUCs up-regulated the expression of epithelial genes Cdh1, Krt8, and Krt18 following 5-aza-CdR treatment, and the relative expression levels of studied genes were related to 5-aza-CdR doses.
B. Quantification PCR showed normalized relative expressions of Cdh1 and Krt18 of 4 μM 5-aza-CdR-treated MUCs were 13.64 ± 0.05 and 13.57 ± 3.68 fold higher than those of control MUCs respectively, which is statistically significant (P<0.05; Student’s t-test).
C. Nested-Methylation Specific PCR (Nested-MSP) showed that the normalized expression of methylated Cdh1 in control MUCs was higher than unmethylated Cdh1. After 4 μM 5-aza-CdR treatment, the normalized expression of methylated Cdh1 was significantly down-regulated and lower than the expression of unmethylated Cdh1. The normalized relative expression of methylated Cdh1 was remarkably decreased following 5-aza-CdR treatment.
We performed Nested-Methylation Specific PCR (Nested-MSP) study to further examine the effect of demethylation on Cdh1 expression following 5-aza-CdR treatment. The bisulfite converted genomic DNA of control and 4 μM 5-aza-CdR treated MUCs served as the template in the Nested-MSP study, which was amplified using both methylated and unmethylated Cdh1 primer. In electrophoresis analysis, expressions of unmethylated Cdh1 in control and 5-aza-CdR treatment groups were used as references for normalizing methylated Cdh1 expressions in order to compare relative expressions of methylated Cdh1. It was shown that the expression of methylated Cdh1 was significantly higher than unmethylated Cdh1 in control MUCs, while in 5-aza-CdR-treated MUCs the expression of methylated Cdh1 was significantly lower than unmethylated Cdh1. In relative expression comparison, the expression of methylated Cdh1 was remarkably decreased following 5-aza-CdR treatment (Fig. 4C). The decreased relative expression of methylated Cdh1 in 5-aza-CdR-treated MUCs is able to lead to gene transcription, which may be related to the up-regulation of Cdh1 expression following 5-aza-CdR treatment.
Discussion
In this study, we used DNA methyltransferase inhibitor 5-aza-CdR to study whether DNA demethylation is able to stimulate epithelial and hair cell gene expression in stem/progenitor cells. We found that 5-aza-CdR stimulated the expression of epithelial genes including Cdh1, Krt8, Krt18, and Dsp in prosensory-like MUCs at 72 hr after treatment. The expressions of studied hair cell genes were increased in response to 5-aza-CdR treatment. We also found that after 5-aza-CdR treatment the genome-wide methylation level of MUCs was significantly reduced. Nested-MSP result of epithelial gene Cdh1 indicated that the methylation of Cdh1 was significantly decreased after 5-aza-CdR treatment. Additionally, gene expression changes following 5-aza-CdR treatment may be related to treatment time, which could be an independent study and will be studied in the future.
It is suggested that DNA demethylation is able to activate the expression of silenced genes in normal fibroblast cells and human bladder cancer cells [30, 36]. However, whether DNA demethylation is able to stimulate epithelial gene expression in stem/progenitor cells has not been determined. In this study, we found that DNA methyltransferse inhibitor 5-aza-CdR stimulated stem/progenitor-like MUCs to up-regulate the expression of epithelial genes that were not expressed in control MUCs, including Cdh1, Krt8, Krt18, and Dsp. In addition to activate the silenced genes, our study suggested that 5-aza-CdR was able to increase the expression level of genes that showed weak expression prior to the treatment. For example, prosensory gene Lfng was expressed in prosensory-like MUCs. After 4μM 5-aza-CdR treatment, the expression of Lfng in 5-aza-CdR-treated MUCs was 13.32 ± 1.32 fold higher than that in control MUCs. This observation was also reported in cancer cell lines [37, 38]. For instance, the follicular lymphoma cell line exhibited a very low level of Lfng that was caused by hypermethylation in DNA sequence. After the treatment of 5-aza-CdR, the transcription of Lfng was restored and its expression was up-regulated (Bennett et al, 2009). In addition to Lfng, our study shows that other prosensory genes including Jag1, Hes1, Isl1, Bmp4, and Sox2 increased their expression levels following 5-aza-CdR treatment. Our observations indicate that 5-aza-CdR treatment is able to up-regulate the expression of genes that are previously completely silenced or at a low expression level prior to 5-aza-CdR treatment.
Previous studies have suggested that 5-aza-CdR exerts concentration-dependent demethylation effects on a variety of cell lines, including a normal fibroblast cell line [30] and cancer cell lines [39, 40]. In this study, we found that 5-aza-CdR induced concentration-dependent effects on studied genes, including epithelial genes (Cdh1, Krt8, and Krt18), prosensory genes (Jag1, Hes1, Sox2, Dlx5, Bmp4, Lfng, P27kip1, and Pax2), and hair cell genes (Myo7a and Myo6). First, epithelial gene such as Krt18 was not expressed in MUCs before 5-aza-CdR treatment. After 72 hr of treatment, a weak expression of Krt8 was observed in 1 μM 5-aza-CdR treatment, while a relatively higher expression was detected with 2 μM treatment. In the treatment of 4 μM 5-aza-CdR, Krt8 expression level was remarkably increased comparing to control, 1, and 2 μM groups. Second, prosensory gene, taking Sox2 as an example, was weakly expressed in control MUCs. After 1 μM 5-aza-CdR treatment, MUCs had an increased Sox2 expression, and this increased expression of Sox2 obtained its relatively highest expression at 4 μM treatment. Third, hair cell genes including Myo7a and Myo6 were hardly detected in control MUCs, but they expressed along with 5-aza-CdR treatment and exhibited the highest expression level in the 4 μM group. Together, our research suggests that studied genes including Cdh1, Krt8, Krt18, Jag1, Hes1, Sox2, Dlx5, Bmp4, Lfng, P27kip1, Pax2, Myo7a, and Myo6 showed gene expression changes in a concentration-dependent manner following 5-aza-CdR treatment.
5-aza-CdR is a commonly used DNA methyltransferase inhibitor for a genome-wide demethylation, which is able to exert its demethylation effects via at least two patterns: 5′-CpG island specific or none specific. First, 5-aza-CdR is able to stimulate the expression of methylation-silenced genes, which possess methylated CpG islands in their promoter sequences at 5′ region [41]. For example, a study on a gastric cancer cell line found that 5-aza-CdR-induced genome-wide demethylation up-regulated the expression of silenced genes that had methylated CpG islands at their 5′-promoter regions [42]. In line with this observation, we found that genes with CpG islands at their 5′-promoter region, including epithelial genes Cdh1 and Krt18, prosensory genes Jag1, Hes1, Dlx5, Lfng, and P27kip1, and hair cell gene Myo6, had significantly up-regulated their expressions in response to 5-aza-CdR treatment. Second, 5-aza-CdR treatment also induces demethylation that is not 5′-CpG island specific. For instance, 5-aza-CdR leads to demethylation of the human bladder tumor cell line T24, in which approximately 60% of the stimulated genes did not have CpG islands in their 5′ regions [36]. This result indicates that 5-aza-CdR may be able to induce genome-wide demethylation that is not 5′ CpG island specific. In our observation, we investigated the locations of 5′ CpG islands of the studied epithelial, prosensory, and hair cell genes. We found that Krt8, Sox2, Bmp4, and Myo7a did not have CpG islands in their 5′ region, which includes 1000bp before the transcription start codon as well as the first exon. These genes exhibited increased gene expression in response to 5-aza-CdR treatment, which suggests that 5-aza-CdR may cause gene expression changes independent of 5′-CpG island methylation in prosensory-like MUCs. In general, our and other observations suggest that 5-aza-CdR is able to induce a genome-wide demethylation and stimulate gene expression in both 5-CpG island dependent and non-dependent ways.
Whether DNA methylation play a role in inducing stem/progenitor cells to stimulate epithelial gene expression has not been determined. In leukemia cell line studies, Paul Corn and his research team have reported that the silence of Cdh1 was caused by DNA methylation on its CpG islands located around Cdh1 transcription start codon, including the CpG islands in its promoter and first exon sequence [32]. In our study of prosensory-like MUCs, RT-PCR results showed that expressions of epithelial genes, including Cdh1, Krt8, Krt18, and Dsp were stimulated following 5-aza-CdR treatment. These epithelial genes demonstrated a significantly higher expression level following 4 μM 5-aza-CdR treatment, which was confirmed by our quantitative PCR results. Nested-MSP of Cdh1 showed that the normalized expression of methylated Cdh1 had a higher expression level than unmethylated Cdh1 in the control MUCs. However, after 5-aza-CdR treatment, the normalized expression of methylated Cdh1 decreased and was significantly lower than the unmethylated Cdh1. The change of relative expression of methylated Cdh1 suggests that DNA demethylation may be a possible mechanism involved in activating the expression of epithelial gene Cdh1 in prosensory-like MUCs. Moreover, we found that prosensory-like MUCs exhibited an increased expression of hair cell genes Myo6 and Myo7a following 5-aza-CdR treatment, which indicates that DNA methylation may play a role in the induction of differentiated genes. Overall, our and previous studies indicate that DNA methyltransferase inhibitor is able to induce a genome-wide demethylation and up-regulated the expression of epithelial and hair cell genes in prosensory-like MUCs. Therefore, our study may provide a cell model to study epithelial hair cell regeneration via epigenetic approaches.
In summary, DNA methyltransferase inhibitor 5-aza-CdR induced a remarkable genome-wide demethylation in stem/progenitor cells, which was determined by quantifying the percentage of global methylated DNA of prosensory-like MUCs in the control and treatment groups. In response to DNA demethylation, expressions of prosensory genes including Jag1, Hes1, Sox2, Dlx5, Bmp4, Lfng, P27kip1, and Pax2, and hair cell genes Myo7 and Myo6 were up-regulated following 5-aza-CdR treatment. Concurrently, MUCs acquired an activation of epithelial genes, which was demonstrated by the increased expression of Cdh1, Krt8, Krt18, and Dsp in the treatment group. The methylation/demethylation pattern of Cdh1 determined by the Nested-MSP study indicated that 5-aza-CdR-induced demethylation may play a role in stimulating the expression of Cdh1 in MUCs. Our study suggests that an epigenetic approach is able to modify gene expression in stem/progenitor cells, which also provides a cell model for studying stem cell differentiation via epigenetic modifications.
Acknowledgments
The authors thank Neelkumar Patel for his technical support and Jue Wang, Fei Nei, and Xiaoyang Li for valuable comments to the manuscript. The study is supported by NIDCD/NIH (1 R01 DC013275) and the Grants Plus Program from the Wayne State University.
Footnotes
Conflict of interest
The authors indicate no potential conflicts of interest.
References
- 1.Shi F, Edge AS. Prospects for replacement of auditory neurons by stem cells. Hear Res. 2013;297:106–112. doi: 10.1016/j.heares.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu Z, Ulfendahl M. The potential of stem cells for the restoration of auditory function in humans. Regen Med. 2013;8(3):309–318. doi: 10.2217/rme.13.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groves AK, Zhang KD, Fekete DM. The genetics of hair cell development and regeneration. Annu Rev Neurosci. 2013;36:361–381. doi: 10.1146/annurev-neuro-062012-170309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ronaghi M, Nasr M, Heller S. Concise review: Inner ear stem cells--an oxymoron, but why? Stem Cells. 2012;30(1):69–74. doi: 10.1002/stem.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotanche DA, Kaiser CL. Hair cell fate decisions in cochlear development and regeneration. Hear Res. 2010;266(1–2):18–25. doi: 10.1016/j.heares.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okano T, Kelley MW. Stem cell therapy for the inner ear: recent advances and future directions. Trends Amplif. 2012;16(1):4–18. doi: 10.1177/1084713812440336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Liu H, Heller S. Pluripotent stem cells from the adult mouse inner ear. Nat Med. 2003;9(10):1293–1299. doi: 10.1038/nm925. [DOI] [PubMed] [Google Scholar]
- 8.Oshima K, Grimm CM, Corrales CE, et al. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J Assoc Res Otolaryngol. 2007;8(1):18–31. doi: 10.1007/s10162-006-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White PM, Doetzlhofer A, Lee YS, Groves AK, Segil N. Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature. 2006;441(7096):984–987. doi: 10.1038/nature04849. [DOI] [PubMed] [Google Scholar]
- 10.Mizutari K, Fujioka M, Hosoya M, et al. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron. 2013;77(1):58–69. doi: 10.1016/j.neuron.2012.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley MW, Talreja DR, Corwin JT. Replacement of hair cells after laser microbeam irradiation in cultured organs of corti from embryonic and neonatal mice. J Neurosci. 1995;15(4):3013–3026. doi: 10.1523/JNEUROSCI.15-04-03013.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Hu Z. Sensory epithelial cells acquire features of prosensory cells via epithelial to mesenchymal transition. Stem Cells Dev. 2012;21(10):1812–1821. doi: 10.1089/scd.2011.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oesterle EC, Campbell S, Taylor RR, Forge A, Hume CR. Sox2 and JAGGED1 expression in normal and drug-damaged adult mouse inner ear. J Assoc Res Otolaryngol. 2008;9(1):65–89. doi: 10.1007/s10162-007-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batts SA, Shoemaker CR, Raphael Y. Notch signaling and Hes labeling in the normal and drug-damaged organ of Corti. Hear Res. 2009;249(1–2):15–22. doi: 10.1016/j.heares.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7(11):837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Hu Z. Sensory Epithelial Cells Acquire Features of Prosensory Cells Via Epithelial to Mesenchymal Transition. Stem Cells Dev. 2012;21(10):1812–1821. doi: 10.1089/scd.2011.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barald KF, Kelley MW. From placode to polarization: new tunes in inner ear development. Development. 2004;131(17):4119–4130. doi: 10.1242/dev.01339. [DOI] [PubMed] [Google Scholar]
- 18.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 19.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 20.Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55(3):451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 22.Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002;289(1–2):41–48. doi: 10.1016/s0378-1119(02)00469-9. [DOI] [PubMed] [Google Scholar]
- 23.Ghoshal K, Datta J, Majumder S, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25(11):4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Auclair G, Weber M. Mechanisms of DNA methylation and demethylation in mammals. Biochimie. 2012;94(11):2202–2211. doi: 10.1016/j.biochi.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Phillips T. The Role of Methylation in Gene Expression. Nature Education. 2008;1(1):116. [Google Scholar]
- 27.Sigalotti L, Fratta E, Coral S, et al. Epigenetic drugs as pleiotropic agents in cancer treatment: biomolecular aspects and clinical applications. J Cell Physiol. 2007;212(2):330–344. doi: 10.1002/jcp.21066. [DOI] [PubMed] [Google Scholar]
- 28.Sigalotti L, Fratta E, Coral S, Maio M. Epigenetic drugs as immunomodulators for combination therapies in solid tumors. Pharmacol Ther. 2014;142(3):339–350. doi: 10.1016/j.pharmthera.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 29.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 30.Mossman D, Kim KT, Scott RJ. Demethylation by 5-aza-2′-deoxycytidine in colorectal cancer cells targets genomic DNA whilst promoter CpG island methylation persists. BMC Cancer. 2010;10:366. doi: 10.1186/1471-2407-10-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patra A, Deb M, Dahiya R, Patra SK. 5-Aza-2′-deoxycytidine stress response and apoptosis in prostate cancer. Clin Epigenetics. 2011;2(2):339–348. doi: 10.1007/s13148-010-0019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corn PG, Smith BD, Ruckdeschel ES, Douglas D, Baylin SB, Herman JG. E-cadherin expression is silenced by 5′ CpG island methylation in acute leukemia. Clin Cancer Res. 2000;6(11):4243–4248. [PubMed] [Google Scholar]
- 33.Ling ZQ, Li P, Ge MH, et al. Hypermethylation-modulated down-regulation of CDH1 expression contributes to the progression of esophageal cancer. Int J Mol Med. 2011;27(5):625–635. doi: 10.3892/ijmm.2011.640. [DOI] [PubMed] [Google Scholar]
- 34.Lin SL. Concise review: Deciphering the mechanism behind induced pluripotent stem cell generation. Stem Cells. 2011;29(11):1645–1649. doi: 10.1002/stem.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan X, Ehnert S, Culmes M, et al. 5-azacytidine improves the osteogenic differentiation potential of aged human adipose-derived mesenchymal stem cells by DNA demethylation. PLoS One. 2014;9(3):e90846. doi: 10.1371/journal.pone.0090846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang G, Gonzales FA, Jones PA, Orntoft TF, Thykjaer T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2′-deoxycytidine. Cancer Res. 2002;62(4):961–966. [PubMed] [Google Scholar]
- 37.Bennett LB, Schnabel JL, Kelchen JM, et al. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes Chromosomes Cancer. 2009;48(9):828–841. doi: 10.1002/gcc.20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almstedt M, Blagitko-Dorfs N, Duque-Afonso J, et al. The DNA demethylating agent 5-aza-2′-deoxycytidine induces expression of NY-ESO-1 and other cancer/testis antigens in myeloid leukemia cells. Leuk Res. 2010;34(7):899–905. doi: 10.1016/j.leukres.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58(1):95–101. [PubMed] [Google Scholar]
- 40.Mund C, Hackanson B, Stresemann C, Lubbert M, Lyko F. Characterization of DNA demethylation effects induced by 5-Aza-2′-deoxycytidine in patients with myelodysplastic syndrome. Cancer Res. 2005;65(16):7086–7090. doi: 10.1158/0008-5472.CAN-05-0695. [DOI] [PubMed] [Google Scholar]
- 41.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19(11):7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita S, Tsujino Y, Moriguchi K, Tatematsu M, Ushijima T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2′-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006;97(1):64–71. doi: 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]