Abstract
A successful HIV-1 vaccine must elicit immune responses that impede mucosal virus transmission, though functional roles of protective HIV-1 Envelope (Env)-specific mucosal antibodies remain unclear. Colostrum is a rich source of readily accessible mucosal B cells that may help define the mucosal antibody response contributing to prevention of postnatal HIV-1 transmission. To examine the HIV-1 Env-specific colostrum B cell repertoire, single B cells were isolated from 17 chronically HIV-infected, lactating women, producing 51 blood and 39 colostrum HIV-1 Env-specific B cell antibodies. All HIV-1 Env-specific colostrum-derived antibodies were IgG1 isotype and had mean heavy chain complementarity-determining region 3 (CDR3) lengths and mutation frequencies similar to those isolated from blood. However, variable heavy chain (VH) gene subfamily 1~69 usage was higher among colostrum than blood HIV-1 Env-reactive antibodies (49% versus 20%, p = 0.006, Fisher’s exact test). Additionally, more HIV-1 Env-specific colostrum antibodies were gp120-specific than those isolated from blood (44% versus 16%, p = 0.005, Fisher’s exact test). One cross-compartment HIV-1 Env-specific clonal B cell lineage was identified. These unique characteristics of colostrum B cell antibodies suggest selective homing of HIV-1-specific IgG1-secreting memory B cells to the mammary gland and have implications for targeting mucosal B cell populations by vaccination.
Keywords: HIV-1, colostrum, B cell, antibodies
Introduction
The HIV-1 epidemic continues to be fueled by 2.3 million new infections annually,1 highlighting the critical need for an effective HIV-1 vaccine. As HIV-1 transmission occurs mainly at mucosal surfaces, effective anti-HIV-1 antibody responses at the portal of viral entry may reduce the risk of virus acquisition. Yet, it remains unclear whether effective HIV-1 vaccination necessitates protective mucosal antibody responses. Furthermore, whether mucosal antibody responses were potentially responsible for the moderate protection observed in the RV144 vaccine trial is unknown.2 Determining the role of HIV-1 Env-specific mucosal IgG and IgA antibodies in preventing mucosal HIV-1 transmission is therefore an essential step towards HIV-1 vaccine design and evaluation.
With recent advances in recombinant antibody technology, numerous HIV-1-neutralizing monoclonal antibodies (mAbs) have been produced from B lymphocytes isolated from peripheral blood of HIV-1-infected individuals.3, 4, 5, 6 However, the specificity and function of HIV-1-specific mAbs produced by mucosal B cell populations have yet to be explored, primarily due to insufficient lymphocyte content of mucosal secretions and difficulties in obtaining mucosal tissue specimens. Importantly, colostrum is a rich source of mucosal B lymphocytes that are thought to originate in the gastrointestinal-associated lymphoid tissue (GALT) and produce local antibodies that effectively protect infants against neonatal pathogens (“gut-mammary axis”).7 As a result, colostrum is an ideal source from which HIV-1 Env-specific antibodies can be isolated from mucosal B cell populations, allowing characterization of the spectrum of HIV-1 Env-specific antibodies produced at mucosal surfaces.
Breastfeeding transmission accounts for over one third of the 260,000 infant HIV-1 infections that occur annually.8, 9 Remarkably, without antiretroviral prophylaxis, only ten percent of HIV-exposed breastfeeding infants become infected despite chronic mucosal HIV-1 exposure.10 This relatively low transmission rate suggests that the immunologic components in breast milk contribute to protecting the majority of infants exposed to HIV-1 via breastfeeding. Interestingly, despite high levels of secretory IgA in milk,7, 11 the anti-HIV-1 humoral response seems to be primarily mediated by IgG antibodies.9, 12, 13 Prior work has further shown that HIV-1-specific antibodies with distinct anti-viral functions in milk may contribute to the low rate of virus transmission.13, 14, 15 In addition, we previously isolated two HIV-1-neutralizing IgG1 mAbs from colostrum, providing initial insight into the characteristics of Env-specific mAbs produced by colostrum B cells.16
We now describe the specificity, variable gene usage, and maturation of a large number of human monoclonal antibodies (mAbs) isolated from colostrum from a cohort of 17 HIV-infected, lactating women. This study of colostrum HIV-1 Env-specific mucosal B cell responses has identified characteristics of colostrum antibodies that suggest selective homing of IgG1-producing HIV-1 Env-specific B cells to the lactating mammary gland.
Results
Clinical characteristics of the cohort of 17 chronically HIV-1-infected, lactating Malawian women
The median baseline CD4+ T cell count of the chronically HIV-1-infected, lactating women included in this study was 275 cells/μl (range, 80 – 592 cells/μl), and the median baseline plasma viral load was 5,207 RNA copies/ml (range, 200 – 174,976 RNA copies/ml) (Table 1).13 The median breast milk viral load at four to six weeks postpartum was 247 RNA copies/ml (range, 20 – 144,750 RNA copies/ml). Fifteen subjects were untreated during pregnancy with the exception of single dose nevirapine during labor, and two began highly active anti-retroviral therapy (HAART) during pregnancy. Three women transmitted HIV-1 to their infant via breastfeeding.
Table 1.
Clinical characteristics and mAbs isolated from blood (BLD) and colostrum (CLM) of 17 chronically HIV-1-infected lactating women from Malawi (CHAVI009).
| Subject ID | Peripheral CD4+ T cell countd | Plasma Viral Loadd | Breast Milk Viral Loade | Treatment | BLD B cell mAbs (% Env-reactive) | CLM B cell mAbs (% Env-reactive) | B Cell Sort Method |
|---|---|---|---|---|---|---|---|
| CH0608 | 275 | 200 | 20 | HAARTb | 24 (4%) | 0 | Total B, Con-S gp120 |
| CH3601 | 452 | 60,335 | f | sdNVPc | 3 (33%) | 22 (9%) | Con-S gp120, Total memory |
| CH4403a | 208 | 100,892 | 144,750 | sdNVP | 0 | 1 (0%) | Con-S gp120 |
| CH8700 | 123 | 200 | 285 | HAART | 14 (0%) | 0 | Con-S gp140 |
| CH8802 | 382 | 4,269 | 120 | sdNVP | 9 (0%) | 6 (17%) | Con-S gp120 |
| CH8908 | 178 | 9,246 | 120 | sdNVP | 0 | 1 (100%) | Con-S gp140 |
| CH9105 | 266 | 5,207 | 120 | sdNVP | 12 (83%) | 14 (100%) | Con-S gp140 |
| CH9208 | 447 | 200 | 120 | sdNVP | 6 (33%) | 8 (100%) | Con-S gp140 |
| CH9300 | 415 | 91,717 | 254 | sdNVP | 35 (23%) | 5 (40%) | Con-S gp120, Con-S gp140 |
| CH9606 | 558 | 5,158 | 180 | sdNVP | 11 (18%) | 1 (100%) | Con-S gp140 |
| CH9701 | 217 | 63,707 | 4,250 | sdNVP | 5 (25%) | 3 (0%) | Total B, Total memory, Con-S gp140 |
| CH9803 | 592 | 1,678 | 863 | sdNVP | 6 (100%) | 2 (100%) | Con-S gp140 |
| CH0009 | 429 | 1,256 | 120 | sdNVP | 6 (83%) | 2 (50%) | Con-S gp140 |
| CH0305 | 198 | 200 | 240 | sdNVP | 3 (100%) | 0 | Con-S gp140 |
| CH0404a | 80 | 22,600e | 29,425 | sdNVP | 19 (58%) | 10 (70%) | Con-S gp140 |
| CH0301 | 406 | 75,934 | 790 | sdNVP | 6 (0%) | 3 (0%) | Total B |
| CH1209a | 153 | 174,976 | 26,818 | sdNVP | 14 (7%) | 2 (0%) | Total B |
Subject transmitted HIV-1 postnatally to infant.
Subject received Highly Active Anti-Retroviral Therapy (HAART) during pregnancy and breastfeeding.
Subject and infant received single dose Nevirapine (sdNVP) at delivery.
Measured during third trimester of pregnancy.
Measured four to six weeks postpartum.
Four to six week milk sample unavailable.
High frequency of memory B cells in colostrum compared to blood
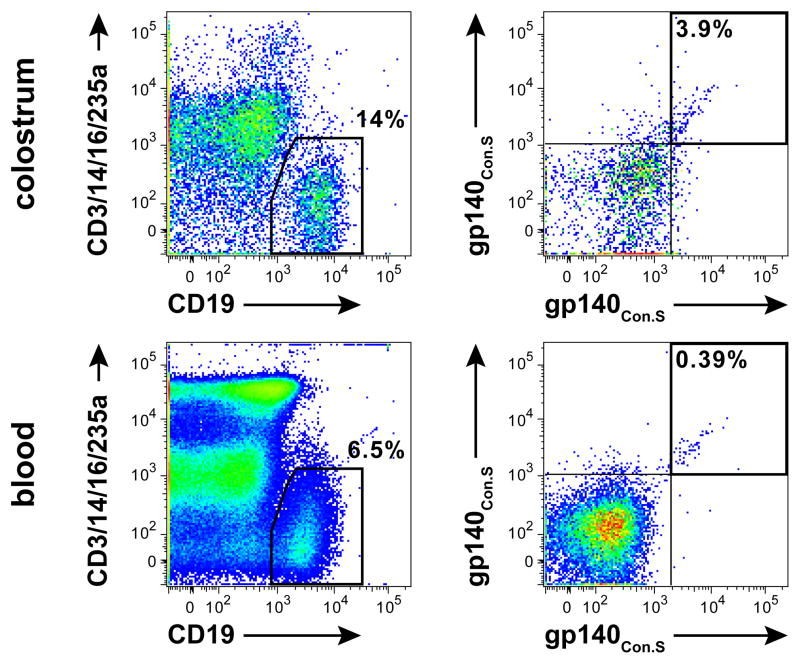
The proportion of total CD19+ B cells ranged from 0.6 to 22% and 3 to 11% of total live cells isolated from colostrum and blood, respectively. Representative flow plots illustrating total B cell and Env-specific phenotype distribution of the colostrum and blood B cell populations from one subject are shown in Figure 1. In both blood and colostrum, most B cells were IgD– memory, yet colostrum had a significantly higher proportion of IgD– memory B cells than blood (median: 96% versus 62%, respectively, p < 0.0001, Wilcoxon matched-pairs signed rank test; Table 2). The proportions of CD27+ memory B cells (IgD–/CD27+) were similar in colostrum and blood (median: 34% versus 28%, respectively). Moreover, the median frequencies of plasmablasts and plasma cells (IgD–/CD38Hi) were the same between colostrum and blood (both, 4%).17, 18 Colostrum and blood had similar median frequencies of B cells reactive with consensus HIV-1 Env constructs, including a group M consensus (Con-S) gp120 (0% and 0.1%, respectively, n = 5) and gp140 (2% versus 0.2%, respectively; n = 12).
Figure 1. Total B cell and Con-S gp140 HIV-1 Env-specific B cell populations in colostrum (top) and blood (bottom) of an HIV-1-infected, lactating woman.
Total B cells were gated as CD3–/CD14–/CD16–/CD235a– and CD19+ (left). HIV-1 Env-specific B cells were gated from the total B cell population as Con-S gp140 AF647+/Con-S gp140 BV421+ (right).
Table 2.
Colostrum and blood B lymphocyte phenotype.
| % IgD– (range) (n = 17) | % IgD–/CD27+ (range) (n = 17) | %IgD–/CD38Hi (range) (n = 17) | % IgD-/Con-S gp120++ (range) (n = 5) | % IgD-/Con-S gp140++ (range) (n = 12) | |
|---|---|---|---|---|---|
| Colostrum | 96a (69–100) | 34 (0–83) | 4 (0–39) | 0 (0–4.0) | 2 (0.2–6) |
| Blood | 62 (23–98) | 28 (9–82) | 4 (0.6–17) | 0.1 (0–0.2) | 0.2 (0–6) |
|
| |||||
| p-valueb | <0.0001 | 0.13 | 0.33 | 0.38 | 0.06 |
Median.
Wilcoxon matched-pairs signed rank test comparing colostrum and blood; p < 0.05 in bold.
HIV-1 Con-S gp140-specific sorting efficiently isolates HIV-1 Env-specific antibody-secreting B cells
Colostrum-derived cell samples showed substantial variation in cell viability, necessitating optimization of the sort gating method. Initial samples were gated for total B cells (CD19+, CD3–/CD14–/CD16–/CD235a–) and total memory B cells (CD19+, CD3–/CD14–/CD16–/CD235a–, IgD–, CD38+), yielding few HIV-1 Env-specific mAbs defined by PCR amplification, transient transfection, and ELISA screening (two HIV-1 Env-specific mAbs isolated per 807 colostrum B cells amplified, 0.2%). We then used antigen-specific sorting of B cells with HIV-1 Con-S gp120 with no increase in the yield of HIV-1 Env-specific antibodies from colostrum (1/409, 0.2%). Finally, we used a HIV-1 Con-S gp140 probe for antigen-specific sorting to target gp41-reactive and conformationally-dependent antibodies. This strategy increased the yield of HIV-1 Env-reactive antibodies (36/1109, 3%), showing that HIV-1 Con-S gp140 was more specific than Con-S gp120 for isolating B cells from colostrum. From blood-derived B cells, there was a 5% yield by ELISA of HIV Env-reactive antibodies sorted using the HIV-1 Con-S gp140 probe versus 4% from Con-S gp120-specific sorting.
Correlation between the proportion of HIV-1 Env-specific B cells in colostrum and HIV-1 Env-specific IgG response in whole breast milk
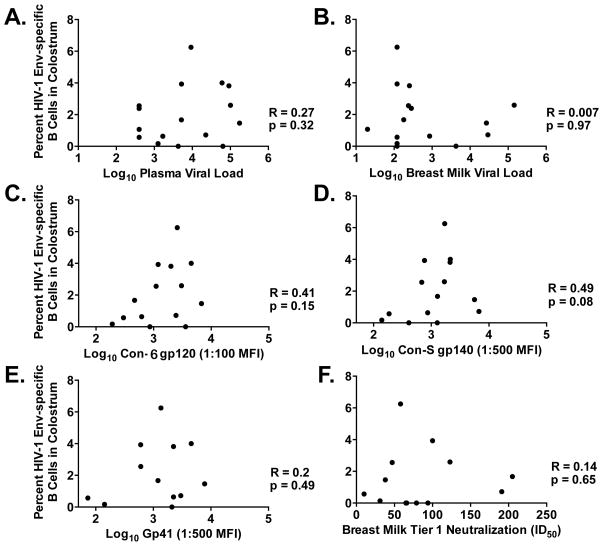
The frequency of HIV-1 Env-specific colostrum B cells identified by antigen-specific flow cytometry (Con-S gp120, n = 5 and Con-S gp140, n = 12) was compared to plasma virus load during the third trimester of pregnancy and breast milk viral load at four to six weeks postpartum. No correlations were found between the colostrum Env-specific B cell frequency and plasma or milk viral load (Figure 2A and B, respectively). Furthermore, no correlation was observed between the proportion of HIV-1 Env-specific B cells and the consensus Con-6 gp120-binding (R = 0.41; p = 0.15) or Con-S gp140-binding (R = 0.49; p = 0.08) IgG response in milk (both, Spearman rank correlation; Figure 2C and 2D). With B cells isolated by gp120-specific sorting excluded, the colostrum Env-specific B cell frequency and the gp41-binding IgG response in milk were not associated (Figure 2E). Similarly, there was no correlation between the colostrum Env-specific B cell frequency and milk neutralization response against a tier 1 neutralization-sensitive clade-matched virus (C.MW965; Figure 2F).
Figure 2. Correlation of the proportion of HIV-1 Env-specific B cells isolated from colostrum with virus load and humoral responses in milk.
There is no association between the proportion of HIV-1 Env-specific B cells in colostrum and (A) plasma viral load during pregnancy or (B) breast milk viral load at four to six weeks postpartum. There is no correlation between the proportion of HIV-1 Env-specific B cells in colostrum and (C) breast milk HIV-1 Env-specific Con-6 gp120-binding (R = 0.41, p = 0.15) or Con-S gp140-binding IgG response (R = 0.49, p = 0.08; both, Spearman Rank Correlation). The proportion of HIV-1 Env-specific B cells in colostrum is not correlated with (E) gp41-binding IgG response or (F) breast milk tier 1 HIV-1 (C.MW956) neutralization potency.
IgG1 predominance of HIV-1 Env-specific mAbs isolated from colostrum B cells
From blood samples of 17 women, we isolated 1495 single-sorted B cells, of which 243 yielded a pair of VHDJH and VLJL genes. Seventy of these antibodies were excluded from analysis due to duplicate or indeterminate heavy chain sequences, and 51 of the 173 remaining blood antibodies were Env-reactive by ELISA (29%). We isolated 2919 colostrum B cells from the same cohort, yielding 161 antibodies, of which 80 were excluded as above, and 39 of the remaining 81 colostrum antibodies were HIV-1 Env-reactive by ELISA (48%).
Nonreactive antibodies produced by blood B cells were primarily IgM isotype (71%), while most nonreactive antibodies in colostrum were IgG1 isotype (55%; Figure 3). A small number of IgA (9%) and IgD (2%) nonreactive antibodies were isolated from blood B cells. Similarly, we found few colostrum B cells producing IgA antibodies (7%), and none were HIV-1 Env-specific. Interestingly, Env-reactive antibodies in colostrum were all IgG1 isotype (100%), consistent with our prior isolation of only IgG1 HIV-1 Env-specific colostrum antibodies.16 In contrast, IgA, IgG1, IgG3, IgD, and IgM Env-reactive antibodies were isolated from blood.
Figure 3. HIV-1 Env-reactive monoclonal antibodies isolated from colostrum are IgG1 isotype.
The isotype distribution of HIV-1 Env-reactive (left) and nonreactive (right) antibodies isolated from colostrum (top) and blood (bottom) of chronically HIV-1-infected, lactating women is shown by color. The number of antibodies in each population is indicated in the center of each donut.
Restricted VH gene usage in HIV-1 Env-specific mAbs produced by colostrum B cells
Monoclonal antibodies isolated from blood and colostrum used a total of 30 and 27 different VH gene subfamilies, respectively (Figure 4). The identified HIV-1 Env-specific blood and colostrum antibodies used 20 and 12 different VH gene subfamilies, respectively. HIV-1 Env-specific antibodies from both compartments employed VH 1~2, 1~69, 3~15, 3~23, 3~30, 3~30-3, 3~49, and 5~51 alleles. The only identified unique VH gene subfamily used by Env-reactive colostrum B cell antibodies was 3~43. Notably, VH 1~69 constituted 49% of the gene usage of HIV-1 Env-specific colostrum antibodies, compared to 12% in nonreactive colostrum antibodies (p = 0.0005, Fisher’s exact test). Similarly, VH 1~69 was the most commonly used gene subfamily in HIV-1 Env-specific antibodies from blood (20%), significantly higher than in nonreactive blood antibodies (6%, p = 0.01, Fisher’s exact test). Yet, VH 1~69 was used at a significantly higher rate in HIV-1 Env-specific antibodies isolated from colostrum than blood (p = 0.006, Fisher’s exact test).
Figure 4. Distinct VH gene usage in HIV-1 Env-reactive antibodies isolated from colostrum.
These charts show the VH gene usage in HIV-1 Env-reactive (left) and nonreactive (right) antibodies isolated from colostrum (top) and blood (bottom). The number of antibodies in each population is indicated in the center of each donut. Subfamilies contributing at least 6% of the total VH gene usage in one antibody population are shown on all donuts in color. The VH1~69 gene subfamily was used at a significantly higher rate in HIV-1 Env-specific antibodies from colostrum compared to those from blood (p = 0.006, Fisher’s exact test).
CDR3 lengths and somatic hypermutation frequencies of colostrum HIV-1 Env-specific IgG B cell mAbs are similar to those isolated from peripheral B cells
As broadly HIV-1-neutralizing antibodies often display long CDR3 lengths and high somatic hypermutation rates,19 we examined these parameters in blood and colostrum Env-specific mAbs. The mean CDR3 lengths of nonreactive IgG antibodies in blood and colostrum were similar (15 and 17 amino acids, respectively), as were the mean CDR3 lengths of HIV-1 Env-reactive IgG antibodies in blood and colostrum (16 and 17 amino acids, respectively; Table 3). Somatic hypermutation rates were also similar between Env-reactive IgG antibodies from blood (6%) and colostrum (7%), as well as between nonreactive blood and colostrum antibodies (both, 6%).
Table 3.
Somatic hypermutation frequencies and CDR3 lengths are not significantly different between HIV-1 Env-reactive IgG antibodies in blood and colostrum.
| Nonreactive | HIV-1 Env-reactive | p-value | ||
|---|---|---|---|---|
| Mean CDR3 length (amino acids) | Blood | 15 | 16 | 0.83 |
| Colostrum | 17 | 17 | 0.94 | |
|
|
||||
| p-valuea | 0.12 | 0.13 | ||
|
| ||||
| Mean mutation frequency (%) | Blood | 6 | 6 | 0.97 |
| Colostrum | 6 | 7 | 0.24 | |
|
|
||||
| p-value | 0.90 | 0.21 | ||
Analyses performed using linear mixed model; p > 0.05 is not significant.
High rate of gp120-directed IgG1 antibodies in the colostrum HIV-1 Env-specific B cell mAb repertoire
The majority of the neutralizing epitopes identified on the HIV-1 Env protein complex are contained within the gp120 protein,20 whereas the gp41 protein has only a single identified neutralizing site, the membrane proximal external region (MPER).21 However, the predominant early target of acute HIV-1 Env antibody responses in plasma and the genital mucosa is gp41.22, 23, 24 Therefore, we compared the distribution of gp120 and gp41-directed antibodies produced by blood and colostrum B cells. Strikingly, while the majority of HIV-1 Env-reactive antibodies isolated from blood were gp41-specific (65%), only 21% of the colostrum HIV-1 Env-reactive antibodies were gp41-directed (p < 0.0001, Fisher’s exact test; Table 4). Rather, more HIV-1 Env-reactive antibodies isolated from colostrum were gp120-specific than in blood (44% versus 16%, respectively, p = 0.005, Fisher’s exact test). In addition, significantly more colostrum than blood antibodies were multi-Env-specific (bind gp120 and gp41 antigens; 33% versus 12%, respectively, p = 0.02, Fisher’s exact test). If only those antibodies produced by Con-S gp140-specific-sorted B cells were analyzed, differences in antigen specificity between Env-reactive colostrum and blood antibodies persisted (Table 4). Finally, gp120-specific and gp-41-specific antibodies isolated from colostrum used VH 1~69 gene at a relatively higher frequency than those in blood (59% versus 25% for gp120 and 50% versus 18% for gp41).
Table 4.
Predominance of gp120-directed HIV-1 Env-specific antibodies isolated from colostrum B cells.
| Env-specific mAbs isolated from all Sort Methods | gp120 | gp41 | gp140 | multi-Env |
|---|---|---|---|---|
| Blood (n = 51) | 8 (16%) | 33 (65%) | 4 (8%) | 6 (12%) |
| VH1~69 usage | 2 (25%) | 6 (18%) | 0 | 2 (33%) |
|
| ||||
| Colostrum (n = 39) | 17 (44%) | 8 (21%) | 1 (3%) | 13 (33%) |
| VH1~69 usage | 10 (59%) | 4 (50%) | 0 | 5 (38%) |
|
| ||||
| Env-specific mAbs isolated from Con-S gp140-Specific Sorting | gp120 | gp41 | gp140 | multi-Env |
|
| ||||
| Blood (n = 40) | 8 (20%) | 24 (60%) | 4 (10%) | 4 (10%) |
| VH1~69 usage | 2 (25%) | 4 (17%) | 0 | 1 (25%) |
|
| ||||
| Colostrum (n = 36) | 14 (39%) | 8 (22%) | 1 (3%) | 13 (36%) |
| VH1~69 usage | 9 (64%) | 4 (50%) | 0 | 5 (38%) |
Nonreactive and gp41-directed B cell mAbs of HIV-1-infected, lactating mothers who transmitted HIV-1 postnatally to their infant
We aimed next to compare the HIV-1 Env-specific colostrum B cell repertoire of the postnatally transmitting and nontransmitting women included in this cohort, though our analysis was limited by the low number of transmitting women (three) and low colostrum antibody recovery from this group. The women who transmitted HIV-1 to their infant via breastfeeding had either only nonreactive colostrum B cell antibodies isolated (CH4403, n = 1; CH1209, n = 2) or predominately gp41-specific colostrum B cell antibodies (CH0404, n = 10: 50% gp41-specific, 10% gp120-specific, 10% multi-Env-specific, 30% nonreactive; Figure S2). Although colostrum B cells of transmitting subject CH0404 produced predominately gp41-specific antibodies, this subject’s blood had equal numbers of gp41- and gp120-specific antibodies (both, n = 5). Of the 14 subjects who did not transmit HIV-1 to their infant postnatally, one or more HIV-1 Env-reactive colostrum antibodies were isolated from seven subjects. Their HIV-1 Env-reactive colostrum B cell antibody populations consisted primarily of gp120-binding antibodies, with the exception of CH9300 (gp140, n=1; gp120, n=1) and CH9803 (gp41, n=2; Figure S2).
Cross-compartment clonal IgG1-secreting B cell lineage identified
Six groups of clonally-related HIV-1 Env-reactive B cell antibodies were identified in blood and/or milk of three HIV-1-infected subjects: CH9105, CH9208, and CH0404 (Table 5). In addition, one cross-compartment clonal set was identified which contained two gp120-specific colostrum antibodies and one gp120-specific blood antibody (Figure 5), indicating systemic trafficking of colostrum HIV-1 Env-specific B cells. Like the Env-reactive antibodies isolated from colostrum, all clonally related antibodies were IgG1 isotype. This finding was surprising given the previously reported prevalence of IgA-secreting cells in colostrum7, 11, 25 and suggests that class switching to IgA is not required for B cells to home to the lactating mammary gland.
Table 5.
Single and cross-compartment HIV-1 Env-reactive clonal B cell lineages isolated from blood (BLD) and colostrum (CLM).
| Clonal Lineagea | Compartment (Number of mAbs) | Env Specificity (Number of mAbs) | Ig Isotype | VH | VK or VL |
|---|---|---|---|---|---|
| CH9105 | |||||
| DH279, DH280 | CLM (2) | gp120 (2) | IgG1 | 1~69 | 1~12 |
| DH281, DH282 | CLM (2) | gp41 (2) | IgG1 | 1~2 | 1~12 |
| DH276, DH277, DH278 | CLM (2)/BLD (1) | gp120 (3) | IgG1 | 1~69 | 3~20 |
| CH9208 | |||||
| DH283, DH284 | CLM (2) | gp120 (2) | IgG1 | 3~49 | 3~20 |
| DH285, DH286, DH287 | CLM (3) | gp120 (3) | IgG1 | 1~69 | 3~20 |
| CH0404 | |||||
| DH288, DH289 | CLM (2) | gp41 (2) | IgG1 | 1~69 | 3~11 |
The clonal lineages are named by their member antibody numbers.
Figure 5. Cross-compartment gp120-specific, IgG1-secreting B cell clonal lineage tree from subject CH9105.

One cross-compartment clonal set containing two gp120-specific colostrum antibodies and one blood antibody is illustrated. The inferred UCAs are at the roots, the intermediates are at the branches, and the antibody heavy chain sequences are at the leaves. The legend indicates the scale of evolutionary distance. All clones are reactive to Con-S gp120 by ELISA, and mutation frequencies of intermediate and antibody sequences relative to the inferred UCA are shown as percentages.
Discussion
Breastfeeding remains a significant mode of HIV-1 transmission in many developing countries, but the protective role of HIV-1 Env-specific maternal antibodies has not been consistently established.8, 9 Several studies have demonstrated a predominately IgG anti-HIV-1 Env response in breast milk,9, 12, 13 despite the high concentration of total IgA in breast milk and other mucosal compartments.7, 11 Furthermore, the HIV-1 Env-specific IgG responses in breast milk correlate to those in blood, albeit at a two log lower magnitude, suggesting that IgG transudate from plasma to the breast milk compartment is the predominant breast milk anti-HIV-1 antibody response.13 Yet, there is also some evidence of distinct anti-HIV-1 IgG antibody specificities and functions between the two compartments.15 Moreover, ADCC response of breast milk HIV-1 Env-specific IgG was recently associated with protection against postnatal HIV-1 transmission.14 Thus, our study provides a characterization of the genetic qualities and specificity of breast milk HIV-1 Env-specific B cells, facilitating better strategies to target an effective local antibody response through maternal vaccination.
Maternal B cells in breast milk, specifically IgA-secreting and potentially IgG-secreting B cells, appear to home to the lactating mammary gland from mesenteric lymph nodes in GALT along the “gut-mammary axis,” and many are specific for gut microflora.7, 26, 27, 28 Because the lymphocyte content is high in colostrum compared to mature milk,10 this fluid provided a unique opportunity to examine the HIV-1 Env-specific B cells that traffic to this mucosal compartment. In our B cell repertoire analysis, we observed that the colostrum of chronically HIV-1-infected women contained a significantly greater proportion of memory B cells than blood (p < 0.0001), corresponding with previous data showing that breast milk B cells are primarily class-switched memory B cells.12 Importantly, the HIV-1 Env-specific colostrum antibodies had reduced isotype breadth compared to those from blood, as all were IgG1 isotype, consistent with a previous report of predominately IgG1-producing HIV-1 Env-specific B cells in breast milk.12 Furthermore, our identification of a HIV-1 Env-reactive cross-compartment clonal IgG1-secreting B cell lineage indicates systemic trafficking of IgG1-producing B cells to the lactating mammary gland.
The absence of a correlation between plasma or milk virus load and the colostrum HIV-1 Env-specific B cell frequency in this study was unsurprising given the evidence that systemic virus populations continually repopulate the breast milk compartment virus pool.29 Furthermore, the administration of nevirapine at delivery likely interrupted any relationship between the local B cell responses and milk viral load. Prior studies have demonstrated that only low levels of virus neutralization responses are detectable in breast milk, and these whole milk responses did not correlate with breast milk viral load.13 The lack of correlation between the proportion of HIV-1 Env-specific B cells and HIV-1 Env-specific IgG binding responses or viral neutralization in milk signifies that a locally generated colostrum B cell IgG1 antibody response may only contribute to a small portion of the total milk anti-HIV-1 IgG response. Importantly, however, locally produced antibodies may be able to interfere with transmission of breast milk virus within the infant GI tract. The predominance of IgG1-producing antigen-specific B cells in colostrum also has important implications for the traditional paradigm that chiefly IgA-secreting B cells are trafficked within the gut-mammary axis.7
While there is a strong correlation between the total HIV-1 Env-specific IgG responses in blood and milk, the HIV-1 Env-specific IgA responses in milk and plasma are not associated.13 A detectable HIV-1-specific IgA response in milk of HIV-1-infected women has been demonstrated at 15 days postpartum in fewer individuals and at lower magnitude compared to the milk HIV-1 Env-specific IgG response.9 In our colostrum B cell analysis, the few IgA colostrum antibodies isolated were non-reactive. Our findings lend support to the idea that HIV-1 Env-specific IgA-secreting B cells may be primarily located within mammary tissue, not in the secretory fluid. HIV-1 Env-specific IgA-producing B cells have not been easily isolated from mucosal compartments; thus, it remains unclear whether mucosal IgA responses have a role in protecting the mucosal surface against HIV-1 infection.
The HIV-1 Env-specific mAbs isolated from colostrum in our study did not have a particularly high mean somatic hypermutation rate or long CDR3 length, characteristics of broadly neutralizing antibodies isolated from blood.3, 4, 5, 6, 19, 21, 30, 31 However, colostrum and blood HIV-1 Env-specific B cell mAbs were distinguishable by different VH gene usage. HIV-1 Env-reactive B cell colostrum mAbs demonstrated a higher rate of VH1~69 usage than nonreactive colostrum antibodies or HIV-1 Env-reactive B cell blood mAbs (p = 0.0005 and p = 0.006, respectively, Fisher’s exact test). Previous data has suggested that VH1~69 gene usage may facilitate direct heavy chain CDR2 interaction with the CD4-inducible epitope ons gp120.30 Therefore, the higher frequency of VH1~69 gene subfamily usage may relate to the distinct specificities of Env-specific antibodies in colostrum compared to blood. Conversely, the antigen-specific sorting employed in this study could have skewed gene usage by better targeting specific B cell populations. However, both gp41-specific and gp120-specific colostrum antibodies used VH1~69 at a somewhat higher frequency than those from blood, indicating that high VH1~69 usage by colostrum B cells may not be solely driven by antigen binding properties.
The proportions of gp120-, multi-Env-, and gp41-specific antibodies isolated from the systemic and mucosal compartments in this study are not biologically representative, as our use of antigen-specific B cell sorting biased the results by selecting for HIV-1 Env-specific antibodies that bind to the antigen employed. Given the small numbers of colostrum Env-specific B cells, slight differences in the affinity of the gp140 versus gp120 protein for the B cell receptor may explain the superior ability of the gp140 epitope to isolate antigen-specific B cells. However, as Con-S gp140-specific sorting is effective in isolating both gp41-specific and gp120-specific antibodies, it is an unlikely source of bias towards one specificity.22 Our data therefore indicate that gp120-binding mAbs are more frequently produced by colostrum than blood HIV-1 Env-specific B cells, and whether they are more common in chronically HIV-infected lactating women who do not transmit HIV-1 to their infant postnatally should be studied in more detail. As there were only three postnatally transmitting women in the entire CHAVI009 cohort, all with low colostrum mAb recovery, an association between gp120-specific B cell predominance in colostrum and no postnatal transmission of HIV-1 cannot be concluded with our current panel of antibodies, yet highlights the need for further investigation.
A primarily gp120- and multi-Env-specific B cell response in the lactating mammary gland contrasts with the gp41-specific antibody predominance of the acute systemic and mucosal B cell response to HIV-1.22, 24 The mechanism of distinct antigen-specific B cell homing to the mammary gland is unclear but could be hormonally or receptor-mediated.7, 26, 32 Gp120-directed antibodies may have more neutralizing capability than gp41-directed antibodies, as neutralizing gp41-directed antibodies are rare and have only been found to target a single epitope (MPER). Selective trafficking of HIV-1 Env-specific B cells with distinct VH gene usage and high rates of gp120-binding antibodies to the lactating mammary gland could thus produce a more effective antigen-specific response that contributes to the protection of the majority of HIV-exposed breastfed infants against postnatal HIV-1 transmission. Moreover, this distinct B cell population in the colostrum compartment is a potential target of future maternal vaccine candidates developed to reduce postnatal HIV-1 transmission.
Methods
Ethics statement
This study (CHAVI009) was approved by the Division of DAIDS; NIAID; NIH; the College of Medicine Research and Ethics Committee in Malawi (P.06/06/440); and the institutional review board at Duke University (Pro00030437). Subjects signed a written consent.
Subjects and colostrum processing
Colostrum and blood samples were obtained from a total of 77 chronically HIV-1 infected, lactating women enrolled between 2008 and 2009 in the CHAVI009 study in Malawi, 17 of which were included in this study based on high colostrum cell number to maximize yield of antigen-specific B cells.13, 29 The subjects were followed throughout pregnancy and breastfeeding. All subjects and infants received a single dose of nevirapine during the peripartum period. Infant HIV-1 status was determined by whole blood HIV-1 DNA PCR.33 Postnatal HIV-1 transmission was defined by negative infant whole blood HIV-1 DNA PCR at birth and at four to six weeks of age, followed by a positive whole blood HIV-1 DNA PCR at three months of age or later.33
Baseline plasma viral load and peripheral CD4 T cell count were measured during the subjects’ third trimester of pregnancy. Breast milk and blood samples were collected from the subjects within one week following delivery and again at four to six weeks postpartum. Breast milk collected within seven days of delivery was defined as “colostrum.” Breast milk viral load was measured at four to six weeks postpartum due to the limited amount of colostrum able to be collected. The milk samples were processed within four hours of collection and separated into cellular and skim milk fractions by low speed centrifugation as previously described.34 Colostrum cells (CLM) and PBMC were then cryopreserved within six hours of collection and stored viably in liquid nitrogen.
Flow cytometry panel antibodies and isolation of B cells from colostrum and blood
CLM and PBMC isolated from EDTA-anticoagulated blood were thawed in a 37 degree water bath. Right and left breast CLM were combined if both available. The samples were then immediately re-suspended in 14 milliliters pre-warmed RPMI/10% FBS and afterward centrifuged at 250xg for 10 minutes. Following removal of the supernatant, the cells were re-suspended and washed in R10, once to remove residual DMSO from the freezing media and a second time for cell counting. After a count was obtained, the cells were washed and re-suspended with PBS/2% FBS prior to labeled antibody staining.
Thawed CLM and PBMC were stained with the following panel of antibodies for flow cytometry: anti-IgM-fluorescein isothiocyanate [FITC] (G20-127, BD Biosciences, San Jose, CA), anti-IgD-phycoerythrin [PE] (IA6-2, BD Biosciences), anti-CD10-electron coupled dye [ECD] (ALB1, BD Biosciences), anti-CD3-PE-Cy5 (HIT3a, BD Biosciences), anti-CD14-PE-Cy5 (RMO52, Beckman Coulter, Brea, CA), anti-CD16-PE-Cy5 (3G8, BD Biosciences), anti-CD235a-PE-Cy5 (GA-R2, BD Biosciences), anti-CD27-PE-Cy7 (323, eBiosciences, San Diego, CA), anti-CD38-APC-Alexa Fluor [AF] 700 (LS198-4-3, Beckman Coulter), and anti-CD19-allophycocyanin [APC]-Cy7 (SJ25C1, BD Biosciences). CLM and PBMC were stained with either HIV-1 consensus clade C gp120 Env (Con-S gp120) or HIV-1 group M consensus gp140 Env (Con-S gp140) for antigen-specific B cell sorts. Con-S gp120 was labeled with Pacific Blue (PacBlue) and AF647 (both, Invitrogen, Carlsbad, CA). The Con-S gp140 protein included an AviTag that was then biotinylated via biotin ligase. The biotinylated protein was then tetramerized to Streptavidin-BV421 (Sony Biotechnology, Inc., Champaign, IL) and Streptavidin-AF647 (Invitrogen, Carlsbad, CA). Aqua Vital Dye was used to distinguish live from dead cells (Invitrogen, Carlsbad, CA). Controls consisted of matching isotype antibodies.
B cells from blood and colostrum were isolated and sorted according to four methods. Total B cells were gated as viable (Aqua Vital Dye –), CD3–/CD14–/CD16–/CD235a–, and CD19+. Total memory B cells were further selected by gating as IgD– and CD38+ from the total B cell population. For antigen-specific sorts, total B cells were gated as Con-S PacBlue + and Con-S AF647 + (Con-S gp120-specific), or Con-S BV421 + and Con-S AF647 + (Con-S gp140-specific). The B cells were individually sorted into 96 well plates pre-charged with a RNA stabilization cocktail and cryopreserved as described previously.6, 35
Phenotyping of isolated B cells
Flow cytometry data were collected on the FACSAria2 instrument (BD Biosciences) with FACSDiVa software and analyzed by manual gating with FlowJo software. The complete gating strategy for the B cell phenotype analysis is shown in Figure S1A. A broad lymphocyte gate was used to incorporate larger plasma cells and activated B cells which may be somewhat granular, as well as those associated with fat globules in colostrum due to its high lipid content.17, 26 Side scatter and forward scatter width versus height geometric gating excluded doublets and clumps. All Aqua Vital Dye + (dead) cells were omitted, and the total B cell population was defined as CD3–/CD14–/CD16–/CD235a– and CD19+ live cells. This gate was determined for the blood B cell population of each subject and copied to the colostrum B cell population. B cell phenotype was further identified as IgD– (memory and antibody-secreting B cells), IgD–/CD27+ (class-switched memory B cells), IgD–/CD38Hi (plasmablasts or plasma cells), and Con-S gp120- or Con-S gp140-specific.17
To ensure that inadequate compensation was not responsible for the observed diagonal of antigen-specific B cells, the compensation matrix was artificially adjusted to increase the compensation for each antigen-specific reagent fluorochrome (Figure S1B). Additionally, we stained PBMC and colostrum cells from HIV-1 uninfected women to determine if there was a background level of staining with the reagents; no staining of memory B cells was observed in these samples (Figure S1C).
Immunoglobulin (Ig) gene amplification, sequencing, and screening for HIV-1 Env-reactivity
After colostrum B cell sorting, the expressed Ig VH and variable light chain (VL) genes were amplified by reverse transcription (RT) and nested PCR as previously described.35, 36 The PCR products were purified and sequenced.35 Ig isotype was determined by sequence homology, and somatic hypermutation frequencies, inferred V(D)J rearrangement, and CDR3 length were determined by Somatic Diversification Analysis (SoDA), a web-based programming algorithm.37 Overlapping PCR was performed to co-express the variable region PCR products with full length IgG1 (for heavy chain) and kappa or lambda (for light chain) cassettes, and the PCR products were transiently transfected in 293T cells without the need for a cloning step as described previously.23, 35, 36 The supernatants of transfected cells were screened for reactivity against the following HIV-1 Env proteins by enzyme-linked immunoassay (ELISA)22, 23, 35: a transmitted/founder virus 1086.C gp120 and gp14038; CH505 gp12039; a consensus clade C gp140 Env with deletions in the gp41 cleavage site, fusion domain and immunodominant region [CFI]; Con-S gp140 CFI; gp41 sp400 peptide (RVLAVERYLRDQQLLGIWGCSGKLICTTAVPWNASWSNKSLNKI); gp41 sp62 peptide (GGG-QQEKNEQELLELDKWASLWN); gp41 MPER21 (CPC Scientific); resurfaced stabilized core-3 (RSC3)5 produced by Drs. Hua-Xin Liao and Barton Haynes; and HIV-1 MN recombinant gp41 (Immunodiagnostics).
HIV-1 Env-reactive antibodies were defined as having IgG quantitation > 0 in small scale transfection and an Env protein binding optical density (OD) ≥ 0.13 by ELISA, with background OD < 0.06.22, 23, 35 Percent reactivity for the blood and colostrum antibody repertoire of each individual was calculated as percent antibodies reactive by ELISA divided by the total number of antibodies produced from the compartment. Env specificities were classified as follows: gp41 (reactive against gp41, sp62, sp400, and/or MPER; or gp41 and any gp140), gp120 (reactive against any gp120 protein, gp120 and any gp140, and/or RSC3), gp140 (reactive only against any gp140 protein), or multi-Env (reactive against both gp41 and gp120 proteins).
Breast milk supernatant HIV-1 neutralization and Env-specific IgG binding
Heterologous virus neutralization was performed on breast milk samples collected four to six weeks postpartum from all untreated subjects except CH3601, whose four to six week sample was not collected. The assays were performed in TZM-bl reporter cells with HIV-1 MW965.26, a clade C tier 1 virus, by methods described previously and are reported as 50% inhibitory dilution.13, 40
HIV-1 Env-specific IgG levels were quantified from breast milk samples collected within one week of delivery for subject CH3601 and four to six weeks postpartum for all remaining untreated subjects using a customized HIV-1 binding antibody multiplex assay as described previously.13, 22 HIV-1 Env-specific IgG responses were measured against a recombinant HIV-1 gp41 Env (Immunodiagnostics), multiclade consensus gp120 Env (Con-6 gp120),41 and Con-S gp140 CFI (from H.X. Liao) at a dilution of 1:500 (gp41 and gp140) or 1:100 (gp120). 2F5 was used as positive control for gp41 and 2G12 as positive control for gp120 and gp140. The coefficient of variation between duplicates was ≤20% and a minimum of 100 beads was counted per sample. Milk Env-specific IgG responses are reported as mean florescent intensity (MFI) after blank beads subtraction.
Clonal B Cell Lineage Analysis
Clonal relatedness among antibodies was assessed in two stages. First, candidate relatives were determined by shared V and J gene usage, and by a statistical test testing the hypothesis of a shared single CDR3 sequence, conditional on the mutation frequencies observed in the region encoded by the V gene. Second, confirmation was obtained through manual inspection for evidence suggesting independent rearrangements. The sequences were aligned using BioEdit software, and the inferred unmutated common ancestor (UCA) and intermediate heavy chain V(D)J sequences were determined by Antigen Receptor Probabilistic Parser (ARPP) software and analyzed as described previously.36, 39, 42 Clonal lineage trees were created with FigTree software.
Statistics
Comparisons of B cell phenotypes in colostrum and blood were performed using the nonparametric Wilcoxon matched-pairs signed rank test. Correlations between subjects’ plasma or milk viral load, milk tier 1 neutralization, milk HIV-1 Env-specific IgG binding, and the proportion of HIV-1 Env-specific colostrum B cells were performed using Spearman’s rank correlation. The threshold of inclusion of colostrum Env-specific B cell frequency in these correlations was a total live B cell population of greater than 100 cells. Rates of variable gene usage (VH 1~69 versus other genes) and antigen specificity (gp41, gp120, or multi-Env versus other Env) were compared between colostrum and blood antibodies using 2 × 2 contingency tables with Fisher’s exact test. Somatic hypermutation frequencies and CDR3 lengths were compared between Env-reactive and nonreactive IgG antibodies in blood and colostrum using linear mixed models. Antibodies with identical heavy chains (duplicates) and heavy chains unable to be analyzed into constituent gene segments by SoDA (indeterminate sequences) were excluded from all statistical analyses. A p value of less than 0.05 (two-tailed) was considered significant for all analyses.
Supplementary Material
Acknowledgments
We acknowledge the following individuals for their contributions and support: Josh Amos, Sheri Chen, Annie Hogan, James Holland, Jennifer Kirchherr, Krissey Lloyd, Robert Parks, Christina Stolarchuk, Ashley Trama, Wilton Williams, and Yi Yang. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health R01 AI106380 (Permar), the Howard Hughes Medical Institute (Sacha), the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery, grant number UM1-AI100645-01, and the Duke University Center for AIDS Research (CFAR), an NIH funded program (5P30 AI064518). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors have no conflicts of interest.
Disclosure
We have no conflicts of interest to disclose.
References
- 1.UNAIDS. Fact Sheet: New HIV Infections. 2013 [cited]Available from: http://www.unaids.org/en/resources/campaigns/globalreport2013/factsheet/
- 2.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 3.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458(7238):636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 4.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329(5993):856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris L, Chen X, Alam M, Tomaras G, Zhang R, Marshall DJ, et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PloS one. 2011;6(9):e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roux ME, McWilliams M, Phillips-Quagliata JM, Weisz-Carrington P, Lamm ME. Origin of IgA-secreting plasma cells in the mammary gland. The Journal of experimental medicine. 1977;146:1311–1322. doi: 10.1084/jem.146.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kourtis AP, Butera S, Ibegbu C, Belec L, Duerr A. Breast milk and HIV-1: vector of transmission or vehicle of protection? Lancet Infect Dis. 2003;3(12):786–793. doi: 10.1016/s1473-3099(03)00832-6. [DOI] [PubMed] [Google Scholar]
- 9.Perre PVd, Simonon A, Hitimana D-G, Dabis F, Msellati P, Mukamabano B, et al. Infective and anti-infective properties of breastmilk from HIV-1-infected women. The Lancet. 1993;341:914–918. doi: 10.1016/0140-6736(93)91210-d. [DOI] [PubMed] [Google Scholar]
- 10.Rousseau CM, Nduati RW, Richardson BA, Steele MS, John-Stewart GC, Mbori-Ngacha DA, et al. Longitudinal analysis of human immunodeficiency virus type 1 RNA in breast milk and of its relationship to infant infection and maternal disease. The Journal of infectious diseases. 2003;187(5):741–747. doi: 10.1086/374273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandtzaeg P. The secretory immune system of lactating human mammary glands compared with other exocrine organs. Ann N Y Acad Sci. 1979;409:353–382. doi: 10.1111/j.1749-6632.1983.tb26883.x. [DOI] [PubMed] [Google Scholar]
- 12.Tuaillon E, Valea D, Becquart P, Al Tabaa Y, Meda N, Bollore K, et al. Human milk-derived B cells: a highly activated switched memory cell population primed to secrete antibodies. J Immunol. 2009;182(11):7155–7162. doi: 10.4049/jimmunol.0803107. [DOI] [PubMed] [Google Scholar]
- 13.Fouda GG, Yates NL, Pollara J, Shen X, Overman GR, Mahlokozera T, et al. HIV-specific functional antibody responses in breast milk mirror those in plasma and are primarily mediated by IgG antibodies. J Virol. 2011;85(18):9555–9567. doi: 10.1128/JVI.05174-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mabuka J, Nduati R, Odem-Davis K, Peterson D, Overbaugh J. HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS pathogens. 2012;8(6):e1002739. doi: 10.1371/journal.ppat.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becquart P, Hocini H, Garin B, Sépou A, Kazatchkine MD, Bélec L. Compartmentalization of the IgG immune response to HIV-1 in breast milk. AIDS. 1999;13(11):1323–1331. doi: 10.1097/00002030-199907300-00008. [DOI] [PubMed] [Google Scholar]
- 16.Friedman J, Alam SM, Shen X, Xia SM, Stewart S, Anasti K, et al. Isolation of HIV-1-neutralizing mucosal monoclonal antibodies from human colostrum. PloS one. 2012;7(5):e37648. doi: 10.1371/journal.pone.0037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski DA, Wei C, Qian Y, Rosenberg AF, Sanz I. Advances in human B cell phenotypic profiling. Frontiers in immunology. 2012;3(302) doi: 10.3389/fimmu.2012.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanz I, Wei C, Lee FE, Anolik J. Phenotypic and functional heterogeneity of human memory B cells. Seminars in immunology. 2008;20(1):67–82. doi: 10.1016/j.smim.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37(3):412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haynes BF, Moody MA, Liao HX, Verkoczy L, Tomaras GD. B cell responses to HIV-1 infection and vaccination: pathways to preventing infection. Trends in molecular medicine. 2011;17 (2):108–116. doi: 10.1016/j.molmed.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol. 1993;67(11):6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82(24):12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao HX, Chen X, Munshaw S, Zhang R, Marshall DJ, Vandergrift N, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. The Journal of experimental medicine. 2011;208(11):2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yates NL, Stacey AR, Nolen TL, Vandergrift NA, Moody MA, Montefiori DC, et al. HIV-1 gp41 envelope IgA is frequently elicited after transmission but has an initial short response half-life. Mucosal immunology. 2013;6(4):692–703. doi: 10.1038/mi.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz-Jouanen E, Williams RC. T and B lymphocytes in human colostrum. Clin Immunol Immunopathol. 1979;3(2):248–255. doi: 10.1016/0090-1229(74)90011-7. [DOI] [PubMed] [Google Scholar]
- 26.Butler J, Kehrli M. Immunoglobulins and immunocytes in the mammary gland and its secretions. In: Mestecky JF, Beinenstock J, Lamm M, Mayer L, McGhee JR, Strover W, editors. Mucosal Immunol. 3. Academic Press; San Diego: 2004. [Google Scholar]
- 27.Permar SR, Wilks AB, Ehlinger EP, Kang HH, Mahlokozera T, Coffey RT, et al. Limited contribution of mucosal IgA to Simian immunodeficiency virus (SIV)-specific neutralizing antibody response and virus envelope evolution in breast milk of SIV-infected, lactating rhesus monkeys. J Virol. 2010;84(16):8209–8218. doi: 10.1128/JVI.00656-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belec L, Kourtis AP. B lymphocyte-derived humoral immune defenses in breast milk transmission of the HIV-1. Adv Exp Med Biol. 2012;743:139–160. doi: 10.1007/978-1-4614-2251-8_10. [DOI] [PubMed] [Google Scholar]
- 29.Salazar-Gonzalez JF, Salazar MG, Learn GH, Fouda GG, Kang HH, Mahlokozera T, et al. Origin and evolution of HIV-1 in breast milk determined by single-genome amplification and sequencing. J Virol. 2011;85(6):2751–2763. doi: 10.1128/JVI.02316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. PNAS. 2004;101(9):2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janeway C. The mucosal immune system. In: Murphy K, Travers P, Walport M, editors. Janeway's Immunobiology. 7. Garland Science; New York, NY: 2008. pp. 459–495. [Google Scholar]
- 33.Fouda GG, Mahlokozera T, Salazar-Gonzalez JF, Salazar MG, Learn G, Kumar SB, et al. Postnatally-transmitted HIV-1 Envelope variants have similar neutralization-sensitivity and function to that of nontransmitted breast milk variants. Retrovirology. 2013;10:3. doi: 10.1186/1742-4690-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Permar SR, Kang HH, Carville A, Mansfield KG, Gelman RS, Rao SS, et al. Potent SIV-specific cellular immune responses in the breast milk of SIV-infected, lactating rhesus monkeys. J Immunol. 2008;181(5):3643–3650. doi: 10.4049/jimmunol.181.5.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. Journal of virological methods. 2009;158(1–2):171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85(19):9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volpe JM, Cowell LG, Kepler TB. SoDA: implementation of a 3D alignment algorithm for inference of antigen receptor recombinations. Bioinformatics. 2006;22(4):438–444. doi: 10.1093/bioinformatics/btk004. [DOI] [PubMed] [Google Scholar]
- 38.Go EP, Liao HX, Alam SM, Hua D, Haynes BF, Desaire H. Characterization of host-cell line specific glycosylation profiles of early transmitted/founder HIV-1 gp120 envelope proteins. Journal of proteome research. 2013;12(3):1223–1234. doi: 10.1021/pr300870t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496(7446):469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montefirori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005 Jan;Chapter 12(Unit 12.11) doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 41.Gao F, Weaver EA, Lu Z, Li Y, Liao H-X, Ma B, et al. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol. 2005;79(2):1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kepler TB. Reconstructing a B-cell clonal lineage: statistical inference of unobserved ancestors. F1000 Research. 2013;2(103) doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.