Abstract
Anatomical structures and mechanisms linking genes to neuropsychiatric disorders are not deciphered. Reciprocal copy number variants at the 16p11.2 BP4-BP5 locus offer a unique opportunity to study the intermediate phenotypes in carriers at high risk for autism spectrum disorder (ASD) or schizophrenia (SZ). We investigated the variation in brain anatomy in 16p11.2 deletion and duplication carriers. Beyond gene dosage effects on global brain metrics, we show that the number of genomic copies negatively correlated to the gray matter volume and white matter tissue properties in cortico-subcortical regions implicated in reward, language and social cognition. Despite the near absence of ASD or SZ diagnoses in our 16p11.2 cohort, the pattern of brain anatomy changes in carriers spatially overlaps with the well-established structural abnormalities in ASD and SZ. Using measures of peripheral mRNA levels, we confirm our genomic copy number findings. This combined molecular, neuroimaging and clinical approach, applied to larger datasets, will help interpret the relative contributions of genes to neuropsychiatric conditions by measuring their effect on local brain anatomy.
Introduction
Copy number variants (CNVs) are major contributors to common neuropsychiatric disorders and cognitive deficits.1 Investigations of cohorts with specific CNVs allow the characterization of endophenotypes2 associated with neuropsychiatric disorders, such as autism spectrum disorder (ASD) and schizophrenia (SZ). The 16p11.2 CNV (breakpoint 4–5, BP4-BP5, 29.6-30.2 Mb- Hg19) phenotypes are characterized by both reciprocal and overlapping deficits that include energy imbalance, language impairment, ASD and SZ.3, 4, 5, 6, 7, 8 Notably, both 16p11.2 deletion and duplication have been associated with ASD, while only the duplication is enriched in SZ cohorts.9, 10 Deletion carriers present with increased head circumference6, 8 and body mass index (BMI),3, 7 while duplication carriers are at risk for microcephaly and being underweight.5 Manipulations of zebrafish embryos and mouse models suggest a close relationship between gene dosage at this locus and brain anatomy. Over- and under-expression of the KCDT13 gene in zebrafish induces mirror alterations interpreted as correlates of micro- and macrocephaly.11 Murine models mimicking 16p11.2 deletion and duplication demonstrate reciprocal regional brain volume changes.12
The aim of this study is to identify imaging endophenotypes in a group of 16p11.2 CNV carriers at high risk for ASD and SZ. We investigate the effects of gene dosage, defined as the number of genomic copies at the 16p11.2 locus on brain structure using the state-of-the-art structural magnetic resonance imaging (MRI). We find a correlation between gene dosage and alterations in brain structure with diametrically opposite changes in both global and local brain volumes that parallel specific changes in tissue microstructure. The anatomical areas affected by gene dosage are also key areas involved in ASD, SZ and obesity, supporting the notion that common molecular mechanisms may be involved in these conditions.
Materials and Methods
Participants
The study was reviewed and approved by the local Ethics committee and signed consents were obtained from the participants or legal representatives prior to the investigation. Participants (Table 1) were taking part in a larger phenotyping project on the deletion/duplication of the 16p11.2 region. Carriers were referred to the study by the clinical geneticist who had initially established the genetic diagnosis in the context of a neurodevelopmental disorder. Inclusion criteria: participants were selected based on the presence of a 16p11.2 deletion or duplication comprising the BP4-BP5 region. Controls were non-carriers in the same families. Exclusion criteria: none beside an age <6 years. Seventeen participants were unable to complete the scan because of incompliance related to moderate or severe intellectual disability, anxiety, significant behavioral issues or extreme BMIs with waist circumference beyond the limit of scanning safety standards.
Table 1. Population characteristics.
| Deletion n=14 | Controls n=23 | Duplication n=17 | |
|---|---|---|---|
| Age in years (mean±s.d.) | 24.6±13.3 | 33±12.3 | 35.7±12.5 |
| Gender (M/F) | 8/6 | 11/12 | 10/7 |
| Handedness (L/R) | 3/11 | 0/23 | 3/14 |
| BMI z-score (mean ±s.d.) | 2.37±2.14a | 0.37±1.07 | −0.56±1.61 |
| FSIQ (mean±s.d.) | 74±14b | 98±13 | 72±16b |
| NVIQ (mean±s.d.) | 79±11b | 102±13 | 73±15b |
Abbreviations: BMI, body mass index; F, female; FSIQ, full-scale intellectual quotient; M, male; NVIQ, non-verbal intellectual quotient; L, left; R, right.
Significantly different from the two other groups, ANCOVA, post-hoc group comparisons, P<0.05 Bonferroni corrected.
Significantly different from the control group, ANCOVA, post-hoc group comparisons, P<0.05, Bonferroni corrected.
All had whole genome arrays confirming either a recurrent deletion or duplication of the BP4-BP5 region. The larger project aims at phenotyping a European cohort of 16p11.2 rearrangement carriers. It includes neuropsychological and behavioral assessments, medical, psychiatric and neurological examinations.
Anthropometric measures, psychiatric and cognitive assessment
We collected anthropometric data such as height and weight to calculate the BMI. Obesity is defined as BMI>30 kg m-2 in adults and >2 standard deviations in children.13 Underweight is considered significant <18.5 kg m2 and <2 standard deviations in children.13 Z-scores were computed for all data using gender, age and geographically matched reference population as previously described in Zufferey et al.8 Overall cognitive functioning was measured using the Wechsler Intelligence scales for children (WISC-IV)14 as well as the Wechsler Intelligence scale for adults (WAIS-III).15 All assessments were performed by a board-certified neuropsychologist. Diagnostic and Statistical Manual of Mental Disorders, Fourth edition-text revision16 diagnoses were made by licensed psychologist and psychiatrist using history, parent report as well as the Diagnostic Interview for Genetic Studies.17 An additional assessment was performed to investigate prodroms of SZ using the Schizophrenia Proneness Instrument Adult version.18 The diagnosis of ASD was established by a certified clinician using the Autism Diagnostic Interview-Revised19 and the Autism Diagnostic Observation Schedule.20 Of note, only two duplication carriers were on medication: lithium, aripiprazol and valproate.
Quantitative RT-PCR
For quantitative polymerase chain reaction, 100 ng of high-quality total RNA isolated from Epstein–Barr virus transformed lymphoblastoid cell lines was converted to cDNA using Superscript VILO (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Primers were designed using PrimerExpress 2.0 software (Applied Biosystems, Foster City, CA, USA), with default parameters except for the primer- and minimal amplicon lengths, which were set at 17–26 bp and 60 bp respectively. The amplification factor of each primer pair was tested using a cDNA dilution series and only assays with amplification factors between 1.75 and 2.00 were retained. A representative set of samples was tested for genomic contamination. Quantitative polymerase chain reaction experiments were performed in triplicates using SYBR-Green (Roche, Basel, Switzerland) as reporter. The reaction mixtures were prepared in 384-well plate using a Freedom Evo robot (Tecan, Männedorf, Switzerland) and run in an ABI 7900HT sequence detection system (Applied Biosystems) using the following conditions: 50 °C for 2 min, 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and then 60 °C for 1 min, after which dissociation curves were established. Applicable normalization genes were included in each experiment to enable compensation for fluctuations in expression levels between experiments. Using SDS v2.4 software (Applied Biosystems) the threshold and baseline values were adjusted when necessary to obtain raw cycle threshold values. The cycle threshold values were further analyzed using qBase plus software (Biogazelle, Zwijnaarde, Belgium), which calculates relative expression values per sample per tested gene upon designation of the normalization genes and corrects for the amplification efficiency of the performed assay.
MRI Data acquisition and processing
All participants were examined on a 3T whole body scanner (Magnetom TIM Trio, Siemens Healthcare, Erlangen, Germany) using a 12-channel RF receive head coil and RF body transmit coil. Participants presenting with moderate to severe clinical phenotype were unable to undergo the scanning procedure. A total of 57 participants underwent the MRI protocol, which included T1-weighted (T1w) and diffusion-weighted (DWI) data acquisition. Anatomical images were acquired using a multi-echo magnetization prepared rapid gradient echo sequence (ME-MPRAGE: 176 slices; 256 × 256 matrix; echo time (TE): TE1: 1.64 ms, TE2: 3.5 ms, TE3: 5.36 ms, TE4: 7.22 ms; repetition time (TR): 2530 ms; flip angle 7°). The DWI protocol consisted of 2 mm contiguous slices covering the whole brain (TE=83 ms, TR=9020 ms) along 60 spherically distributed gradient directions with b-value=700 s mm-2 with 10 reference images with no diffusion weighting (b-value=0 s/mm2). Due to movement of artifacts, 3 out of 57 T1w images and 5 DWI out of 45 who were able to complete the DWI protocol were excluded.
Multi-echo T1w images were averaged then classified into probability of belonging to gray matter (GM), white matter (WM) or cerebrospinal fluid using Gaussian mixture model within the ‘unified segmentation' framework.21 Images were transformed non-linearly to standard Montreal Neurological Institute space using the diffeomorphic spatial registration algorithm implemented in SPM8.22 GM probability maps were subsequently ‘modulated' by the Jacobian determinants of the deformations to account for local compression and expansion because of linear and non-linear transformation.23 Finally, GM probability maps were smoothed with an isotropic Gaussian kernel of 8 mm full-width at half-maximum.
Cortical surface extraction was performed on the averaged multi-echo T1w images using the default settings of the Freesurfer software (Freesurfer, Boston, MA, USA)24 (http://surfer.nmr.mgh.harvard.edu/). Individual images were examined for potential defects of surface reconstruction. Mean cortical thickness and cortex surface area were then computed for each subject.
DWI images were corrected for Eddy current and motion artifacts with the Artefact correction in diffusion MRI (ACID toolbox).25 Diffusion tensor based indices—fractional anisotropy and mean diffusivity, were computed with the Camino Diffusion MRI toolkit (Camino Diffusion MRI toolkit, University College of London, UK).26 Fractional anisotropy and mean diffusivity maps were aligned to the T1w images using affine transformation. In order to enhance the specificity for a particular tissue class and to avoid diffusion indices value changes because of Gaussian smoothing kernel during the spatial registration to Montreal Neurological Institute space, we applied a previously described combined weighting/smoothing procedure.27
For voxel-based statistical analysis of gene dosage-dependent regional effects we used a linear regression model in the General Linear Model framework of SPM8. Age effects were analyzed separately for GM and WM sub-space by creating two corresponding design matrices. Explicit masking using binary masks of GM and WM ensured inclusion of the same number of voxels in all analyses. All GM/WM data were included in the same model with gender, total intracranial volume (TIV) (the sum of GM, WM and cerebrospinal fluid volume) and first-order polynomial expansion of age as regressors. We tested for overall cognitive functioning (full-scale intellectual quotient), but as results did not show significant effect on whole-brain or regional structures, we did not add it as a covariate in the statistical parametric mapping analyses. Statistical thresholds were applied at P<0.05 after family-wise error correction for multiple comparisons over the whole volume of the GM/WM mask. Trends were assessed by using an auxiliary uncorrected voxel threshold of P<0.001.28
Multivariate linear models analysis
Multivariate linear model was used to discover high-order correlation mapping between the two datasets: gene expression and brain volume. The multivariate approach takes into account the full dimensionality of both data sets (all voxels and all mRNA levels for all individuals). The method simultaneously determines the best representations in each datasets in order to explain maximum covariance between gene expression and volume. The representation in the brain space is called eigenimage and show the level contribution (either positive or negative) for each voxel to the correlation mapping. Similarly, the gene loadings (positive or negative) show the contribution of each gene to the correlation mapping. The number of mappings and the significance of the mapping is assessed with Wilks' Lambda statistic.28, 29
Statistical analyses
Statistical analyses were performed using Matlab and R 3.0.2. Plots and heat maps have been generated using R libraries ggplot2 and gplots, respectively.
Results
Study participants and MRI measures
We acquired data on fourteen 16p11.2 BP4-BP5 deletion and seventeen duplication carriers as well as twenty-three intrafamilial controls. The median age of participants was 34 years (range 7–58). The three groups were not significantly different in terms of age, gender or handedness (Table 1). Overall cognitive functioning (full-scale intellectual quotient) in deletion and duplication carriers was 2 standard deviations (~27 points) below that in the control group, which is consistent with previous studies.5, 8 Two duplication carriers met diagnostic criteria for ASD and none of the participants had clinical signs of SZ (all clinical data and neuro-radiological findings are presented in Supplementary Tables 1 and 2).
In an unbiased whole-brain approach, we analyzed regionally derived structural MRI estimates of GM and WM volume, cortical thickness and surface area, and also TIV.24, 30 We also investigated brain microstructure using an independent data set of DWI images from the same subjects, from which we computed fractional anisotropy and mean diffusivity as indices of local tissue integrity.31 Gene dosage effects on brain anatomy were tested explicitly over the cohorts of deletion carriers, controls and duplication carriers using differential contrasts and conjunction analyses.
Global brain differences
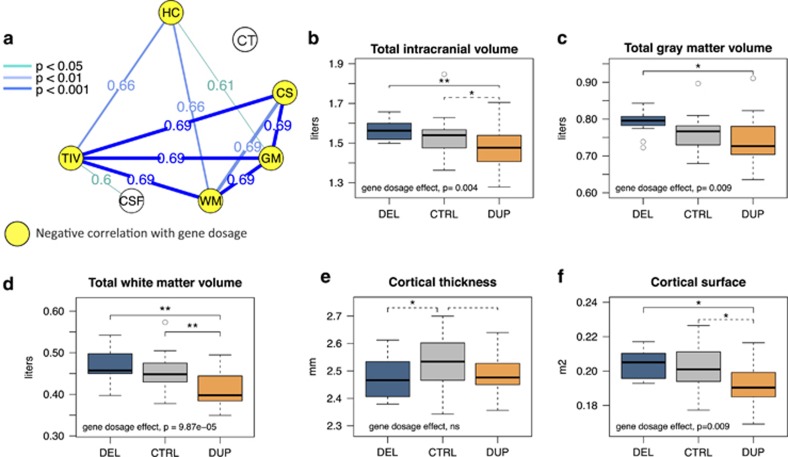
We first tested the sensitivity of our whole-brain imaging approach to detect global effects of gene dosage since early brain overgrowth is a feature common to both ASD and the 16p11.2 deletion.8, 32 We confirm the strong correlation between head circumference and gene dosage in the studied group (P=0.0001)5, 7 and demonstrate the negative correlation between TIV and number of 16p11.2 copies (P=0.004, Figure 1b). Both GM (P=0.009) and WM volume (P=9.8 e-05) contribute to the observed effect on TIV (Figures 1c and d). There is no significant correlation between gene dosage and cerebrospinal fluid volume suggesting that there are no compensatory cerebrospinal fluid changes secondary to brain atrophy. The analysis of cortical anatomy shows correlation between gene dosage and global cortical surface area (P=0.009, Figure 1f). As opposed to global measures of volume and surface, which are all inter-correlated (Figure 1a) and modulated by gene dosage, only cortical thickness is decreased in both deletions and duplication carriers (Figures 1a–e). This may be related to previously reported relationship between intellectual disability and cortical thickness.33, 34 Of note, adjusting for intellectual quotient does not affect our findings (Supplementary Figure 1). Analyses on global metrics are detailed in Supplementary Table 3.
Figure 1.
Effects of gene dosage on global brain metrics. (a): Correlation between global brain metrics in 16p11.2 CNV carriers and controls. The intensity of blue lines represents positive r Pearson's coefficient correlations at different statistical thresholds. All global measures are inter-correlated except for CT and CSF. (b–f): Boxplots representing TIV, GM, WM volume, cortical surface area and cortical thickness adjusted for age and gender in deletion, duplication carriers and intrafamilial controls. **P⩽0.001; *P⩽0.05 uncorrected. Gene dosage effect is estimated in a linear regression analysis using the number of copies (1, 2 or 3), and including age, gender as covariates. Significant differences between groups after Bonferroni correction (threshold at P<0.01) are represented by solid black lines, trends—by dashed lines. HC, head circumference; TIV, total intracranial volume; GM, gray matter; WM, white matter; CSF, cerebrospinal fluid; CT, cortical thickness; CS, cortical surface area; DEL, deletion carriers; CTRL, intrafamilial controls; DUP, duplication carriers.
There is a strong correlation between BMI and head circumference in 16p11.2 CNV carriers.8 We also find a negative correlation between BMI and gene dosage (P<0.001, Table 1). Total GM volume (P=0.008) and TIV (P=0.019) are correlated to BMI (Supplementary Table 4) and this relationship becomes borderline significant (P=0.06) after adjusting for the copy number effect on BMI.
Regional volume brain differences
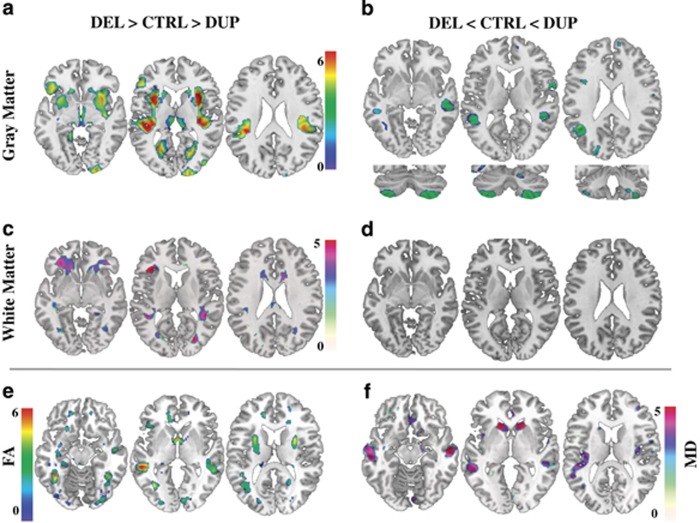
Using the same statistical design and voxel-based morphometry, we analyzed local GM and WM changes beyond global volume effects. The spatial pattern of gene dosage-associated changes overlaps with key areas of the reward system including the medio-dorsal thalamus, insula, ventral striatum, orbito-frontal cortex and WM corresponding to fronto-striatal projections (Figures 2a and c). The language circuitry, comprising the left-middle temporal gyrus, bilateral supramarginal and superior temporal gyri and adjacent WM connections, also shows strong gene dosage dependence (Figures 2a–c and Supplementary Table 5). We further observe gene dosage effects on cerebellar anatomy, including lobules VIIb, VIII and crus II bilaterally (Figure 2b). Subsequent gender-by-gene dosage interaction analysis demonstrates that this effect is mainly driven by decreased volume in male deletion carriers (Supplementary Figure 2). The analysis of an independent dataset of DWI images sensitive to water diffusion properties of brain tissue in a subset of 45 participants confirms a reciprocal gene dosage effect on reward and language circuit associated regions—striatum, middle and superior temporal gyrus (Figures 2e and f). The changes of diffusion-tensor-derived indices (fractional anisotropy and mean diffusivity) suggest a dosage-dependent effect on brain microstructure beyond the volume changes already described.35
Figure 2.
Effects of gene dosage on local brain volume and tissue properties. Results of voxel-based whole-brain general linear analyses showing: (a) negative gene dosage effect (DEL>CTRL>DUP) on GM volume in ventral striatum, thalamus, superior temporal region, fusiform, precuneus, insula and calcarine sulci bilaterally as well as in right occipital region. (b) Positive gene dosage effect (DEL<CTRL<DUP) on GM volume in the middle temporal gyrus and in cerebellar lobule VIII, VIIb and crus II. (c) Negative gene dosage effect on WM volume within fronto-striatal projections, anterior thalamic and superior longitudinal fasciculus. (d) Absence of significant positive gene dosage effect on WM volume. (e and f) FA and MD changes overlapping with GM changes in superior temporal gyrus and caudate bilaterally. For representation purposes, results significant at a voxel level at threshold of P<0.05 family-wise error corrected for multiple comparisons are displayed at significance threshold of P<0.001 uncorrected at voxel level in standard Montreal Neurological Institute space. Color bars represent T scores. DEL, deletion carriers; CTRL, intrafamilial controls; DUP, duplication carriers; GM, gray matter; WM, white matter; FA, fractional anisotropy; MD, mean diffusivity.
We further investigated the contribution of the deletion and duplication to the gene dosage analysis (deletion>control>duplication) results on the brain structure. We performed a conjunction analysis testing the intersection of the two differential contrasts: (deletion>control)∩(control>duplication) (Supplementary Figure 3). This stringent analysis shows that both the deletion and duplication contribute to the negative correlation between gene dosage and putamen volume. The two differential contrasts suggest that changes in the reward system are driven by the duplication, while deletion carriers contribute to modifications in language and social cognition networks (Supplementary Figure 3 and Supplementary Table 5). Due to the stringency of the conjunction analysis, we cannot exclude rejecting modest brain changes following the gene dosage-dependent pattern. A larger sample size would be required to identify brain anatomy changes specific to either deletion or duplication carriers.
Regional cortical thickness and surface area differences
We observed cortical thickness changes mainly driven by deletion carriers in the fronto-temporal regions, particularly in insula, supramarginal and superior temporal gyrus (Supplementary Table 6 and Supplementary Figure 4). These regions overlap spatially with the cortical volume changes derived in the whole-brain voxel-based morphometry analysis. For measures of cortical surface area, the overlap is restricted to the frontal pole.
16p11.2 dosage-related brain alterations common to idiopathic ASD and SZ
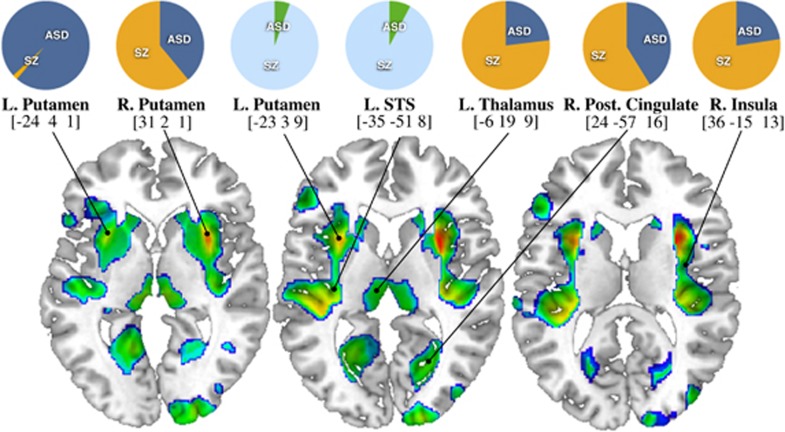
The 16p11.2 CNVs that confer high risk for ASD and SZ, exemplify the concept of shared genetic factors in psychiatric disorders. We formally tested the spatial overlap between the described brain patterns and the results of a recent meta-analysis in ASD and SZ brain morphometry36 (Figure 3). Areas subject to strong 16p11.2 gene dosage effects overlap with six out of the eight brain structures most commonly affected in ASD and SZ—left and right putamen, insula, posterior cingulate, thalamus and superior temporal gyrus, but with none of the SZ-specific regions36 (Figure 3 and Supplementary Table 7).
Figure 3.
Spatial mapping of meta-analysis data in autism spectrum disorder and schizophrenia on to 16p11.2 gene dosage brain pattern. Projection of meta-analysis data in ASD and SZ36 on the statistical map of 16p11.2 gene dosage effects on gray matter volume in deletion and duplication carriers (DEL>CTRL>DUP). Pie charts represent previously published data on the relative contribution of ASD and SZ to brain volume change at particular location.36 Orange-blue pie charts indicate gray matter decreases; cyan-green pie charts show gray matter increases in ASD and SZ. Coordinates correspond to Montreal Neurological Institute standard space. ASD, autism spectrum disorder; SZ, schizophrenia; STS, superior temporal sulcus.
mRNA expression levels and brain anatomy
16p11.2 BP4-BP5 CNVs either delete or duplicate 28 genes. We therefore tested whether the effects of gene dosage on brain anatomy are mediated by changes in gene expression measured in lymphoblastic cell lines of twenty-seven of our participants. We investigated in a linear regression analysis, mRNA levels of eighteen genes mapping within and one gene (SH2B1) outside the BP4-BP5 interval. While SH2B1 expression is not affected by changes in copies of the BP4-BP5 CNVs, mRNA levels for all eighteen genes within the interval are correlated to the number of genomic copies and twelve of these genes show strong correlation (Pearson r>0.75) with each other (Supplementary Figure 5).
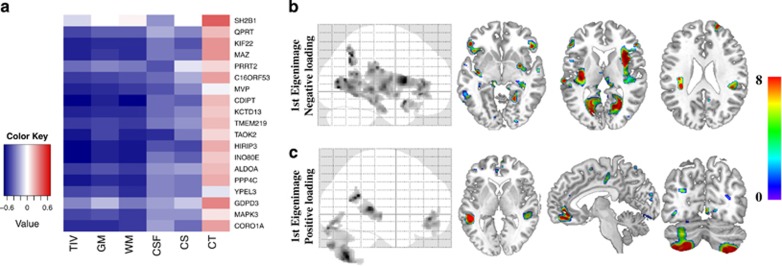
mRNA levels of all assessed genes within the BP4-BP5 interval except GDPD3 and PRRT2 show negative correlation with global metrics of brain volume (Figure 4a and Supplementary Table 8). To characterize the differential contributions of highly correlated gene expression levels on brain structure, we performed whole-brain multivariate analyses based on singular value decomposition. This method identifies the linear combination of brain voxels—eigenimages—that are best predicted by a linear mixture of gene expression levels. Despite the reduced subject sample size, this analysis replicated the anatomical pattern described by categorical analyses of gene dosage effects (Figures 2 and 4). The first eigenimage is characterized by negative loadings on the striatum, fusiform gyrus and thalamus bilaterally (Figure 4b), as well as positive loadings on both cerebellar hemispheres (Figure 4c). Put simply, voxels with negative loadings indicate that low mRNA levels are associated with increased GM volumes while those with positive loadings with GM volume reduction. In our cohorts, the first three eigenimages explain more than 30% of variance in brain anatomy, and are mainly driven by the number of genomic copies (P=0.003, 0.01 and 0.07 for the first three eigenvariates, respectively). The correlation between gene expression and the number of genomic copies (Supplementary Figure 5) as well as the high level of shared variance between genes at this locus preempts identification of the contributions of individual gene to specific anatomical patterns (Supplementary Table 9 and Figure 6).
Figure 4.
Effects of mRNA levels on global and local metrics of brain anatomy. Brain anatomy changes explained by mRNA levels of 18 genes within the 16p11.2 BP4-BP5 interval in a subset of 27 participants. (a) Matrix correlation between mRNA levels and global measures of brain volume. The color key represents the Pearson correlation coefficient. Most mRNA levels, except for SH2B1, PRRT2 and GDPD3 are correlated to TIV, GM and WM. There are no correlations with cortical thickness and CSF. Statistical P values for all correlations are detailed in Supplementary Table 7. (b-c) Eigenimages represent the statistical parametric maps resulting from singular value decomposition analysis. This method simultaneously determines the best combination in each dataset (gene expression and voxels) in order to explain maximum covariance between gene expression and local brain volume. Negative loading indicate that low mRNA levels are associated with increased GM volumes while positive loading represents the inverse effect. TIV, total intracranial volume; GM, gray matter; WM, white matter; CSF, cerebrospinal fluid; CS, cortical surface area; CT, cortical thickness.
Discussion
Using an unbiased whole-brain approach, we demonstrate that genomic copy number at the 16p11.2 BP4-BP5 locus is associated with brain anatomy changes in a dosage-dependent manner, and that these structural changes are present in the absence of either an ASD or SZ diagnosis. Our findings including areas implicated in reward, language and social cognition allow generating new hypotheses on how gene dosage results in reciprocal and overlapping phenotypes observed in 16p11.2 deletion and duplication carriers. This provides a general framework to study the effects of CNV on cognition and behavior in common neuropsychiatric disorders such as ASD and SZ. This approach circumvents a number of confounders such as the effects of ascertainment, medication and clinical symptoms2, 37 and has recently been used to investigate predefined anatomical regions related to early psychosis in 15q11.2 CNVs carriers.38
There is a strong correspondence between the changes in brain anatomy patterns and the phenotypic traits characterizing 16p11.2 deletion or duplication carriers. In particular, opposing volume changes in key nodes of the reward circuitry—striatum, medio-dorsal thalamus, orbito-frontal cortex and insula—which are associated with eating behavior,39, 40, 41 may explain the mirror BMI phenotype in 16p11.2 CNVs carriers. Similarly, the reciprocal changes in the language areas—middle, superior temporal gyrus and caudate42 may underlie the language deficits reported in deletion but not in duplication carriers. Our results showing involvement of the striatum have recently been corroborated by the findings of Portmann et al.43 in a mouse model of 16p11.2 deletion syndrome.
Whole genome studies have identified genetic factors, including 16p11.2 CNVs, shared between ASD and SZ. This led to the assumption that the two disorders may represent opposite manifestations of the same underlying mechanism or trait.44 There is a large overlap in the reported patterns of gene dosage-dependent brain anatomy changes with well-established structural signatures of ASD and SZ. Notably, alterations in reward system structure, influenced by 16p11.2 gene dosage, is also the main structural change shared by ASD and SZ.36, 45, 46 This finding supports the notion of a common abnormal mechanism underlying these two conditions. None of the studied participants met diagnostic criteria for SZ and only two did so for ASD (Supplementary Table 1), which suggests that the observed brain modifications are not the consequence of a long standing ASD or SZ diagnosis, but that they may be considered as intermediate phenotypes. This is also in keeping with previous findings of brain modifications predating the onset of psychosis.47
Obesity is a well-known comorbidity of ASD or SZ.48 It has been hypothesized that the reward system, and the striatum in particular, may underlie the frequent co-occurrence of metabolic and psychiatric manifestations.37, 39, 41, 49 This idea is in line with our findings and suggests that modulation of the reward system by genes at the 16p11.2 locus is driving a group of disorders as opposed to the notion that different genes lead to different symptoms in CNV carriers.
Analyses of cortex anatomy showed cortical thickness reduction in 16p11.2 deletion carriers and a similar trend in duplication carriers. This shared feature may be explained by decreased intellectual quotient and risk for ASD in both deletion and duplication carriers. However, both cortical thickness and surface area are correlated to intellectual quotient and this relationship changes with age.50 Of note, SZ is also associated with widespread cortical thining.51 The dissociation between cortical surface and thickness with regards to 16p11.2 gene dosage supports the notion of distinct genetic mechanisms regulating these cortical anatomy features.52, 53 As shown in the general population and ASD, it is most likely that the 16p11.2 gene dosage effects on brain volume are related to changes in total cortical surface.52, 54, 55 Therefore, we refrain from drawing conclusions on topology overlap and causal links between regional thickness/surface changes and brain volume alterations.54
In agreement with the known effect of gender in ASD and in carriers of 16p11.2 CNVs,5, 56 we observe a volume reduction in both cerebellar hemispheres restricted to male deletion carriers (Supplementary Figure 2). This finding supports the notion of differential neurodevelopmental effects of genetic variants in males and females as well as mounting evidence for cerebellar involvement in intellectual disability, language impairment and ASD.57, 58
The reciprocal effects of gene dosage on global metrics of brain volume and cortical surface area corroborate previous reports on head size measurements in humans and zebrafish.5, 11 In the absence of gene dosage-dependent cortical thickness changes, we interpret the effects of 16p11.2 CNVs on brain volume and cortical surface area as evidence of abnormal neurogenesis.59 TAOK2, MAPK3, MVP, KIF22, ALDOA and KCTD13, are 16p11.2 genes previously linked to neurogenesis and/or apoptosis,11, 60, 61 and could represent candidate genes implicated in the control of brain growth.
Whole brain analysis using continuous measures of mRNA levels further validates results using the number of genomic copies (1, 2 or 3), and both approaches identify that the same anatomical structures are involved. Given the significant amount of shared variance between mRNA levels of the studied genes at the 16p11.2 BP4-BP5 locus we refrain from drawing conclusions about potential differential contribution of single genes on brain anatomy.
One of the anticipated limitations of the study is the inability to acquire imaging data in participants with significant behavioral deficits and BMI/waist circumference beyond limits of MRI scanning safety standards. Results might not generalize to other 16p11.2 CNV carriers with extreme obesity or a full-blown clinical symptoms of ASD or SZ. Nevertheless, with respect to global cognition, our cohort is highly representative of 16p11.2 with intellectual quotient measures identical to what has been previously published.4, 8
Conclusion
In this study, we demonstrate that gene dosage at the 16p11.2 locus modulates specific neural circuitry including foremost the reward system. The patterns of brain anatomy changes in fronto-subcortical networks could be interpreted as endophenotypes of ASD, SZ and obesity associated with 16p11.2 rearrangements in the absence of diagnostic criteria for ASD and SZ. The complementary analysis using peripheral measures of gene expression levels brings further evidence for the correlation between gene dosage and brain structure. This combined approach applied to larger datasets should allow dissection of the relative contributions of genes to human behavior and cognition through a fine-grained analysis of human brain anatomy.
Acknowledgments
We thank the participants, families and referring providers (see Supplementary Table 1) for their contribution. We are very grateful to Richard S. Frackowiak for his valuable comments and edits on the manuscript. We acknowledge Stéphanie Selmoni, Nathalie Isidor and Ana Machado for their contribution in the recruitment of the participants as well as Guillaume Sierro and Melody Laser for their help scanning the participants. This work was supported by the Leenaards Foundation Prize (SJ, AR and NH), a Swiss National Science Foundation (SNSF) Sinergia grant CRIS FN CRSII33-133044 (AR, SJ), and the Simons Foundation Autism Research Initiative SFARI274424 (AR) the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 604102. BD was supported by the SNSF (NCCR Synapsy, project grant Nr 320030_135679 and SPUM 33CM30_140332/1), Foundation Parkinson Switzerland, Foundation Synapsis, Novartis Foundation for medical–biological research and Deutsche Forschungsgemeinschaft (Kfo 247). LREN is supported by the Roger de Spoelberch and the Partridge Foundations. SJ was supported by a professorship grant from the SNSF PP00P3_144902/2. KM is a grantee of a scholarship from the Swiss Scientific Exchange NMS Program and MZ by a fellowship from the doctoral school of the Faculty of Biology and Medicine, University of Lausanne. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Participants were scanned at the ‘Centre d'Imagerie BioMédicale' (CIBM) which is a research initiative of the following partners: University of Lausanne, Swiss Federal Institute of Technology Lausanne, University of Geneva, Centre Hospitalier Universitaire Vaudois, Hôpitaux Universitaires de Genève and the Leenaards and the Jeantet Foundations. We thank the participants and their families for their contribution. 16p11.2 European Consortium contributing members: M-C Addor, Service of Medical Genetics, Centre Hospitalier Universitaire Vaudois, Lausanne University, Lausanne. J Andrieux, Institut de Génétique Médicale, Hopital Jeanne de Flandre, Lille. B Arveiler, Univ. Bordeaux, Maladies Rares: Génétique et Métabolisme, Service de Génétique Médicale, CHU-Bordeaux, Bordeaux. G Baujat, Centre de Référence Département de Génétique, Hôpital Necker-Enfants malades, Paris. F Béna, Service of Genetic Medicine, University Hospitals of Geneva, Geneva. D Bonneau, Service de génétique médicale, CHU-Angers, Angers. S Bouquillon, Institut de Génétique Médicale, Hopital Jeanne de Flandre, Lille. O Boute, Hôpital Jeanne de Flandre, CHRU de Lille, Lille. A Brusco, University of Torino, Department of Medical Sciences, Turin. D Campion, Service de psychiatrie, Centre hospitalier de Rouvray, Sotteville lès Rouen. A David, Service de Génétique Médicale, CHU-Nantes, Nantes. M-A Delrue, Univ. Bordeaux, Maladies Rares: Génétique et Métabolisme, Service de Génétique Médicale, CHU-Bordeaux, Bordeaux. M Doco-Fenzy, Service de Génétique, CHU, EA3801 SFR-CAP Santé Reims. C Fagerberg, Department of Clinical Genetics, Odense University Hospital, Odense. L Faivre, Hôpital d'Enfants, CHU Dijon. F Forzano, SSD Genetica Medica, EO Ospedali Galliera, Genova. D Giachino, Genetica Medica, Azienda Ospedaliera Universitaria San Luigi Gonzaga Orbassano, Torino. A Guichet, Service de génétique médicale, CHU-Angers. O Guillin, Service de psychiatrie, Centre hospitalier de Rouvray, Sotteville lès Rouen. D Héron, Département de Génétique, Hôpital Pitié-Salpêtrière, Université Pierre et Marie Curie, Paris. B Isidor, Service de Génétique Médicale, CHU-Nantes, Nantes. A Jacquette, Département de Génétique, groupe hospitalier Pitié-Salpétrière, Paris. H Journel, Génétique médicale, Centre Hospitalier Bretagne Atlantique, Vannes. B Keren, Département de Génétique Cytogénétique et Embryologie AP-HP, Hôpital Pitié-Salpêtrière, Université Pierre et Marie Curie, Paris. D Lacombe, Univ. Bordeaux, Maladies Rares: Génétique et Métabolisme, Service de Génétique Médicale, CHU-Bordeaux, Bordeaux. C Le Caignec, Service de Génétique Médicale, CHU Nantes, Nantes. M-P Lemaître, Service de Neuropédiatrie, Centre Hospitalier Régional Universitaire, Lille. J Lespinasse, Service génétique médicale, CH Chambéry. G Mandrile, Genetica Medica, Azienda Ospedaliera Universitaria San Luigi Gonzaga Orbassano, Torino. M Mathieu-Dramard, Service de Génétique Clinique, CHU Amiens, Amiens. C Mignot, Service de Génétique Clinique, CHU La Pitié Salpêtrière, France; Service de Neuropédiatrie, APHP, Hôpital Armand Trousseau. F Petit, Hopital Jeanne de Flandre, CHRU de Lille, Lille. G Plessis, Service de Génétique, CHU Clémenceau, Caen. F Prieur, Service de génétique médicale, CHU St Etienne C Rooryck, Univ. Bordeaux, Maladies Rares: Génétique et Métabolisme, Service de Génétique Médicale, CHU-Bordeaux, Bordeaux. D Sanlaville, Service de Cytogénétique Constitutionnelle, Hospices Civils de Lyon, CHU de Lyon and Centre de Recherche en Neuroscience de Lyon, Lyon. MM Van Haelst, Department of Medical Genetics, University Medical Center, Utrecht. L Van Maldergem, Centre de Génétique humaine, CHU-Besançon.
The authors declare no conflict of interests.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson E, Bernier R, Porche K, Jackson FI, Goin-Kochel RP, Snyder LG, et al. The Cognitive and behavioral phenotype of the 16p11.2 deletion in a clinically ascertained population Biol Psychiatry(in press) [DOI] [PMC free article] [PubMed]
- Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, Kutalik Z, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey F, Sherr EH, Beckmann ND, Hanson E, Maillard AM, Hippolyte L, et al. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J Med Genet. 2012;49:660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. New Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horev G, Ellegood J, Lerch JP, Son YE, Muthuswamy L, Vogel H, et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci USA. 2011;108:17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obesity: preventing and managing the global epidemic. Report of a WHO consultation World Health Organ Tech Rep Ser 2000894i–xii.1-253. [PubMed] [Google Scholar]
- Wechsler D. WISC-IV Echelle d'intelligence de Wechsler pour enfants et adolescents: Quatrième édition. Les Editions du Centre de Psychologie Appliquée: Paris; 2005. [Google Scholar]
- Wechsler D. WAIS-III Echelle d'intelligence de Wechsler pour adultes. les Éditions du Centre de psychologie appliquée: Paris; 2008. [Google Scholar]
- Association, A.P Diagnostic and Statistical Manual of Mental DisordersFourth Edition. DSM-IV-TR®American Psychiatric Association; 2000 [Google Scholar]
- Preisig M, Fenton BT, Matthey ML, Berney A, Ferrero F. Diagnostic interview for genetic studies (DIGS): inter-rater and test-retest reliability of the French version. Eur Arch Psychiatr Clin Neurosci. 1999;249:174–179. doi: 10.1007/s004060050084. [DOI] [PubMed] [Google Scholar]
- Schultze-Lutter F, Ruhrmann S, Klosterkötter J. Shizophrenia, Proneness Instrument (SPI-A) Giovanni Fioriti: Rome; 2007. [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH, Jr, Leventhal BL, DiLavore PC, et al. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi S, Moller HE, Kugel H, Muller DK, Deppe M. Correcting eddy current and motion effects by affine whole-brain registrations: evaluation of three-dimensional distortions and comparison with slicewise correction. Magn Reson Med. 2010;64:1047–1056. doi: 10.1002/mrm.22501. [DOI] [PubMed] [Google Scholar]
- Cook PA, Bai Y, Nedjati-Gilani S, Seunarine KK, Hall MG, Parker GJ, Alexander DC. Camino: Open-Source Diffusion-MRI reconstruction and processing. 14th Scientific Meeting of the International Society for Magnetic Resonance in Medicine. 2003. p. P 2759.
- Draganski B, Ashburner J, Hutton C, Kherif F, Frackowiak RS, Helms G, et al. Regional specificity of MRI contrast parameter changes in normal ageing revealed by voxel-based quantification (VBQ) Neuroimage. 2011;55:1423–1434. doi: 10.1016/j.neuroimage.2011.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ. Testing for anatomically specified regional effects. Hum Brain Mapp. 1997;5:133–136. doi: 10.1002/(sici)1097-0193(1997)5:2<133::aid-hbm7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Kawasaki Y, Suzuki M, Kherif F, Takahashi T, Zhou SY, Nakamura K, et al. Multivariate voxel-based morphometry successfully differentiates schizophrenia patients from healthy controls. Neuroimage. 2007;34:235–242. doi: 10.1016/j.neuroimage.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Ashburner J. Computational anatomy with the SPM software. Magn Reson Imaging. 2009;27:1163–1174. doi: 10.1016/j.mri.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. 1996. J Magn Reson. 2011;213:560–570. doi: 10.1016/j.jmr.2011.09.022. [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, et al. Early brain overgrowth in autism associated with an increase in cortical surface area before age 2 years. Arch Gen Psychiatry. 2011;68:467–476. doi: 10.1001/archgenpsychiatry.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh S, Bansal R, Xu D, Hao X, Liu J, Peterson BS. Neuroanatomical correlates of intellectual ability across the life span. Dev Cogn Neurosci. 2011;1:305–312. doi: 10.1016/j.dcn.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu Y, Zhu M, Wang C, Wang J, Zhang Y, et al. Reduced cortical thickness in mental retardation. PloS One. 2011;6:e29673. doi: 10.1371/journal.pone.0029673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha L, Livy DJ, Beaulieu C, Wheatley BM, Gross DW. In vivo diffusion tensor imaging and histopathology of the fimbria-fornix in temporal lobe epilepsy. J Neurosci. 2010;30:996–1002. doi: 10.1523/JNEUROSCI.1619-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C, Yu K, Fung G, Leung M, Wong C, Li Q, et al. Autistic disorders and schizophrenia: related or remote? An anatomical likelihood estimation. PloS One. 2010;5:e12233. doi: 10.1371/journal.pone.0012233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm O, Heinz A, Walter H, Kirsch P, Erk S, Haddad L, et al. Striatal Response to Reward Anticipation: Evidence for a Systems-Level Intermediate Phenotype for Schizophrenia. JAMA Psychiatry. 2014;71:531–539. doi: 10.1001/jamapsychiatry.2014.9. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2013;505:361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- Kenny PJ. Reward mechanisms in obesity: new insights and future directions. Neuron. 2011;69:664–679. doi: 10.1016/j.neuron.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raji CA, Ho AJ, Parikshak NN, Becker JT, Lopez OL, Kuller LH, et al. Brain structure and obesity. Hum Brain Mapp. 2010;31:353–364. doi: 10.1002/hbm.20870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannacciulli N, Del Parigi A, Chen K, Le DS, Reiman EM, Tataranni PA, et al. Brain abnormalities in human obesity: a voxel-based morphometric study. Neuroimage. 2006;31:1419–1425. doi: 10.1016/j.neuroimage.2006.01.047. [DOI] [PubMed] [Google Scholar]
- Price CJ. A review and synthesis of the first 20 years of PET and fMRI studies of heard speech, spoken language and reading. Neuroimage. 2012;62:816–847. doi: 10.1016/j.neuroimage.2012.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portmann T, Yang M, Mao R, Panagiotakos G, Ellegood J, Dolen G, et al. Behavioral abnormalities and circuit defects in the basal ganglia of a mouse model of 16p11.2 deletion syndrome. Cell Reports. 2014;7:1077–1092. doi: 10.1016/j.celrep.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi BJ, Crofts HJ. Association testing of copy number variants in schizophrenia and autism spectrum disorders. J Neurodev Disord. 2012;4:15. doi: 10.1186/1866-1955-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolphs R. Cognitive neuroscience of human social behaviour. Nat Rev Neurosci. 2003;4:165–178. doi: 10.1038/nrn1056. [DOI] [PubMed] [Google Scholar]
- Nickl-Jockschat T, Habel U, Michel TM, Manning J, Laird AR, Fox PT, et al. Brain structure anomalies in autism spectrum disorder—a meta-analysis of VBM studies using anatomic likelihood estimation. Human Brain Mapp. 2012;33:1470–1489. doi: 10.1002/hbm.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelis C, Velakoulis D, McGorry PD, Wood SJ, Suckling J, Phillips LJ, et al. Neuroanatomical abnormalities before and after onset of psychosis: a cross-sectional and longitudinal MRI comparison. Lancet. 2003;361:281–288. doi: 10.1016/S0140-6736(03)12323-9. [DOI] [PubMed] [Google Scholar]
- Heiskanen T, Niskanen L, Lyytikainen R, Saarinen PI, Hintikka J. Metabolic syndrome in patients with schizophrenia. J Clin Psychiatry. 2003;64:575–579. doi: 10.4088/jcp.v64n0513. [DOI] [PubMed] [Google Scholar]
- Lopresti AL, Drummond PD. Obesity and psychiatric disorders: commonalities in dysregulated biological pathways and their implications for treatment. Prog Neuro-psychopharmacol Biol Psychiatry. 2013;45:92–99. doi: 10.1016/j.pnpbp.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Schnack HG, van Haren NE, Brouwer RM, Evans A, Durston S, Boomsma DI, et al. Changes in thickness and surface area of the human cortex and their relationship with intelligence Cereb Cortex(in press). [DOI] [PubMed]
- Rimol LM, Nesvåg R, Hagler DJ, Jr, Bergmann O, Fennema-Notestine C, Hartberg CB, et al. Cortical volume, surface area, and thickness in schizophrenia and bipolar disorder. Biol Psychiatry. 2012;71:552–560. doi: 10.1016/j.biopsych.2011.11.026. [DOI] [PubMed] [Google Scholar]
- Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, et al. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53:1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Sabuncu MR, Yeo BT, Fischl B, Greve DN, Kochunov P, et al. Measuring and comparing brain cortical surface area and other areal quantities. Neuroimage. 2012;61:1428–1443. doi: 10.1016/j.neuroimage.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Ginestet C, Feng Y, Johnston P, Lombardo MV, Lai MC, et al. Brain surface anatomy in adults with autism: the relationship between surface area, cortical thickness, and autistic symptoms. JAMA Psychiatry. 2013;70:59–70. doi: 10.1001/jamapsychiatry.2013.265. [DOI] [PubMed] [Google Scholar]
- Im K, Lee JM, Lyttelton O, Kim SH, Evans AC, Kim SI, et al. Brain size and cortical structure in the adult human brain. Cereb Cortex. 2008;18:2181–2191. doi: 10.1093/cercor/bhm244. [DOI] [PubMed] [Google Scholar]
- Jacquemont S, Coe BP, Hersch M, Duyzend MH, Krumm N, Bergmann S, et al. A higher mutational burden in females supports a "female protective model" in neurodevelopmental disorders. Am J Human Genet. 2014;94:415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Aldinger KA, Ashwood P, Bauman ML, Blaha CD, Blatt GJ, et al. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. 2012;11:777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BT. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:2322–2345. doi: 10.1152/jn.00339.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poluch S, Juliano SL.Fine-tuning of neurogenesis is essential for the evolutionary expansion of the cerebral cortex Cereb Cortexe-pub ahead of print 22 August 2013; doi: 10.1093/cercor/bht232 [DOI] [PMC free article] [PubMed]
- Blaker-Lee A, Gupta S, McCammon JM, De Rienzo G, Sive H. Zebrafish homologs of genes within 16p11.2, a genomic region associated with brain disorders, are active during brain development, and include two deletion dosage sensor genes. Dis Mod Mech. 2012;5:834–851. doi: 10.1242/dmm.009944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anda FC, Rosario AL, Durak O, Tran T, Gräff J, Meletis K, et al. Autism spectrum disorder susceptibility gene TAOK2 affects basal dendrite formation in the neocortex. Nat Neurosci. 2012;15:1022–1031. doi: 10.1038/nn.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.