Abstract
Purpose
The cell-cycle checkpoint kinase 2 (CHEK2) is an important signal transducer of cellular responses to DNA damage, whose defects has been associated with increased risk for breast cancer. The CHEK2 1100delC mutation has been reported to confer a twofold increased risk of breast cancer among carriers. The frequency of the mutation varies among populations. The highest frequency has been described in Northern and Eastern European countries. However, the 1100delC mutation has been investigated in different case-control studies and none in Moroccan population. The aim of this study was to evaluate the prevalence of this variant and determine its contribution to the development of breast cancer in sporadic cases and also in members of breast cancer families who tested negative or positive for a deleterious mutation in BRCA1/BRCA2.
Methods
In this case-control study we performed the CHEK2 1100delC mutation analysis by ASO-PCR in 134 breast cancer patients and 114 unaffected control individuals. Most of these families had several cases of breast cancer or ovarian cancer (or both).
Results
No CHEK2 1100delC mutations were detected in any of 134 individuals, including 59 women diagnosed with breast cancer at an early age (<40 years), 10 women with bilateral breast cancer, and 6 women with ovarian cancer.
Conclusion
Our preliminary genetic analysis are consistent with the reported very low frequency of CHEK2 1100delC mutation in North American populations (compared with Northern Europe), rendering CHEK2 1100delC such as an unlikely to be major breast cancer susceptibility genes.
Introduction
Cell-cycle checkpoint kinase 2 (CHEK2 [MIM 604373]) is a tumor suppressor gene widely researched as a strong candidate gene for breast cancer susceptibility (Iniesta et al. 2010; Vahteristo et al. 2002; CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004; Mateus Pereira et al. 2004; Offit et al. 2003; Zhang et al. 2008; Rashid et al. 2005; Kleibl et al. 2005; Cybulski et al. 2007; Friedrichsen et al. 2004; Bernstein et al. 2006; Sodha et al. 2002; McInerney et al. 2009; Dufault et al. 2004; Einarsdottir et al. 2006; Weischer et al. 2007; Baeyens et al. 2005; Fletcher et al. 2009). The CHEK2 gene has been shown to play a major role in deoxyribonucleic acid (DNA) repair, apoptosis and cell cycle regulation. Indeed, in response to DNA damage, the ATM/CHEK2/p53 pathway is activated. The ATM (Ataxia Telangiectasia Mutated) phosphorylates CHEK2, which in turn phosphorylates p53 leading to cell cycle arrest and apoptosis (Iniesta et al. 2010). In addition, CHEK2 protein regulates BRCA1 in response to DNA damage (Oldenburg et al. 2007).
The CHEK2 1100delC protein truncating variant is situated in exon 10 of the functional gene on chromosome 22q. This variant is caused by the deletion of a single cytosine at position 1100 resulting in the introduction of a stop codon after amino acid 380, inducing a loss of activity of the CHEK2 kinase (Iniesta et al. 2010).
The CHEK2 1100delC variant was found in women suffering from breast cancer with familial Li-Fraumeni syndrome (Bell et al. 1999). In 2004, CHEK2-Breast Cancer Consortium did a collaborative analysis with 10 studies from 5 western countries, which involved 10 860 breast cancer cases and 9 065 controls (CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004). The Consortium found the frequency of CHEK2 1100delC to be 1.9% and 0.7% in cases and controls respectively, and confirmed that this gene variant could potentially increase the risk of breast cancer. The CHEK2 1100delC variant has been reported to be a low-penetrance breast cancer susceptibility allele (Vahteristo et al. 2002; CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004; Oldenburg et al. 2003; Kuusisto Kirsi et al. 2011). It results in an approximately two-fold risk of breast cancer in women and a ten-fold risk in men (van der Groep et al. 2011). It has been detected in 5% of breast cancer patients from non-BRCA1 and BRCA2 families (Vahteristo et al. 2002; Meijers-Heijboer et al. 2002). The frequency of CHEK2 1100delC allele varies among different populations. Indeed, high mutation rates are seen in Northern and Eastern European countries (Vahteristo et al. 2002; CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004; Meijers-Heijboer et al. 2002) although its frequency is much lower in North America (Mateus Pereira et al. 2004; Offit et al. 2003), whereas the mutation does not seem to be a triggering factor to breast cancer in Poland (Kwiatkowska et al. 2006; Cybulski et al. 2004) and some multiple-case breast cancer families from Australia (Jekimovs et al. 2005). Likewise, the frequency of CHEK2 1100delC seems to be very low in Southern Europe, Italy (Caligo et al. 2004), Spain (Osorio et al. 2004; Bellosillo et al. 2005) and rare in Brazil (Zhang et al. 2008).
Moreover, another quantitative synthesis was done by Weischer et al. (Weischer et al. 2007). Combined with 16 studies, it showed that CHEK2 1100delC heterozygotes rate was 3- to 5-fold higher in the breast cancer group than the control group. However, this widely discussed variant of CHEK2 – which seemed clearly associated with the predominance of breast cancer in western countries – was rarely detected in Asian populations, such as the Chinese (Song et al. 2006), Koreans (Choi et al. 2008), Japanese (Bell et al. 2007), Singaporeans (Lee & Ang 2008), Malaysians (Thirthagiri et al. 2009) and South Indians (Rajkumar et al. 2003).
In Morocco, according to the Greater Casablanca Cancer Registry, breast cancer seems to be the first female cancer with a standardized incidence of 36.4 per 100,000 women for an average age of 49.5 years (Bouchbika et al. 2013). Although this incidence appears higher compared to the other Maghreb countries (Hamdi Cherif et al. 2010; Ben Abdallah et al. 2009; El Mistiri et al. 2010), it remains low compared to developed countries (Marrett et al. 2008; Botha et al. 2003; Jemal et al. 2011). In the greater area of Casablanca, 57 % of the cases were under 50 years of age (Bouchbika et al. 2013) and 7% of cases registered at the city of Rabat are younger than 35 years (Tazi et al. 2013).
In addition, Morocco is a country of northwestern Africa composed predominantly of Berber and Arab ethnic groups. The contribution of these ethnic groups to the genetic diversity of Moroccan population is evident. Therefore, it is crucial to improve our understanding, both on the subject of genetic susceptibilities and environmental risk factors. In the present study, we examined DNA from Moroccan patients who have been screened and found not to carry BRCA1/2 mutations. The purpose of the study was to determine the frequency of CHEK2 1100delC mutation and the implication of CHEK2 as a breast cancer susceptibility gene in the Moroccan population.
Materials and methods
Subjects
DNA samples were collected from 134 patients. Among them, 113 were recruited from Mohammed VI Center for Cancer Treatment of Ibn Rochd University Hospital of Casablanca during 2009–2010 and 21 patients were kindly provided by the Oncology Unit of the Littoral Clinic during 2013.
Data were collected from medical records and clinico-pathological reports. The group of sporadic cases (n = 75) presented a mean age of 36.9 years with an age range of 23–59 years. The group of familial breast and/or ovarian cases (n = 59) presented a mean age of 44.8 years with an age range of 25–67 years, and included women with specific family-history criteria (Figure 1):
Three or more first or second degree relatives with breast cancer diagnosed in the same familial branch, at any age
Two first degree relatives with breast cancer, with at least one early onset breast cancer case (≤40 years) or male breast cancer case or ovarian cancer case.
Figure 1.
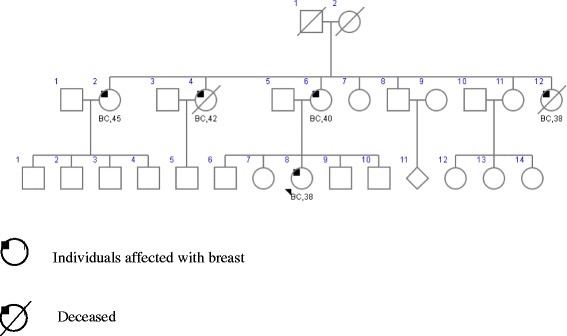
Pedigree corresponding to one of the families included in the present study of CHEK2 1100delC mutation in hereditary breast cancer. The index case is indicated with an arrow.
The control group included a total of 114 matched healthy women with no personal history of cancer diseases selected from DNA bank volunteers of the Genetics and Molecular Pathology Laboratory with a median age of 36.4 years and an age range of 20–77 years.
The study was approved by our institutional ethics committee and written informed consent for genetic testing was obtained from all subjects.
DNA isolation and PCR amplification
Genomic DNA was extracted from peripheral blood leukocytes using the salting out method (Miller et al. 1988). Genomic DNA was dissolved in TE (10 mM Tris–HCl and 0.1 mM EDTA, pH 8.0), confirmed by agarose gel electrophoresis and then quantified using the Nanovue TM Plus spectrophotometer.
ASO-PCR analysis was performed to determine the 1100delC mutation of CHEK2 gene, as described previously (Rashid et al. 2005).The CHEK2 1100delC mutation genotyping was performed with forward primer CHEK2ex10f (5′-GCAAAATTAAATGTCCTAACTTGC-3′) in combination with previously described reverse primers CHEK2ex10r andCHEK2delCr primers (Cybulski et al. 2004).
CHEK2 gene was amplified in a final volume of 25 μl containing: 10× reaction buffer, 25 mM MgCl2, 5 mM dNTPs, 5 μM primers, 500 U HotStarTaq DNA polymerase and 150 ng genomic DNA. Touchdown PCR conditions were as follows: 15 min at 95°C, 10 cycles of 20 s at 94°C, 25 s at annealing temperature 68–55°C touchdown (1.4°C/cycle), and 30 s at 72°C followed by 36 cycles of 20 s at 94°C, 25 s at 55°C, and 35 s at 72°C, and then a final extension step of 10 min at 72°C.
The PCR product was separated by electrophoresis in a 1% agarose gel containing ethidium bromide and visualized under UV illumination. Wild type allele resulted in 537 bp fragment and variant allele resulted in 200 bp fragment.
Results
A total of 134 Moroccan subjects were selected for our study. General characteristic of the subjects including distribution of tumor characteristic such as histological grade and location of cancer were obtained from patients’ medical records and listed in Table 1. In total, 134 breast cancer cases and 114 controls were successfully screened for the 1100delC variant of the CHEK2 gene using the ASO-PCR technique. None of the 134 Moroccan breast cancer patients carried the CHEK2 1100delC mutation. Moreover among the 134 cases, 40 patients had been screened for BRCA1 and BRCA2 mutations (Tazzite et al. 2012). Thus, in the 30 patients who were non-BRCA mutation carriers, the analysis of 1100delC mutation was also found negative.
Table 1.
Characteristics of individuals with breast cancer at time of diagnosis, screened for the CHEK2 1100delC mutation
| Characteristics | Samples |
|---|---|
| Cases/Controls | 134 / 114 |
| Age at diagnosis, mean ± SD (years) | 41 ± 11 |
| Range (years) | 23– 67 |
| Menopausal status | No. (%) |
| Premenopausal | 76(56.71) |
| Postmenopausal | 57(42.53) |
| Missing | 1(0.74) |
| Estrogen receptor | |
| Positive | 100 (74.62) |
| Negative | 24(17.91) |
| Missing | 10 (7.46) |
| Progesterone receptor | |
| Positive | 98 (73.13) |
| Negative | 30(22.38) |
| Missing | 6 (4.47) |
| Estrogen/Progesterone receptor | |
| ER + /PR + | 66 (49.25) |
| ER + /PR − | 15 (11.19) |
| ER − /PR + | 10 (7.46) |
| ER − /PR − | 43 (32.08) |
| Tumor size | |
| <2 cm | 20 (14.92) |
| >2 cm | 56 (41.79) |
| >5 cm | 31(23.13) |
| Tumor of any size with extension | 27 (20.14) |
| Histological grade | |
| 1 | 8 (5.97) |
| 2 | 87 (64.92) |
| 3 | 39 (29.10) |
| Lymph node status | |
| Negative | 56(41.79) |
| Positive | 78 (58.20) |
| Distant metastases | |
| Negative | 106(79.10) |
| Positive | 28 (20.89) |
This finding suggests that this mutation is probably not present or is present at an extremely low frequency in Moroccan population.
Discussion
In our present molecular study, we evaluated the involvement of the low-penetrance susceptibility 1100delC allelic variant in the CHEK2 gene in the Moroccan population. For this purpose, we performed a screening of this mutation in 134 Moroccan breast cancer patients and 114 healthy controls. Our results have shown that none of the 248 analyzed samples carried the CHEK2 1100delC mutation, suggesting that the frequency of this mutation is extremely low (or not present) in the Moroccan population.
On the other hand, our results are in line with some previous findings showing that there was no association between CHEK2 1100delC mutation and risk for Breast Cancer (Zhang et al. 2008; Caligo et al. 2004; Osorio et al. 2004; Bellosillo et al. 2005; Rajkumar et al. 2003; Sodha et al. 2007; Gonzalez-Hormazabal et al. 2008) (Table 2). Nonetheless, it should be noted that in Northern and Eastern European (Vahteristo et al. 2002; CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004; Meijers-Heijboer et al. 2002) CHEK2 1100delC mutation is associated with an increased risk of Breast Cancer (on the basis of age at diagnosis and family history of breast cancer).
Table 2.
Frequency of CHEK2 1100delC mutation in breast cancer cases and controls by ethnicity
| Overall | Overall | Carriers n (frequency of carriers, %) | Carriers n (frequency of carriers, %) | ||||
|---|---|---|---|---|---|---|---|
| Study | Population | Year | Case | Control | Case | Control | Reference |
| América | |||||||
| 1. Meijers-Heijboer and al | USA | 2002 | 264 | 166 | 6 (2.3) | 1(0.6) | (Meijers-Heijboer et al. 2002) |
| 2. Offit KPH and al | USA | 2003 | 67 | 569 | 0 (0) | 2 (0.4) | (Offit et al. 2003) |
| 3. Pereira and al | USA | 2004 | 829 | 859 | 9 (1.1) | 4(0.5) | (Mateus Pereira et al. 2004) |
| 4. Friedrichsen and al | USA | 2004 | 506 | 459 | 6 (1.2) | 2 (0.4) | (Friedrichsen et al. 2004) |
| 5. Bernstein and al | CANADA | 2006 | 1199 | 496 | 18 (1.34) | 1 (0.2) | (Bernstein et al. 2006) |
| 6. Novak and al | CANADA | 2008 | 149 | 141 | 3 (2.01) | 1 (0.7) | (Novak et al. 2008) |
| 7. Shiyu Zhang and al | CANADA | 2008 | 560 | 6460 | 4 (0.7) | 19 (0.3) | (Zhang et al. 2008) |
| 8. Gonzalez-Hormazabal and al | CHILE | 2008 | 196 | 1124 | 0 (0) | 0 (0) | (Gonzalez-Hormazabal et al. 2008) |
| 9. Shiyu Zhang and al | BRAZIL | 2008 | 155 | 377 | 1 (0.7) | 0 (0) | (Zhang et al. 2008) |
| 10. Bell DW | LATINAS | 2007 | 362 | 384 | 1 | 0 (0) | (Sodha et al. 2007) |
| Europe | |||||||
| Sodha and al | UNITED KINGDOM | 2002 | 68 | 300 | 3(4.4) | 0(0) | (Sodha et al. 2002) |
| Check2 Consortium | UNITED KINGDOM | 2004 | 2886 | 3749 | 35(1.2) | 20 (0.53) | (CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004) |
| Meijers-Heijboer and al | UNITED KINGDOM | 2002 | 564 | 288 | 7(1.3) | 1(0.35) | (Meijers-Heijboer et al. 2002) |
| Meijers-Heijboer and al | THE NETHERLANDS | 2002 | 79 | 460 | 2(2.5) | 6(1.2) | (Meijers-Heijboer et al. 2002) |
| Oldenburg and al | THE NETHERLANDS | 2003 | 237 | 212 | 27(11.4) | 6(2.8) | (Oldenburg et al. 2003) |
| Check2 Consortium | THE NETHERLANDS | 2004 | 1706 | 184 | 65(3.8) | 3(1.6) | (CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004) |
| De Jong and al | THE NETHERLANDS | 2005 | 962 | 367 | 28(2.9) | 5(1.4) | (De Jong et al. 2005) |
| Offit KPH and al | ASHKENAZI JEWISH | 2003 | 33 | 1096 | 1(3) | 3(0.3) | (Offit et al. 2003) |
| Shiyu Zhang and al | JEWISH | 2008a | 320 | 180 | 4(1.3) | 0(0) | (Zhang et al. 2008) |
| McInerney and al | IRELAND | 2009 | 903 | 1016 | 5(0.5) | 1(0.1) | (McInerney et al. 2009) |
| Martinez-Bouzas and al | BASQUE COUNTRY | 2007 | 214 | 120 | 2(0.93) | 0(0) | (Martınez-Bouzas et al. 2007) |
| Cybulski and al | POLAND | 2004a | 1017 | 4000 | 5(0.5) | 10(0.25) | (Cybulski et al. 2004) |
| Cybulski and al | POLAND | 2007b | 4454 | 5496 | 20(0.4) | 12(0.2) | (Cybulski et al. 2007) |
| Kleibl and al | CZECH REPUBLIC | 2005 | 1046 | 730 | 4(0.38) | 2(0.27) | (Kleibl et al. 2005) |
| Check2 Consortium | GERMANY | 2004 | 985 | 401 | 11(1.1) | 1(0.25) | (CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004) |
| Dufault and al | GERMANY | 2004 | 516 | 1315 | 8(1.6) | 6(0.5) | (Dufault et al. 2004) |
| Rashid and al | GERMANY | 2005 | 613 | 651 | 5(0.82) | 6(0.92) | (Rashid et al. 2005) |
| Einarsdottir and al | SWEDEN | 2006 | 1510 | 1334 | 20(0.7) | 8(0.4) | (Einarsdottir et al. 2006) |
| Margollin and | SWEDEN | 2007 | 450 | 760 | 10(2.2) | 5(0.7) | (Margolin et al. 2007) |
| Weischer and al | DENMARK | 2007 | 1088 | 4643 | 13(1.1) | 22(0.5) | (Weischer et al. 2007) |
| Vahteristo and al | FINLAND | 2002 | 1035 | 1885 | 21(2.1) | 26 (1.4) | (Vahteristo et al. 2002) |
| Check2 Consortium | FINLAND | 2004 | 464 | 447 | 13(2.9) | 5(1.1) | (CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004) |
| Kirsi and al | FINLAND | 2011 | 82 | 380 | 3(0.037) | 6(0.016) | (Kuusisto Kirsi et al. 2011) |
| Osorio and al | SPAIN | 2004 | 456 | 400 | 0(0) | 0(0) | (Osorio et al. 2004) |
| Caligo and al | ITALY | 2004 | 939 | 334 | 1(0.1) | 0(0) | (Caligo et al. 2004) |
| Baeyens and al | BELGIUM | 2005 | 178 | 100 | 4(2.24) | 0(0) | (Baeyens et al. 2005) |
| Asia | |||||||
| Shiyu Zhang and al | FILIPINO | 2008 | 342 | 7 | 0(0) | 0(0) | (Zhang et al. 2008) |
| Chekmariova and al | RUSSIA | 2006 | 815 | 448 | 22(5.2) | 1(0.2) | (Chekmariova et al. 2006) |
| Bell DW | JAPANE | 2007 | 428 | 378 | 0(0) | 0(0) | (Sodha et al. 2007) |
| Rajkumar T and al | SOUTH INDIA | 2003 | 22 | 1 | 0(0) | 0(0) | (Rajkumar et al. 2003) |
| Africa | |||||||
| Present study | MOROCCO | 2013 | 134 | 114 | 0(0) | 0(0) | Present study |
A rational explanation for these discrepancies may be due to ethnic or geographic variations. As reported in several studies, it is evident that the contribution of CHEK2 1100delC mutation to the burden of cancer varies according to the ethnic group, and from country to country (Antoni et al. 2007; Gonzalez-Hormazabal et al. 2008; Martınez-Bouzas et al. 2007) (Table 2). For instance, CHEK2 1100delC mutation was frequently observed in some Western and Northern Europe (CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004), but it was very rare in the Central Europe (Kleibl et al. 2005; Kwiatkowska et al. 2006), Southern Europe (Italy and Spain) (Caligo et al. 2004; Osorio et al. 2004) and Australia (Jekimovs et al. 2005). In Basque Country, Martinez-Bouzas et al. reported the 1100delC mutation in 0.93% of the cases with breast cancer, and in none of the control populations. Therefore, the authors raise the hypothesis of the existence of a 1100delC frequency gradient from the North-West to the South-East of Europe, caused by an ancestral common origin in the Northern Europe (Osorio et al. 2004; Gonzalez-Hormazabal et al. 2008; Martınez-Bouzas et al. 2007). Likewise, this variant is very low in North America (CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004; Offit et al. 2003), and rare in Brazil (Zhang et al. 2008). The CHEK2 1100delc mutation is not present in Chilean families with familial breast (Gonzalez-Hormazabal et al. 2008), and it was rarely detected in Asian populations, such as the Chinese (Song et al. 2006; Chen et al. 2008), Koreans (Choi et al. 2008), Japanese (Sodha et al. 2007), Singaporeans (Lee & Ang 2008), Malysians (Thirthagiri et al. 2009), South Indians (Rajkumar et al. 2003), and Philippines (Zhang et al. 2008).
Antoniou et al. (2002) suggested that susceptibility to breast cancer in non-carriers of BRCA1 and BRCA2 mutations may be mainly due to a “polygenic” model, with a large number of susceptibility alleles, each conferring a small increase in risk. Further evaluation of these interactions will be required to identify and analyze other susceptibility genes. Moreover, Martınez-Bouzas et al. (2007) detected a 0,93% prevalence among 214 Basque Country non BRCA1/2 patients and Gutiérrez-Enríquez et al. (2008)) reported 0,3% frequency in 331 non BRCA1/2 families from Basque Country and Catalonia. On the other hand, Zhang et al. (2008) found no CHEK2 mutation in 307 White women with breast cancer and a BRCA mutation. It is unlikely that women with a BRCA mutation will be found to harbor a CHEK2 mutation.
However, it is possible that other CHEK2 variants will confer susceptibility to breast cancer in other countries. Therefore, many studies screened the full coding sequence of CHEK2. For example, a splice-site mutation in CHEK2 IVS2 + G > A has been found to be associated with breast cancer susceptibility in Poland (Cybulski et al. 2007), and the S428F allele of the CHEK2 gene increases breast cancer risk in Ashkenazi Jewish women (Shaag et al. 2005). Likewise, several previous studies have suggested that CHEK2 I157T variant may contribute to inherited breast cancer predisposition (Cybulski et al. 2007; Kilpivaara et al. 2004; Bogdanova et al. 2005).
A limitation of our study was the reliance on family members’ reports of cancer in their relatives. Secondly, the sample size was limited and it is important to confirm our findings in a larger study. Thirdly, this study only focused on single gene without taking into consideration SNP-SNP and gene–gene interactions, or the possibility of linkage disequilibrium between polymorphisms, which may affect individual susceptibility to breast cancer.
Conclusion
Since its discovery as a BC susceptibility allele, the occurrence of CHEK2 1100delC mutation depends on the geographical area and/or ethnical characteristics of populations (CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer 2004). Thus, the absence or rarity of the CHEK2*1100delC heterozygosity among patients with breast cancer in our population underlines the importance of considering ethnic background before offering a genetic test. Further studies need to expand and elaborate on the putative contribution of this variant in the Moroccan population.
Acknowledgments
We thank all the patients and their families for their participation in this study. We also thank all the staff of the Oncology Department of Ibn Rochd University Hospital and the Genetic and Molecular Pathology Laboratory for their collaboration. Our gratitude goes also to Dr. Omar Hajji and all the staff of Oncology department of Littoral Clinic for their assistance in data and sample collection. We gratefully acknowledge Dr. Nadia Said for her excellent assistance to draft the manuscript.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CM carried out the molecular genetic studies, recruited the patients and drafted the manuscript. BD helped to draft the manuscript. OH, AT, HJ and AB participated in the design of the study and the recruitment of patients. SN conceived the study, participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Chaymaa Marouf, Email: maroufchaymaa@gmail.com.
Omar Hajji, Email: c.marouf@hotmail.fr.
Brehima Diakité, Email: staouil@bmcebank.co.ma.
Amal Tazzite, Email: tresverre11@hotmail.com.
Hassan Jouhadi, Email: taouil.salah@gmail.com.
Abdellatif Benider, Email: s.nadia.10@hotmail.fr.
Sellama Nadifi, Email: m.saramarouf@gmail.com.
References
- Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy – two sides of the same coin? Nature. 2007;7:925–936. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Pharoah PD, McMullan G, Day NE, Stratton MR, Peto J. A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer. 2002;86(1):76–83. doi: 10.1038/sj.bjc.6600008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeyens A, Claes K, Willems P, De Ruyck K, Thierens H, Vral A. Chromosomal radiosensitivity of breast cancer with a CHEK2 mutation. Cancer Genet Cytogenet. 2005;163:106–12. doi: 10.1016/j.cancergencyto.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, Birch JM, Li FP, Garber JE, Haber DA. Heterozygous germ line hCHEK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–31. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- Bell DW, Kim SH, Godwin AK, Schiripo TA, Harris PL, Haserlat SM, Wahrer DC, Haiman CA, Daly MB, Niendorf KB, Smith MR, Sgroi DC, Garber JE, Olopade OI, Le Marchand L, Henderson BE, Altshuler D, Haber DA, Freedman ML. Genetic and functional analysis of CHEK2 (CHK2) variants in multiethnic cohorts. Int J Cancer. 2007;121:2661–7. doi: 10.1002/ijc.23026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellosillo B, Tusquets I, Longaron R, Pérez-Lezaun A, Bellet M, Fabregat X, Serrano S, Sole F. Absence of CHEK2 mutations in Spanish families with hereditary breast cancer. Cancer Genet Cytogenet. 2005;161:93–95. doi: 10.1016/j.cancergencyto.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Ben Abdallah M, Zehani S, Maalej M, Hsairi M, Hechiche M, Ben Romdhane K, Boussen H, Saadi A, Achour N, Ben Ayed F. Cancer du sein en Tunisie: caractéristiques épidémiologiques et tendance évolutive de l’incidence. Tunis Med. 2009;87:417–25. [PubMed] [Google Scholar]
- Bernstein JL, Teraoka SN, John EM, Andrulis IL, Knight JA, Lapinski R, Olson ER, Wolitzer AL, Seminara D, Whittemore AS, Concannon P. The CHEK2*1100delC allelic variant and risk of breast cancer: Screening results from the breast cancer family registry. Cancer Epidemiol Biomark Prev. 2006;15:348–352. doi: 10.1158/1055-9965.EPI-05-0557. [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Enben-Dubrowinskaja N, Feshchenko S, Lazjuk GI, Rogov YI, Dammann O, Bremer M, Karstens JH, Sohn C, Dork T. Association of two mutations in the CHEK2 gene with breast cancer. Int J Cancer. 2005;116:263–266. doi: 10.1002/ijc.21022. [DOI] [PubMed] [Google Scholar]
- Botha JL, Bray F, Sankila R, Parkin DM. Breast cancer incidence and mortality trends in 16 European countries. Eur J Cancer. 2003;39:1718–29. doi: 10.1016/S0959-8049(03)00118-7. [DOI] [PubMed] [Google Scholar]
- Bouchbika Z, Haddad H, Benchakroun N, Eddakaoui H, Kotbi S, Megrini A, Bourezgui H, Sahraoui S, Corbex M, Harif M, Benider A. Cancer incidence in Morocco: report from Casablanca registry 2005–2007. Pan Afr Med J. 2013;16:31. doi: 10.11604/pamj.2013.16.31.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligo MA, Agata S, Aceto G, Crucianelli R, Manoukian S, Peissel B, Scaini MC, Sensi E, Veschi S, Cama A, Radice P, Viel A, D’Andrea E, Montagna M. The CHEK2 c.1100delC mutation plays an irrelevant role in breast cancer predisposition in Italy. Hum Mutat. 2004;24:100–101. doi: 10.1002/humu.20051. [DOI] [PubMed] [Google Scholar]
- CHEK2 Breast Cancer Case–control Consortium CHEK2*1100delC and susceptibility to breast cancer A collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74:1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekmariova EVSA, Buslov KG, Iyevleva AG, Ulibina YM, Rozanov ME, Mitiushkina NVTA, Matsko DE, Voskresenskiy DA, Chagunava OL, Devilee P, Cornelisse CSV, Imyanitov EN. CHEK2 1100delC mutation is frequent among Russian breast cancer patients. Breast Cancer Res Treat. 2006;100:99–102. doi: 10.1007/s10549-006-9227-7. [DOI] [PubMed] [Google Scholar]
- Chen W, Yurong S, Liansheng N. Breast cancer low-penetrance allele 1100delC in the CHEK2 gene: not present in the Chinese familial breast cancer population. Adv Ther. 2008;25:496–501. doi: 10.1007/s12325-008-0057-3. [DOI] [PubMed] [Google Scholar]
- Choi DH, Cho DY, Lee MH, Park HS, Ahn SH, Son BH, Haffty BG. The CHEK2 1100delC mutation is not present in Korean patients with breast cancer cases tested for BRCA1 and BRCA2 mutation. Breast Cancer Res Treat. 2008;112:569–73. doi: 10.1007/s10549-007-9878-z. [DOI] [PubMed] [Google Scholar]
- Cybulski C, Gorski B, Huzarski T, Masojc’ B, Mierzejewski M, Debniak T, Teodorczyk U, Byrski T, Gronwald J, Matyjasik J, Złowocka E, Lenner M, Grabowska E, Nej K, Castaneda J, Medrek K, Szymanska A, Szymanska J, Kurzawski G, Suchy J, Oszurek O, Witek A, Narod SA, Lubinski J. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75:1131–1135. doi: 10.1086/426403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C, Wokolorczyk D, Huzarski T, Byrski T, Gronwald J, Górski B, Debniak T, Masojć B, Jakubowska A, van de Wetering T, Narod SA, Lubiński J. A deletion in CHEK2 of 5,395 bp predisposes to breast cancer in Poland. Breast Cancer Res Treat. 2007;102:119–122. doi: 10.1007/s10549-006-9320-y. [DOI] [PubMed] [Google Scholar]
- De Jong MM, Nolte IM, Te Meerman GJ, Van der Graaf WTA, Oosterom E, Bruinenberg M, Van der Steege G, Oosterwijk JC, Van der Hout AH, Boezen HM, Schaapveld M, Kleibeuker JH, De Vries EGE. No increased susceptibility to breast cancer from combined CHEK2 1100delC genotype and the HLA class III region risk factors. Eur J Cancer. 2005;41:1819–1823. doi: 10.1016/j.ejca.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Dufault MR, Betz B, Wappenschmidt B, Hofmann W, Bandick K, Golla A, Pietschmann A, Nestle-Kramling C, Rhiem K, Huttner C, Von Lindern C, Dall P, Kiechle M, Untch M, Jonat W, Meindl A, Scherneck S, Niederacher D, Schmutzler RK, Arnold N. Limited relevance of the CHEK2 gene in hereditary breast cancer. Int J Cancer. 2004;110:320–325. doi: 10.1002/ijc.20073. [DOI] [PubMed] [Google Scholar]
- Einarsdottir K, Humphreys K, Bonnard C, Palmgren J, Iles MM, Sjolander A, Li Y, Seng Chia K, Liu ET, Hall P, Liu J, Wedren S. Linkage disequilibrium mapping of CHEK2: Common variation and breast cancer risk. PLoS Med. 2006;3:e168. doi: 10.1371/journal.pmed.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mistiri M, Pirani M, El Sahli N, El Mangoush M, Attia A, Shembesh R, Habel S, El Homry F, Hamad S, Federico M. Cancer profile in eastern Libya: incidence and mortality in the year 2004. Ann Oncol. 2010;21:1924–6. doi: 10.1093/annonc/mdq334. [DOI] [PubMed] [Google Scholar]
- Fletcher O, Johnson N, Dos Santos SI, Kilpivaara O, Aittomäki K, Blomqvist C, Nevanlinna H, Wasielewski M, Meijers-Heijerboer H, Broeks A, Schmidt MK, Van’t Veer LJ, Bremer M, Dörk T, Chekmariova EV, Sokolenko AP, Imyanitov EN, Hamann U, Rashid MU, Brauch H, Justenhoven C, Ashworth A, Peto J. Family history, genetic testing, and clinical risk prediction: pooled analysis of CHEK2*1100delC in 1,828 bilateral breast cancers and 7,030 controls. Cancer Epidemiol Biomarkers Prev. 2009;18:230–4. doi: 10.1158/1055-9965.EPI-08-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Malone KE, Doody DR, Daling JR, Ostrander EA. Frequency of CHEK2 mutations in a population based, patient case–control study of breast cancer in young women. Breast Cancer Res. 2004;6:R629–R635. doi: 10.1186/bcr933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Hormazabal P, Castro VG, Blanco R, Gomez F, Peralta O, Waugh E, Bravo T, Reyes JM, Jara L. Absence of CHEK2 1100delC mutation in familial breast cancer cases from a South American population. Breast Cancer Res Treat. 2008;110:543–545. doi: 10.1007/s10549-007-9743-0. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Enríquez S, Balmaña J, Baiget M, Díez O. Detection of the CHEK2 1100delC mutation by MLPA BRCA1/2 analysis: a worthwhile strategy for its clinical applicability in 1100delC low-frequency populations? Breast Cancer Res Treat. 2008;107:455–7. doi: 10.1007/s10549-007-9555-2. [DOI] [PubMed] [Google Scholar]
- Hamdi Cherif M, Zaidi Z, Abdellouche D, Hamdi S, Lakhdari N, Djema BA. Registre du cancer de Sétif (Algérie): incidence, tendance et survie, 1986–2005. J Afr Cancer. 2010;2:245–58. doi: 10.1007/s12558-010-0075-3. [DOI] [Google Scholar]
- Iniesta MD, Gorin MA, Chien LC, Thomas SM, Milliron KJ, Douglas JA, Merajver SD. Absence of CHEK2*1100delC mutation in families with hereditary breast cancer in North America. Cancer Genet Cytogenet. 2010;202:136–140. doi: 10.1016/j.cancergencyto.2010.07.124. [DOI] [PubMed] [Google Scholar]
- Jekimovs CR, Chen X, Arnold J, Gatei M, Richard DJ, Investigators kConFab. Spurdle AB, Khanna KK, Chenevix-Trench G. Low frequency of CHEK2 1100delC allele in Australian multiple-case breast cancer families: functional analysis in heterozygous individuals. Br J Cancer. 2005;92:784–790. doi: 10.1038/sj.bjc.6602381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kilpivaara O, Vahteristo P, Falck J, Syrjakoski K, Eerola H, Easton D, Bartkova J, Lukas J, Heikkila P, Aittomaki K, Holli K, Blomqvist C, Kallioniemi OP, Bartek J, Nevanlinna H. CHEK2 variant I157t may be associated with increased breast cancer risk. Int J Cancer. 2004;111:543–547. doi: 10.1002/ijc.20299. [DOI] [PubMed] [Google Scholar]
- Kleibl Z, Novotny J, Bezdickova D, Malik R, Kleiblova P, Foretova L, Petruzelka L, Ilencikova D, Cinek P, Pohlreich P. The CHEK2 c.1100delC germline mutation rarely contributes to breast cancer development in the Czech Republic. Breast Cancer Res Treat. 2005;90:165–167. doi: 10.1007/s10549-004-4023-8. [DOI] [PubMed] [Google Scholar]
- Kuusisto Kirsi M, Aleksandra B, Vihinen M, Schleutker J, Sallinen SL. Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals. Breast Cancer Res. 2011;13:R20. doi: 10.1186/bcr2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska E, Skasko E, Niwinska A, Wojciechowska-Lacka A, Rachtan J, Molong L, Nowakowska D, Konopka B, Janiec-Jankowska A, Paszko Z, Steffen J. Low frequency of the CHEK2*1100delC mutation among breast cancer probands from three regions of Poland. Neoplasma. 2006;53:305–308. [PubMed] [Google Scholar]
- Lee AS, Ang P. CHEK2*1100delC screening of Asian women with a family history of breast cancer is unwarranted. J Clin Oncol. 2008;16:4335. doi: 10.1200/JCO.2008.16.4335. [DOI] [PubMed] [Google Scholar]
- Margolin S, Eiberg H, Lindblom A, Bisgaard ML. CHEK2 1100delC is prevalent in Swedish early onset familial breast Cancer. BMC Cancer. 2007;7:163. doi: 10.1186/1471-2407-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrett LD, De P, Airia P, Dryer D. Cancer in Canada in 2008. CMAJ. 2008;179:1163–70. doi: 10.1503/cmaj.080760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martınez-Bouzas CBE, Guerra I, Gorostiaga J, Mendizabal JL, De-Pablo JLG-AE, Sanz-Parra A, Tejada MI. CHEK2 1100delC is present in familial breast cancer cases of the Basque Country. Breast Cancer Res Treat. 2007;103:111–113. doi: 10.1007/s10549-006-9351-4. [DOI] [PubMed] [Google Scholar]
- Mateus Pereira LH, Sigurdson AJ, Doody MM, Pineda MA, Alexander BH, Greene MH, Struewing JP. CHEK2:1100delC and female breast cancer in the United States. Int J Cancer. 2004;112:541–543. doi: 10.1002/ijc.20439. [DOI] [PubMed] [Google Scholar]
- McInerney NM, Miller N, Rowan A, Colleran G, Barclay E, Curran C, Kerin MJ, Tomlinson IP, Sawyer E. Evaluation of variants in the CHEK2, BRIP1 and PALB2 genes in an Irish breast cancer cohort. Breast Cancer Res Treat. 2009;121:203–210. doi: 10.1007/s10549-009-0540-9. [DOI] [PubMed] [Google Scholar]
- Meijers-Heijboer H, Van den Ouweland A, Klijn J, Wasielewski M, De Snoo A, Oldenburg R, Hollestelle A, Houben M, Crepin E, Van Veghel-Plandsoen M, Elstrodt F, Van Duijn C, Bartels C, Meijers C, Schutte M, McGuffog L, Thompson D, Easton DF, Sodha N, Seal S, Barfoot R, Mangion J, Chang-Claude J, Eccles D, Eeles R, Evans DG, Houlston R, Murday V, Narod S, Peretz T, Peto J, Phelan C, Zhang HX, Szabo C, Devilee P, Goldgar D, Futreal PA, Nathanson KL, Weber BL, Rahman N, Stratton MR. Low-penetrance susceptibility to breast cancer due to CHEK2*1100delC in non-carriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak DJ, Chen LQ, Ghadirian P, Hamel N, Zhang P, Rossiny V, Cardinal G, Robidoux A, Tonin PN, Rousseau F, Narod SA, Foulkes WD. Identification of a novel CHEK2 variant and assessment of its contribution to the risk of breast cancer in French Canadian women. BMC Cancer. 2008;8:239. doi: 10.1186/1471-2407-8-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offit KPH, Kirchhoff T, Kolachana P, Rapaport B, Gregersen P, Johnson S, Yossepowitch OHH, Satagopan J, Robson M, Scheuer L, Nafa K, Ellis N. Frequency of CHEK2*1100delC in New York breast cancer cases and controls. BMC Med Genet. 2003;4:1. doi: 10.1186/1471-2350-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg RA, Kroeze-Jansema K, Kraan J, Morreau H, Klijn JGM, Hoogerbrugge N, Ligtenberg MJL, van Asperen CJ, Vasen HFA, Meijers C, Meijers-Heijboer H, de Bock TH, Cornelisse CJ, Devilee P. The CHEK2*1100delC variant acts as a breast cancer risk modifier in non-BRCA1/BRCA2 multiple-case families. Cancer Res. 2003;63:8153–8157. [PubMed] [Google Scholar]
- Oldenburg RA, Meijers-Heijboer H, Cornelisse CJ, Devilee P. Genetic susceptibility for breast cancer: how many more genes to be found? Oncol Hematol. 2007;63:125–149. doi: 10.1016/j.critrevonc.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Osorio A, Rodriguez-Lopez R, Diez O, de la Hoya M, Martinez JI, Vega A, Esteban-Cardenosa E, Alonso C, Caldes T, Benitez J. The breast cancer low-penetrance allele 1100delC in the CHEK2 gene is not present in Spanish familial breast cancer population. Int J Cancer. 2004;108:54–56. doi: 10.1002/ijc.11414. [DOI] [PubMed] [Google Scholar]
- Rajkumar T, Soumittra N, Nancy NK, Swaminathan R, Sridevi V, Shanta V. BRCA1, BRCA2 and CHEK2 (1100 del C) germline mutations in hereditary breast and ovarian cancer families in South India. Asian Pac J Cancer Prev. 2003;4:203–208. [PubMed] [Google Scholar]
- Rashid MU, Jakubowska A, Justenhoven C, Harth V, Pesch B, Baisch C, Pierl CB, Bruning T, Ko Y, Benner A, Wichmann HE, Rauch H, Hamann U. German populations with infrequent CHEK2*1100delC and minor associations with early-onset and familial breast cancer. Eur J Cancer. 2005;41:2896–2903. doi: 10.1016/j.ejca.2005.04.049. [DOI] [PubMed] [Google Scholar]
- Shaag A, Walsh T, Renbaum P, Kirchhoff T, Nafa K, Shiovitz S, Mandell JB, Welcsh P, Lee MK, Ellis N, Offit K, Levy-Lahad E, King MC. Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Hum Mol Genet. 2005;14:555–563. doi: 10.1093/hmg/ddi052. [DOI] [PubMed] [Google Scholar]
- Sodha N, Bullock S, Taylor R, Mitchell G, Guertl-Lackner B, Williams RD, Bevan S, Bishop K, McGuire S, Houlston RS, Eeles RA. CHEK2 variants in susceptibility to breast cancer and evidence of retention of the wild type allele in tumours. Br J Cancer. 2002;87:1445–1448. doi: 10.1038/sj.bjc.6600637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodha DW, Kim SH, Godwin AK, Schiripo TA, Harris PL, Haserlat SM, Wahrer DCR, Haiman CA, Daly MB, Niendorf KB, Smith MR, Sgroi DC, Garber JE, Olopade OI, Marchand LL, Henderson BE, Altshuler D, Haber DA, Freedman ML. Genetic and functional analysis of CHEK2 (CHK2) variants in multiethnic cohorts. Int J Cancer. 2007;121:2661–2667. doi: 10.1002/ijc.23026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CG, Hu Z, Yuan WT, Di GH, Shen ZZ, Huang W, Shao ZM. CHEK2 c.1100delC may not contribute to genetic background of hereditary breast cancer from Shanghai of China. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23:443–5. [PubMed] [Google Scholar]
- Tazi MA, Er-Raki A, Benjaafar N. Cancer incidence in Rabat, Morocco: 2006–2008. Ecancer. 2013;7:338. doi: 10.3332/ecancer.2013.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazzite A, Jouhadi H, Nadifi S, Aretini P, Falaschi E, Collavoli A, Benider A, Caligo MA. BRCA1 and BRCA2 germline mutations in Moroccan breast/ovarian cancer families: novel mutations and unclassified variants. Gynecol Oncol. 2012;125:687–692. doi: 10.1016/j.ygyno.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Thirthagiri E, Cheong LS, Yip CH, Teo SH. CHEK2*1100delC does not contribute to risk to breast cancer among Malay, Chinese and Indians in Malaysia. Fam Cancer. 2009;8:355–358. doi: 10.1007/s10689-009-9244-x. [DOI] [PubMed] [Google Scholar]
- Vahteristo P, Bartkova J, Eerola H, Syrjakoski K, Ojala S, Kilpivaara O, Tamminen A, Kononen J, Aittomaki K, Heikkila P, Holli K, Blomqvist C, Bartek J, Kallioniemi OP, Nevanlinna H. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71:432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Groep P, van der Wall E, van Diest PJ. Pathology of hereditary breast cancer. Cell Oncol. 2011;34:71–88. doi: 10.1007/s13402-011-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischer M, Bojesen SE, Tybjaerg-Hansen A, Axelsson CK, Nordestgaard BG. Increased risk of breast cancer associated, with CHEK2*1100delC. J Clin Oncol. 2007;25:57–63. doi: 10.1200/JCO.2005.05.5160. [DOI] [PubMed] [Google Scholar]
- Zhang S, Phelan CM, Zhang P, Rousseau F, Ghadirian P, Robidoux A, Foulkes W, Hamel N, McCready D, Trudeau M, Lynch H, Horsman D, Lourdes Leon De Matsuda M, Aziz Z, Gomes M, Magalhaes Costa M, Liede A, Poll A, Sun P, Narod SA. Frequency of the CHEK2 1100delC Mutation among Women with Breast Cancer: An International Study. Cancer Res. 2008;68:7. doi: 10.1158/0008-5472.CAN-07-5187. [DOI] [PubMed] [Google Scholar]


