Abstract
Aim
Heart disease is recognized as a consequence of dysregulation of cardiac gene regulatory networks. Previously, unappreciated components of such networks are the long non-coding RNAs (lncRNAs). Their roles in the heart remain to be elucidated. Thus, this study aimed to systematically characterize the cardiac long non-coding transcriptome post-myocardial infarction and to elucidate their potential roles in cardiac homoeostasis.
Methods and results
We annotated the mouse transcriptome after myocardial infarction via RNA sequencing and ab initio transcript reconstruction, and integrated genome-wide approaches to associate specific lncRNAs with developmental processes and physiological parameters. Expression of specific lncRNAs strongly correlated with defined parameters of cardiac dimensions and function. Using chromatin maps to infer lncRNA function, we identified many with potential roles in cardiogenesis and pathological remodelling. The vast majority was associated with active cardiac-specific enhancers. Importantly, oligonucleotide-mediated knockdown implicated novel lncRNAs in controlling expression of key regulatory proteins involved in cardiogenesis. Finally, we identified hundreds of human orthologues and demonstrate that particular candidates were differentially modulated in human heart disease.
Conclusion
These findings reveal hundreds of novel heart-specific lncRNAs with unique regulatory and functional characteristics relevant to maladaptive remodelling, cardiac function and possibly cardiac regeneration. This new class of molecules represents potential therapeutic targets for cardiac disease. Furthermore, their exquisite correlation with cardiac physiology renders them attractive candidate biomarkers to be used in the clinic.
Keywords: Myocardial infarction, Heart failure, Transcriptome, Long non-coding RNAs, Next-generation sequencing
Introduction
Coronary artery disease is the most frequent cardiovascular disorder and typically leads to acute myocardial infarction (MI) and ultimately heart failure (HF).1,2 Despite continued advances, HF is rapidly evolving into a major global epidemic requiring novel therapeutic approaches. In the light of this, the elucidation of novel regulatory mechanisms involved in HF pathogenesis holds the promise of identifying new avenues for this prevalent deadly disease. In the adult heart, stress-dependant pathological haemodynamic and neurohormonal signals induce a maladaptive remodelling response, a process characterized by cardiomyocyte (CM) hypertrophy, interstitial fibrosis, and ultimately cellular dysfunction resulting in contractile and functional failure.2 Importantly, the hallmark of pathological remodelling in the adult heart is a global transcriptional reprogramming, resulting in the reactivation of a foetal cardiac gene programme.3 At the molecular level, these signals activate a network of interacting cardiac signal transduction cascades that converge on evolutionary conserved cardiac transcription factors (TFs).2–4 These core TFs (e.g. SRF, NKX2.5, MEF2c, GATA4, and Tbox factors) interact in a combinatorial manner to elicit specific temporal and spatial gene expression programmes that are ultimately responsible for pathological remodelling.
In this context, the notion of gene regulatory networks (GRNs) being primarily protein-based regulatory systems has been somewhat premature.5 A number of recent studies have demonstrated that GRN activity is under the control of a myriad of interleaved networks of non-coding RNAs (ncRNAs). Non-coding RNAs control every aspect of GRN activity including transcriptional control, post-transcriptional processing, and epigenetic targeting.6 The best-characterized ncRNAs in the heart are the small microRNAs (miRNAs), which adjust entire functional networks of mRNAs via post-transcriptional gene silencing, implicating miRNAs as important stress-dependant modulators.7,8 In addition to small ncRNAs, global transcriptomic screens have identified other functional classes of transcripts, which are larger than 200 nucleotides, collectively known as long non-coding RNAs (lncRNAs).9–11 The functions of most lncRNAs remain unknown; however, many have been shown to exert non-redundant roles in a diverse array of biological processes including chromosome X inactivation,12 imprinting,13 splicing,14 and transcriptional regulation.15 In particular, lncRNAs appear to be important for the global modulation of cell-specific epigenomic states via directing chromatin modification complexes to their sites of action.16 Furthermore, mammalian lncRNAs appear to be expressed in a highly cell-type and context-specific manner.17–19 Considering the functionality of these transcripts, lncRNAs may represent an important class of regulatory mediators of cardiogenic lineage-specific commitment during development and of specialized cellular functions involved in maintaining cardiac integrity. Accordingly, the majority of cardiogenic lncRNAs functionally characterized to date regulates developmental processes.20,21 However, their potential role controlling mature tissue homoeostasis and adaptation to stress remains largely unexplored.
The intrinsic -cis and -trans activating and epigenomic orchestrating properties of lncRNAs warrants the need to explore and generate catalogues of cardiac-specific lncRNAs in diseased adult tissues. In this study, we set out to characterize the cardiac long non-coding transcriptome and in particular the dynamically modulated fraction post-MI (Supplementary material online, Figure S1). We coupled deep RNA-sequencing with ab initio transcript reconstruction, and integrated genome-wide data sets to systematically identify and annotate novel heart-specific lncRNAs. We show that these lncRNAs are highly cardiac and context specific, correlating with cardiac physiology, suggesting a role as modulators of the pathological response. Using functional inference based on developmental chromatin state transitions, we functionally annotated these novel lncRNAs demonstrating that they are predominantly implicated with cardiac developmental, structural, and functional gene programmes. In particular, novel lncRNAs are predominantly associated with active enhancer states. We validated several novel lncRNAs in developmental and pathological models in vitro and in vivo, and demonstrate that the expression of specific novel lncRNAs is closely associated with individual cardiac patho-physiological traits. Finally, we identified hundreds of predicted human orthologues and validated their expression in human samples. A number of these validated human orthologues were differentially expressed in human heart disease, supporting conserved roles in cardiac remodelling. Collectively, we have described a novel class of mammalian heart-specific lncRNAs with unique regulatory and functional characteristics, relevant to maladaptive pathological remodelling, cardiac function, and potentially regeneration. Further characterization of these novel lncRNAs could provide unprecedented opportunities for diagnosis and therapeutic intervention.
Methods
Cardiac injury models: microsurgery
Ligation of the left anterior descending artery: MI in mice (C57/BL6; males; 12 weeks of age) was induced, as previously described. See Supplementary material online, extended experimental procedures.
Echocardiography
Transthoracic echocardiographies were performed using a 30-MHz probe and the Vevo 770 Ultrasound machine (VisualSonics, Toronto, ON, Canada).
RNA purification and selection of representative samples for RNA sequencing
Total RNA was isolated from mouse hearts using the RNeasy isolation kit (Qiagen) 2 weeks after infarction. Expression of selected cardiac markers, i.e. Nppa, Nppb, Myh6, Mhy7, Col1a, Postn, and Tgfb1, was determined in sham-operated and infarcted heart samples. Four samples in each group were selected for RNA-sequencing based on expected patterns of gene expression known to be representative of normal vs. pathological hearts, i.e. significant increased expression of Nppa, Nppb, Mhy7, Col1a, Postn, and Tgfb1, and decreased expression of Myh6 in infarcted heart when compared with sham-operated hearts. Samples were also selected on echocardiographic parameters, in particular control hearts demonstrated normal ejection fraction (EF) (>45%), whereas infarcted heart showed decreased EF (<15%).
RNA-sequencing and analysis
Sequencing libraries were prepared according to Illumina RNA Seq library kit instructions with Poly(A) selection. Libraries were sequenced with the Illumina HiSeq2000 (2 × 100 bp). Paired-end RNAseq reads were mapped and analysed as described in Supplementary material online, extended experimental procedures.
Primary cell cultures and transfection
Please refer to Supplementary material online, extended experimental procedures.
Embryonic stem cell differentiation
Mouse embryonic stem (ES) cells were differentiated into CMs as described previously.22
RNA expression
RNA was isolated using the RNeasy Kit (Qiagen) according to manufacturer's instructions, using on column DNase treatment. Complimentary DNA was generated using the SuperScript III kit (Invitrogen) with random hexamer primers. Quantitative real-time PCR (qRT–PCR) was carried out using the Applied Biosystems SYBR Green and TaqMan PCR kit and an ABI Prism 7500 cycler and analysed using the ΔΔCt method.
Chromatin-based integrated analysis
For details see Supplementary material online, extended experimental procedures.
Human samples
For details see Supplementary material online, extended experimental procedures.
Statistical analysis
Data throughout the paper are expressed as means ± SEM. One-way ANOVA was used to test significance of data comparisons between experimental groups, with P-values <0.05 were considered significant (95% confidence interval).
Accession numbers
Data sets have been deposited in the Gene Expression Omnibus Database under accession number GSE52313.
Results
Global identification of long non-coding RNAs expressed in the heart and regulated during myocardial infarction
We first set out to characterize global transcriptional regulation during myocardial adaptation to stress for both the coding and non-coding transcriptomes. We utilized a well-characterized pathophysiological model of cardiac stress in the mouse, namely MI obtained by left anterior descending artery ligation. Fourteen days post-infarction, the myocardium was characterized by intense remodelling, decreased cardiac function and induction of cardiac markers of stress (Nppa, Nppb, Myh7, Col1a1, Postn, and Tgfb1) (Supplementary material online, Table S1). We then identified lncRNAs expressed in the infarcted adult mouse heart via RNA-sequencing and ab initio transcriptome reconstruction of carefully selected control (sham-operated hearts) and infarcted samples based on those best representing the maladaptive remodelling response of a large group (n = 17, data not shown) (Supplementary material online, Figure S2A). Massive parallel sequencing was used to obtain paired-end reads of experimental libraries from the border zone (BZ) of four infarcted hearts and four corresponding regions of sham-operated non-infarcted hearts, and Cufflinks23 was utilized to perform ab initio transcript assembly on mapped paired-end reads. Immediately, post-MI the mammalian heart gradually evolves into three distinct regions, the infarct, BZ, and remote zone (RZ) (Supplementary material online, Figure S2B and C). Of these regions the BZ, which is located between infarct and RZ, is the location of significant biological processes including inflammation and fibrosis, and is thought to be of importance for the immediate adaptive response and subsequent long-term remodelling that ultimately leads to HF. We therefore sequenced the transcriptome of the BZ to ensure the identification of transcripts involved in these key remodelling processes.
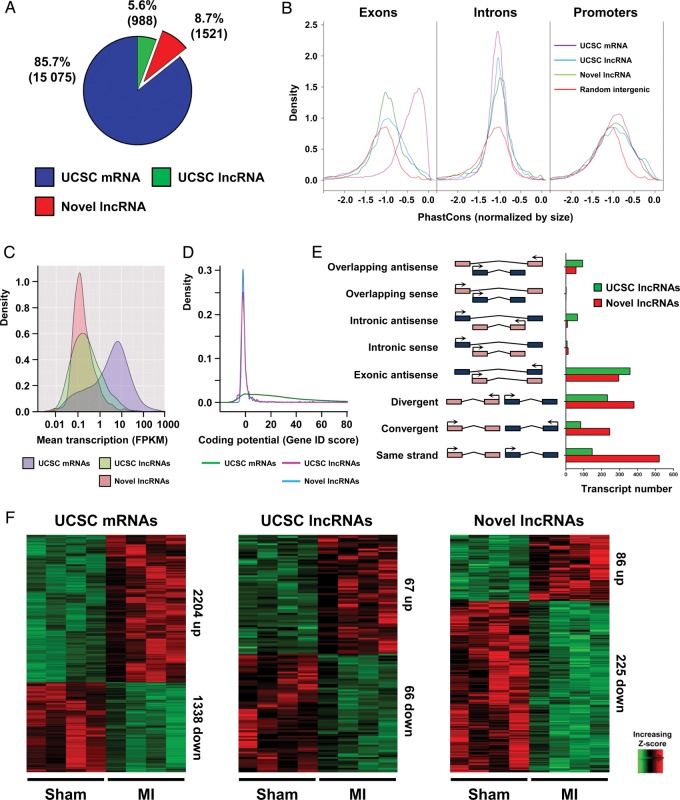
This analysis reconstructed 17 584 multi-exonic transcripts, of which 15 075 (2204 up-regulated and 1338 down-regulated) correspond to University of California Santa Cruz (UCSC)-annotated protein-coding genes (Figure 1A–F). Our lncRNA annotation pipeline identified 2509 multi-exonic lncRNAs (>200 bp). There were 988 (67 up-regulated and 66 down-regulated) UCSC-annotated lncRNAs and 1521 (86 up-regulated and 225 down-regulated) novel unannotated lncRNAs, encompassing all known lncRNA locus-types (Figure 1E). To verify the non-coding nature of our novel lncRNA candidates, we used the GeneID-coding potential score and found that these novel transcripts have minimal protein-coding potential, comparable with UCSC-annotated lncRNAs (Figure 1D). Furthermore, novel lncRNAs and UCSC lncRNAs were expressed at significantly lower levels than coding genes (Figure 1C). Novel and UCSC lncRNA exons were less conserved than coding exons although promoters were equally conserved (Figure 1B).
Figure 1.
Global identification of long non-coding RNAs expressed in the heart during myocardial infarction . RNA-Seq during myocardial infarction was performed on border zone and sham operated samples to characterize the infarction associated transcriptome. (A) Pie chart showing composition of PolyA+ transcriptome, UCSC mRNAs (blue) UCSC long non-coding RNAs (green) and novel long non-coding RNAs (red). Transcript numbers in each group are indicated in brackets (B) Kernel density plots of phastCons score distribution of UCSC mRNA, UCSC long non-coding RNA, novel long non-coding RNA, and random intergenic sequence for exons, introns, and promoters. (C) Kernel density plot of the transcript abundance [fragments per kilobases per million reads (FPKM)] of UCSC mRNAs, UCSC long non-coding RNAs and novel long non-coding RNAs. (D) Kernel density plot of coding potential (Gene ID score). (E) Number of long non-coding RNA genes in orientations relative to nearby coding genes (F) Heatmaps showing hierarchical clustering of differentially expressed transcripts within the three RNA classes after myocardial infarction.
We further characterized the basic features of novel lncRNAs, comparing them with UCSC-coding genes and lncRNAs where appropriate. The size of the novel lncRNA transcripts was comparable with coding genes and slightly greater than annotated lncRNAs even if they had fewer exons than both annotated transcript classes (Supplementary material online, Figure S2H–J). Approximately 70% of all novel lncRNAs have three exons or less (40% having only two exons). To validate that the cDNA libraries of the Sham and MI samples represent a transcriptome associated with infarction-dependant remodelling, we first analysed well-characterized stress marker expression in the RNA-Seq data and showed their appropriate modulation (Supplementary material online, Figure S2D). In addition, several novel lncRNAs demonstrated changes in expression (Supplementary material online, Figure S2D and K). Furthermore, globally, the cardiac transcriptome correlated well within Sham samples, and to an independent ENCODE adult heart RNA-Seq data set (Supplementary material online, Figure S2E and F).24 Unsupervised hierarchical clustering of all UCSC coding, lncRNA, and novel lncRNA transcripts identified two distinct clusters corresponding to the Sham and MI cDNA libraries demonstrating that the transcriptome was sensitive to and representative of the phenotypic adaptation observed in the heart post-MI (Supplementary material online, Figure S2G).
Long non-coding RNAs are potent post-transcriptional modulators of gene expression via their ability to act as competitive endogenous RNAs (ceRNAs) for miRNAs.25 To investigate whether the cardiac lncRNA transcriptome may be involved in cardiac-specific ceRNA networks, we selected three miRNAs known to be modulated in the heart after infarction and determined the putative target lncRNAs and mRNAs. We generated miRNA–lncRNA–mRNA correlation networks with modulated miRNAs and target transcripts visualized in Hive plots, where miRNAs, mRNAs, and lncRNAs are presented on three axes, and the arcs connecting the axes represent positive (red) and negative (blue) correlation between nodes (Supplementary material online, Figure S2L). It was particularly striking that lncRNAs correlated positively with these miRNAs, whereas mRNAs correlated negatively. Moreover, the miRNAs appeared to target common lncRNAs and mRNAs in a synergistic manner (Supplementary material online, Figure S2M). These findings are consistent with the implication of lncRNAs as decoys for miRNAs.
Novel long non-coding RNAs are heart specific and proximal to cardiac developmental genes
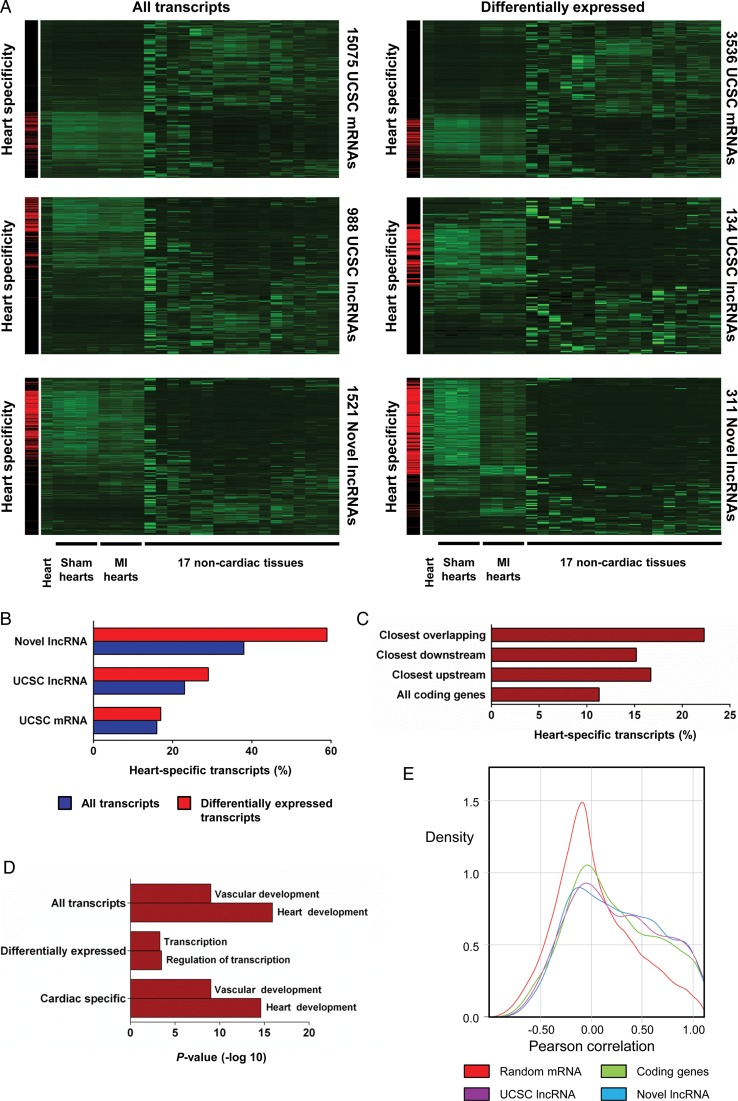
The majority of lncRNAs identified in our analysis represents novel lncRNAs that have previously escaped annotation. This could reflect in part the high levels of cell and tissue specificity typically associated with lncRNAs. We, therefore, computationally realigned 17 mouse non-cardiac ENCODE RNA-Seq data sets,24 and found that 16% of UCSC mRNAs and 23% of UCSC lncRNAs were classified as heart specific (Figure 2A and B, see Supplementary material online for a detailed definition of the heart specificity score). In contrast, 38% of the novel lncRNAs were heart-specific, a significant enrichment vs. UCSC mRNAs and lncRNAs (novel lncRNA vs. mRNA: P< 2.2 × 10−16, novel lncRNA vs. UCSC lncRNA: P< 6.4 × 10−9). Furthermore, differentially expressed novel lncRNAs were significantly more heart-specific than all transcript classes, with 60% of these novel lncRNAs being classified as heart specific (novel lncRNA vs. mRNA: P< 2.2 × 10−16, novel lncRNA vs. UCSC lncRNA: P< 2.2 × 10−16) (Figure 2A and B, Supplementary material online, Figure S3A). Heart specificity of selected candidates was confirmed by qRT–PCR (Supplementary material online, Figure S3B). This increased heart specificity is likely a consequence of the association of novel lncRNAs with enhancer sequences active in the heart. Long non-coding RNAs have been shown to regulate coding gene expression both in cis and in trans. If cis-regulation was common, one would expect proximal coding genes to also be more heart specific. In support of this, we found that overlapping, proximal upstream or downstream coding genes were significantly more heart-specific than the entire coding gene collection (three categories vs. all other genes: 6.4 × 10−8) (Figure 2C). Furthermore, gene ontology (GO) analysis of these proximal coding genes revealed significant enrichment in GO biological process categories associated with heart development, cardiac function, and transcriptional regulation (Figure 2D). Interestingly, differentially expressed novel lncRNAs were more associated with transcriptional control, suggesting that modulated novel lncRNAs may be implicated in the transcriptional reprogramming observed in the remodelling heart.
Figure 2.
Tissue specificity of novel long non-coding RNAs. Novel long non-coding RNAs are more heart-specific and proximal to cardiac developmental genes. (A) Clustering of all and differentially expressed UCSC mRNAs, UCSC long non-coding RNAs and novel long non-coding RNAs across Sham-operated hearts, infarcted hearts, ENCODE heart and 17 non-cardiac mouse tissues. Left-hand panels highlight cardiac-specific transcripts in red. (B) Heart specificity of all and differentially expressed UCSC mRNAs, UCSC long non-coding RNAs and novel long non-coding RNAs. (C) Heart specificity of all UCSC coding genes compared with UCSC coding genes proximal to novel overlapping downstream and upstream long non-coding RNAs. (D) Enriched gene ontology terms for genes closest to all, differentially expressed and heart-specific novel long non-coding RNAs. (E) Kernel density plot displaying correlation of RNA expression for random gene pairs, neighbouring coding gene pairs, UCSC long non-coding RNA coding gene pairs and novel long non-coding RNA coding gene pairs during myocardial infarction.
The enrichment of specific cardiac gene functions in coding genes adjacent to novel lncRNAs raised the possibility that novel lncRNAs acted in cis, and directly affected the expression of their chromosomal neighbours. To test this, we studied the expression of all transcript classes with their neighbouring coding genes post-MI, and identified many examples of highly correlated novel lncRNA-coding gene pairs (Supplementary material online, Table S2 and Figure S3C). Novel lncRNAs and their neighbouring coding genes were more correlated to each other than random gene pairs. However, coding gene pairs and UCSC lncRNA neighbours were also more correlated than random coding gene pairs (Figure 2E). Together these data suggest that novel lncRNAs are not more correlated to their coding gene neighbours post-infarction than expected for any given pair of neighbouring loci.
The cardiac transcriptome is highly correlated with cardiac physiological traits
Quantitative expression profiling using RNA sequencing has a number of significant advantages over hybridization-based array methods. Arguably, the greatest advantage is the greater dynamic range in transcript abundance determination. These unique characteristics lend global transcriptome data sets amenable to direct correlation profiling with continuous traits of choice. The heart itself represents a unique organ that can be physiologically characterized in vivo in terms of functional contractile and remodelling parameters.
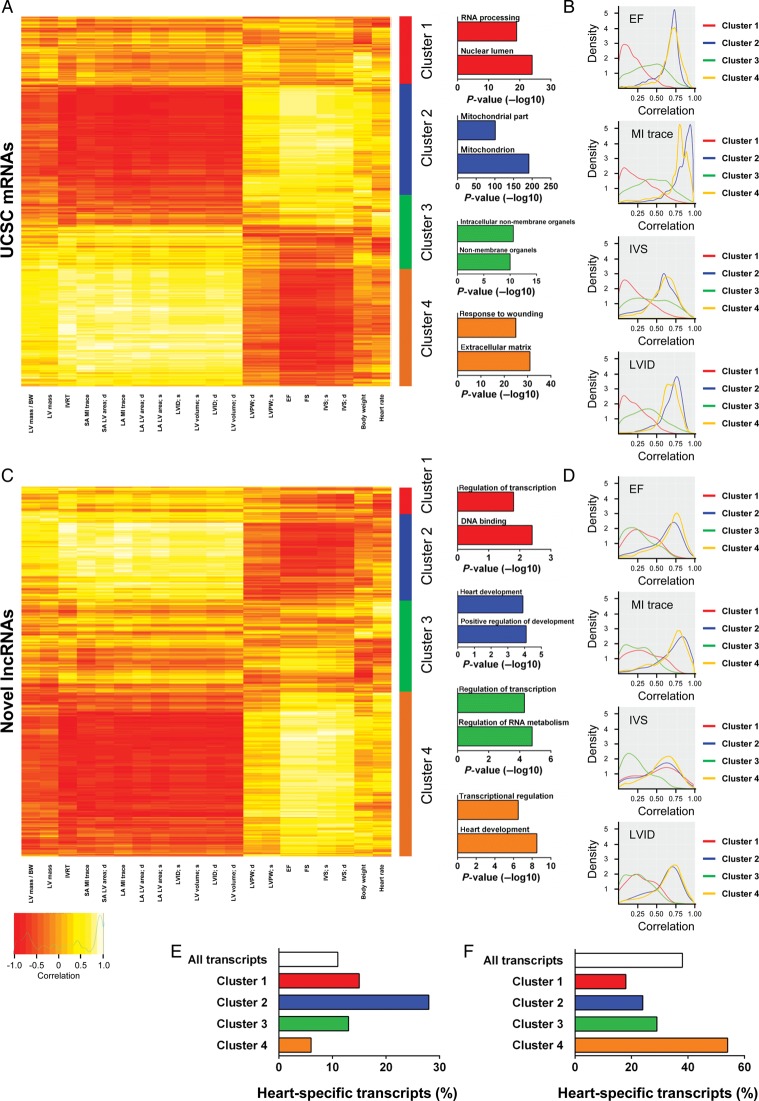
We therefore correlated the cardiac transcriptome with physiological traits measured by echocardiography during MI (Sham and MI samples were used for correlation analysis). Both the coding and non-coding transcriptome correlated well with parameters of cardiac dimensions and function (Figure 3); however, globally novel lncRNAs were better correlated than UCSC lncRNAs with all physiological traits assessed (Supplementary material online, Figure S4A). To gain a deeper molecular insight and potentially identify molecular pathways associated with physiological traits, we executed unsupervised clustering and further downstream analysis of UCSC-coding genes and novel lncRNAs. We identified four clusters for both coding (Figure 3A) and novel lncRNA (Figure 3C) transcripts. In each case, these consisted of one cluster that correlated positively with cardiac function and negatively with remodelling parameters, one cluster with the inverse of these correlations and two clusters with non-specific intermediate correlations. Gene ontology and heart-specificity analyses were performed on individual clusters. With respect to novel lncRNAs, GO analysis was executed on the most proximal coding genes. In the coding gene group, the most heart-specific cluster was Cluster 2 (Figure 3E), which was positively correlated with cardiac functional traits and associated with genes involved in mitochondrial biology (Figure 3A). The least heart-specific cluster (Cluster 4) was positively correlated with remodelling and associated with genes involved in wound healing and extracellular matrix (Figure 3A). Within novel lncRNAs, the most heart-specific cluster, i.e. Cluster 4 (Figure 3F), was again positively correlated with cardiac function associated traits. Proximal coding genes to novel lncRNAs in Cluster 4 were enriched with heart development associated processes (Figure 3C). Since novel lncRNAs that correlated strongly with particular physiological traits were likely to be involved in biological processes associated with those traits, these findings indicated that novel lncRNAs within this cluster could represent a class of cardiac-specific regulators of developmental gene programmes, which was reactivated in the damaged myocardium. The least heart-specific clusters were clusters one and two, which was positively correlated with remodelling traits.
Figure 3.
Physiological clustering of the cardiac transcriptome. The cardiac transcriptome is highly correlated with cardiac physiological traits. (A) Clustering of UCSC mRNAs based on correlation of expression with echocardiography-derived physiological traits after myocardial infarction (Sham-operated and myocardial infarction samples were used for correlations). Clusters are illustrated right of the heatmap with top-enriched gene ontology terms for coding genes in each cluster displayed. (B) Kernel density plots of correlation of mRNA expression with selected physiological traits (ejection fraction, myocardial infarction trace, interventricular septal, and left ventricular internal diameter) within each cluster. (C) Clustering based on correlation of novel long non-coding RNA expression with echocardiography-derived physiological traits. Clusters are illustrated right of the heatmap with top-enriched terms for coding genes closest to novel long non-coding RNA. (D) Kernel density plots of correlation of long non-coding RNA expression with selected physiological traits (ejection fraction, myocardial infarction trace, interventricular septal, and left ventricular internal diameter) within each cluster. Heart specificity of UCSC mRNAs (E) and novel long non-coding RNAs (F) in clusters one to four.
These data demonstrated that unsupervised clustering of transcripts was able to distinguish physiological traits. In addition, it indicated that lncRNAs could represent specific markers of particular physiological traits. To test this, we compared correlation distributions for each UCSC-coding gene and novel lncRNA cluster, with each of the following traits; EF, interventricular septal thickness at systole (IVS), MI trace, and left ventricular internal diameter at systole (LVID) (Figure 3B and D). UCSC-coding gene Clusters 2 and 4 strongly correlated with all these traits when compared with non-specific clusters (Clusters 1 and 3) (Figure 3B). A similar pattern of correlation was observed with novel lncRNA Clusters 2 and 4 (Figure 3D). Interestingly, novel lncRNA Cluster 1 exhibited poor correlation with LVID, EF, and MI trace but correlated well with IVS, even though this trait is typically associated with EF in our model (Supplementary material online, Figure S4B). This unique characteristic is likely a consequence of the exquisite context and cell-type specific expression of lncRNAs, and has intriguing implications for the utilization of novel lncRNAs as biomarkers.
Novel long non-coding RNAs are associated with specific chromatin states in the adult heart
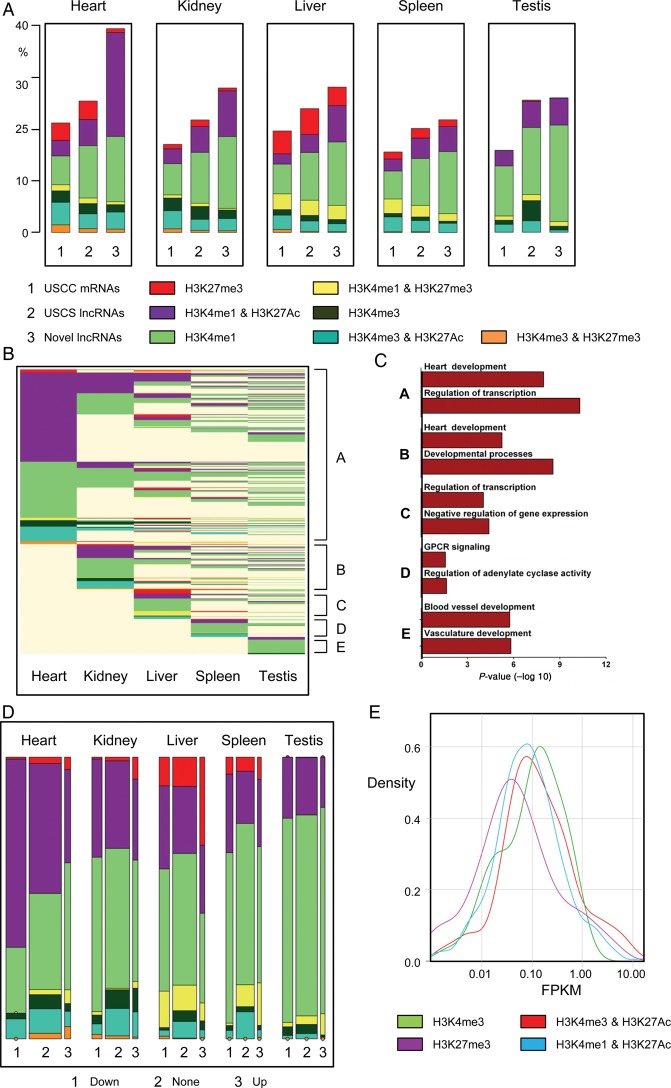
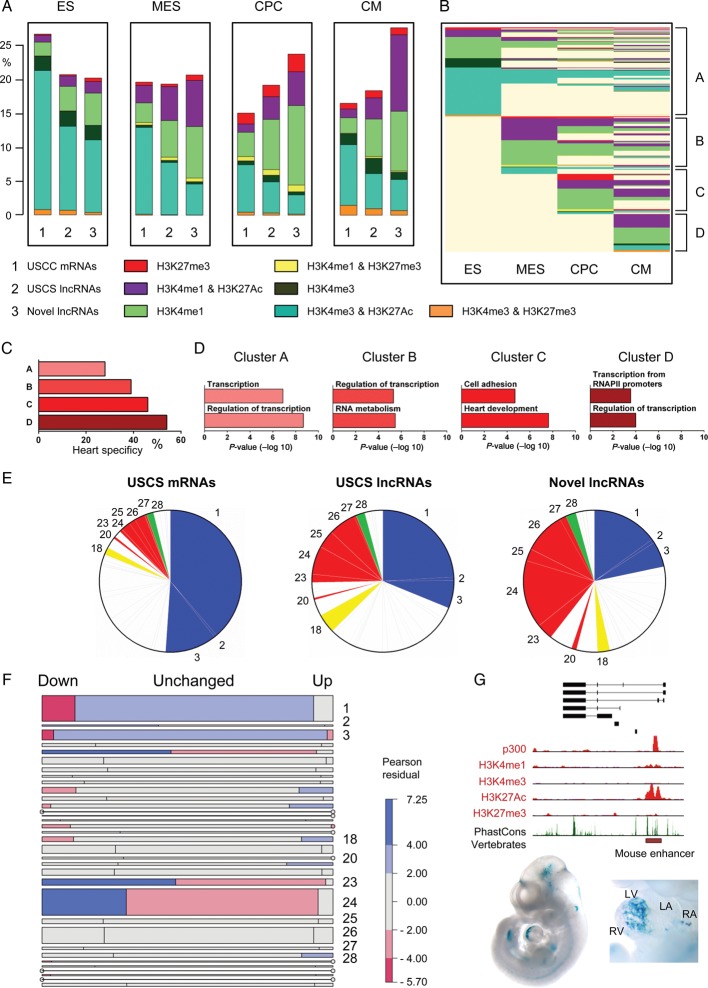
Distinct chromatin states can be used to annotate genomic sequence, and can demarcate regulatory and transcribed genomic sequences.10 We investigated chromatin states at the novel lncRNAs and compared with those associated with UCSC-coding genes and lncRNAs utilizing publically available ChIP-Seq catalogues.24 We produced frequency plots for chromatin states around the transcriptional start sites (TSS) of novel lncRNAs and UCSC transcripts in five adult mouse tissues (heart, kidney, liver, spleen, and testis). Modifications included H3K27me3 and H3K4me3 (associated with inactive and active promoters, respectively) and H3K4me1 and H3K27Ac (associated with poised and active promoters and enhancers). Additionally, combinations of these modifications are associated with specific functions, for example, H3K4me1 in combination with H3K27Ac is associated with active enhancers while H3K27me3 and H3K4me3 together are associated with ‘bivalent’ domains which are implicated in developmental processes.26–30 We calculated the frequencies of each chromatin state in promoters of all transcripts specifically in the adult mouse heart. Novel lncRNAs were more frequently associated with active enhancer states (H3K4me1 and H3K27Ac) when compared with UCSC-coding and lncRNA genes (Figure 4). Considering the increased heart specificity of novel lncRNAs, it is interesting to observe that they are significantly enriched with active enhancer sequences, the most tissue-specific functional elements within the genome. In agreement with this observation, it has recently been shown that enhancer-associated lncRNAs are typically more cell/tissue-specific than promoter associated lncRNAs.31 Conversely, novel lncRNAs were significantly less associated with the polycomb repressed mark (H3K27me3) or bivalent domain (H3K4me3 and H3K27me3). There were no readily discernible differences in frequency of modifications associated with active canonical promoters (H3K4me3) or poised promoters and enhancers (H3K4me1). To explore whether these differences in chromatin states were tissue specific, we extended our analysis to non-cardiac tissues. The enrichment of novel lncRNAs with active enhancer-specific marks was less pronounced in non-cardiac tissues, suggesting that a large fraction of the active enhancer states were cardiac specific (Figure 4A). Novel lncRNAs were sorted based on their chromatin states across individual tissues (Clusters A to E) (Figure 4B). Most novel lncRNAs were associated with a distinct chromatin states in the heart, compared with all other tested tissues. Analysis of novel lncRNAs with active states only in one particular tissue demonstrated that novel lncRNAs with heart-specific chromatin states were both more cardiac-specific in their expression (Supplementary material online, Figure S5) and more likely to be associated with proximal coding genes involved in heart developmental processes (Figure 4C).
Figure 4.
Novel long non-coding RNAs are associated with specific chromatin states in the adult heart. Novel long non-coding RNAs are predominantly associated with adult heart-specific active enhancers (A) Bar charts of frequencies of UCSC mRNAs (1), UCSC long non-coding RNAs (2) and novel long non-coding RNAs (3) marked by H3K27me3, H3K4me3, H3K4me1, H3K4me3/H3K27Ac, H3K4me1/H3K27me3, H3K4me3/H3K27Ac, or H3K4me3/H3K27me3 in adult mouse heart, kidney, liver, spleen, and testis. (B) Epigenetic state map of all novel long non-coding RNAs marked by one of the above markers in at least one adult tissue including heart, kidney, liver, spleen, and testis. Rows are recursively clustered by marks (clusters A–E) in heart, kidney, liver, spleen, and testis. (C) Gene ontology terms significantly enriched in clusters A to E. (D) Mosaic plots showing the frequencies of transcripts, by differential expression category (vertical blocks) and by chromatin states (coloured cells within vertical blocks). Area of each coloured cell is proportional to frequency across all tissues; width of vertical bars is proportional to frequency of differential expression category; height of cell is proportional to frequency of chromatin state within expression category. (E) Enhancer-associated novel long non-coding RNAs (H3K4me1/H3K27Ac) tend to be more lowly expressed when compared with novel long non-coding RNAs with canonical promoter signatures (H3K4me3).
We further analysed chromatin state frequencies at the promoters of differentially expressed transcripts, either up or down-regulated post-infarction. Coding genes and UCSC lncRNAs exhibited no obvious enrichments in chromatin states associated with the up- or down-regulated transcripts in the five tissues assessed (data not shown). In contrast, novel lncRNAs exhibited a heart-specific enrichment of active enhancer states associated with transcripts that were down-regulated post infarction (Figure 4D). This is likely to be of functional importance with respect to the global reprogramming of gene expression observed post-infarction, much of which is under the control of differentially activated and repressed cell and tissue specific enhancers.
Finally, methylation and acetylation of histone lysine residues are known to be critical determinants of transcriptional activity.10,29 For coding genes, H3K4me3 enrichment at the TSS correlates with active transcription while H3K27me3 is associated with a repressed transcriptional state. To determine whether novel lncRNAs exhibited a similar correlation between chromatin state and transcription, we integrated ChIP and RNA-Seq data sets from the adult heart. Novel lncRNAs with H3K4me3 enrichment exhibited higher expression than those marked by the polycomb repressing mark, H3K27me3 (Figure 4E). Interestingly, novel lncRNAs associated with an exclusively active enhancer signature were less abundant than those with a canonical active promoter mark, suggesting that novel lncRNAs derived from active cardiac enhancers were less expressed compared with those with a canonical promoter signature.
Inferring functions for novel long non-coding RNAs based on developmental chromatin state patterns
Pathological cardiac remodelling is associated with the global reactivation of the foetal gene programme. We reasoned that many novel lncRNAs likely represent ‘foetal’ genes with important roles during cardiogenesis. To investigate this, we utilized ChIP-Seq data generated in a directed differentiation system that recapitulated the step-wise differentiation of mouse ES cells to differentiated CMs.32 Reminiscent of our findings in the adult heart vs. other tissues, novel lncRNAs were enriched with active enhancer states in cardiac precursor cells (CPCs) and differentiated CM when compared with UCSC-annotated transcripts (Figure 5A). Considering that novel lncRNAs were discovered in the adult heart, it makes it more likely for them to be associated with enhancers that are active in more differentiated cell lineages (i.e. CPCs and CMs) during cardiogenic differentiation of embryonic stem cells in vitro.
Figure 5.
Inferring functions for novel long non-coding RNAs based on developmental chromatin state patterns. Novel long non-coding RNAs are more typically associated with chromatin state patterns linked to cardiac development and function. (A) Bar charts of frequencies of UCSC mRNAs (1), UCSC long non-coding RNAs (2) and novel long non-coding RNAs (3) marked by H3K27me3, H3K4me3, H3K4me1, H3K4me3/H3K27Ac, H3K4me1/H3K27me3, H3K4me3/H3K27Ac, and H3K4me3/H3K27me3 in embryonic stem cells, mesodermal precursors, cardiac precursor cells, and cardiomyocytes. (B) Epigenetic state map of all novel long non-coding RNA marked by one of the above markers in at least one lineage. Rows are recursively clustered by marks in embryonic stem, mesodermal precursors, cardiac precursor cell and cardiomyocyte (clusters A–D). (C) Heart specificity of chromatin-based clusters A–D. (D) Gene ontology terms significantly enriched in clusters A–D. (E) Pie charts illustrating associations of UCSC mRNAs, UCSC long non-coding RNAs and novel long non-coding RNAs with pre-determined chromatin state patterns according to Wamstad et al.32. (F) Mosaic plot showing frequencies of transcripts by chromatin state cluster (horizontal blocks) and by differential expression status (cells within horizontal blocks). Area of each coloured cell is proportional to frequency across all lineages. Height of horizontal bar is proportional to frequency of cluster; width of cell is proportional to frequency of differential expression status within cluster. Cells are shaded according to the Pearson residual, providing a measure of enrichment within each cluster. Red shading indicates that the observed frequency is lower than expected while blue indicates greater than expected frequency. (G) The novlnc6 genomic locus is associated with a bonafide developmental enhancer (red box). Lower panel is a Lac-Z stained embryo and isolated heart with in vivo enhancer activity at E11.5. Stained embryo images were obtained from http://enhancer.lbl.gov/.
Clustering novel lncRNAs according to their chromatin state at each stage of cardiac differentiation showed that novel lncRNAs exhibited stage-specific chromatin state transitions (Figure 5B). This suggested that lncRNAs likely had key roles in the developmental transitions at which the chromatin state transitions occurred. For example, novel lncRNAs with exclusively active enhancer states in mesodermal precursors (MES) represent ideal candidates for molecules involved in cardiac mesoderm specification, whereas novel lncRNAs that gain active marks during the CPCs to CM transition are likely involved in CM differentiation. Supporting this, we found that novel lncRNAs with active chromatin states at the CP to CM stages were more heart-specific and associated with coding genes implicated in heart development (Figure 5C and D).
A previous study demonstrated that co-expressed genes during cardiac differentiation could be functionally grouped based on different chromatin state patterns.32 Each subgroup of genes appeared to be involved in distinct biological processes, including signalling, metabolism, and cardiac muscle contraction. We reasoned that novel lncRNAs that shared specific chromatin patterns as those described for coding genes were likely to be involved in comparable biological processes, thus providing a novel powerful unbiased chromatin-based proxy to functionally annotate novel lncRNAs. We mapped the novel lncRNAs and UCSC annotated genes to pre-determined cumulative patterns of chromatin transitions during ES cell differentiation (ChIP clusters 1–34). We classified the novel lncRNAs based on which chromatin pattern they were associated with, and inferred a biological function based on the coding genes and biological processes previously linked to each cluster (Supplementary material online, Table S4).32 For clarity, we present 11 ChIP clusters and inferred biological processes associated with each of our transcript classes. ChIP clusters 1–3 are associated with ubiquitous housekeeping and non-cardiac developmental processes. UCSC-coding genes and lncRNAs were enriched within these clusters while novel lncRNAs were depleted (Figure 5E and Supplementary material online, Figure S6A). On the other hand, novel lncRNAs were enriched in ChIP-cluster 20 and clusters 23–27, which are associated with cardiac developmental and functional processes including muscle contraction. Furthermore, novel lncRNAs associated with these inferred functions were significantly more heart-specific, validating our functional inference approach (Supplementary material online, Figure S6B).
We also identified ChIP-clusters that were enriched or depleted in up- and down-regulated novel lncRNAs post-MI, providing a functional insight into the roles of novel lncRNAs in this response (Figure 5F). Novel lncRNAs in ChIP-clusters 23 and 24 were enriched in down-regulated lncRNAs post-infarction. These clusters are associated with CM maturation and sarcomeric genes (e.g. Myoz2 and Myl2). The enrichment of ChIP-cluster 23 and 24 novel lncRNAs in down-regulated lncRNAs could be indicative of the re-activation of the foetal gene programme in the BZ post-infarction and/or a loss of mature CMs. Furthermore, ChIP-clusters enriched in up-regulated novel lncRNAs included cluster 18, which is associated with immune and inflammatory responses (e.g. IL17b), and cluster 28, which is associated with calcium homoeostasis and G-protein-coupled receptor signalling (e.g. Gnb3), processes that are typically activated in the BZ of the infarcted heart. Giving further support to the notion that our novel lncRNAs may be cardiac developmental associated transcripts, we mapped them to a list of bonafide in vivo validated enhancers active specifically within the E11.5 mouse heart.33 We found that seven of our novel lncRNAs map to validated cardiac enhancers (Supplementary material online, Table S3) including novlnc6 (see below), which maps to mm74, a cardiac enhancer specifically active within the embryonic left ventricle (Figure 5G).
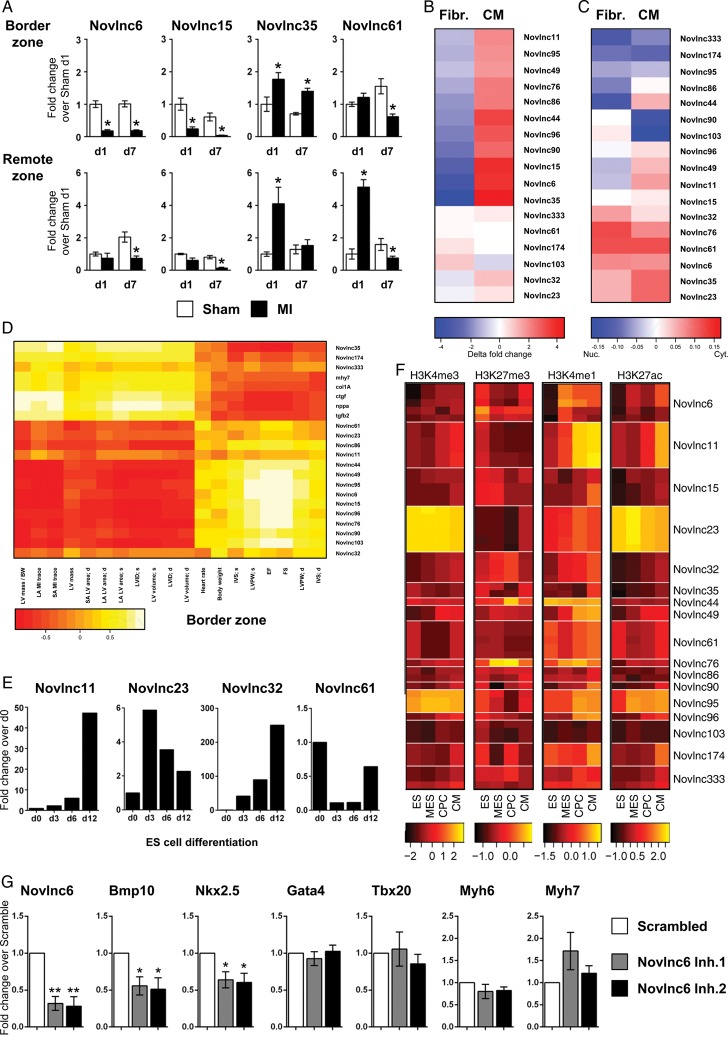
Validation of novel long non-coding RNAs
To gain confidence in our transcript nominations, we validated multiple unannotated novel transcripts by qRT–PCR. Seventeen high priority novel candidates were selected based on their association with specific parameters (Supplementary material online, Table S5), and their expression was quantified in the BZ and RZ, 1 and 7 days after infarction. Cardiac function and morphology was stereotypically perturbed with markers of cardiac stress differentially expressed as expected (Supplementary material online, Figure S7A and B). The novel lncRNAs exhibited various kinetics of expression in both the BZ and RZ during the acute and chronic phases (Figure 6A and Supplementary material online, Figure S7C). Many lncRNAs were down-regulated, e.g. Novlnc6 and 15, some were transiently induced at Day 1 in both BZ and RZ (Novlnc35), while others were gradually increased in BZ and RZ (Novlnc174). These distinct kinetic and spatial patterns of expression demonstrate that novel lncRNAs are dynamically regulated in response to MI, and suggest that they likely play important roles in the adaptive process.
Figure 6.
Selection and validation of novel long non-coding RNAs. High priority candidates were selected based on their association with unique and specific characteristics. See Supplementary material online, Table S5 (A) Quantitative RT–PCR analysis of selected novel long non-coding RNAs in Sham-operated (white bar) and infarcted heart (black bar), in the border and remote zones at 1 and 7 days post-infarction. (B) Heatmap representation of qRT–PCR analysis of long non-coding RNAs in cardiomyocytes and fibroblasts isolated from neonatal mouse hearts. (C) Heatmap representation of qRT–PCR analysis of long non-coding RNAs in the cytoplasmic or nuclear fractions of cardiomyocytes and fibroblasts from neonatal mouse hearts. Blue indicates nuclear enrichment and red cytoplasmic enrichment. (D) Heatmap representation of correlation between validated novel long non-coding RNA expression in the border zone and echocardiography-derived physiological traits after infarction. (E) Expression of selected novel long non-coding RNA in embryonic stem, mesodermal precursors, cardiac precursor cell and cardiomyocyte as measured by qRT–PCR. (F) Heatmaps of ChIP enrichment values of different chromatin modification at TSS region of validated novel long non-coding RNA in embryonic stem, mesodermal precursors, cardiac precursor cell, and cardiomyocyte. Colour scale is based on log10 of the ChIP signal. (G) Mouse neonatal cardiomyocytes were transfected with GapmeRs targeting a novlnc6 or random scrambled sequence. Cells were harvested 48 h post-transfection and assayed for Novlnc6, Bmp10, Nkx2–5, GATA4, Tbx20, Myh6, and Myh7 expression by qRT–PCR. Bars represent means ± SEM (n = 6 independent experiments). **P < 0.001; *P < 0.05.
The two major cell types within the adult heart are CMs and cardiac fibroblasts (FBs) with both being important in maladaptive remodelling. To better characterize the novel lncRNAs, we quantified their expression in CMs and FBs isolated from the neonatal mouse heart. The selected lncRNAs were either highly CM-specific (Novlnc35), equally expressed in both cell types (Novlnc61) or primarily expressed in FBs (Novlnc103) (Figure 6B). LncRNA function is also dependant on subcellular localization. Enhancer-associated lncRNAs tend to be more enriched in the nucleus, whereas lncRNAs involved in post-transcriptional and translational processes tend to be more cytoplasmic. Nuclear and cytoplasmic RNA fractions were isolated from neonatal CMs and FBs (Figure 6C and Supplementary material online, Figure S7D). Validated lncRNAs were either enriched in nuclear (Novlnc174) or cytoplasmic (Novlnc61) fractions, in addition to being equally present in both (Novlnc15). Some lncRNAs interestingly displayed differential nuclear vs. cytoplasmic enrichment in CMs and FBs (Novlnc90, −103, −49, −11). This may be of functional relevance to roles in these different cell types. We also correlated the expression of these validated lncRNAs with physiological traits in Day 1 and Day 7 control and MI tissue samples (Figure 6D and Supplementary material online, Figure S7E). The majority of the lncRNAs correlated well with physiological traits both in the BZ and RZ. Interestingly, some of our novel lncRNAs were better correlated than canonical stress genes with cardiac function and remodelling (Supplementary material online, Figure S7F).
Many of the validated novel lncRNAs exhibited changes in chromatin states during cardiogenesis in ES cells, suggesting that they may have roles as ‘foetal’ developmental genes (Figure 6F). To confirm this, mouse ES cells were differentiated through embryoid body (EB) formation using the hanging drop model recapitulating embryonic cardiac development in vitro. Novel lncRNAs were modulated during cardiac differentiation with expression correlating with the dynamic changes in chromatin states observed at their promoters (Figure 6E and F). Some lncRNAs were induced late during differentiation at the CM stage (Novlnc44, −41, −11, −32, Figure 6E and F; Supplementary material online, Figure S7G), and are likely involved in terminal CM differentiation and maturation. Novel lncRNAs exhibiting this profile were mapped to ChIP-clusters 24 and 25, which are predicted to be associated with heart development, and particularly z-disk and sarcomere function, which corresponds to terminal cardiac maturation. Other lncRNAs were maximally expressed at MES and CPC stages (Novlnc49, −333) and are likely to be involved in more transient developmental processes.
To evaluate whether novel lncRNAs could be associated with specific functions involving regulation of cardiac protein coding genes, we focused on Novlnc6, which fulfils several criteria and unique features prototypical of lncRNAs including low-coding potential (Gene ID score: −0.42; median for UCSC mRNA, UCSC lncRNAs, and Novel lncRNAs is 22.64, −1.01 and −0.76, respectively). Furthermore Novlnc6 shares a chromatin pattern in differentiating ES cells with the key cardiac signalling ligand BMP10 (ChIP-cluster 25) (Supplementary material online, Table S5, suggesting that Novlnc6 could be involved in similar regulatory pathways. As an experimental model, we used primary isolated neonatal mouse CMs expressing high levels of Novlnc6. Cells were transfected with modified anti-sense oligonucleotides (GapmeRs) targeting Novlnc6 (Figure 6G). Key cardiac TFs and downstream cardiac target genes involved in stress signalling, contractile apparatus and BMP10 signalling were examined. This screen identified Nkx2.5 and BMP10 as potential targets of Novlnc6-mediated regulation. Nkx2.5 encodes a core cardiac TF, high in the regulatory hierarchy of the cardiac GRN and critical for the regulation of other cardiac TFs and downstream cardiac differentiation, structural, and maturation genes.34 Furthermore Nkx2–5 has been shown to be downstream of BMP10 signalling during cardiac development.35 Our data support therefore the notion that novel lncRNAs includes functionally important regulatory transcripts.
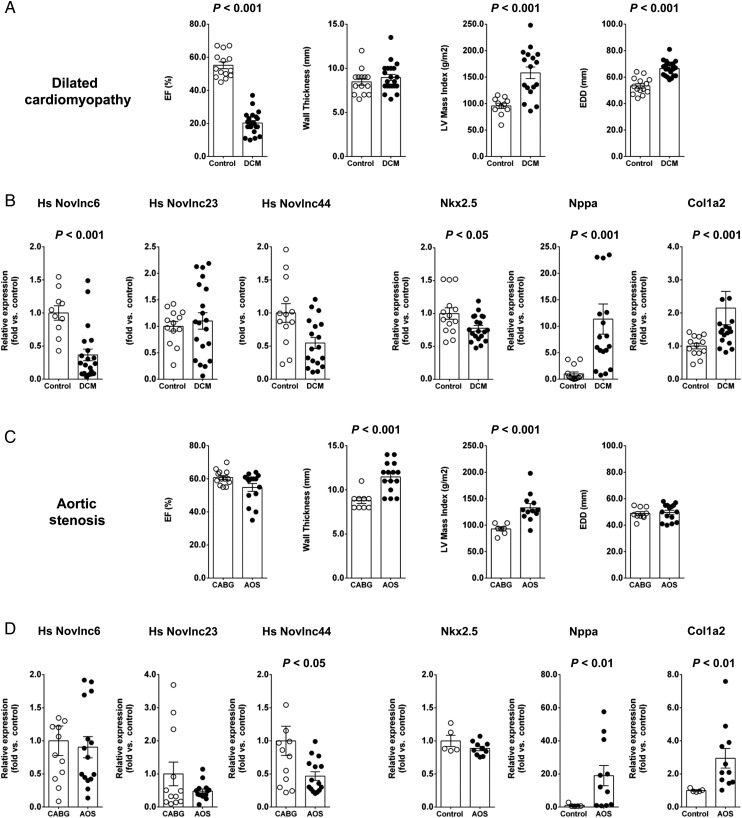
Dysregulation of human orthologues in cardiac pathology
Considering the unique characteristics associated with the novel lncRNAs, we searched for human orthologues. We mapped our novel lncRNA catalogue to the human genome using TransMap, a cross species mRNA alignment tool. TransMap maps our novel mouse lncRNAs sequences across the human genome using syntenic BLASTZ alignments that consider conserved gene order and synteny. Of the 1521 novel lncRNAs, ∼73% were mapped to the human genome (Supplementary material online, Table S6). To validate and characterize predicted orthologues, we designed primers encompassed within the putative exons of three human orthologues, corresponding to mouse Novlnc6, −23 and −44 (Supplementary material online, Figure S8B–D). Quantitative RT–PCR was executed on RNA isolated from the left ventricle of a healthy male. All three putative human orthologues were readily amplified and expressed at relatively high levels (Supplementary material online, Figure S8A). To determine the potential roles of these orthologues in cardiac pathology, we examined their cardiac expression in two independent human heart pathologies. Patients with idiopathic dilated cardiomyopathy (DCM) and with aortic stenosis (AOS) were assessed. These two patient groups presented with perturbed cardiac functions and associated maladaptive remodelling as expected for such pathologies (Figure 7A and C and Supplementary material online, Table S7). Furthermore, cardiac stress marker genes were also differentially expressed (Figure 7B and D). In patients with DCM, novlnc6 was significantly modulated (Figure 7B). Interestingly, the predicted target gene of Novlnc6, i.e. the key cardiac TF Nkx2-5, was also significantly down-regulated in patients with DCM. In contrast to DCM, patients with AOS were not associated with differential expression of Novlnc6, or the predicted target gene of Novlnc6, Nkx2-5. Novlnc44, however, was significantly down-regulated (Figure 7D). These data suggested that some candidate novel lncRNA human orthologues were expressed at significant levels in the human heart, and furthermore, are differentially modulated in human cardiac pathologies.
Figure 7.
Dysregulation of human long non-coding RNA orthologues in cardiac pathology. Human novel long non-coding RNA orthologues were detected and differentially modulated in pathological cardiac states. (A) Physiological parameters of the dilated cardiomyopathy patient cohorts. (B) Expression of Novlnc6, Novlnc23, Novlnc44, Nkx2.5, Nppa, and Col1a2 in heart samples from dilated cardiomyopathy patients. (C) Physiological parameters of the aortic stenosis patient cohorts. (D) Expression of Novlnc6, Novlnc23, Novlnc44, Nkx2.5, Nppa, and Col1a2 in heart samples from aortic stenosis patients.
Discussion
Long non-coding RNAs are emerging as key regulatory components of GRNs. However, little is known about the roles of these molecules in disease-relevant organs such as the heart especially during the adaptive response to stress. To address this we have systematically identified and characterized the mouse long non-coding transcriptome after MI. Hundreds of novel heart-specific lncRNAs with unique functional and regulatory characteristics have been identified for the first time. We integrated genome-wide approaches to associate novel lncRNAs with cardiac cell types, developmental processes, physiological traits, and human disease states. By integrating global chromatin state maps, we identified novel lncRNAs with potential roles in both cardiac development and pathological cardiac remodelling. Using chromatin-based inference, we generated biologically meaningful functional annotations for the novel lncRNAs. We also identified human orthologues and demonstrated that they are dysregulated in human pathological states. These studies open up new avenues to study and functionally characterize the roles of novel heart-specific lncRNAs in pathological cardiac remodelling, physiological function, and ultimately therapeutic reprogramming in the context of regenerative therapies.
Novel long non-coding RNAs as potential heart specific modulators of cardiac gene programmes
The newly identified lncRNAs exhibit a striking tissue-specific pattern of expression. This is in agreement with recent analysis of lncRNAs in other tissues such as pancreatic beta cells and neuronal cells.17–19 We, therefore, propose that novel lncRNAs described here are major heart-specific sensors and/or effectors of the cardiac stress response post-MI. We present a number of pieces of evidence to support this conclusion. First, coding genes that are proximal to novel lncRNAs are associated with heart developmental processes and global transcriptional control. This directly implicates novel lncRNAs in the transcriptional reprogramming that underlies the reactivation of the foetal gene programme in the damaged heart. In agreement with this, novel lncRNAs correlate positively in expression with physiological traits associated with cardiac function and negatively with traits associated with remodelling. At the chromatin level, novel lncRNAs that are down-regulated post-infarction are highly associated with active heart-specific enhancer states. Considering that lncRNAs have emerged as key functional components required for enhancer activity, it suggests that novel lncRNAs underpin the global heart-specific transcriptional reprogramming post-MI. Furthermore, chromatin state transition-derived functional inferences demonstrate that large fractions of the novel lncRNAs are associated with ChIP-clusters linked to very specific cardiac structural and functional processes. Additionally, modulated lncRNAs are overrepresented in ChIP clusters associated with biological processes implicated in the pathological stress response. For example, up-regulated lncRNAs are enriched in inflammatory and calcium signalling-associated ChIP clusters while down-regulated lncRNAs are enriched in ChIP clusters linked to negative regulation of growth and to force generating contractile proteins. One such prototypical novel lncRNA, novlnc6, is associated with a bonafide cardiac developmental enhancer and appears to modulate the expression of Nkx2.5 , a master cardiac TF, critical for the modulation of gene programmes involved in cardiogenic differentiation, maturation, homoeostasis.34 Interestingly, BMP10, a key signalling ligand for cardiogenesis, which maintains Nkx2.5 expression during development,35 shares chromatin transition patterns with Novlnc6 during ES cell cardiac differentiation. BMP10 is significantly down-regulated upon Novlnc6 knockdown, suggesting that Nxk2.5 modulation occurs secondary to BMP10 change in expression or can be regulated independently by novlnc6. Finally, novel lncRNAs were also significantly associated with cardiac miRNA networks that have previously been shown to be post-transcriptional modulators of the cardiac stress response, namely miR-133a, −499, and 30c. This suggests that cardiac lncRNAs could modulate gene programmes at multiple regulatory levels, including post transcriptionally via interaction with miRNAs.
Novel long non-coding RNAs as potential biomarkers of cardiac form and function
RNA-seq-based expression profiling allowed us to correlate the cardiac transcriptome with continuous numerical traits of cardiac dimensions and function. Analysis of clustered coding genes revealed that genes associated with mitochondrial biology positively correlated with cardiac function and negatively with remodelling. Free fatty acid oxidation is decreased during the transition to HF due to repressed activity of mitochondrial fatty acid oxidation enzymes.36,37 Our data support this metabolic shift in the mitochondria. Conversely, coding genes that correlated positively with remodelling were associated with wound healing, extracellular matrix production, and fibrosis, processes that are activated during the physio-pathological response to stress.2 More importantly, Novel lncRNAs correlated significantly better than annotated lncRNAs with all physiological traits. This superior correlation with physiological traits exhibited by novel lncRNAs is likely as a consequence of their unique functional and regulatory characteristics, including increased heart specificity, increased association with cardiac-specific enhancer states and enrichment within functional clusters linked to cardiac function. Clustering of novel lncRNAs identified specific clusters that, like coding genes, correlated either positively, or negatively with functional and remodelling-associated traits. This allows us to functionally annotate our novel lncRNAs based on their correlations with physiological traits in the heart post-injury. Novel lncRNAs that correlate positively with remodelling traits are likely, akin to coding genes, to be involved in fibrotic, remodelling, and hypertrophic associated processes. Supporting this, we show that novel lncRNAs that correlate best with cardiac function are in fact the most heart-specific cluster and associated with proximal coding genes implicated in heart development.
Although novel lncRNAs and coding genes are generally comparable in their correlations with physiological traits, we identified one major distinguishing feature that dissociated their clustering with individual traits. Novel lncRNAs contain a single cluster that correlated specifically with IVS and not the linked EF. This specificity suggests the existence of unique functional and regulatory attributes associated with novel lncRNAs and therefore they could represent powerful biomarkers of such traits. This would be especially relevant in the context of cardiac pathologies where cardiac morphology is affected independently of cardiac function, such as heart failure with preserved ejection fraction (HFpEF).38 Patients with HFpEF are difficult to diagnose; however, this pathology is characterized with remodelling of the IVS. Novel lncRNAs in Cluster 1 could therefore be ideal biomarkers for this pathology as they are specifically associated with IVS independently of the linked functional trait, EF. Further emphasizing the sensitivity of novel lncRNAs, we demonstrate that individual novel lncRNAs correlated better with physiological traits than genes encoding prototypical cardiac stress markers. Furthermore, a recent study has demonstrated that human cardiac lncRNA expression signatures, and not mRNA or miRNAs, were able to distinguish left ventricular tissue samples from patients suffering with ischaemic and non-ischaemic cardiomyopathies—pre- and—post-left ventricular assist device (LVAD) unloading.39 These data complement our data demonstrating that cardiac lncRNAs represent exquisite sensors of cardiac form, function, and ultimately pathology. Recently, specific lncRNAs in the blood have been demonstrated to be predictive of various cancer states.40 It will be important to determine whether the novel lncRNAs identified here can also be detectable in the plasma, and whether their expression correlates with cardiac pathological states.
Cardiac novel long non-coding RNAs are associated with specific chromatin state transitions
A powerful approach for the functional annotation of novel lncRNAs is to characterize their underlying chromatin states in relevant biological situations. Chromatin state maps were previously used for the identification of the intergenic class of long non-coding transcripts (lincRNAs),11 which were characterized based on the presence of an active canonical promoter signature at their TSS (H3K4me3) and which regulate genes primarily in -trans. Other classes of lncRNAs are derived from active enhancers, as recognized by the presence of H3K4me1 and H3K27ac marks and primarily regulate genes in -cis.41–43 Globally, novel lncRNAs identified here are enriched with chromatin states marking active enhancers while relatively few are associated with canonical promoter states typically found at intergenic lincRNAs.11 A number of recent studies have implicated ncRNAs as key functional components required for enhancer activity and chromatin looping.41 We suspect that our novel lncRNAs are likely involved in global heart-specific enhancer dependant cis-regulation and thus modulators of the transcriptional reprogramming required for the reactivation of the foetal gene programme. This is supported by the observation that proximal coding genes to novel lncRNAs with active states exclusive to the adult heart are associated with heart development (foetal gene programme) and transcriptional control (global transcriptional programming). Moreover, enhancer-associated novel lncRNA expression correlates with proximal coding gene expression post-MI. Interestingly, novel lncRNAs associated with active enhancer states are significantly less expressed vs. novel lncRNAs associated with canonical active promoters. It has previously been shown that enhancer derived transcripts are typically less stable than other lncRNAs, which may contribute to this difference in expression.44 Additionally, this may also reflect cis-regulatory action of enhancer-associated lncRNAs at their genomic site of transcription, which potentially requires lower copy number to elicit their regulatory effects31 Furthermore, the higher association with active enhancer states likely explains the increased heart specificity of novel lncRNAs when compared with UCSC lncRNAs. Indeed, Novel lncRNAs derived from tissue-specific enhancers are typically more tissue-specific in their expression, correlating with the unique tissue specificity of the enhancers that generate them.31
We established a novel approach for lncRNA functional annotation that utilized patterns of chromatin state transitions during the differentiation of ES to CMs allowing us to assign novel lncRNAs to a series of pre-determined chromatin pattern clusters corresponding to distinct sets of functionally related genes32. We found that novel lncRNAs were associated with chromatin clusters linked to very specific cardiac processes, including heart developmental and contractile genes, while mRNAs and UCSC lncRNAs were more associated with non-cardiac specific housekeeping processes. This demonstrates that novel lncRNAs are enriched in clusters with inferred cardiac functions during cardiac differentiation. This also provides a novel powerful paradigm for the functional annotation of distinct classes of lncRNAs that can be adopted in other appropriate contexts. In addition, lncRNAs with unique specific chromatin states involved in developmental specification and differentiation represent interesting regulatory molecules as potential inductive and/or reprogramming factors. For instance, a role in reprogramming has been reported in recent work showing that a single lncRNA can promote induction of pluripotent cells from somatic cells.45 Long non-coding RNAs are emerging as global modulators of the epigenome via their ability to interact with and genomically target ubiquitously expressed chromatin readers, writers, and erasers.16 The stress pathways activated post-MI are associated with dynamic remodelling of chromatin states,46 leading to net regulatory output control of multiple, simultaneously activated pathological networks. Novel lncRNAs described here are likely integral to context and cardiac-specific global modulation of these states and targeting them could provide a powerful therapeutic approach to abrogate pathological gene expression and HF progression.
Novel long non-coding RNAs and human cardiac pathology
The identification of novel human lncRNAs will provide a new framework to study human cardiac patho-physiology. Our study has revealed hundreds of human orthologues of novel lncRNAs, of which a few candidates were shown to be modulated in two human cardiac disease states, namely dilated cardiomyopathy and aortic stenosis. One particular novel lncRNA, novlnc6, was significantly down-regulated in dilated cardiomyopathy. Importantly, Novlnc6 knockdown in CMs results in a concomitant down-regulation of BMP10 and Nkx2.5, two important regulators of CM identity. Novel human lncRNAs identified thus represent candidate regulators of protein coding genes implicated in human cardiac pathology. More, generally, these results set the stage for more focused studies to dissect the potential roles of inherited and acquired defects in novel lncRNA genes in human cardiac disease. Furthermore, considering that novel lncRNAs were identified in a MI model, it would be of interest to characterize the expression of human orthologues in patients following infarction. Moreover, it is likely that novel lncRNAs might be useful at identifying subgroups within affected individuals. In this context, future studies should investigate the value of novel lncRNAs as phenotypic biomarkers. Characterization of these novel lncRNAs could therefore provide unprecedented opportunities for understanding disease progression, and for diagnosis and intervention.
Supplementary material
Supplementary material is available at European Heart Journal online.
Funding
This work is supported by a grant from the Swiss National Science Foundation (grant no 406340-128129) within the frame of the National Research Programme 63 on ‘Stem cells and Regenerative Medicine’. Funding to pay the Open Access publication charges for this article was provided by the University of Lausanne Medical School.
Conflict of interest: none declared.
Acknowledgements
We are grateful to Keith Harshman and Genomic Technologies Facility, University of Lausanne, Switzerland facility for support and sequencing. We thank Benoit Bruneau, Gladstone Institute, San Francisco, CA for providing access to custom ChIP-Seq tracks. In the process of this analysis, we took advantage of CvDC data produced part as the Bench-to-Bassinet Programme. We thank Len A. Pennacchio, Lawrence Berkeley National Laboratory, for access to the mouse transgenic enhancer database (http://enhancer.lbl.gov/).
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB American Heart Association Statistics C, Stroke Statistics S. Heart disease and stroke statistics--2012 update: a report from the American Heart Association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 3.van Berlo JH, Maillet M, Molkentin JD. Signaling effectors underlying pathologic growth and remodeling of the heart. J Clin Invest. 2013;123:37–45. doi: 10.1172/JCI62839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 6.Ounzain S, Crippa S, Pedrazzini T. Small and long non-coding RNAs in cardiac homeostasis and regeneration. Biochim Biophys Acta. 2013;1833:923–933. doi: 10.1016/j.bbamcr.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumarswamy R, Thum T. Non-coding RNAs in cardiac remodeling and heart failure. Circ Res. 2013;113:676–689. doi: 10.1161/CIRCRESAHA.113.300226. [DOI] [PubMed] [Google Scholar]
- 9.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y Consortium F, Group RGER, Genome Science G. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 10.Consortium EP, Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Dunham I, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Khatun J, Kheradpour P, Kundaje A, Lassmann T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LA, Adams LB, Kelly CJ, Zhang J, Wexler JR, Green ED, Good PJ, Feingold EA, Bernstein BE, Birney E, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Gerstein M, Giddings MC, Gingeras TR, Green ED, Guigo R, Hardison RC, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Snyder M, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Khatun J, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Giddings MC, Bernstein BE, Epstein CB, Shoresh N, Ernst J, Kheradpour P, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Ward LD, Altshuler RC, Eaton ML, Kellis M, Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena HP, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Risk BA, Robyr D, Ruan X, Sammeth M, Sandu KS, Schaeffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Hayashizaki Y, Harrow J, Gerstein M, Hubbard TJ, Reymond A, Antonarakis SE, Hannon GJ, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR, Rosenbloom KR, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kent WJ, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Furey TS, Song L, Grasfeder LL, Giresi PG, Lee BK, Battenhouse A, Sheffield NC, Simon JM, Showers KA, Safi A, London D, Bhinge AA, Shestak C, Schaner MR, Kim SK, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Birney E, Iyer VR, Lieb JD, Crawford GE, Li G, Sandhu KS, Zheng M, Wang P, Luo OJ, Shahab A, Fullwood MJ, Ruan X, Ruan Y, Myers RM, Pauli F, Williams BA, Gertz J, Marinov GK, Reddy TE, Vielmetter J, Partridge EC, Trout D, Varley KE, Gasper C, Bansal A, Pepke S, Jain P, Amrhein H, Bowling KM, Anaya M, Cross MK, King B, Muratet MA, Antoshechkin I, Newberry KM, McCue K, Nesmith AS, Fisher-Aylor KI, Pusey B, DeSalvo G, Parker SL, Balasubramanian S, Davis NS, Meadows SK, Eggleston T, Gunter C, Newberry JS, Levy SE, Absher DM, Mortazavi A, Wong WH, Wold B, Blow MJ, Visel A, Pennachio LA, Elnitski L, Margulies EH, Parker SC, Petrykowska HM, Abyzov A, Aken B, Barrell D, Barson G, Berry A, Bignell A, Boychenko V, Bussotti G, Chrast J, Davidson C, Derrien T, Despacio-Reyes G, Diekhans M, Ezkurdia I, Frankish A, Gilbert J, Gonzalez JM, Griffiths E, Harte R, Hendrix DA, Howald C, Hunt T, Jungreis I, Kay M, Khurana E, Kokocinski F, Leng J, Lin MF, Loveland J, Lu Z, Manthravadi D, Mariotti M, Mudge J, Mukherjee G, Notredame C, Pei B, Rodriguez JM, Saunders G, Sboner A, Searle S, Sisu C, Snow C, Steward C, Tanzer A, Tapanari E, Tress ML, van Baren MJ, Walters N, Washieti S, Wilming L, Zadissa A, Zhengdong Z, Brent M, Haussler D, Kellis M, Valencia A, Gerstein M, Raymond A, Guigo R, Harrow J, Hubbard TJ, Landt SG, Frietze S, Abyzov A, Addleman N, Alexander RP, Auerbach RK, Balasubramanian S, Bettinger K, Bhardwaj N, Boyle AP, Cao AR, Cayting P, Charos A, Cheng Y, Cheng C, Eastman C, Euskirchen G, Fleming JD, Grubert F, Habegger L, Hariharan M, Harmanci A, Iyenger S, Jin VX, Karczewski KJ, Kasowski M, Lacroute P, Lam H, Larnarre-Vincent N, Leng J, Lian J, Lindahl-Allen M, Min R, Miotto B, Monahan H, Moqtaderi Z, Mu XJ, O'Geen H, Ouyang Z, Patacsil D, Pei B, Raha D, Ramirez L, Reed B, Rozowsky J, Sboner A, Shi M, Sisu C, Slifer T, Witt H, Wu L, Xu X, Yan KK, Yang X, Yip KY, Zhang Z, Struhl K, Weissman SM, Gerstein M, Farnham PJ, Snyder M, Tenebaum SA, Penalva LO, Doyle F, Karmakar S, Landt SG, Bhanvadia RR, Choudhury A, Domanus M, Ma L, Moran J, Patacsil D, Slifer T, Victorsen A, Yang X, Snyder M, White KP, Auer T, Centarin L, Eichenlaub M, Gruhl F, Heerman S, Hoeckendorf B, Inoue D, Kellner T, Kirchmaier S, Mueller C, Reinhardt R, Schertel L, Schneider S, Sinn R, Wittbrodt B, Wittbrodt J, Weng Z, Whitfield TW, Wang J, Collins PJ, Aldred SF, Trinklein ND, Partridge EC, Myers RM, Dekker J, Jain G, Lajoie BR, Sanyal A, Balasundaram G, Bates DL, Byron R, Canfield TK, Diegel MJ, Dunn D, Ebersol AK, Ebersol AK, Frum T, Garg K, Gist E, Hansen RS, Boatman L, Haugen E, Humbert R, Jain G, Johnson AK, Johnson EM, Kutyavin TM, Lajoie BR, Lee K, Lotakis D, Maurano MT, Neph SJ, Neri FV, Nguyen ED, Qu H, Reynolds AP, Roach V, Rynes E, Sabo P, Sanchez ME, Sandstrom RS, Sanyal A, Shafer AO, Stergachis AB, Thomas S, Thurman RE, Vernot B, Vierstra J, Vong S, Wang H, Weaver MA, Yan Y, Zhang M, Akey JA, Bender M, Dorschner MO, Groudine M, MacCoss MJ, Navas P, Stamatoyannopoulos G, Kaul R, Dekker J, Stamatoyannopoulos JA, Dunham I, Beal K, Brazma A, Flicek P, Herrero J, Johnson N, Keefe D, Lukk M, Luscombe NM, Sobral D, Vaquerizas JM, Wilder SP, Batzoglou S, Sidow A, Hussami N, Kyriazopoulou-Panagiotopoulou S, Libbrecht MW, Schaub MA, Kundaje A, Hardison RC, Miller W, Giardine B, Harris RS, Wu W, Bickel PJ, Banfai B, Boley NP, Brown JB, Huang H, Li Q, Li JJ, Noble WS, Bilmes JA, Buske OJ, Hoffman MM, Sahu AO, Kharchenko PV, Park PJ, Baker D, Taylor J, Weng Z, Iyer S, Dong X, Greven M, Lin X, Wang J, Xi HS, Zhuang J, Gerstein M, Alexander RP, Balasubramanian S, Cheng C, Harmanci A, Lochovsky L, Min R, Mu XJ, Rozowsky J, Yan KK, Yip KY, Birney E. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, Aumayr K, Pasierbek P, Barlow DP. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 14.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakic N, Garcia-Hurtado J, Rodriguez-Segui S, Pasquali L, Sauty-Colace C, Beucher A, Scharfmann R, van Arensbergen J, Johnson PR, Berry A, Lee C, Harkins T, Gmyr V, Pattou F, Kerr-Conte J, Piemonti L, Berney T, Hanley N, Gloyn AL, Sussel L, Langman L, Brayman KL, Sander M, McCarthy MI, Ravassard P, Ferrer J. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramos AD, Diaz A, Nellore A, Delgado RN, Park KY, Gonzales-Roybal G, Oldham MC, Song JS, Lim DA. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013;12:616–628. doi: 10.1016/j.stem.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Wu SM. Isolation and functional characterization of pluripotent stem cell-derived cardiac progenitor cells. Curr Protoc Stem Cell Biol. 2010 doi: 10.1002/9780470151808.sc01f10s14. Chapter 1:Unit 1F 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouse EC, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, Giste E, Johnson A, Zhang M, Balasundaram G, Byron R, Roach V, Sabo PJ, Sandstrom R, Stehling AS, Thurman RE, Weissman SM, Cayting P, Hariharan M, Lian J, Cheng Y, Landt SG, Ma Z, Wold BJ, Dekker J, Crawford GE, Keller CA, Wu W, Morrissey C, Kumar SA, Mishra T, Jain D, Byrska-Bishop M, Blankenberg D, Lajoie BR, Jain G, Sanyal A, Chen KB, Denas O, Taylor J, Blobel GA, Weiss MJ, Pimkin M, Deng W, Marinov GK, Williams BA, Fisher-Aylor KI, Desalvo G, Kiralusha A, Trout D, Amrhein H, Mortazavi A, Edsall L, McCleary D, Kuan S, Shen Y, Yue F, Ye Z, Davis CA, Zaleski C, Jha S, Xue C, Dobin A, Lin W, Fastuca M, Wang H, Guigo R, Djebali S, Lagarde J, Ryba T, Sasaki T, Malladi VS, Cline MS, Kirkup VM, Learned K, Rosenbloom KR, Kent WJ, Feingold EA, Good PJ, Pazin M, Lowdon RF, Adams LB. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 31.Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol. 2013;14:R131. doi: 10.1186/gb-2013-14-11-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruneau BG. Transcriptional regulation of vertebrate cardiac morphogenesis. Circ Res. 2002;90:509–519. doi: 10.1161/01.res.0000013072.51957.b7. [DOI] [PubMed] [Google Scholar]
- 35.Huang J, Elicker J, Bowens N, Liu X, Cheng L, Cappola TP, Zhu X, Parmacek MS. Myocardin regulates BMP10 expression and is required for heart development. J Clin Invest. 2012;122:3678–3691. doi: 10.1172/JCI63635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bugger H, Schwarzer M, Chen D, Schrepper A, Amorim PA, Schoepe M, Nguyen TD, Mohr FW, Khalimonchuk O, Weimer BC, Doenst T. Proteomic remodelling of mitochondrial oxidative pathways in pressure overload-induced heart failure. Cardiovasc Res. 2010;85:376–384. doi: 10.1093/cvr/cvp344. [DOI] [PubMed] [Google Scholar]
- 37.Sack MN, Rader TA, Park S, Bastin J, McCune SA, Kelly DP. Fatty acid oxidation enzyme gene expression is downregulated in the failing heart. Circulation. 1996;94:2837–2842. doi: 10.1161/01.cir.94.11.2837. [DOI] [PubMed] [Google Scholar]
- 38.Borlaug BA, Paulus WJ. Heart failure with preserved ejection fraction: pathophysiology, diagnosis, and treatment. Eur Heart J. 2011;32:670–679. doi: 10.1093/eurheartj/ehq426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL, Nerbonne JM. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNA in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129:1009–1021. doi: 10.1161/CIRCULATIONAHA.113.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, Fujiwara H, Okamoto K, Otsuji E. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–3193. [PubMed] [Google Scholar]
- 41.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, de Laat W, Agami R. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 43.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, Manos PD, Datta S, Lander ES, Schlaeger TM, Daley GQ, Rinn JL. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKinsey TA, Olson EN. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J Clin Invest. 2005;115:538–546. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]