Mutations in calreticulin (CALR) were recently described to be present in the majority of patients with a JAK2-unmutated myeloproliferative neoplasm (MPN).1,2 This discovery has had rapid clinical impact, and testing for CALR has been embedded in national and international diagnostic guidelines.3, 4, 5 However, a human MPN-derived cell line harboring a CALR mutation has not been reported and the mechanisms by which mutated-CALR results in an MPN remain unclear.
To begin to investigate the pathogenetic consequences of mutant CALR, we searched for patient-derived cell lines harboring CALR mutations. None were identified by exome sequencing of 1015 cell lines, including 37 derived from hematopoietic neoplasms.1 We therefore looked for cell lines derived from patients with leukemic transformation of a preceding MPN. Given that CALR and JAK2 mutations are almost completely mutually exclusive,1,2,6 we focused on four such lines known to lack a JAK2 mutation (MONO-MAC-6, MARIMO, GDM-1 and ELF-153), and also tested a further 52 other predominantly myeloid cell lines (Supplementary Table 1). Mutation screening was done by Sanger sequencing as previously described,1 and details of other methods are in the Supplementary Information.
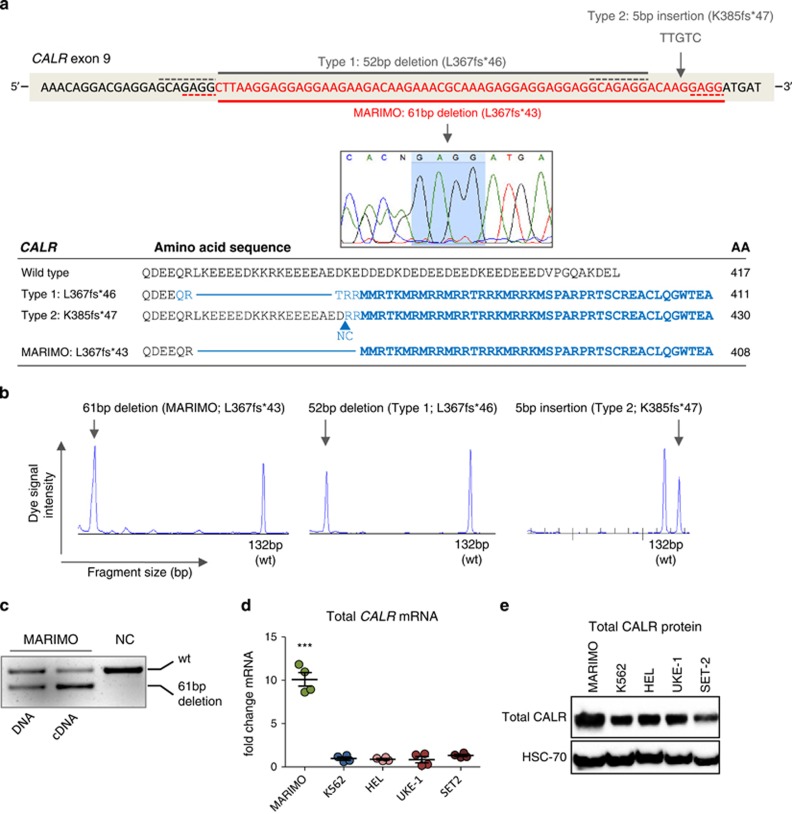
The only cell line found to harbor a CALR mutation was MARIMO, originally derived from a 68-year-old female with AML-M2, and an antecedent history of ET.7 MARIMO is negative for JAK2V617F and MPL exon 10 mutations (data not shown) and carries a heterozygous 61-basepair (bp) deletion in CALR exon 9 (c.1099_1159del; L367fs*43), which, like all other reported CALR mutations, results in a +1- bp shift in the reading frame and thus generates a novel C terminus (Figure 1a). In patients, the commonest two CALR mutations, accounting for 85% of cases, are a 52-bp deletion (type 1; c.1099_1150del; L367fs*46) and a 5-bp insertion (type 2; c.1154_1155_ins; K385fs*47).8 Both the type 1 deletion and the MARIMO deletion are immediately preceded by a nucleotide sequence identical to that at the 3' end of the deletion (Figure 1a). The 61-bp MARIMO deletion is readily detected by fragment analysis and represents a useful positive control for diagnostic clinical testing (Figure 1b).
Figure 1.
Identification of a CALR-mutated human cell line. (a) Top panel shows the mutated region in CALR exon 9 (red bases). The commonest CALR mutations are shown above the DNA sequence. Solid gray line shows type 1 (52- bp deletion; c.1099_1150del; L367fs*46) and gray arrow shows type 2 (5- bp insertion; c.1154_1155_ins; K385fs*47 mutations). The CALR mutation in human cell line MARIMO is shown below the DNA sequence. Solid red line and capillary sequencing image show a heterozygous 61-bp deletion (c.1099_1159del; L367fs*43) in MARIMO. Dashed gray and red lines represent the homologous sequence flanking the deleted regions in type 1 and MARIMO mutations, respectively, also highlighted in the capillary sequencing image (pale blue) for MARIMO. Lower panel shows the predicted protein sequence of the commonest CALR mutations and of MARIMO with total protein sizes. Amino acids (AA) in the new reading frame are shaded blue and the common novel peptide sequence shared by the different CALR variants are in bold blue. (b) PCR amplification of CALR exon 9 followed by fragment size analysis, as used for diagnostic testing for CALR mutations. Vertical heights of peaks represent dye signal intensity and horizontal position of peaks reflect the fragment size of the PCR amplicon. Wild type (wt) peak occurs at 132- bp. Left panel shows wt and mutated alleles of MARIMO (61-bp separation in peaks), middle panel shows Type 1/L367fs*46 with peak separation of 52 bp and right panel shows Type 2/K385fs*47 peaks separated by 5 bp. (c) Agarose gel image showing wt (upper band) and mutated-CALR (lower band) in MARIMO DNA and cDNA. (d) Quantitative real-time PCR of total CALR mRNA levels expressed as a fold change relative to house-keeping RPLP0 levels, for the cell lines MARIMO, the BCR-ABL1+ CML cell line K562, and the JAK2V617F+ cell lines HEL, UKE-1 and SET-2. Graph depicts all data points generated in two independent experiments performed in duplicate. ***P<0.001 (e) Western blot showing total CALR protein levels of MARIMO and four other myeloid cell lines.
Allele-specific PCR demonstrated expression of the mutant CALR allele (Figure 1c). Compared with other cell lines derived from patients with JAK2V617F (HEL, UKE-1 and SET-2) or CML (K562) total CALR mRNA levels were 10-fold higher in MARIMO (Figure 1d) and total CALR protein levels were also increased albeit more modestly (Figure 1e).
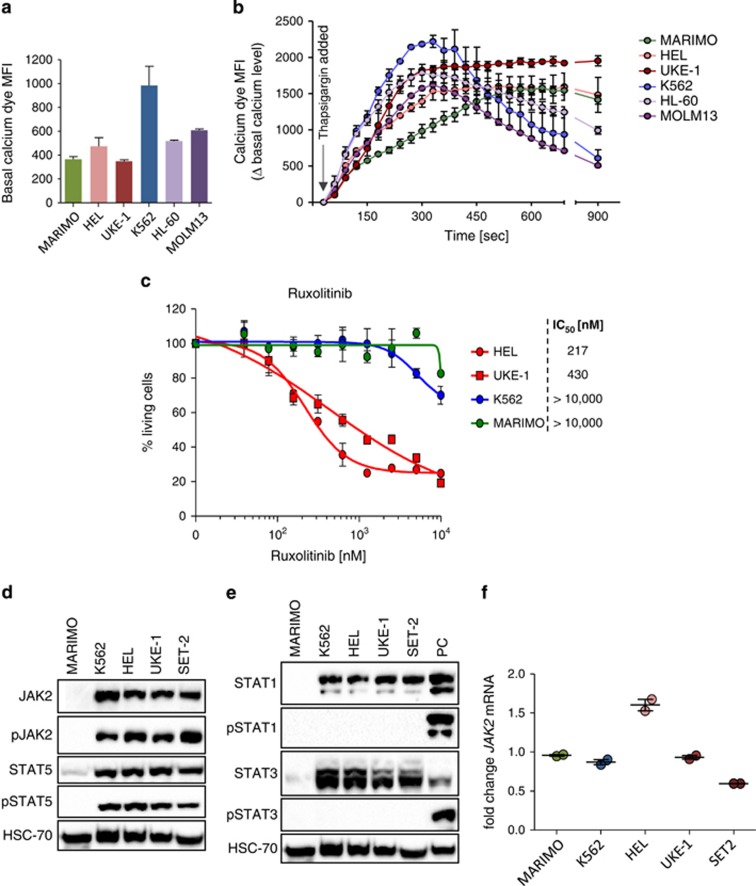
MARIMO cells expressed cell surface marker CD15 but not other progenitor or lineage-affiliated markers (Supplementary Table 2). The proliferation and cell cycle status of MARIMO was unremarkable compared with other myeloid lines (Supplementary Figure 1). We next analysed cellular calcium stores since CALR has an important role in endoplasmic reticulum (ER) mediated calcium homeostasis9 and mutant CALR protein lacks variable numbers of calcium binding sites present in the wild-type C terminus. No significant differences in basal cytoplasmic calcium levels were found amongst the six cell lines tested (Figure 2a). Cell lines were then treated with 1 μM thapsigargin, which blocks ER Ca2+-ATPase channels resulting in ER calcium depletion and increased cytosolic calcium levels.10 MARIMO cells showed the slowest rate of increase of cytoplasmic calcium levels upon addition of thapsigargin (Figure 2b), consistent with the concept that mutant CALR alters ER dependent calcium homeostasis.
Figure 2.
Characterization of the cell line MARIMO. (a and b) Basal cytoplasmic calcium level (a) and changes in cytoplasmic calcium levels over time upon addition of thapsigargin (b) in MARIMO and five other leukemic cell lines. (c) Dose response curves for the JAK2-inhibitor Ruxolitinib in the cell lines MARIMO, K562, HEL and UKE-1. (d and e) Western blots showing protein levels of the inactive and phosphorylated forms of JAK2 and STAT5 (d) and STAT1 and STAT3 (e). PC, positive control. (f) JAK2 mRNA levels expressed as fold changes relative to RPLP0 in MARIMO and four myeloid cell lines.
The mutual exclusivity of JAK2 and CALR mutations argues that they may share pathogenetic mechanisms and has been used to suggest that CALR mutations may activate JAK2/STAT5 signaling. This concept is supported by expression profiling of patient-derived granulocytes11 together with a report that expression of CALR in Ba/F3 cells confers interleukin-3 independence and is accompanied by increased STAT5 phosphorylation.2 However other studies have reported distinct transcriptional signatures in JAK2V617F-mutated and JAK2V617F-unmutated MPNs.12,13 Interpretation of these apparently conflicting results is complicated by several issues including limitations of overexpression systems, the uncertain relevance of granulocytes to disease pathogenesis and difficulties inherent to studies of signaling in primary cells containing variable proportions of mutant cells. To circumvent some of these issues, and to gain insight into the consequences of CALR mutations, we explored the properties of MARIMO cells.
The dependence of MARIMO cells on JAK signaling was initially assessed using the JAK inhibitors Tofacitinib (a JAK2/3 inhibitor) and JAK-inhibitor-I (a pan-JAK inhibitor) (Supplementary Figure 2). MARIMO cells were more resistant to both inhibitors than seven cell lines harboring mutant JAK2 or JAK3. Dose response studies using the clinically approved JAK-inhibitor Ruxolitinib (INCB018424, a JAK1/2 inhibitor) showed that HEL and UKE-1 (both JAK2V617F positive) had IC50 values of 217 and 430 nm, respectively (Figure 2c). In marked contrast the IC50 value for MARIMO was greater than 10 000 nm, demonstrating that MARIMO was not dependent on JAK2 signaling. Consistent with these data, western blot analysis showed that, compared with JAK2-mutant cells, MARIMO cells contained markedly reduced levels of JAK2, phosphorylated-JAK2 (pJAK2), STAT5 and pSTAT5 (Figure 2d). The lack of JAK2-STAT5 signaling was not accompanied by a compensatory increase in STAT1 or STAT3 phosphorylation (Figure 2e). JAK2 transcript levels in MARIMO were similar to other cell lines (Figure 2f), suggesting either decreased translation or increased degradation of JAK2.
Together, our data demonstrate that the MARIMO cell line harbors a CALR mutation and yet is not dependent on JAK/STAT signaling, in marked contrast to JAK2-mutated cell lines. Our results therefore raise the possibility that mutations of CALR and JAK2 may share activation of pathways other than the STATs. Superficially our data appear to contrast with reports that JAK2-unmutated and CALR-mutated MF patients respond to ruxolitinib.14,15 However in JAK2V617F-positive patients studies of mutant allele burden show that Ruxolitinib has a minimal effect on the mutant clone.15 It is therefore likely that the clinical responses to Ruxolitinib (reduced splenomegaly and improved constitutional symptoms) do not reflect a cytotoxic effect of the drug on the neoplastic clone, but instead are at least in part due to down-modulation of pro-inflammatory signaling cascades.16
Acknowledgments
We are grateful to Hitoshi Kiyoi (Nagoya University) for provision of cell line MARIMO. We thank Barbara Newman, Haemato-Oncology Diagostic Service, Addenbrooke's Hospital, Cambridge, UK. Work in the Green lab is supported by Leukemia and Lymphoma Research, Cancer Research UK, the NIHR Cambridge Biomedical Research Centre, the Cambridge Experimental Cancer Medicine Centre, and the Leukemia and Lymphoma Society of America. WW is supported by the Austrian Science Foundation (J 3578-B21). JN is supported by a Kay Kendall Leukaemia Clinical Fellowship.
Author Contributions
KK performed western blots and reverse transcriptase–PCR; WW performed calcium release-, inhibitor-, proliferation- and cell cycle assays; JN performed cell line screen; HQ and HD prepared cell lines; AB, EB performed clinical assays. All authors wrote and reviewed the manuscript. ARG directed the research.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369:2391–2405. doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369:2379–2390. doi: 10.1056/NEJMoa1311347. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Thiele J, Vannucchi AM, Barbui T. An overview on CALR and CSF3R mutations and a proposal for revision of WHO diagnostic criteria for myeloproliferative neoplasms. Leukemia. 2014;28:1407–1413. doi: 10.1038/leu.2014.35. [DOI] [PubMed] [Google Scholar]
- Reilly JT, McMullin MF, Beer PA, Butt N, Conneally E, Duncombe AS, et al. Use of JAK inhibitors in the management of myelofibrosis: a revision of the British Committee for Standards in Haematology Guidelines for Investigation and Management of Myelofibrosis 2012 Br J Haematol 2014. e-pub ahead of print 25 June 2014; doi: 10.1111/bjh.12985 [DOI] [PubMed]
- Harrison CN, Butt N, Campbell P, Conneally E, Drummond M, Green AR, et al. Modification of British Committee for Standards in Haematology diagnostic criteria for essential thrombocythaemia Br J Haematol 2014. e-pub ahead of print 17 June 2014; doi: 10.1111/bjh.12986 [DOI] [PubMed]
- Tefferi A, Lasho TL, Finke CM, Knudson RA, Ketterling R, Hanson CH, et al. CALR vs JAK2 vs MPL-mutated or triple-negative myelofibrosis: clinical, cytogenetic and molecular comparisons. Leukemia. 2014;28:1472–1477. doi: 10.1038/leu.2014.3. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Kondo M, Ichihashi T, Hashimoto N, Inazawa J, Ohno R, et al. A novel myeloid cell line, Marimo, derived from therapy-related acute myeloid leukemia during treatment of essential thrombocythemia: consistent chromosomal abnormalities and temporary C-MYC gene amplification. Cancer Genet Cytogenet. 1998;100:21–24. doi: 10.1016/s0165-4608(97)00017-4. [DOI] [PubMed] [Google Scholar]
- Guglielmelli P, Nangalia J, Green AR, Vannucchi AM. CALR mutations in myeloproliferative neoplasms: hidden behind the reticulum. Am J Hematol. 2014;89:453–456. doi: 10.1002/ajh.23678. [DOI] [PubMed] [Google Scholar]
- Michalak M, Groenendyk J, Szabo E, Gold LI, Opas M. Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem J. 2009;417:651–666. doi: 10.1042/BJ20081847. [DOI] [PubMed] [Google Scholar]
- Wictome M, Henderson I, Lee AG, East JM. Mechanism of inhibition of the calcium pump of sarcoplasmic reticulum by thapsigargin. Biochem J. 1992;283:525–529. doi: 10.1042/bj2830525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel J-P, Mermel CH, et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood. 2014;123:e123–e133. doi: 10.1182/blood-2014-02-554634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmers S, Will B, Waller CF, Abdulkarim K, Johansson P, Andreasson B, et al. JAK2V617F-negative ET patients do not display constitutively active JAK/STAT signaling. Exp Hematol. 2007;35:1695–1703. doi: 10.1016/j.exphem.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigdecanet E, Espinet B, Lozano JJ, Sumoy L, Bellosillo B, Arenillas L, et al. Gene expression profiling distinguishes JAK2V617F-negative from JAK2V617F-positive patients in essential thrombocythemia. Leukemia. 2008;22:1368–1376. doi: 10.1038/leu.2008.112. [DOI] [PubMed] [Google Scholar]
- Passamonti F, Caramazza D, Maffioli M. JAK inhibitor in CALR-mutant myelofibrosis. N Engl J Med. 2014;370:1168–1169. doi: 10.1056/NEJMc1400499. [DOI] [PubMed] [Google Scholar]
- Verstovsek S, Mesa RA, Gotlib J, Levy RS, Gupta V, DiPersio JF, et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med. 2012;366:799–807. doi: 10.1056/NEJMoa1110557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 Inhibitor, in myelofibrosis. N Engl J Med. 2010;363:1117–1127. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.