SUMMARY
Mitochondrial division is essential for mitosis and metazoan development, but a mechanistic role in cancer biology remains unknown. Here, we examine the direct effects of oncogenic RASG12V mediated cellular transformation on the mitochondrial dynamics machinery and observe a positive selection for dynamin related protein 1 (DRP1), a protein required for mitochondrial network division. Loss of DRP1 prevents RASG12V-induced mitochondrial dysfunction, and renders cells resistant to transformation. Conversely, in human tumor cell lines with activating MAPK mutations, inhibition of these signals leads to robust mitochondrial network reprogramming initiated by DRP1 loss resulting in mitochondrial hyper-fusion and increased mitochondrial metabolism. These phenotypes are mechanistically linked by ERK1/2 phosphorylation of DRP1 serine 616; DRP1S616 phosphorylation is sufficient to phenocopy transformation-induced mitochondrial dysfunction, and DRP1S616 phosphorylation status dichotomizes BRAFWt from BRAFV600E positive lesions. These findings implicate mitochondrial division and DRP1 as crucial regulators of transformation with unexpected leverage in chemotherapeutic success.
Keywords: apoptosis, DRP1, MAPK, metabolism, mitochondria, mitochondrial dynamics, oncogenes, targeted therapeutics
INTRODUCTION
Mitochondria are endo-symbiotic organelles that created an evolutionary advantage for nascent eukaryotes that arose in an increasingly oxidizing environment billions of years ago (Katz, 2012). This advantage is hypothesized to center on the notion that mitochondria provided energy and macromolecular precursors in exchange for a more stable environment. The complex signaling pathways shared between a cell and its mitochondrial network has only recently become apparent with perturbations directly contributing to aging, metabolic diseases, and neurodegeneration (Schapira and Tolosa, 2010; Wallace, 2010). While cancer is a disease characterized by aberrations directly linked to numerous pathways involving mitochondria (Hanahan and Weinberg, 2011), little is mechanistically understood about how cancer associated mutations directly affect mitochondrial biology, and how this impacts upon cancer cell phenotypes.
A commonly mutated pathway in cancer is the mitogen activated protein kinase (MAPK) cascade, which is responsible for orchestrating signaling events at the cell surface leading to a series of gene expression changes in the nucleus (Brose et al., 2002; Davies et al., 2002; McCubrey et al., 2007; Young et al., 2009; Yuen et al., 2002). In the wild type state, this pathway is mediated by a series of kinases, phosphatases, and exchange factors that carefully control and balance cellular signaling, proliferation, and cell death (McCubrey et al., 2007). However, when a hyper-activating oncogenic mutation occurs within this pathway (e.g., RASG12V or BRAFV600E), the cellular phenotype is aberrant signaling, uncontrolled proliferation, and silencing of the cell death machinery (Montagut and Settleman, 2009).
A hallmark feature of cancer cells with oncogenic MAPK signaling mutations is the metabolic shift away from oxidative phosphorylation towards anaerobic glycolysis, which is termed the Warburg effect (Warburg, 1956). Several studies indicate that oncogenic RASG12V signaling promotes mitochondrial dysfunction and subsequent metabolic reprogramming to favor increased glycolytic flux and glutaminolysis (Baracca et al., 2010; Son et al., 2013; Ying et al., 2012), however there is no known mechanism directly linking oncogenic MAPK signaling to mitochondrial dysfunction in primary cells.
In this study, we provide evidence that RASG12V expression and transformation selects for dynamin related protein 1 (DRP1), a large GTPase required for mitochondrial division. Genetic or pharmacological loss of DRP1 prevents RASG12V-induced mitochondrial dysfunction, and renders cells resistant to transformation and colony formation. Conversely, in human tumor cell lines with oncogenic MAPK mutations, inhibition of these signals leads to robust mitochondrial network reprogramming initiated by reduced DRP1 phosphorylation - a key event that offers novel prognostic and chemotherapeutic potential.
RESULTS
RASG12V-induced transformation selects for increased DRP1 function and coincident mitochondrial fragmentation
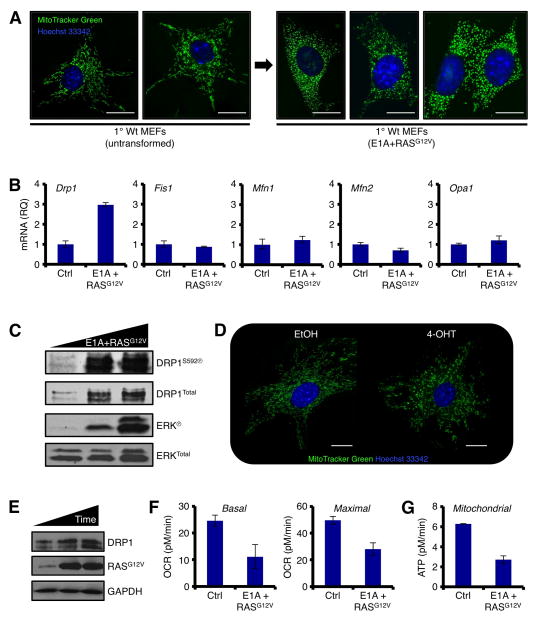
Primary mouse embryonic fibroblasts (MEFs) infected with E1A and the oncogenic form of RAS (RASG12V) undergo rapid immortalization and transformation, which is defined by avoidance of the Hayflick limit, clonogenic survival, and the loss of contact inhibition (Hanahan and Weinberg, 2011; Land et al., 1983; Ruley, 1983). To identify changes in mitochondrial network shape during transformation, we infected primary MEFs with E1A+RASG12V and monitored the shape of the mitochondrial network using live cell fluorescent microscopy. Uninfected primary MEFs displayed a highly dynamic and interconnected mitochondrial network (Fig. 1A, Movie S1). In contrast, the introduction of E1A+RASG12V led to marked mitochondrial division (a.k.a. mitochondrial fission) and a loss in network dynamics (Fig. 1A, Movie S2).
Figure 1.
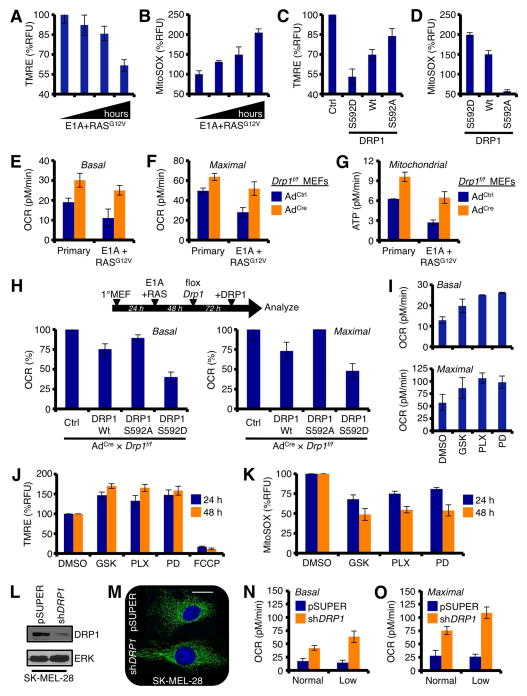
RASG12V-induced transformation selects for increased DRP1 function and co-incident mitochondrial fragmentation. (A) Primary Wt MEFs were infected with E1A+RASG12V and cultured. Cells were loaded with MitoTracker Green and Hoechst 33342 (nuclei), and imaged before and after transformation. (B) qPCR analyses for the mitochondrial dynamics machinery before and after transformation, normalized to 18S and Gapdh. (C) Same as B, but SDS-PAGE and western blot. (D) Primary Wt MEFs were infected with ERTam-regulated RASG12V, treated with 100 nM 4-OHT for 24 h, and imaged as in A. (E) Cell lysates (0, 12, 24 h 4-OHT treatment) from D were subjected to SDS-PAGE and western blot. (F) Basal and maximal mitochondrial oxygen consumption rates (OCR) were determined by Seahorse XF96 analyses before and after transformation. (G) Mitochondrial ATP generation was determined by Seahorse XF96 analyses before and after transformation.
Mitochondrial network division is the result of either enhanced function of the mitochondrial fission machinery (e.g., DRP1, Fis1), or the inhibition of mitochondrial fusion proteins (e.g., Mitofusin 1 and 2, Mfn1/2; Optic atrophy 1, OPA1). To gain mechanistic insights explaining the mitochondrial division phenotype following the introduction of E1A+RASG12V, we screened the mitochondrial fission and fusion components for E1A+RASG12V dependent changes. As shown in figure 1B, Drp1 mRNA expression was specifically induced following E1A+RASG12V; and this correlated with increased DRP1 protein and activation via serine 592 phosphorylation (Fig. 1C). All other components of the mitochondrial dynamics machinery remained essentially unchanged by qPCR and western blot analyses (Figs. 1B, and data not shown). The expression of E1A alone did not result in mitochondrial network or protein changes (data not shown). To determine if RASG12V was sufficient to promote mitochondrial division and enhanced DRP1 expression, we infected primary MEFs with a 4-hydroxytamoxifen (4-OHT) inducible form of RASG12V, added 4-OHT, and visualized the mitochondrial network. Indeed, the addition of 4-OHT led to rapid mitochondrial division, and expression of DRP1 (Figs. 1D–E). While RASG12V activation is sufficient for these phenotypes, cellular transformation requires the addition of E1A. Therefore, E1A+RASG12V will be used throughout our study. Together, these observations suggest that E1A+RASG12V promotes rapid mitochondrial division, potentially through the induction of DRP1, a pro-fission protein.
Next, we hypothesized that E1A+RASG12V mediated mitochondrial division could compromise mitochondrial function. Therefore, we examined the consequences of E1A+RASG12V on mitochondrial oxygen consumption and ATP generation by Seahorse analyses. Indeed, the introduction of E1A+RASG12V was sufficient to decrease basal and maximal rates of oxygen consumption (Fig. 1F), and this paralleled a marked decrease in mitochondrial ATP generation (Fig. 1G).
DRP1 expression and activity are required for RASG12V-induced cellular transformation
The above observations propose that enhanced Drp1 expression is responsible for E1A+RASG12V induced mitochondrial division and compromised function, and these phenotypes potentially contribute to the transformation process. To determine the requirement for DRP1 in transformation, we evaluated the loss of DRP1 function in three systems: (i) Drp1 RNAi, (ii) genetic removal of floxed Drp1 alleles, and (iii) pharmacological inhibition of DRP1.
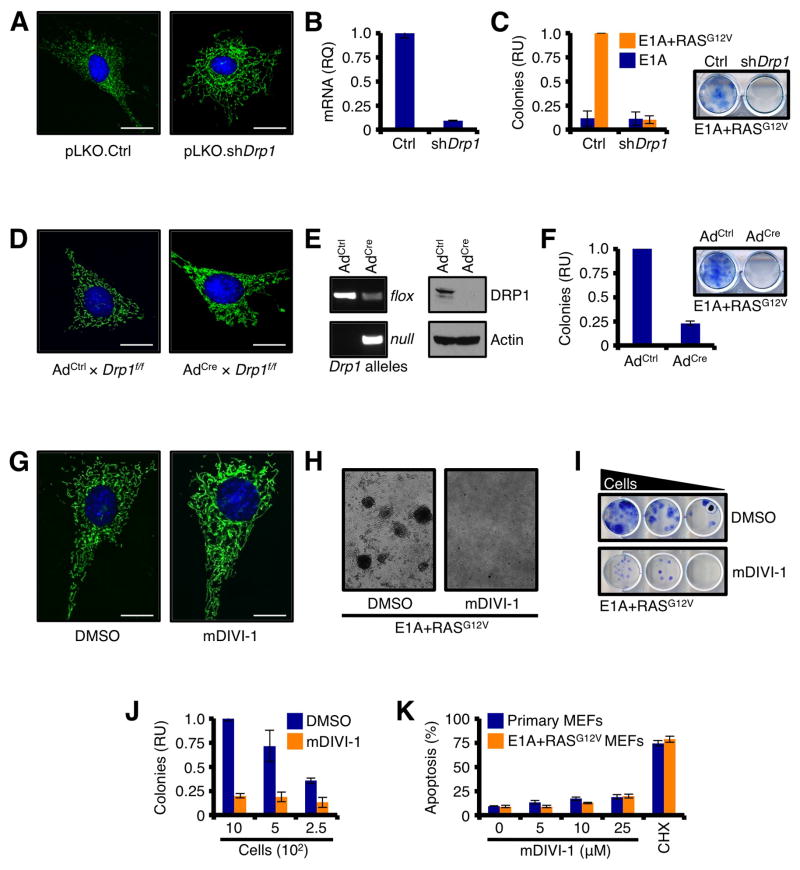
Primary MEFs expressing Drp1 shRNA were generated, and displayed a highly connected mitochondrial network along with an ~ 90% decrease in Drp1 mRNA (Figs. 2A–B). These cells were then infected with E1A+RASG12V, and allowed to form colonies. Control shRNA cells formed colonies when infected with E1A+RASG12V, but Drp1 shRNA cells failed to undergo transformation and generate colonies (Fig. 2C). A similar result was obtained using floxed primary Drp1f/f MEFs. While the addition of adenoviral Cre recombinase efficiently removed Drp1 alleles flanked by LoxP sites, reduced DRP1 protein, and resulted in a highly connected mitochondrial network, the removal of Drp1 led to a significant reduction in E1A+RASG12V mediated transformation and colony formation (Figs. 2D–F). Finally, primary MEFs were induced to undergo transformation with E1A+RASG12V in the presence of a small molecule inhibitor to DRP1, named mDIVI-1 (or DMSO) (Cassidy-Stone et al., 2008). Treatment with mDIVI-1 rapidly produced a highly connected mitochondrial network (Fig. 2G), and abolished E1A+RASG12V mediated transformation and clonogenic survival (Figs. 2H–J). SV40-mediated cellular transformation, which leads to in situ RAS mutations, was also blocked by mDIVI-1 (Figs. S1A–B, and data not shown). As control, mDIVI-1 treatment alone had minimal effects on the survival of primary or transformed cells (Figs. 2K). These studies indicate that DRP1 expression and function are required for E1A+RASG12V mediated transformation.
Figure 2.
DRP1 expression and activity are required for RASG12V-induced cellular transformation. (A) Primary Wt MEFs stably expressing pLKO.shDrp1 (or control vector) were loaded with MitoTracker Green and Hoechst 33342 (nuclei) and imaged. (B) Drp1 knockdown was verified by qPCR analysis, and normalized to 18S and Gapdh. (C) Primary Wt MEFs stably expressing pLKO.shDrp1 (or control vector) were infected with E1A or E1A+RASG12V, and the resulting colonies were quantified. Representative E1A+RASG12V induced colonies ± shDrp1 are shown. (D) Primary Drp1f/f MEFs were treated with control adenovirus (AdCtrl) or Cre recombinase-expressing adenovirus (AdCre), and imaged 96 h later. (E) The floxing of Drp1 was confirmed by PCR of genomic DNA, and western blot. (F) Primary Drp1f/f MEFs were infected with E1A+RASG12V and the resulting colonies were quantified. Representative E1A+RASG12V induced colonies ± AdCre are shown. (G) Primary Wt MEFs were treated with mDIVI-1 (25 μM; or DMSO) for 24 h and imaged. (H) Primary Wt MEFs were treated with mDIVI-1 (25 μM; or DMSO) for 24 h, infected with E1A+RASG12V, and the resulting colonies were imaged by bright field microscopy. (I–J) Dilutions of primary Wt MEFs were treated with mDIVI-1 (25 μM; or DMSO) for 24 h, infected with E1A+RASG12V, and the resulting colonies were stained (I) and quantified (J). (K) Primary and E1A+RASG12V transformed Wt MEFs were treated with indicated concentrations of mDIVI-1 for 24 h and analyzed by AnnexinV.
ERK1/2 phosphorylates DRP1 at serine 616, which is permissive for RASG12V-induced cellular transformation
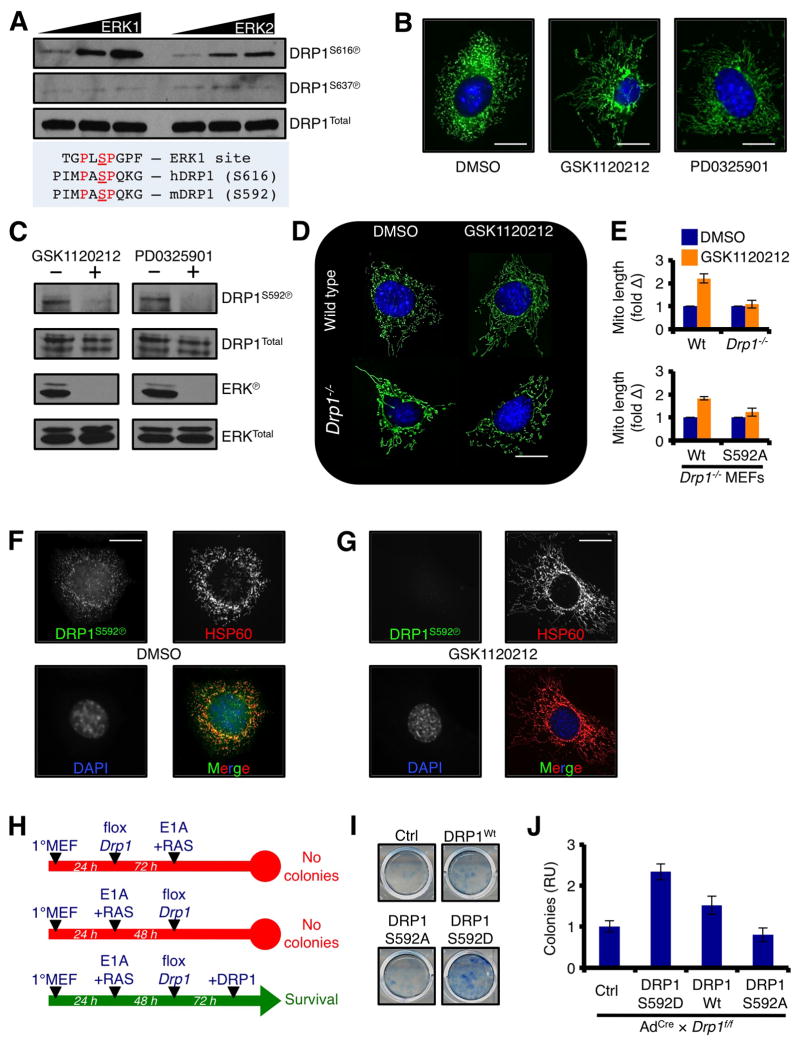
In order for mitochondrial division to occur in a DRP1-dependent manner, human DRP1 protein must be activated by phosphorylation at serine 616 (n.b., equivalent to murine DRP1S592; similar to Fig. 1C). We next determined if the oncogenic RAS pathway directly regulated DRP1 serine 616 phosphorylation (DRP1S616Ⓟ), and if this residue impacted on RASG12V-induced transformation. Recombinant, functional, full-length human DRP1 was subjected to an in vitro kinase assay with active ERK1 and ERK2, and both ERK1 and ERK2 promoted dose-dependent DRP1S616Ⓟ, whereas DRP1S637 was unaffected (Fig. 3A). These in vitro ERK1/2 results suggest that oncogenic MAPK signaling may promote DRP1S592Ⓟ in cells. Therefore, we treated freshly transformed E1A+RASG12V MEFs with a GSK1120212 or PD0325901, two small molecule inhibitors to MEK that potently block RASG12V signaling, and analyzed mitochondrial network shape and DRP1S616Ⓟ status. GSK1120212 or PD0325901 treatment led to marked mitochondrial fusion (Fig. 3B) and rapid loss of DRP1S592Ⓟ (Fig. 3C).
Figure 3.
DRP1 is directly phosphorylated by ERK1/2 at serine 616, which is permissive for RASG12V-induced cellular transformation. (A) 50 ng of recombinant, full-length human DRP1 was incubated with 0.1 units of recombinant human ERK1 or ERK2 for 30 minutes at 37°C, and subjected to western blot using phospho-specific anti-DRP1 antibodies. (B) E1A+RASG12V transformed Wt MEFs were treated with GSK1120212 (100 nM) or PD0325901 (500 nM) for 5 h, loaded with MitoTracker Green and Hoechst 33342 (nuclei) and imaged. (C) Cells were treated as in B, and lysates were western blotted for indicated proteins. ERKⓅ is shown as a positive control for drug sensitivity. (D) E1A+RASG12V transformed Wt or Drp1−/− MEFs were treated with GSK1120212 (100 nM) for 5 h and imaged. (E) Drp1−/− MEFs were stably reconstituted with DRP1Wt or DRP1S592A, treated with GSK1120212 (100 nM) for 5 h and imaged. Mitochondrial lengths were measured, and presented as fold change. Data from D are also provided for comparison. (F–G) E1A+RASG12V transformed Wt MEFs were treated with GSK1120212 (10 nM, F; or DMSO, G) for 5 h. Cells were fixed and stained for HSP60 (Texas Red), DRP1S592Ⓟ (FITC), and nuclei (DAPI). (H) Summary of Drp1 floxing, E1A+RASG12V mediated transformation, and DRP1 reconstitution (Wt, S592D, or S592A) protocol and results. (I) Representative E1A+RASG12V mediated colony formation assays from floxed Drp1f/f MEFs reconstituted with DRP1 (Wt, S592D, or S592A). (J) Quantification from I.
Despite that very few Drp1−/− MEFs survive E1A+RASG12V mediated transformation, we were able to generate Drp1−/− MEFs reconstituted with either wild type DRP1 (DRP1Wt) or a mutant form of DRP1 that cannot be phosphorylated (DRP1S592A) to examine the requirement for DRP1S592 in GSK1120212 regulated mitochondrial fusion. Wt MEFs treated with GSK1120212 rapidly fuse their mitochondria, whereas Drp1−/− mitochondria do not change their length (Figs. 3D–E). Drp1−/− MEFs reconstituted with DRP1Wt are re-sensitized to MEK regulated mitochondrial fusion; in contrast, Drp1−/− mitochondria reconstituted with DRP1S592A do not respond before or after MEK inhibition. Finally, we determined if ERK1/2 regulated DRP1S592Ⓟ co-localized with mitochondria by immunofluorescence. Indeed, Wt MEFs stained for antibodies specific to DRP1S592Ⓟ revealed a mitochondrial distribution that co-localized with HSP60 (a mitochondrial matrix marker) (Fig. 3F), and this mitochondrial DRP1S592Ⓟ pattern was eliminated following GSK1120212 induced mitochondrial fusion (Fig. 3G).
To directly examine a role for DRP1S592Ⓟ in transformation, we performed a series of E1A+RASG12V induced colony formation assays using Cre treated Drp1f/f MEFs reconstituted with DRP1Wt, DRP1S592D (i.e., a phospho-mimetic allele), or DRP1S592A (i.e., phospho-null allele). Drp1 was necessary before and after the addition of E1A+RASG12V, as almost zero colonies developed when Drp1 was floxed (Fig. 2F; and summarized in Fig. 3H). Reconstitution with DRP1Wt allowed for minimal cellular transformation, and we speculate this is due to the timing required for floxing and re-introduction/expression of the wild type allele. Despite this issue, introduction of DRP1S592D resulted in a marked increase in transformation that was approximately double that of DRP1Wt (Figs. 3I–J). Moreover, DRP1S592A acted similarly to control, suggesting that the phosphorylation of serine 592 is an essential component to E1A+RASG12V induced colony formation (Figs. 3I–J).
The inhibition of oncogenic MAPK signaling promotes rapid loss of DRP1 serine 616 phosphorylation, DRP1 expression, and mitochondrial fission in human cancer cells
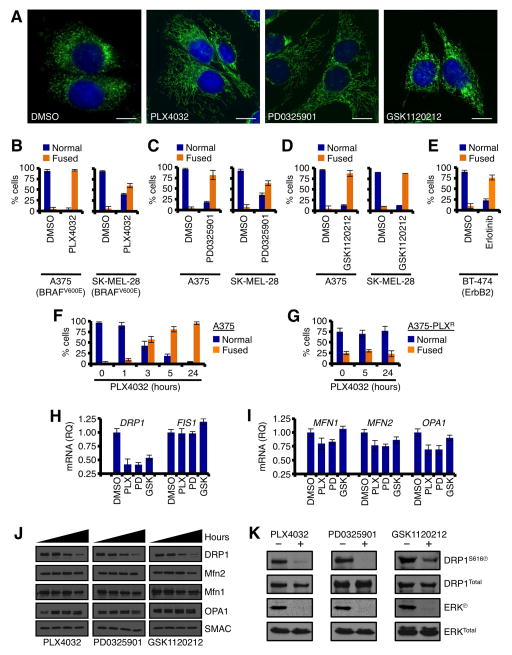
In the previous model systems, we introduced E1A+RASG12V and observed that mitochondrial division and DRP1 expression/function markedly increased due to an oncogenic MAPK signal. To expand and corroborate these findings, we next examined if the inhibition of oncogenic MAPK signaling in human cancer cell lines negatively influenced mitochondrial division and DRP1 expression/function. We selected a panel of human cell lines that harbor oncogenic MAPK signaling including BRAFV600E and ErbB2, as small molecule inhibitors to these mutations (PLX4032 and Erlotinib, respectively) and downstream proteins (PD0325901 and GSK1120212 are MEK inhibitors) are clinically relevant (N.b., as there are no specific RAS inhibitors, we could not evaluate this mutation directly). In the A375 human BRAFV600E melanoma line, inhibition of oncogenic MAPK signaling by PLX4032, PD0325901, or GSK1120212 led to a markedly fused mitochondrial network (Fig. 4A). This phenotype occurred in all human tumor cell lines tested (e.g., A375, SK-MEL-28, BT-474, & HT29), and in response to oncogenic MAPK signaling inhibition by multiple small molecule inhibitors to the pathway (Figs. 4B–E, S2A–C). Importantly, mitochondrial fusion resulting from the inhibition of oncogenic MAPK signaling (e.g., PLX4032) was initiated within one to three hours (Fig. 4F), was dependent upon the on-target effects of the inhibitors as drug-resistant lines failed to fuse their mitochondria (Fig. 4G), independent of mitochondrial biogenesis (Fig. S2E), and was reversible upon drug washout (data not shown).
Figure 4.
The inhibition of oncogenic MAPK signaling promotes rapid mitochondrial fusion, and loss of DRP1 expression and serine 616 phosphorylation in human cancer cells. (A) A375 were treated with PLX4032 (1 μM), PD0325901 (50 nM), or GSK1120212 (10 nM) for 5 h, loaded with MitoTracker Green and Hoechst 33342 (nuclei) before live cell imaging. (B–E) A375, SK-MEL-28, BT-474 were treated with PLX4032 (1 μM), PD0325901 (50 nM), GSK1120212 (10 nM), or Erlotinib (2 μM) for 5 h, loaded with MitoTracker Green and Hoechst 33342 (nuclei) before live cell imaging. Normal verses fused mitochondrial networks in > 300 cells were quantified. (F–G) Time course of PLX4032 induced mitochondrial fusion in parental A375 cells and PLX4032 resistant A375 cells. (H–I) A375 cells were treated with indicated drugs for 2 h, harvested for total RNA, subjected to qPCR for indicate genes, and normalized against actin. (J) The results in H–I were confirmed by western blot at 0, 12, 24, 48 h. SMAC is a mitochondrial loading control. (K) A375 cells were treated as in H, and lysates were western blotted for indicated proteins. ERKⓅ is shown as a positive control for drug sensitivity.
To determine the mechanism explaining how the inhibition of oncogenic MAPK signaling promotes mitochondrial fusion, we screened the mitochondrial fission and fusion components for drug dependent changes. Complementary to the E1A+RASG12V results (Figs. 1–3), all the oncogenic MAPK inhibitors selectively silenced DRP1 mRNA, protein, and DRP1S616Ⓟ (Figs. 4H–K, S2D); and no other mitochondrial dynamics components were affected (Figs. 4H–K, S2D). These data suggest that oncogenic MAPK inhibitors target DRP1 expression and function to promote mitochondrial hyper-fusion.
Oncogenic MAPK signaling reprograms mitochondrial network function via DRP1 serine 616 phosphorylation
DRP1 regulated mitochondrial network remodeling upon initiation (Figs. 1–3) and inhibition (Fig. 4) of oncogenic MAPK signaling suggests that this pathway directly alters mitochondrial function. To examine this, we first determined how cellular transformation impacts upon mitochondrial membrane potential (ΔφM) and mitochondrial reactive oxygen species (mtROS) as respective measures of function and efficiency (Brand and Nicholls, 2011; Sena and Chandel, 2012). Primary MEFs were infected with E1A+RASG12V and cultured for 24 – 96 h, ΔφM and mtROS were then quantified by TMRE (tetramethylrhodamine ethyl ester) and MitoSOX staining, respectively. The introduction of E1A+RASG12V resulted in a marked decrease in ΔφM and a collateral ~100% increase in mtROS generation (Figs. 5A–B), suggesting that cellular transformation rapidly compromises mitochondrial function. As E1A+RASG12V promotes Drp1 expression and DRP1S592Ⓟ, we next measured the direct consequences of exogenous DRP1Wt, DRP1S592D, and DRP1S592A on ΔφM and mtROS in primary Wt MEFs. While DRP1Wt shifted ΔφM and mtROS in the direction observed with E1A+RASG12V, DRP1S592D expression alone was sufficient to remodel ΔφM and mtROS levels similar to those induced by cellular transformation (Figs. 5C–D). Importantly, the phospho-null version of DRP1S592A was similar to the vector control (Figs. 5C–D).
Figure 5.
Oncogenic MAPK signaling reprograms mitochondrial network function via DRP1 serine 616 phosphorylation. (A) Primary Wt MEFs were infected with E1A+RASG12V, cultured up to 96 h (24, 38, 72, 96 h), loaded with TMRE (100 nM), and analyzed by flow cytometry. (B) Same as in A, but MitoSOX (5 μM) was measured. (C–D) Primary Wt MEFs were infected with DRP1 variants, cultured for 96 h, loaded with TMRE or MitoSOX, and analyzed by flow cytometry. (E–F) Primary and E1A+RASG12V infected (48 h post-infection) Drp1f/f MEFs (followed with AdCre or AdCtrl for 72 h) were analyzed for basal and maximal OCRs. (G) Same as E–F, but mitochondrial ATP generation was quantified. (H) Primary Drp1f/f MEFs were infected with E1A+RASG12V, cultured for 48 h, treated with AdCre or AdCtrl for 72 h, and reconstituted with DRP1 (Wt, S592D, or S592A). Basal and maximal OCRs were determined 48 h later. (I) A375 were treated with PLX4032 (1 μM), PD0325901 (50 nM), or GSK1120212 (10 nM) for 48 h, and analyzed for basal and maximal OCRs. (J–K) Same as I, but loaded with TMRE (J) or MitoSOX (K), and analyzed by flow cytometry. (L) SK-MEL-28 were infected with shDRP1 (or pSUPER) for 72 h, and lysates were western blotted for indicated proteins. (M) Same as L, but cells were imaged with MitoTracker Green and Hoechst 33342. (N–O) SK-MEL-28 shDRP1 (or pSUPER) cells were analyzed for basal and maximal OCRs in the presence of normal (35 mM) or low (5 mM) glucose.
The introduction of either DRP1S592D or E1A+RASG12V (which also increases DRP1S592Ⓟ, Fig. 1C) in primary MEFs led to decreased ΔφM and substantially more mtROS (Figs. 5A–D). Next, we investigated how genetic removal of endogenous Drp1 regulated mitochondrial function before and after E1A+RASG12V. Primary Drp1f/f MEFs were treated with adenoviral Cre recombinase (or control adenovirus) for 72 h and examined for mitochondrial function with a Seahorse XF96 analyzer. In parallel, primary Drp1f/f MEFs were infected with E1A+RASG12V for 48 h, followed with adenoviral Cre recombinase (or control adenovirus) for 72 h, and analyzed. The genetic removal of Drp1 was sufficient to increase basal and maximal mitochondrial oxygen consumption rates (OCR) in untransformed Drp1f/f MEFs (Figs. 5E–F); and this also led to collateral increases in metabolic pathways (e.g., glycolysis & TCA) to meet increased mitochondrial substrate demand (Fig. S3A). In contrast, unfloxed primary Drp1f/f MEFs infected with E1A+RASG12V displayed reduced basal and maximal OCRs (Figs. 5E–F), which corroborate the loss of ΔφM and increased mtROS observed earlier in primary Wt MEFs (Figs. 5A–B). The E1A+RASG12V-induced decrease in basal and maximal mitochondrial function was prevented when Drp1 was floxed (Figs. 5E–F), suggesting that DRP1 function is responsible for transformation regulated mitochondrial function. Remarkably, transformation-induced decreases to mitochondrial OCRs and ATP generation returned normal (i.e., levels observed in primary untransformed MEFs) when Drp1 was floxed (Figs. 5E–F). All changes to basal and maximal OCRs also paralleled mitochondrial ATP generation rates (Fig. 5G).
To determine the direct effect of DRP1S592Ⓟ in basal and maximal mitochondrial OCRs, we infected Drp1f/f MEFs with E1A+RASG12V, floxed Drp1, reconstituted the cells with DRP1Wt, DRP1S592D, or DRP1S592A (Figs. S3B–C), and analyzed their mitochondrial function. The expression of DRP1Wt resulted in a ~25% decrease in OCRs (Fig. 5H), which complemented previous observations that Drp1 loss leads to increased OCRs (Figs. 5E–F); likewise, DRP1S592D maximally reduced basal and maximal OCRs by >50% (Fig. 5H). In contrast, the DRP1S592A reconstitution failed to influence mitochondrial function (Fig. 5H).
The above experiments show that oncogenic MAPK signaling inhibitors promote mitochondrial fusion through the regulation of DRP1 expression and function (Figs. 3 – 4). Therefore, we next examined if these inhibitors also altered mitochondrial function. Cells were treated with PLX4032, PD0325901, or GSK1120212 for up to 48 h, and analyzed for basal and maximal OCRs, ΔφM, and mtROS generation. Indeed, the inhibition of oncogenic MAPK signaling resulted in increased basal and maximal OCRs (Fig. 5I), along with time-dependent increases in ΔφM and a reduction in mtROS generation (Figs. 5J–K). In previous experiments, we observed that exposure to these drugs led to rapid DRP1S616/S592 dephosphorylation and down-regulation of DRP1 mRNA and protein. As such, we determined if silencing of DRP1 was sufficient to phenocopy the drug-induced changes to mitochondrial function. Cells were infected with either shDRP1 (or pSUPER), cultured for 72 h, and DRP1 knockdown was determined by western blot (Fig. 5L) and imaging of the mitochondrial network (Fig. 5M). After loss of DRP1 was confirmed in these cells, we analyzed their mitochondrial function. Indeed, DRP1 loss increased basal and maximal mitochondrial OCRs, and the removal of glucose (n.b., this was performed to mimic drug-induced loss of GLUT3 and ECAR, Figs. S4F–G) further enhanced mitochondrial functions (Figs. 5N–O). These observations suggest that DRP1 governs mitochondrial function: (i) before oncogenic signaling is initiated, figures 5E–G; (ii) after E1A+RASG12V transformation, figures 5A–I; and (iii) following the inhibition of oncogenic MAPK signaling, figures 5J–K.
To our knowledge, there is no mechanism to explain a DRP1-regulated decrease in mitochondrial function following transformation with E1A+RASG12V (Figs. 5C–H). Therefore, we screened the upstream mitochondrial respiratory chain complexes before (within 2 days of E1A+RASG12V addition) and after transformation (greater than 3 days following E1A+RASG12V addition when morphological features of transformation and increased cell proliferation were apparent) for changes in activity. As shown in figures S3D-E, complex I demonstrated an E1A+RASG12V and time-dependent decrease in activity, yet complex II remained unchanged in all conditions. We then isolated mitochondrial DNA from these cells, and performed next generation sequencing to analyze components of complex I. Indeed, two genes encoding components of complex 1, NADH dehydrogenase I and II (Nd1 and Nd2), were found to be mutated at several positions (Fig. S3F). Quantitative PCR and western blot analyses were performed to determine the potential influence of these mutations on mRNA and protein. While Nd1 mRNA was unchanged, Nd2 mRNA was reduced below 50% compared to control (Fig. S3G); in contrast, ND1 protein levels were markedly increased as basal levels were undetectable in untransformed cells (Fig. S3H). ND2 protein expression was reduced by ~ 50%, which paralleled the decreases in Nd2 mRNA (Fig. S3G–H). We interpret these data to suggest that these mutations impact either on expression, stability, and/or function of ND1. Importantly, ND1 and ND2 protein changes were not detected in Cre-treated Drp1f/f MEFs (Fig. S3H). We could not interpret changes to mRNA in the absence of Drp1 because these cells rapidly entered senescence and globally reduced the expression of multiple mitochondrial mRNAs (Fig. S3I). Finally, to determine the effect of sustained complex I activity on E1A+RASG12V induced transformation we retrovirally-expressed NDI1, a yeast NADH-ubiquinone oxidoreductase that permits bypass of endogenous complex I function (de Vries and Grivell, 1988). Primary Wt MEFs expressing NDI1 exhibited a three-fold increase in complex I activity, and this was associated with an approximate 60% reduction in E1A+RASG12V mediated colony formation (Figs. S3J–K). We were not able to silence individual components of complex I in Wt MEFs without leading to severely compromised survival or proliferative capacities (data not shown), and therefore could not perform complementary loss-of-function experiments.
DRP1 serine 616 phosphorylation status governs cancer cell survival after the inhibition of oncogenic MAPK signaling
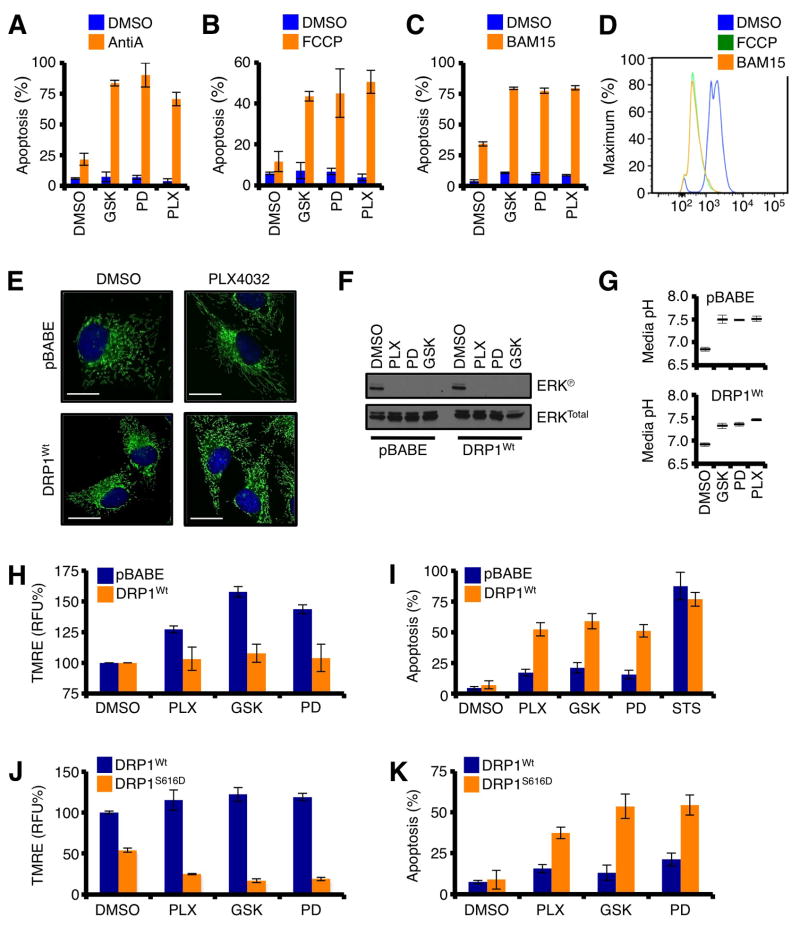
Our observations revealed that the inhibition of oncogenic MAPK signaling promotes DRP1-dependent re-organization of mitochondrial dynamics and enhanced mitochondrial function (Figs. 4 – 5). As the inhibition of oncogenic MAPK signaling leads to substantially reduced carbohydrate metabolites (i.e., glycolysis, TCA, & pentose phosphate pathway), ECAR, and GLUT expression (Figs. S4A–G), enhanced mitochondrial function may be required for energy production and survival. To determine the impact of enhanced mitochondrial function following the inhibition of oncogenic MAPK signaling, we first examined if mitochondrial function was essential to cancer cell survival following treatment with GSK1120212, PD0325901, or PLX4032. A375 cells were pre-treated with PLX4032, PD0325901, or GSK1120212 for 8 h, then treated with either Antimycin A (AntiA; Complex III inhibitor) or FCCP (mitochondrial uncoupler) for 24 h before analyzing survival. While AntiA and FCCP minimally influenced cell survival as single agents, both revealed potent pro-apoptotic effects after the inhibition of oncogenic MAPK signaling (Figs. 6A–B). Off-target toxicity of AntiA and FCCP limits their potential as chemotherapeutic agents, but a novel protonophore mitochondrial uncoupler (named BAM15) with no described in vivo toxicity due to rapid metabolism may provide an opportunity to utilize mitochondrial function as a potential target (Kenwood et al., 2014). Similar to data in figures 6A–B, the combination treatment of BAM15 with GSK1120212, PD0325901, or PLX4032 led to marked apoptotic responses that were very rapid (Fig. 6C); and BAM15 effectively decreased ΔφM similar to FCCP (Fig. 6D). These data suggest that impinging upon DRP1-regulated mitochondrial network function may provide a novel pharmacological target to uniquely eliminate cancer cells.
Figure 6.
DRP1 serine 616 phosphorylation status governs cancer cell survival after the inhibition of oncogenic MAPK signaling. (A–C) A375 cells were pre-treated with PLX4032 (1 μM), PD0325901 (50 nM), or GSK1120212 (10 nM) for 8 h, then treated with either Antimycin A (25 μM, A), FCCP (25 μM, B), or BAM15 (25 μM, C) for 24 h before AnnexinV analysis. (D) A375 cells were treated with FCCP (25 μM) or BAM15 (25 μM) for 2 h, loaded with TMRE (100 nM), and analyzed by flow cytometry. (E) A375-pBABE and A375-DRP1Wt cells were treated with PLX4032 (1 μM) for 24 h, and loaded with MitoTracker Green and Hoechst 33342 (nuclei) before live cell imaging. (F) A375-pBABE and A375-DRP1Wt cells were treated with PLX4032 (1 μM), PD0325901 (50 nM), or GSK1120212 (10 nM) for 1 h, and lysates were western blotted for indicated proteins. (G) Same as in F, but media acidification was measured after 24 h. This is a rapid means to determine ECAR (Figs. S4A–E). (H) A375-pBABE and A375-DRP1Wt cells were treated with PLX4032 (1 μM), PD0325901 (50 nM), or GSK1120212 (10 nM) for 24 h before TMRE staining and flow cytometry. (I) The same cells and drugs as in H, but for 72 h, followed with AnnexinV staining. (J) A375-DRP1Wt and A375-DRP1S616D were treated as in H. (K) The same cells and drugs as in J, but for 48 h, followed with Annexin V staining.
To directly examine this hypothesis, we generated cells with stable expression of DRP1, or vector control (pBABE), and then determined how oncogenic MAPK inhibitors impacted upon mitochondrial responses and survival. First, we characterized these cells for responses to GSK1120212, PD0325901, or PLX4032. While A375-pBABE cells responded to PLX4032 by rapidly fusing their mitochondria, A375-DRP1Wt cells maintained their fragmented mitochondrial network phenotype (Fig. 6E). The lack of mitochondrial fusion in A375-DRP1Wt cells was not due to decreased drug sensitivity because all the drug treatments rapidly decreased ERK phosphorylation and media acidification (Figs. 6F–G). Next, we examined how stable DRP1 expression influenced mitochondrial responses following GSK1120212, PD0325901, or PLX4032 treatment. For these assays we measured ΔφM by TMRE as this allowed for quantitative single cell analysis. As shown previously in figure 5J, GSK1120212, PD0325901, or PLX4032 treatments led to increased ΔφM, and A375-pBABE responded similarly (Fig. 6H); this contrasted with A375-DRP1Wt cells, which failed to promote mitochondrial fusion and increase ΔφM. In parallel, we examined how stable DRP1 expression influenced cell survival following the inhibition of oncogenic MAPK signaling. A375-pBABE cells demonstrated minimal apoptotic responses after 72 h of drug treatment (Fig. 6I), as expected (Serasinghe et al., 2014). In contrast, A375-DRP1Wt cells were sensitized to GSK1120212, PD0325901, and PLX4032-induced apoptosis suggesting that sustained DRP1 over-expression and/or function was detrimental to cancer cell survival following the inhibition of oncogenic MAPK signaling (Fig. 6I). The sensitization to apoptosis was unique to oncogenic MAPK signaling inhibitors, as a general inducer (Staurosporine, STS) of apoptosis was unaffected by stable DRP1 expression (Fig. 6I).
Finally, we examined the impact of DRP1S616Ⓟ in mediating the changes to ΔφM and apoptosis following GSK1120212, PD0325901, or PLX4032 treatments for 48 h (n.b., shorter treatments were needed for DRP1S616D expressing cells, as noted below). A375 cells expressing DRP1Wt minimally increased ΔφM, and this led to weak sensitization to apoptosis after 48 hours (Figs. 6J–K). In contrast, cells expressing the phospho-mimetic allele of human DRP1 (i.e., DRP1S616D) significantly reduced ΔφM after GSK1120212, PD0325901, or PLX4032 treatments (Fig. 6J); and this correlated with a marked sensitization to apoptosis after 48 h. Together these data suggest that failure to promote mitochondrial fusion and collateral increases in mitochondrial function leads to apoptosis following the inhibition of oncogenic MAPK signaling (Fig. 6K).
DRP1S616 phosphorylation status dichotomizes wild type and BRAFV600E positive primary melanoma lesions
To determine if the above signaling mechanisms were observed in human disease, we next evaluated if oncogenic MAPK signaling correlated with a positive DRP1S616Ⓟ status in BRAFV600E positive melanoma. We chose BRAFV600E positive melanoma because our previous data suggest mitochondrial fragmentation is observed in many BRAFV600E melanoma cell lines (Fig. 3), and numerous targeted therapies with implications in melanoma treatment (i.e., PLX4032, GSK1120212, PD0325901) directly regulate mitochondrial fusion and function (Figs. 3–5).
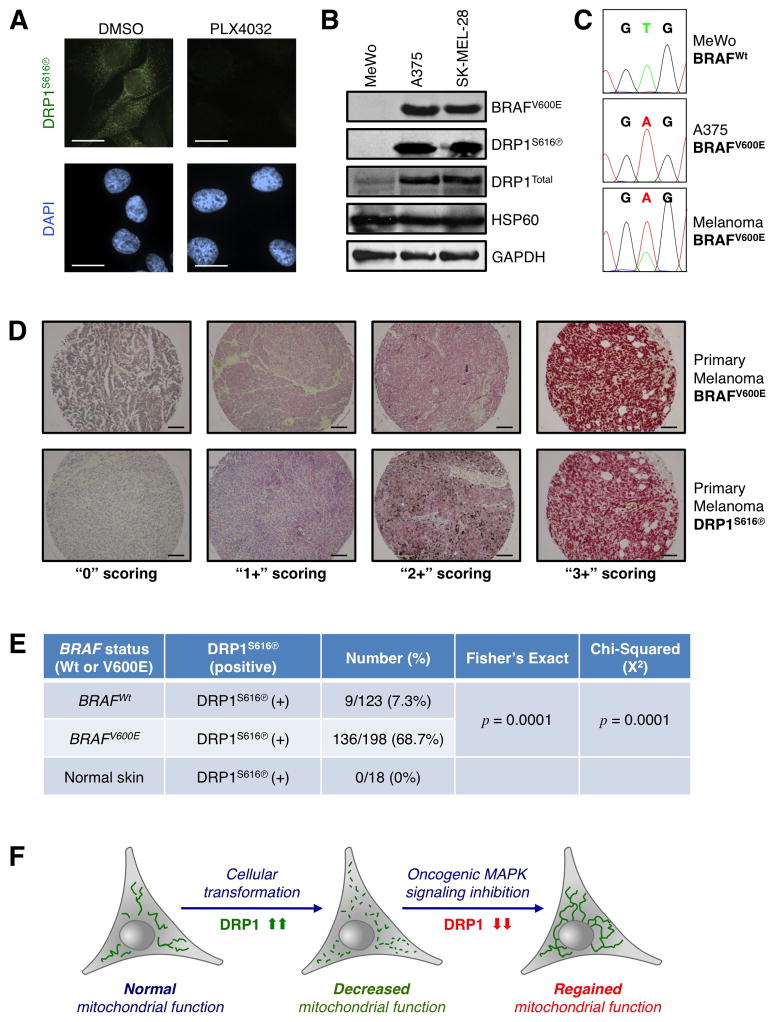
First, we validated that our reagents to detect DRP1S616Ⓟ and BRAFV600E are specific and robust. DRP1S616Ⓟ is decreased by PLX4032 treatment (Fig. 4K), and we corroborated that the DRP1S616Ⓟ antibody immuno-reactivity demonstrated similar regulation in PLX4032 treated A375 cells (Fig. 7A). Using standard human melanoma cell lines, we next assessed the detection of BRAFV600E with a BRAFV600E specific antibody (clone VE1) by western blot, and only mutant BRAF was detected (Fig. 7B). We also confirmed the VE1 western blot results by BRAF exon 15 sequencing (Fig. 7C). These results validated that the antibodies against DRP1S616Ⓟ and BRAFV600E are specific to their targets. Using these antibodies, we performed immunohistochemistry (IHC) with 321 formalin-fixed, paraffin-embedded (FFPE) melanoma tissue sections, and developed BRAFV600E and DRP1S616Ⓟ scoring (0, 1+ = negative; 2+, 3+ = positive) based on standard histopathological analyses within the Mount Sinai Medical Center. Representative BRAFV600E and DRP1S616Ⓟ FFPE section stainings are provided (Fig. 7D, S5A). In 198 cases of BRAFV600E melanoma, we observed DRP1S616Ⓟ in 136 cases (68.7%); in contrast, only 9 of 123 cases of BRAFWt melanoma demonstrated significant DRP1S616Ⓟ staining (Fig. 7E, S5B). Fisher’s Exact (p = 0.0001) and Chi-Squared (p = 0.0001) analyses revealed that these relationships are highly significant. Within our analyses, we also evaluated positive and negative DRP1S616Ⓟ staining in BRAFWt and BRAFV600E lesions (Fig. 7E, S5B), and the detection of DRP1S616Ⓟ in normal skin (0/18 cases; Fig. 7E, S5B). These IHC data suggest that DRP1S616Ⓟ is significantly related to BRAFV600E status in human melanoma. When considered along with our biochemical and cellular data, these results reveal that a deeper understanding of DRP1 expression, function, and regulation will provide fundamental insights into cellular transformation, cancer detection, and therapeutic success.
Figure 7.
DRP1S616 phosphorylation status dichotomizes wild type and BRAFV600E positive primary melanoma lesions. (A) Immunofluorescence for DRP1S616Ⓟ (green) and nuclei (DAPI, blue) was performed on A375 cells treated with PLX4032 (1 μM) for 12 h. (B) Lysates from BRAFWt (MeWo) and BRAFV600E (A375 & SK-MEL-28) cells were western blotted for indicated proteins. The BRAFV600E mutant protein is specifically detected by the BRAF antibody clone VE1. (C) After genomic DNA isolation, BRAF exon 15 sequencing was performed to corroborate BRAFWt (GTG) and BRAFV600E (GAG) codon distinction with VE1. N.b., Wt and mutant BRAF codon 600 alleles are detected in melanoma sections due to both heterozygosity and normal cells within the samples. (D) Immunohistochemistry was performed to detect the status of BRAFV600E and DRP1S616Ⓟ; and examples of tumor scoring are shown. (E) 321 FFPE human melanoma biopsy sections were stained and scored for BRAFV600E and DRP1S616Ⓟ status; results were analyzed by a Fisher’s Exact Test (p = < 0.0001) and Chi-Squared Test (p = < 0.0001) for statistical significance. Normal skin is shown for negative controls. (F) Proposed model of how DRP1 is a key regulator of mitochondrial network shape and function before and after cellular transformation; along with subsequent to the inhibition of oncogenic MAPK signaling in cancer cells.
DISCUSSION
These data reveal a critical role for DRP1 and DRP1S616Ⓟ in mediating mitochondrial function, transformation potential, and apoptosis upon initiation and inhibition of oncogenic MAPK signaling. In brief, we show that DRP1 mediated mitochondrial division is required for RAS-induced transformation (Figs. 1 – 2); inhibition of oncogenic MAPK signaling rapidly mediates mitochondrial fusion via DRP1 (Figs. 3 – 4); DRP1S616 phosphorylation actuates mitochondrial dysfunction and cancer metabolism (Figs. 5 – 6); and DRP1S616 phosphorylation status dichotomizes wild type and BRAFV600E positive primary melanoma lesions (Fig. 7). Over the past decade, intense investigation has focused on exploring the molecular mechanisms of mitochondrial fission (Santel & Frank, 2008; Youle et al., 2005; Chan, 2006), yet minimal mechanistic and therapeutic information linking the mitochondrial fission machinery to cancer promoting pathways has been discovered (Corrado et al., 2012; Grandemange et al., 2009; Rehman et al., 2012). Here, we provide a transformative concept that DRP1, and potentially other members of the mitochondrial dynamics machinery warrant evaluation as direct regulators of oncogenic MAPK signaling and cancer related phenotypes.
Data presented here suggest that mitochondrial function is linked to DRP1 upon initiation and inhibition of oncogenic MAPK signaling, and that glycolysis and TCA respond quickly to the genetic removal of Drp1 to supply mitochondria with substrates for ATP generation (Figs. 1 – 5). Motivation to explore mitochondrial function after the inhibition of oncogenic MAPK signaling originated from clinical results suggesting that while BRAFV600E melanoma patients display nearly 100% metabolic responses (i.e., complete loss of glucose uptake by positron emission tomography) to PLX4032 treatment, decreases in tumor cell survival and burden varied considerably suggesting that cancer cells must reorganize their metabolic functions to survive (Bollag et al., 2010; McArthur et al., 2012). Indeed, alternative mechanisms for enhanced mitochondrial function have been described following BRAFV600E inhibition, but these pathways did not consider the direct regulation of the mitochondrial dynamics machinery by oncogenic MAPK signaling pathways and subsequent changes to mitochondrial function (Haq et al., 2012). Furthermore, direct collaborations between oncogenic MAPK signaling and DRP1-regulated mitochondrial function are supported by the results in figures 6H-K, where cell fate after the inhibition of oncogenic MAPK was governed by the expression and function of DRP1. DRP1Wt expression was sufficient to prevent increased ΔφM and this led to sensitization to apoptosis. In contrast, DRP1S616D expression alone reduced ΔφM, and this decrease was further evident after the inhibition of oncogenic MAPK signaling, presumably due to decreased endogenous DRP1S616Ⓟ, and the remaining exogenous DRP1S616D is constitutively activated leading to a pathologically segregated mitochondrial network that fails to generate sufficient ATP to ensure survival --- indeed, these cells die rapidly when oncogenic MAPK signaling is inhibited. The IHC data presented in figure 7 also suggest that DRP1S616Ⓟ status is directly related to oncogenic MAPK signaling as DRP1Total was observed in 95 melanoma samples (73/95, 76.8%; Fig. S5C), yet DRP1S616Ⓟ status was observed more frequently in BRAFV600E (68.7%) than DRP1Total (40%). A recent IHC study showed that DRP1Total expression related to the metastatic potential of breast carcinoma (Zhao et al., 2012); while no mechanism was provided, it is possible that increased DRP1 expression may also be sufficient to promote mitochondrial division and metastasis in the absence of oncogenic MAPK signaling. Future studies by our laboratory focus on exploring if the status of individual mitochondrial dynamics machinery components can be utilized to dichotomize benign from malignant lesions.
In the past, transformed Drp1−/− MEFs were generated to study the requirement for DRP1 in mitochondrial division and cell death, but these cells have circumvented the requirement for DRP1 in mitochondrial division through unknown mechanisms (Ishihara et al., 2009; Smirnova et al., 2001). Likewise, our work suggests that these cells may also harbor unknown complexities as the majority of primary Drp1−/− MEFs fail to transform in culture when infected with E1A+RASG12V or SV40 (n.b., SV40 requires stochastic RAS mutations to transform a cell) (Fig. 2). While a role for DRP1 in mitochondrial division during the cell cycle is well established (Kashatus et al., 2011; Mitra et al., 2009; Qian et al., 2013), re-evaluation of DRP1 in apoptosis using floxed Drp1 may be required to reveal mechanisms contributing to cell death in the absence of unknown compensatory mechanisms (Youle & Karbowski, 2005).
An additional interesting aspect to our work that warrants future investigations is the development of mitochondrial DNA mutations and the aberrations in ND1 and ND2 expression that are regulated by E1A+RASG12V, and the complementary observation that sustained complex I activity is sufficient to reduce transformation (Figs. S3D–K). Multiple mutations in complex I are described in various solid tumors, yet the contributions of these mutations to tumorigenesis is not well understood (Chatterjee et al., 2006). It appears that mitochondrial dysfunction contributes to cancer-related metabolic and survival phenotypes along with chemotherapeutic sensitivity, and that the pharmacological abrogation of complex I activity is detrimental to cancer cell survival (Birsoy et al., 2014; Gaude et al., 2014; Wheaton et al., 2014). Together, these studies suggest that the development of cancer model systems that permit examination of both experimental and patient-derived complex I mutations is necessary to reveal the mechanistic contributions to disease and, potentially, treatment.
In conclusion, our work provides novel fundamental insights into the relationship of oncogenic MAPK signaling and DRP1-mediated mitochondrial division (Fig. 7F). Moreover, these data reveal that DRP1S616Ⓟ status may be a useful clinical biomarker to corroborate BRAFV600E status, or potentially any tumor with an oncogenic MAPK signaling mutation. A remaining question is how does mitochondrial division via oncogenic RAS negatively impact upon mitochondrial function? Evidence from our laboratory suggests that mitochondrial DNA mutations rapidly occur in a mitochondrial division dependent manner; how these mutations are dealt with to re-organize mitochondrial function after the inhibition of oncogenic MAPK signaling remains unknown, but this scenario is surely related to restoring mitochondrial connectivity (Parone et al., 2008). Recent studies also suggest that fragmented mitochondria resist BAX-dependent apoptosis, which may further contribute to cancer development and treatment (Renault et al., 2014). Furthermore, exploring the proteins and pathways that regulate DRP1 expression and function may reveal additional factors contributing to the oncogenic potential of cells. We are hopeful that improving our understanding of mitochondrial division and its relationship to cancer will reveal additional mechanistic insights, biomarkers and prognostic indicators for cancer, along with motivation to generate potent second generation drugs (Lackner & Nunnari, 2010) that target fundamental mitochondrial biology to prevent, detect, and/or treat cancer.
EXPERIMENTAL PROCEDURES
Reagents
All cell culture and transfection reagents were from Invitrogen; and standard reagents were from Sigma-Aldrich or Fisher Scientific. Drugs were from: PLX4032/GSK1120212/PD0325901/Erlotinib (Selleck); and FCCP, Antimycin A, 4-OHT, mDIVI-1 (Sigma-Aldrich). Antibodies: DRP1Total, DRP1S616Ⓟ, DRP1S637Ⓟ, ERKTotal, ERKⓅ (Cell Signaling); RAS (EMD/Millipore Anti pan RAS AB-3); Mfn2 (Abcam); OPA1 (BD Biosciences); Actin, GAPDH, HSP60, Mfn1, ND1, ND2, SMAC (Santa Cruz). MitoTracker Green and Hoechst 33342 are from Invitrogen and Anaspec, respectively.
Cell culture, stable clone generation, apoptosis assays, and clonogenic survival
A375, SK-MEL-28, BT-474, HT-29, MeWo, Wt (primary and immortalized), Drp1−/− MEFs were cultured in DMEM supplemented with 10% FBS, 2 mM L-Glutamine, and antibiotics. Drp1f/f conditional knockout mouse embryonic fibroblasts were cultured in IMDM supplemented with 10% FBS, 2 mM L-Glutamine, and antibiotics. Primary Wt MEFs were isolated from C57Bl/6 ED11-13 embryos using a standard protocol.
pBABE-hDRP1Wt was generated by performing Quickchange mutagenesis (Stratagene) with a pBABE-hDRP1K38A construct (Addgene). The point mutations DRP1S616A and DRP1S616D were also generated by site directed mutagenesis of the pBABE-hDRP1 construct using the above method. The mouse DRP1 construct was generated by cloning DRP1 cDNA in to the Retip-Poly retroviral vector, and the point mutants DRP1S592A and DRP1S592D were generated by performing Quickchange reactions on the Retip-Poly-DRP1Wt. The human pSUPER-shDRP1 plasmid was kindly provided by Dr. David Kashatus, and the mouse pLKO-shDRP1 and pLKO-shFis1 plasmids were purchased from Sigma-Aldrich (MissionR shRNA). The pLKO empty vector and scrambled shRNA constructs were kindly provided by the laboratory of Dr. E. Premkumar Reddy. The 293T cell line was used to produce retroviral and lentiviral particles for the generation of stable cell lines. Virus was harvested at 24 and 48 h, pooled, and 0.45 μm filtered. SK-MEL-28 and A375 and stable clones were generated using puromycin (0.4–0.8 μg/ml).
Adenovirus (2×108 titer) expressing Cre recombinase (AdCre, Vector Biolabs), and a negative control Ad-CMV-Null (AdCtrl) was used for the Drp1f/f studies. Cre mediated recombination efficiency was determined by qPCR and genotyping as described (Wakabayashi et al., 2009).
For cell death studies, cells were seeded for 24 h, treated as described, floating and attached cells harvested, labeled with AnnexinV-FITC in binding buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2), and analyzed by flow cytometry as indicated (Logue et al., 2009).
For clonogenic survival studies, cells were seeded for 24 h, treated as indicated for 24 h before changing the media, and cultured for 10–14 days. Colonies were stained with 0.1% methylene blue and imaged. Colonies were then de-stained (20% methanol in 5% acetic acid), and the supernatant was measured for absorption at 568 nm for relative quantification of colony numbers.
Supplementary Material
Acknowledgments
We would like to thank everyone in the Chipuk Laboratory for assistance and support. Drs. Stuart Aaronson, Emily Bernstein, Mark Lebwohl, Cathie Pfleger, Poulikos Poulikakos, E. Premkumar Reddy, and Garabet Yeretssian (Icahn School of Medicine at Mount Sinai) for critical reagents and/or mentorship; Dr. Cole Haynes and Christopher Fiorese (Memorial Sloan Kettering Cancer Center), and Dr. Navdeep Chandel and Lucas Sullivan (Northwestern University) for assistance with Seahorse Analyzer studies; the Stable Isotope & Metabolomics Core (Albert Einstein College of Medicine); Dr. Jodi Nunnari (University of California, Davis) for recombinant DRP1; Drs. Miriam Birge and Rajendra Singh (Mount Sinai Medical Center) for their dermatopathology expertise; and Dominique Bozec for assistance with mice; and Dr. Sandra Milasta for assistance with the NDI1 studies. This work was supported by: NIH grants CA157740 (to J.E.C.) and GM089853 (to H.S.); a pilot project from NIH P20AA017067 (to J.E.C.), the JJR Foundation (to J.E.C.), the William A. Spivak Fund (to J.E.C.), and the Fridolin Charitable Trust (to J.E.C.). This work was also supported in part by two research grants (5-FY11-74 and 1-FY13-416) from the March of Dimes Foundation (to J.E.C.), an Einstein Research Fellowship (to S.Y.W.), an American Skin Association Medical Students Grant (to S.Y.W.), and an American Cancer Society Research Scholar Award (to J.E.C.). Chromatographic and mass spectrometric development for metabolite profiling was supported by a Diabetes Research and Training Center (DRTC) grant P60DK020541.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baracca A, Chiaradonna F, Sgarbi G, Solaini G, Alberghina L, Lenaz G. Mitochondrial Complex I decrease is responsible for bioenergetic dysfunction in K-ras transformed cells. Biochim Biophys Acta. 2010;1797:314–323. doi: 10.1016/j.bbabio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, Wang T, Chen WW, Clish CB, Sabatini DM. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–12. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Mambo E, Sidranksy D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- Corrado M, Scorrano L, Campello S. Mitochondrial dynamics in cancer and neurodegeneration and neuroinflammatory diseases. Intl J Cell Bio. 2012;2012:1–13. doi: 10.1155/2012/729290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- De Vries S, Grivell LA. Purification and characterization of a rotentone-insensitive NADH: Q6 oxidoreductase from mitochondria of Saccharomyces cerevisiae. Eur J Biochem. 1988;176:377–384. doi: 10.1111/j.1432-1033.1988.tb14292.x. [DOI] [PubMed] [Google Scholar]
- Gaude E, Frezza C. Defects in mitochondrial metabolism and cancer. Cancer Metab. 2014;17(2):10. doi: 10.1186/2049-3002-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandemange S, Herzig S, Martinou JC. Mitochondrial dynamics and cancer. Sem Can Biol. 2009;19:50–56. doi: 10.1016/j.semcancer.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA. Origin and diversification of eukaryotes. Annu Rev Microbiol. 2012;66:411–427. doi: 10.1146/annurev-micro-090110-102808. [DOI] [PubMed] [Google Scholar]
- Kenwood BM, Weaver JL, Bajwa A, Poon IK, Byrne FL, Murrow BA, Calderone JA, Huang L, Divakaruni AS, Tomsig JL, et al. Identification of a novel mitochondrial uncoupler that does not depolarize the plasma membrane. Mol Metab. 2014;3:114–123. doi: 10.1016/j.molmet.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL, Nunnari J. Small molecule inhibitors of mitochondrial division: tools that translate basic biological research into medicine. Chem Biol. 2010;17:578–583. doi: 10.1016/j.chembiol.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Logue SE, Elgendy M, Martin SJ. Expression, purification and use of recombinant annexin V for the detection of apoptotic cells. Nat Protoc. 2009;4:1383–1395. doi: 10.1038/nprot.2009.143. [DOI] [PubMed] [Google Scholar]
- McArthur GA, Puzanov I, Amaravadi R, et al. Marked, homogeneous, and early [18F]fluorodeoxyglucose-positron emission tomography responses to vemurafenib in BRAF-mutant advanced melanoma. J Clin Oncol. 2012;30:1628–1634. doi: 10.1200/JCO.2011.39.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagut C, Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 2009;283:125–134. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 2008;3:e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Choi S, Gibson GA, Watkins SC, Bakkenist CJ, Van Houten B. Mitochondrial hyperfusion induced by loss of the fission protein Drp1 causes ATM-dependent G2/M arrest and aneuploidy through DNA replication stress. J Cell Sci. 2012;125:5745–5757. doi: 10.1242/jcs.109769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Wang J, Van Houten B. The role of dynamin-related protein 1 in cancer growth: a promising therapeutic target? Expert Opin Ther Targets. 2013;17:997–1001. doi: 10.1517/14728222.2013.823160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, Archer SL. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26:2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault TT, Floros KV, Elkholi R, Corrigan KA, Kushnareva Y, Wieder S, Lindtner C, Serasinghe MN, Asciolla JJ, Buettner C, Chipuk JE. Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis. Mol Cell Dec. 2014;3 doi: 10.1016/j.molcel.2014.10.028. pii: S1097-2765(14)00863-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruley HE. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- Santel A, Frank S. Shaping mitochondria: the complex post-translational regulation of the mitochondrial fission protein, DRP1. IUBMB Life. 2008;60:448–455. doi: 10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- Schapira AH, Tolosa E. Molecular and clinical prodrome of Parkinson disease: implications for treatment. Nat Rev Neurol. 2010;6:309–317. doi: 10.1038/nrneurol.2010.52. [DOI] [PubMed] [Google Scholar]
- Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serasinghe MN, Missert DJ, Asciolla JJ, Podgrabinska S, Wieder SY, Izadmehr S, Belbin G, Skobe M, Chipuk JE. Anti-apoptotic BCL-2 proteins govern cellular outcome following B-RAF inhibition and can be targeted to reduce resistance. Oncogene. 2014 doi: 10.1038/onc.2014.21. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, Chandel NS. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;13(3):e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–633. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- Young A, Lyons J, Miller AL, Phan VT, Alarcon IR, McCormick F. Ras signaling and therapies. Adv Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- Yuen ST, Davies H, Chan TL, Ho JW, Bignell GR, Cox C, Stephens P, Edkins S, Tsui WW, Chan AS, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res. 2002;62:6451–6455. [PubMed] [Google Scholar]
- Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, Abel PW, Tu Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.