Abstract
Salinity is a major abiotic stress that limits plant productivity and quality throughout the world. Roots are the sites of salt uptake. To better understand salt stress responses in maize, we performed a comparative proteomic analysis of seedling roots from the salt-tolerant genotype F63 and the salt-sensitive genotype F35 under 160 mM NaCl treatment for 2 days. Under salinity conditions, the shoot fresh weight and relative water content were significantly higher in F63 than in F35, while the osmotic potential was significantly lower and the reduction of the K+/Na+ ratio was significantly less pronounced in F63 than in F35. Using an iTRAQ approach, twenty-eight proteins showed more than 2.0- fold changes in abundance and were regarded as salt-responsive proteins. Among them, twenty-two were specifically regulated in F63 but remained constant in F35. These proteins were mainly involved in signal processing, water conservation, protein synthesis and biotic cross-tolerance, and could be the major contributors to the tolerant genotype of F63. Functional analysis of a salt-responsive protein was performed in yeast as a case study to confirm the salt-related functions of detected proteins. Taken together, the results of this study may be helpful for further elucidating salt tolerance mechanisms in maize.
Introduction
Salinity is a major abiotic stress that affects plant growth and yield throughout the world [1–3]. More than 830 million hectares of land, which account for over 6% of the world’s total land area, have been affected by salinity [4]. Maize (Zea mays L.) plays an important role in global food security and economic development. Unfortunately, maize is not a salt-tolerant crop. Therefore, improving salt tolerance has become important for maize production.
Under salt stress conditions, the establishment of healthy seedlings is extremely important for maize plant subsequent development. Roots are the primary sites of salinity perception, and salt sensitivity in roots limits the productivity of the entire plant [5]. Therefore, obtaining a better understanding of salt-responsive mechanisms in seedling roots is critical for improving plant salt tolerance.
To better understand the molecular mechanisms of plant salt tolerance, large-scale transcriptomic analyses have been employed in the roots of numerous plant species. However, transcriptome profiling has limitations because mRNA levels are not always correlated to those of corresponding proteins due to post-transcriptional and post-translational modifications [6–8]. Elucidating changes at the protein level is essential for studying salt stress responses in plants. Proteomic analysis provides new insights into plant responses to salt stress at the protein level[9,10]. Recent advances in proteomics have made it possible to perform large-scale studies to help elucidate salt tolerance mechanisms in roots. To date, more than 905 salt-responsive proteins have been identified in roots from 14 plant species, such as Arabidopsis, rice, wheat, soybean, tomato, and barley [10]. However, relatively few such studies have been performed in maize. Zörb et al. studied proteomic changes in maize roots after a short-term adjustment to saline growth conditions [11]. A set of phosphoproteins were detected. Nevertheless, the application of 25 mM NaCl in this study appears to be relatively low for salt stress treatment, which may have led to the identification of a small number of salt-responsive proteins.
In the current study, to study maize salt responses at the protein level, we examined two contrasting maize inbred lines that showed significantly different phenotypes and physiology under salt stress. We conducted a comparative proteomic analysis of these two lines under 160mM NaCl treatment for 2 days. In addition, we examined the functions of one salt-responsive protein in yeast. The results of this study may be helpful for further salt tolerance studies in maize.
Materials and Methods
Plant materials and NaCl treatment
A total of 162 maize (Zea Mays L.) inbred lines (S1 Table) were used for salt tolerance screening in both the field and in hydroponic solutions. For field screening, 15 seeds were sown for each inbred line, and three replicates were conducted. Seeds were planted in soil compartment in which the NaCl concentration was adjusted to 0.3% (w/v) in Nanpi County, Hebei province, China. For hydroponic selection, five seedlings were used per line, and three replicates were performed. Seedlings were grown hydroponically in Hoagland’s full-strength nutrient solution until the third leaf was fully developed. The solution was aerated continuously with an electric pump and replaced every 2 days. Then, half of the seedlings were cultured in nutrient solution containing a final concentration of 160 mM NaCl while the remaining (control) samples were grown in solution lacking NaCl. The most salt-tolerant genotype and the most salt-sensitive genotype were selected for further analysis. For proteomic analysis, roots from ten plants were harvested and washed with distilled water for three times before being immersed into liquid nitrogen after 2 days of NaCl treatment from control (untreated) and treated samples; the samples were stored at -80°C for further use. Two independent biological replicates were conducted for proteomic analysis to validate the results.
Measurement of physiological parameters
Physiological responses to salinity stress were evaluated by measuring fresh weight, relative water content (RWC), osmotic potential, and relative electrolyte leakage (REL) in shoots, as well as cation content in both shoots and roots after salt treatment. To examine physiological changes in the seedlings, the average values from ten seedlings were calculated for each genotype, and five independent biological replicates were conducted. Plant shoot fresh weight and RWC were measured once per day for 6 consecutive days after salt treatment. Osmotic potential, REL, and cation content were measured after 2 days of treatment. For RWC, leaf fresh weight was measured immediately after the leaves were cut off the seedlings. Then, the sample was immersed in deionized water and incubated at 4°C overnight. The weight of the sample represented its rehydrated weight. Finally the sample was completely dried in an oven and its dry weight was calculated. RWC was calculated as (fresh weight—dry weight) / (rehydrated weight—dry weight). Leaf osmotic potential was measured with a vapor pressure osmometer (Vapro 5600, Wescor, Logan, UT, USA). Fresh leaves were obtained and soaked into deionized water for 6 h. The surface water was removed, and the leaves were frozen at -20°C for 2 h. The leaves were then thawed and pressed to obtain cell sap, which was subsequently analyzed for osmolarity (Os; mmol kg-1); osmotic potential (Mpa) = -Os × 2.58 × 10–3 [12,13]. The REL assay was conducted according to Liu et al. [13]. To measure cation contents, the roots were washed with distilled water for three times, plant shoots and roots were dried at 80°C and digested with 1% acid mixture (nitric acid: perchloric acid = 4:1). Na+ and K+ contents were analyzed using an Eppendorf flame photometer (Eppendorf, Hamburg, Germany).
Protein extraction
Total proteins were extracted from roots according to Lan et al. [14]. Total protein concentrations were determined using a Bradford Protein Assay Kit (GE Healthcare, Pittsburgh, PA, USA) according to the manufacturer’s instructions and the protein samples were stored at -80°C.
Protein digestion and iTRAQ labeling
Total proteins (100 μg samples) were digested with Trypsin Gold (Promega, Madison, WI, USA) at a ratio of protein: trypsin = 30:1 at 37°C for 16 h. After digestion, peptides were dried by vacuum centrifugation and reconstituted in 0.5 M TEAB. Labeling was performed according to the manufacturer’s protocol for iTRAQ (AB Sciex, Foster City, CA USA) with minor modifications. In brief, one unit of iTRAQ reagent (defined as the amount of reagent required to label 100 μg of protein) was thawed and reconstituted in 70 μL isopropanol. The control replicates were labeled with iTRAQ tags 113 and 114 for the salt-sensitive genotype and, 115 and 116 for the salt-tolerant genotype. The 160 mM NaCl treated replicates were labeled with tags 117 and 118, 119 and 121 for the salt-sensitive and-tolerant genotypes, respectively. The labeling reactions were incubated at room temperature for 2 h. Two technical replicates were performed.
LC-ESI-MS/MS analysis
LC-MS/MS analysis was performed on an LC-20AD nanoHPLC (Shimadzu) connected to an LTQ-Orbitrap Velos hybrid mass spectrometer (Thermo, Bremen, Germany). Each fraction was reconstituted in eluent buffer A (2% ACN, 0.1% FA) and centrifuged at 20 000×g for 10 min. Then, 10 μL supernatant was loaded onto a Shimadzu LC-20AD NanoHPLC (by the autosampler on a 2 cm C18 trap column (inner diameter 200 μm) and the peptides were eluted onto a resolving 10 cm analytical C18 column (inner diameter 75 μm). The samples were loaded at 15 μL/min for 4 min, and a 44 min gradient was then performed at 400 nL/min from 2% to 35% buffer B (98% ACN, 0.1% FA), followed by a 2 min linear gradient to 80%. The sample was maintained at 80% buffer B for 4 min and finally to 2% in 1 min.
The peptides were subjected to Nano electrospray ionization followed by tandem mass spectrometry (MS/MS) in an LTQ Orbitrap Velos coupled with HPLC. Intact peptides were detected in the Orbitrap at a resolution of 60 000. Peptides were selected for MS/MS using the high energy collision dissociation operating mode with a normalized collision energy setting of 45%. Ion fragments were detected using the Orbitrap FT-FT method. The electrospray voltage applied was 1.5 kV. Automatic gain control was used to prevent overfilling of the ion trap; 1×104 ions were accumulated in the ion trap to generate of high energy collision dissociation spectra. For MS scans, the m/z scan range was 350 to 2, 000 Da.
Protein identification and quantification
Mascot software (Matrix Science, London, UK) was used to simultaneously identify and quantify proteins. For protein identification, data files from the LC-ESI-MS/MS were searched against the NCBI Viridiplantae database (932, 602 sequences). The search parameters were as follows: trypsin was chosen as the enzyme with one missed cleavage allowed; fixed modifications of carbamidomethylation at Cys; variable modifications of oxidation of Met and N-term Glu-pyroglutamic acid; peptide mass tolerance was set at 10 ppm and fragment mass tolerance was set at ± 0.05 Da. For relative protein quantification, proteins were selected for further analysis based on the following criteria: at least two confident unique peptides, CV between the replicates smaller than 0.30. For each protein meeting the criteria between the biological and technical replicates, the iTRAQ ratios were averaged. For peptides matching multiple proteins during the database search, we conducted the quantitation using unique peptides. Proteins with average ratios greater than 2.0 were regarded as differentially altered proteins. For the gene ontology term enrichment test, the agriGO web service (http://bioinfo.cau.edu.cn/agriGO/index.php) was used [15].
Functional analysis of salt-responsive proteins in yeast
The corresponding gene sequences of the proteins were obtained by searching the MaizeSequence database (http://ensembl.gramene.org/Zea_mays/Info/Index). The ORFs were amplified by PCR from a maize cDNA library and cloned into the yeast expression vector pYES2, which contained the Ura3 selection marker. The exogenous gene was driven by the GAL1 promoter. The construct was introduced into yeast strain YPH500 (ura3-52 lys2-801 amber ade2-101 ochre trp1-Δ63 his3-Δ200 leu2-Δ1) according to the pYES2 vector kit instructions (Invitrogen, Carlsbad, CA, USA). Yeast salt tolerance assays were performed according to Gao et al. [16]; the NaCl concentrations were 0 M, 3 M, and 5 M. Yeast cells were collected before spotted on the agar plates, and yeast total RNAs were extracted using the RiboPure-Yeast RNA Isolation Kit (Life Technologies, Carlsbad, CA, USA).
Results and Discussion
Phenotypic differences between F63 and F35 under salt stress
To explore variations in salt tolerance, 162 maize inbred lines were preliminarily screened in soil compartments. The results were then validated with a hydroponic system in which seedlings harboring three full leaves were treated with 160 mM NaCl for 10 days. The two approaches produced similar results, i.e., lines F63 and F35 were the most salt-tolerant and salt-sensitive genotypes, respectively (S1 Fig.). Under a hydroponic screening system, F35 seedlings gradually withered and died while F63 seedlings survived during the 10-day of salinity treatment. The results showed that inbred line F63 was much more tolerant to salt stress than F35 at the early seedling stage. We chose these lines for further study.
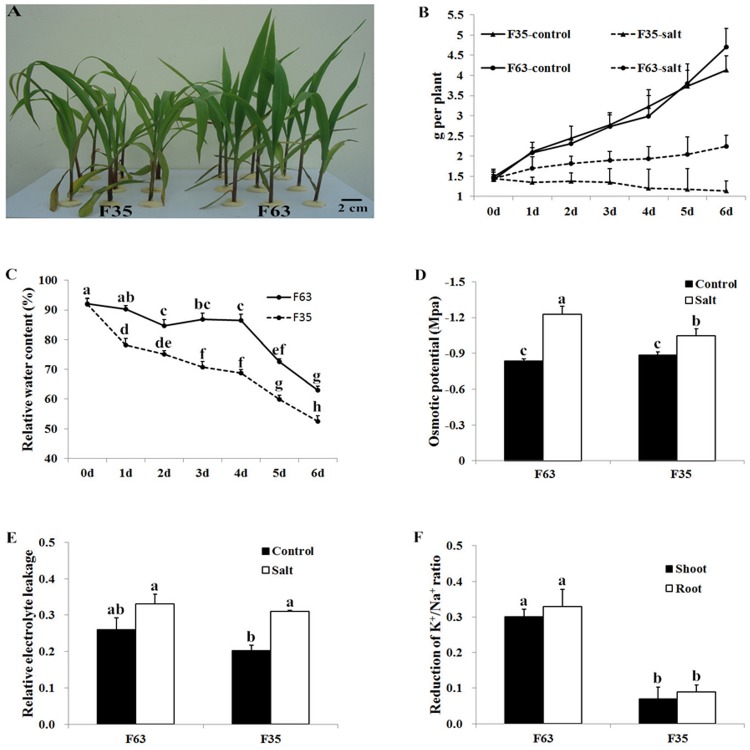
Plant growth is a classic index used to evaluate plant tolerance to abiotic stress [17–19]. Compared with plants grown under normal conditions, plants of both lines treated with 160mM NaCl showed growth retardation. However, F63 plants exhibited better growth status, with straighter and greener leaves than those of F35 (Fig. 1A, Fig. 1B). For F63, the shoot fresh weight increased gradually during the treatment, while for F35, no significant changes were observed during the first 3 days of treatment, followed by a decrease. The differences in shoot fresh weight between F63 and F35 were presumably caused by different levels of water loss in leaves under salt conditions, with F35 exhibiting sharp water loss (Fig. 1B, Fig. 1C).
Fig 1. Morphological and physiological changes in F63 and F35 seedlings under NaCl stress.
(A) Maize inbred lines F63 and F35 were grown hydroponically and treated with 160mM NaCl for 2 days. (B) Shoot fresh weight of F63 and F35 with or without 160mM NaCl treatment. (C) Leaf RWC of F63 and F35. (D) Leaf osmotic potential of F63 and F35 treated with 160mM NaCl for 2 days. (E) Leaf REL of F63 and F35. (F) Reduction of the K+/Na+ ratio after a 2-day NaCl treatment. For each parameter, ten seedlings were selected, and five independent biological replicates were conducted. Bars represent means ± SD (n = 5). Significant differences at P<0.01 according to Tukey’s test are indicated by different letters.
Salt stress can cause both osmotic stress and ion toxicity in plants [4]. To explore the different effects of salinity stress on F63 and F35, we measured the osmotic potential, REL, and K+/Na+ ratios after a 2-day NaCl treatment. The leaf osmotic potential declined in both inbred lines under salt stress, but the rate of decline in F63 was much greater than that of F35 (Fig. 1D). The greater decline of osmotic potential might enable F63 to retain more water in response to salt stress. REL is an indicator of membrane damage under stress conditions [20]. After a 2-day salt treatment, REL increased in both F63 and F35 (Fig. 1E). However, the increase was much greater in F35 than in F63, indicating more severe membrane damage in F35. To further understand the physiological status of the maize seedlings, we determined the concentrations of Na+ and K+ in shoots and roots. Compared to the control, the Na+ concentrations significantly increased in both inbred lines under salt treatment. However, the rate of increase was much greater in the salt-sensitive genotype F35 than in the salt-tolerant genotype F63, indicating that the salt-sensitive genotype accumulated more Na+ ions after salt treatment. Growth retardation of plants under salt stress is primarily caused by the uptake of excess Na+, and greater accumulation of Na+ disruptes K+ absorption and inhibites the activities of many enzymes in the cytoplasm, which impairs metabolism [21,22]. In the current study, the addition of NaCl to the nutrient solution reduced the K+ concentration in the plant, with the greatest decline detected in the roots of F35. Although the results showed that the K+/Na+ ratio in both shoots and roots decreased after treated with 160 mM NaCl, the salt-tolerant genotype F63 exhibited less reduction in this ratio in both shoots and roots compared to F35 (Fig. 1F). Therefore, the high salt tolerance capability of F63 may be closely related to the maintenance of ion homeostasis and membrane integrity under salt stress.
The results revealed that high concentrations of NaCl increased REL and reduced the shoot fresh weight, RWC, osmotic potential, and K+/Na+ ratio in maize. However, the degrees to which maize plants reacted to salt stress were different in F63 and F35. Under NaCl treatment, F63 exhibited a smaller increase in REL and less of reduction in the K+/Na+ ratio than F35. The low osmotic potential of F63 enabled itself to retain more water and a high RWC than F35. Therefore, F63 exhibited a better growth status and higher shoot fresh weight than F35 under salt stress.
Identification of salt stress-responsive proteins by iTRAQ LC-MS
To identify salt stress altered proteins in maize roots, we conducted a comparative proteomic analysis between F35 and F63. We extracted total root proteins from NaCl-treated and untreated seedlings and subjected them to proteomic analysis using a gel-free labeling approach. Two independent biological and technical replicates were performed using iTRAQ labeling followed by HPLC-MS/MS. We detected 856 and 857 proteins from two technical replicates, respectively (S2 and S3 Tables). The overlap of the detected proteins was summarized in S2 Fig. For the 617 reproducibly identified proteins in the replicates, 410 and 415 proteins could be quantified with at least two confident unique peptides and CV between the replicates smaller than 0.30 in F63 and F35, respectively. The detailed information of the quantified proteins was summarized in S4 Table. Compared with the control, treatment with 160 mM NaCl resulted in twenty-four and six differentially responsive proteins (>2.0 fold) in F63 and F35, respectively (Table 1). This result indicates that more proteins were altered in the salt-tolerant inbred line F63 under salt stress, suggesting that a dynamic metabolic process takes place in these plants in response to salt stimulus to help them cope with the osmotic stress and ion toxicity caused by salt stress [4].
Table 1. Classification of the salt-responsive proteins according to their abundance variation under salt treatment.
| F63-increased | F63-decreased | F63-constant | F63-null | Sum | |
|---|---|---|---|---|---|
| F35- increased | 2 | 0 | 3 | 1 | 6 |
| F35- decreased | 0 | 0 | 0 | 0 | 0 |
| F35-constant | 5 | 16 | - | - | 21 |
| F35-null | 1 | 0 | - | - | 1 |
| Sum | 8 | 16 | 3 | 1 | 28 |
Classification of salt-responsive proteins
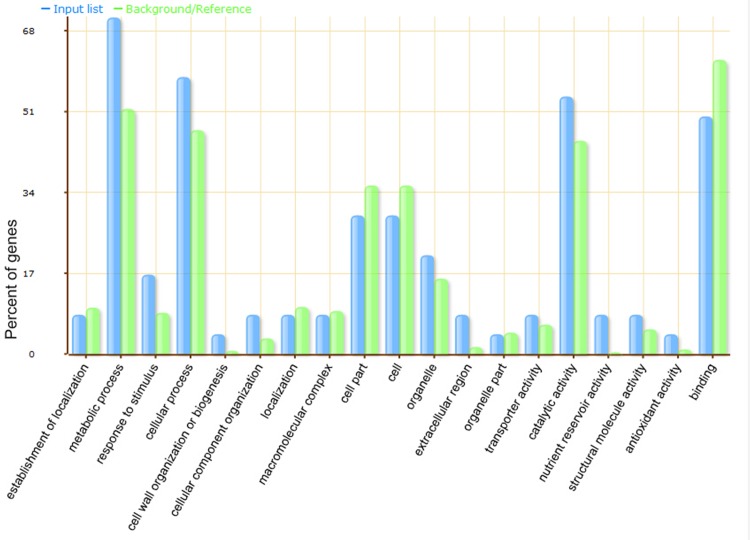
It is of fundamental importance to identify proteins that are differentially altered under salt stress. To further characterize these proteins, we performed gene ontology (GO) analysis using the agriGO web-based program. The salt-responsive proteins were classified into a diverse category (Fig. 2). Proteins involved in carbohydrate metabolic process were specifically enriched in the metabolic process category (p-value = 8.3×10–5, FDR < 0.05, S5 Table). They were exhydrolase II isoform 1 (gi|162463832), fructokinase-2 (gi|162460525), xyloglucan endotransglycosylase homolog (gi|162460193), alpha-1,4-glucan-protein synthase (gi|162463414), fructose bisphosphate aldolase (gi|255645227), lichenase-2 precursor (gi|195629642) and sucrose synthase (gi|459895). No proteins were enriched in the rest of the categories.
Fig 2. Functional classification of differentially-expressed proteins identified in this study.
AgriGO web-based program was used to analyze GO categories. The X-axis is the categories of GO terms. The Y-axis is the percentage of proteins mapped by the categories. The blue column represents input (the 28 differentially-expressed proteins, N = 24). The green column represents background (maize genome reference, N = 39203).
Differentially-regulated salt-responsive proteins in F63
As shown in Table 1, among the twenty-eight salt-responsive proteins, twenty-four were significantly altered in abundance under salt stress in salt-tolerant inbred line F63. Among the twenty-four significantly-altered proteins, twenty-two were specifically regulated in line F63. These differentially-regulated proteins might be the major contributors to the tolerant phenotype of F63.
In plants, the salt overly sensitive (SOS) signaling pathway regulates intracellular sodium ion (Na+) homeostasis and salt tolerance. In Arabidopsis, 14-3-3 proteins could inhibit the SOS signaling pathway by interacting with SOS2 protein for plant adaption to salt stress [23]. In crops, it was reported that the levels of 14-3-3 proteins were decreased in maize and rice [12,24], but increased in wheat and sugar beet under salinity stress conditions [25,26].These results indicated that 14-3-3 proteins may have diverse regulatory effects in plants in response to salt stress [8]. Here, we detected two 14-3-3 like proteins, gi|195635799 and gi|226507586, both of which were salt-reduced in F63. The decreased abundance of these two proteins in F63 might alleviate the inhibition of SOS signaling pathway, which could lead to sodium sequestration from the cytosol [27], and therefore maintain the K+/Na+ ratio better and enhanced the salt-tolerance of F63. Interestingly, the abundance of these two 14-3-3 like proteins was not changed in F35 under salt stress conditions.
Water uptake and flow across the cell membrane are essential for plant growth and development under salt stress. The plasma membrane intrinsic protein (PIP) is a subfamily of aquaporins comprising two subgroups of PIP1 and PIP2; and PIP2 proteins exhibit higher water channel activity [28]. In barley, expressions of several PIPs were down-regulated after 200 mM NaCl treatment, probably to prevent dehydration during salt stress [29]. In maize, ZmPIP2-4 were salt-induced after 2 h of 100 mM NaCl treatment, and salt-reduced under 200 mM NaCl for 24 h [30]. In the present study, the abundance of maize aquaporin PIP2-4 (gi|162459653) protein decreased in F63 under 160 mM NaCl for 2 days, while remained unchanged in F35. As a result, the diffusion of water to the outside of the plasma membrane might be reduced more successfully in F63 than that of in F35 by this change. And this could make F63 exhibit a relatively higher RWC than F35 under salinity condition, and then help the plant resist the physiological drought caused by osmotic stress.
Protein synthesis is of critical importance for plant abiotic stress adaption. The levels of many components of the protein synthesis machinery are altered under salinity conditions. The abundance of most proteins that were involved in protein synthesis is reduced in Arabidopsis roots under salinity treatment [10]. Ribosomal proteins, an important component of protein synthesis machinery, are salt-reduced in Arabidopsis and maize [11,31]. In the present study, two ribosomal-related proteins, ribosomal protein S8 (gi|968902) and 60S ribosomal protein L3-1 (gi|166858), were salt-reduced in the salt-tolerant genotype F63, while they showed inverted trend, even not significant in abundance in the salt-sensitive genotype F35 (Table 2). These results indicated that, under salt stress, the salt-tolerant genotype had the ability to reduce the synthesis of redundant proteins, which may help the plant save energy to battle salt stress.
Table 2. Identification of Salt-Responsive Proteins in Maize Roots.
| Accession‡ | Description | Species | Mass§ | Fold change¶ | CV# | ||
|---|---|---|---|---|---|---|---|
| F63 | F35 | F63 | F35 | ||||
| gi|195635409 | Histone H4 | Zea mays | 24379 | 0.24 | 1.20 | 0.13 | 0.18 |
| gi|162460024 | GST-4 | Zea mays | 27768 | 0.26 | 0.71 | 0.25 | 0.17 |
| gi|21263612 | Formate dehydrogenase | Hordeum vulgare subsp. vulgare | 50265 | 0.28 | 1.02 | 0.23 | 0.11 |
| gi|166858 | 60S ribosomal protein L3–1 | Arabidopsis thaliana | 59670 | 0.31 | 1.42 | 0.19 | 0.10 |
| gi|22160 | Adenine nucleotide translocator | Zea mays | 43216 | 0.33 | 0.81 | 0.07 | 0.08 |
| gi|224031309 | Adenosylhomocysteinase | Zea mays | 65444 | 0.33 | 0.81 | 0.23 | 0.05 |
| gi|293336485 | Heat shock protein 90 | Zea mays | 104965 | 0.37 | 1.01 | 0.12 | 0.04 |
| gi|226500532 | Seed maturation protein PM41 | Zea mays | 56879 | 0.39 | 0.73 | 0.18 | 0.08 |
| gi|162463414 | Alpha-1,4-glucan-protein synthase | Zea mays | 50209 | 0.41 | 1.47 | 0.12 | 0.07 |
| gi|556673 | Heat-shock protein | Secale cereale | 108178 | 0.44 | 1.12 | 0.06 | 0.12 |
| gi|162459653 | Aquaporin PIP2–4 plasma membrane integral | Zea mays | 33877 | 0.44 | 0.59 | 0.26 | 0.08 |
| gi|195635799 | 14-3-3-like protein | Zea mays | 36764 | 0.46 | 0.90 | 0.21 | 0.16 |
| gi|968902 | Ribosomal protein S8 | Oryza sativa Japonica Group | 33047 | 0.47 | 1.31 | 0.02 | 0.24 |
| gi|459895 | Sucrose synthase | Zea mays | 106574 | 0.48 | 0.74 | 0.08 | 0.12 |
| gi|162460525 | Fructokinase-2 | Zea mays | 42551 | 0.48 | 0.58 | 0.11 | 0.15 |
| gi|226507586 | 14-3-3-like protein | Zea mays | 35349 | 0.49 | 0.94 | 0.14 | 0.25 |
| gi|125558097 | Hypothetical protein OsI_25768 | Oryza sativa Indica Group | 18537 | 0.75 | 2.53 | 0.19 | 0.08 |
| gi|162460800 | Peroxidase 42 precursor | Zea mays | 36241 | 0.95 | 2.09 | 0.21 | 0.13 |
| gi|281398970 | Pathogenesis-related protein 10 | Zea mays | 20407 | 1.78 | 2.60 | 0.15 | 0.06 |
| gi|255645227 | Fructose bisphosphate aldolase | Arabidopsis thaliana | 47373 | - | 2.36 | - | 0.12 |
| gi|162463832 | Exhydrolase II isoform 1 | Zea mays | 75783 | 2.54 | 2.01 | 0.14 | 0.19 |
| gi|226508498 | Hypothetical protein | Zea mays | 42827 | 2.78 | 2.23 | 0.11 | 0.12 |
| gi|162460193 | Xyloglucan endotransglycosylase homolog | Zea mays | 27232 | 2.05 | 1.29 | 0.22 | 0.20 |
| gi|215769184 | Unnamed protein product | Oryza sativa Japonica Group | 72725 | 2.07 | - | 0.28 | - |
| gi|226501030 | hypothetical protein LOC100272932 | Zea mays | 27232 | 2.25 | 1.06 | 0.15 | 0.09 |
| gi|76574402 | Cysteine protease Mir1 | Zea diploperennis | 29993 | 2.39 | 0.67 | 0.06 | 0.10 |
| gi|195629642 | Lichenase-2 precursor | Zea mays | 36583 | 3.12 | 0.53 | 0.09 | 0.13 |
| gi|75994608 | Cysteine protease Mir1 | Zea mays subsp. parviglumis | 30325 | 4.62 | 1.34 | 0.04 | 0.15 |
‡ Protein accession number from NCBInr database.
§ Protein molecular weight.
¶ Mean of protein fold changes from salt-treated samples compared with the control.
# Coefficient of variation.
In this study, we also found several proteins do not fall in any known salt stress process. For example, two cysteine proteases (gi|76574402 and gi|75994608) were salt-increased in the salt-tolerant genotype F63, but they showed no significant changes in the salt-sensitive genotype F35. These two proteins are homologs of maize insect resistance 1 (Mir1), a papain-like cysteine protease. It was reported that Mir1 rapidly accumulated in the whorls of insect-resistant maize genotypes in response to feeding by lepidopteran larvae [32,33], indicating that plants may have developed cross-tolerance mechanisms to cope with abiotic and biotic stresses [34].
Functional verification of salt-responsive proteins in yeast
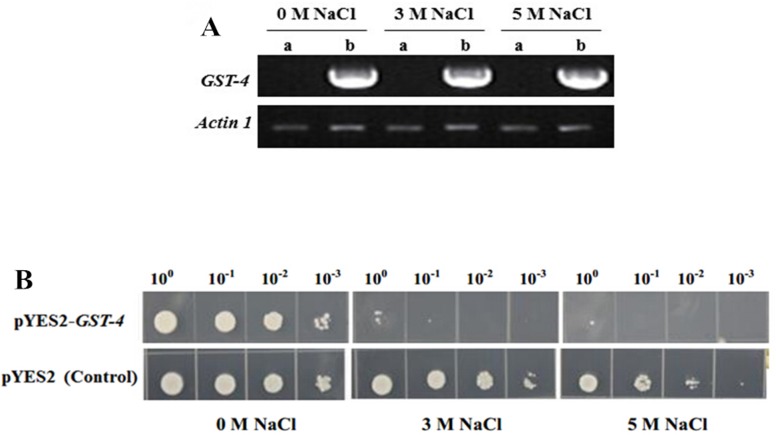
Many cellular processes/mechanisms of NaCl tolerance are conserved in yeast and plant cells [35,36]. In salt-tolerance genotype F63, the abundance of GST-4 (gi|162460024) decreased under salt stress. We over-expressed GST-4 in yeast to test its role in cellular salt tolerance. RT-PCR analysis showed that GST-4 had been transcribed in yeast under 0 M, 3 M and 5 M NaCl treatment (Fig. 3A). Over-expression of this gene did not affect the growth of yeast cells under normal conditions. However, under 3 M or 5 M NaCl treatment, yeast cells harboring pYES2-GST-4 grew more slowly than the control (harboring empty pYES2 vector) (Fig. 3B), indicating that over-expression of GST-4 may have a negative effect under salt stress. The results may be useful for further studies of salt-tolerance in maize.
Fig 3. Yeast cells over-expressing GST-4 showed decreased growth rate under salt stress.
(A) Reverse-trnscript PCR analysis of GST-4 in transgenic yeast after treated with NaCl for 24 h. a: yeast harboring pYES2 vector; b: yeast harboring pYES2-GST-4 vector. (B) Growth of yeast cells harboring pYES2-GST-4 vector or pYES2 vector under NaCl treatment. Yeast cells harboring pYES2-GST-4, or empty pYES2 vector (control) were respectively incubated in SC-ura liquid medium containing with 2% (w/v) galactose for 24 h at 30°C then adjusted to OD600 at 2.0 in 1 mL of medium for the stress experiments. For salt stress treatment, the yeast was resuspended in 0 M, 3 M or 5 M NaCl for 24 h. Serial dilutions were spotted onto SC-ura agar plates and incubated at 30°C for 48 h.
Conclusions
In this study, we identified twenty-eight salt-responsive proteins. Among them, twenty-two were specifically regulated in F63 but remained unchanged in F35. These proteins were mainly involved in signal processing, water conservation, protein synthesis and biotic cross-tolerance. They may contribute to the salt tolerance of this genotype. F63 exhibited a smaller increase in REL and less of reduction in the K+/Na+ ratio under NaCl treatment than F35, demonstrating that this salt-tolerant inbred line maintains membrane integrity and ion homeostasis more successfully than line F35. The low osmotic potential enabled F63 to retain more water and a high RWC in response to salt treatment. Therefore, F63 exhibited a better growth status than F35 under salt stress.
Supporting Information
(A) A total of 162 inbred lines were screened in salt soil pools containing 0.3% (w/v) NaCl in Nanpi County, Hebei Province, China. Fifteen seeds were sown per inbred line, and three replicates were conducted. (B) Maize inbred lines were screened under hydroponic conditions. The salt-tolerant genotype F63 and the salt-sensitive genotype F35 are indicated in the photograph.
(TIF)
Replicate I and replicate II detected 856 and 857 proteins, respectively. 617 proteins were reproducibly identified.
(TIF)
(XLSX)
(XLS)
(XLS)
(XLS)
(XLSX)
Acknowledgments
The authors thank the China National Science Foundation (Grant No. 31201214) and the National Sci-Tech Support program (Grant No. 2013BAD05B01) for providing funds for carrying out this research work.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by grant support from the China National Science Foundation (Grant No. 31201214, http://www.nsfc.gov.cn/publish/portal1/) and the National Sci-Tech Support program (Grant No. 2013BAD05B01, http://program.most.gov.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Parida AK, Das AB, Mittra B (2004) Effects of salt on growth, ion accumulation, photosynthesis and leaf anatomy of the mangrove, Bruguiera parviflora . Trees-Struct Funct 18: 167–174. [Google Scholar]
- 2. Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45: 437–448. [Google Scholar]
- 3. Khan MH, Panda SK (2008) Alterations in root lipid peroxidation and antioxidative responses in two rice cultivars under NaCl-salinity stress. Acta Physiol Plant 30: 81–89. [Google Scholar]
- 4. Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681. 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- 5. Steppuhn H, Asay KH (2005) Emergence, height, and yield of tall, NewHy, and green wheatgrass forage crops grown in saline root zones. Can J Plant Sci 85: 863–875. [Google Scholar]
- 6. Schweppe RE, Haydon CE, Lewis TS, Resing KA, Ahn NG (2003) The characterization of protein post-translational modifications by mass spectrometry. Acc Chem Res 36: 453–461. [DOI] [PubMed] [Google Scholar]
- 7. Canovas FM, Dumas-Gaudot E, Recorbet G, Jorrin J, Mock HP, et al. (2004) Plant proteome analysis. Proteomics 4: 285–298. [DOI] [PubMed] [Google Scholar]
- 8. Zhao Q, Zhang H, Wang T, Chen S, Dai S (2013) Proteomics-based investigation of salt-responsive mechanisms in plant roots. J Proteomics 82:230–253. 10.1016/j.jprot.2013.01.024 [DOI] [PubMed] [Google Scholar]
- 9. Yan S, Tang Z, Su W, Sun W (2005) Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 5: 235–244. [DOI] [PubMed] [Google Scholar]
- 10. Zhang H, Han B, Wang T, Chen S, Li H, et al. (2012) Mechanisms of plant salt response: insights from proteomics. J Proteome Res 11: 49–67. 10.1021/pr200861w [DOI] [PubMed] [Google Scholar]
- 11. Zorb C, Schmitt S, Muhling KH (2010) Proteomic changes in maize roots after short-term adjustment to saline growth conditions. Proteomics 10: 4441–4449. 10.1002/pmic.201000231 [DOI] [PubMed] [Google Scholar]
- 12. Blum A (1989) Osmotic adjustment and growth of Barley genotypes under drought Stress. Crop Sci 29: 230–233. [Google Scholar]
- 13. Liu Y, Du H, He X, Huang B, Wang Z (2012) Identification of differentially expressed salt-responsive proteins in roots of two perennial grass species contrasting in salinity tolerance. J Plant Physiol 169: 117–126. 10.1016/j.jplph.2011.08.019 [DOI] [PubMed] [Google Scholar]
- 14. Lan P, Li W, Wen TN, Shiau JY, Wu YC, et al. (2011) iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol 155: 821–834. 10.1104/pp.110.169508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–70. 10.1093/nar/gkq310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao CQ, Wang YC, Jiang B, Liu GF, Yu LL, et al. (2011) A novel vacuolar membrane H+-ATPase c subunit gene (ThVHAc1) from Tamarix hispida confers tolerance to several abiotic stresses in Saccharomyces cerevisiae. Mol Biol Rep 38: 957–963. 10.1007/s11033-010-0189-9 [DOI] [PubMed] [Google Scholar]
- 17. Munns R, Husain S, Rivelli AR, James RA, Condon AG, et al. (2002) Avenues for increasing salt tolerance of crops, and the role of physiologically based selection traits. Plant Soil 247: 93–105. [Google Scholar]
- 18. Sairam RK, Rao KV, Srivastava GC (2002) Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci 163: 1037–1046. [Google Scholar]
- 19. Ashraf M, Harris PJC (2004) Potential biochemical indicators of salinity tolerance in plants. Plant Sci 166: 3–16. [Google Scholar]
- 20. Pang QY, Chen SX, Dai SJ, Chen YZ, Wang Y, et al. (2010) Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila . J Proteome Res 9: 2584–2599. 10.1021/pr100034f [DOI] [PubMed] [Google Scholar]
- 21. Xiong LM, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14: S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51: 463–499. [DOI] [PubMed] [Google Scholar]
- 23. Zhou H, Lin H, Chen S, Becker K, Yang Y, et al. (2014) Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26:1166–1182. 10.1105/tpc.113.117069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng Y, Qi Y, Zhu Q, Chen X, Wang N, et al. (2009) New changes in the plasma-membrane-associated proteome of rice roots under salt stress. Proteomics 9: 3100–3114. 10.1002/pmic.200800340 [DOI] [PubMed] [Google Scholar]
- 25. Wang MC, Peng ZY, Li CL, Li F, Liu C, et al. (2008) Proteomic analysis on a high salt tolerance introgression strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 8: 1470–1489. 10.1002/pmic.200700569 [DOI] [PubMed] [Google Scholar]
- 26. Yang L, Ma C, Wang L, Chen S, Li H (2012) Salt stress induced proteome and transcriptome changes in sugar beet monosomic addition line M14. J Plant Physiol 169: 839–850. 10.1016/j.jplph.2012.01.023 [DOI] [PubMed] [Google Scholar]
- 27. Zhu J-K (2003) Regulation of ion homeostasis under salt stress. Cur Opin Plant Biol 6: 441–445. [DOI] [PubMed] [Google Scholar]
- 28. Chaumont F, Barrieu F, Jung R, Chrispeels MJ (2000) Plasma membrane intrinsic proteins from maize cluster in two sequence subgroups with differential aquaporin activity. Plant Physiol 122: 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M K (2012) Water transport in plants: from molecules to whole plant. Scientific Conference Proceedings: 781–785.
- 30. Zhu C, Schraut D, Hartung W, Schaffner AR (2005) Differential responses of maize MIP genes to salt stress and ABA. J Exp Bot 56: 2971–2981. [DOI] [PubMed] [Google Scholar]
- 31. Jiang Y, Yang B, Harris NS, Deyholos MK (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58: 3591–3607. [DOI] [PubMed] [Google Scholar]
- 32. Pechan T, Ye LJ, Chang YM, Mitra A, Lin L, et al. (2000) A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other lepidoptera. Plant Cell 12: 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lopez L, Camas A, Shivaji R, Ankala A, Williams P, et al. (2007) Mir1-CP, a novel defense cysteine protease accumulates in maize vascular tissues in response to herbivory. Planta 226: 517–527. [DOI] [PubMed] [Google Scholar]
- 34. Tuteja N (2007) Mechanisms of high salinity tolerance in plants. Methods Enzymol 428: 419–438. [DOI] [PubMed] [Google Scholar]
- 35. Lippuner V, Cyert MS, Gasser CS (1996) Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J Biol Chem 271: 12859–12866. [DOI] [PubMed] [Google Scholar]
- 36. Shin D, Koo YD, Lee J, Lee HJ, Baek D, et al. (2004) Athb-12, a homeobox-leucine zipper domain protein from Arabidopsis thaliana, increases salt tolerance in yeast by regulating sodium exclusion. Biochem Biophys Res Commun 323: 534–540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) A total of 162 inbred lines were screened in salt soil pools containing 0.3% (w/v) NaCl in Nanpi County, Hebei Province, China. Fifteen seeds were sown per inbred line, and three replicates were conducted. (B) Maize inbred lines were screened under hydroponic conditions. The salt-tolerant genotype F63 and the salt-sensitive genotype F35 are indicated in the photograph.
(TIF)
Replicate I and replicate II detected 856 and 857 proteins, respectively. 617 proteins were reproducibly identified.
(TIF)
(XLSX)
(XLS)
(XLS)
(XLS)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.