Abstract
A target protein for nalidixic and oxolinic acids in Escherichia coli, the nalA gene product (Pnal), was purified to homogeneity as judged by gel electrophoresis, using an in vitro complementation assay. It is a dimer of identical 110,000-dalton subunits. A polypeptide of this molecular weight is uniquely induced by a λ nalA transducing phage, thereby showing that the purified Pnal is a product of the nalA gene. Nalidixic and oxolinic acids inhibit DNA gyrase activity and induce formation of a relaxation complex analogue. Treatment of the complex with sodium dodecyl sulfate causes a doublestrand break in the DNA substrate and the resulting linear molecule seems covalently bound to protein. Complex formation, unlike the introduction of supertwists, does not require ATP or relaxed circular DNA and is insensitive to novobiocin. DNA gyrase from a strain with a nalA mutation conferring drug resistance (nalAr) is 1/100 as sensitive to oxolinic and nalidixic acids with respect to inhibition of supertwisting and induction of the pre-linearization complex. Addition of Pnal restores drug sensitivity and stimulates DNA gyrase activity. DNA gyrase preparations and Pnal catalyze a third reaction sensitive to nalidixic and oxolinic acids, the ATP-independent relaxation of supertwister DNA. Relaxation by gyrase from nalAr cells is drug resistant. The nicking-closing activity is distinct from E. coli ω protein in several properties, including the ability to relax positively supertwisted DNA. We postulate that the nalA gene product occurs in two molecular forms, as Pnal and as a gyrase component. Both forms catalyze nicking-closing, and inhibition of this activity by nalidixic and oxolinic acids may account for the inhibition of DNA synthesis by these drugs.
Keywords: oxolinic acid, DNA relaxation, DNA supertwisting, λ transducing phages, relaxation complex
Full text
PDF


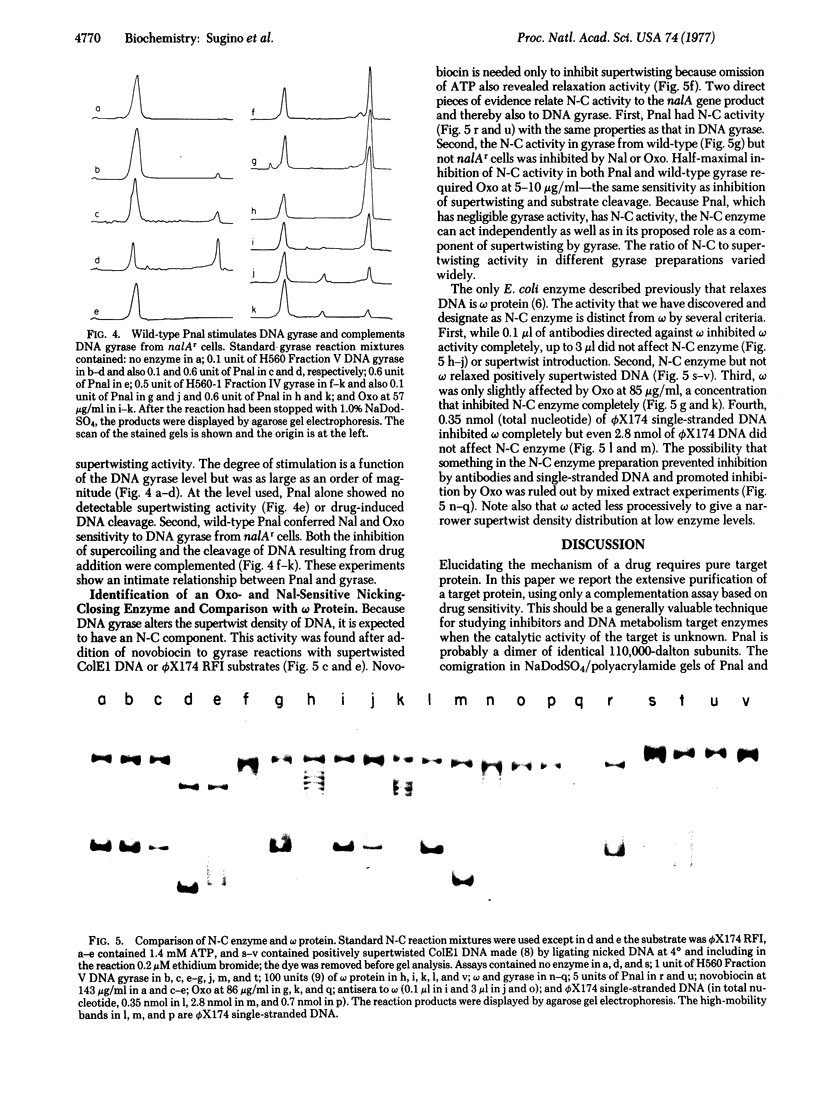

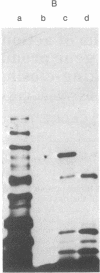
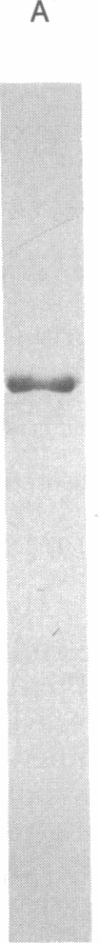
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. W., Baum P. R., Gesteland R. F. Processing of adenovirus 2-induced proteins. J Virol. 1973 Aug;12(2):241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli N. R. The mechanism of action of inhibitors of DNA synthesis. Annu Rev Biochem. 1977;46:641–668. doi: 10.1146/annurev.bi.46.070177.003233. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Itoh T., Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Marians K. J., Ikeda J. E., Schlagman S., Hurwitz J. Role of DNA gyrase in phiX replicative-form replication in vitro. Proc Natl Acad Sci U S A. 1977 May;74(5):1965–1968. doi: 10.1073/pnas.74.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEntee K., Hesse J. E., Epstein W. Identification and radiochemical purification of the recA protein of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3979–3983. doi: 10.1073/pnas.73.11.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. F., Eisenberg S., Bertsch L. L., Kornberg A. A mechanism of duplex DNA replication revealed by enzymatic studies of phage phi X174: catalytic strand separation in advance of replication. Proc Natl Acad Sci U S A. 1977 Jan;74(1):193–197. doi: 10.1073/pnas.74.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. I. Location of the secondary attachment sites and the properties of the lysogens. J Mol Biol. 1972 Feb 14;63(3):483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- Siegel L. M., Monty K. J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966 Feb 7;112(2):346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L. Replication of small plasmids in extracts of Escherichia coli. Mol Gen Genet. 1976 Jun 15;145(3):273–280. doi: 10.1007/BF00325823. [DOI] [PubMed] [Google Scholar]
- Sumida-Yasumoto C., Yudelevich A., Hurwitz J. DNA synthesis in vitro dependent upon phiX174 replicative form I DNA. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1887–1891. doi: 10.1073/pnas.73.6.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderslice R. W., Yegian C. D. The identification of late bacteriophage T4 proteins on sodium dodecyl sulfate polyacrylamide gels. Virology. 1974 Jul;60(1):265–275. doi: 10.1016/0042-6822(74)90384-5. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]