Abstract
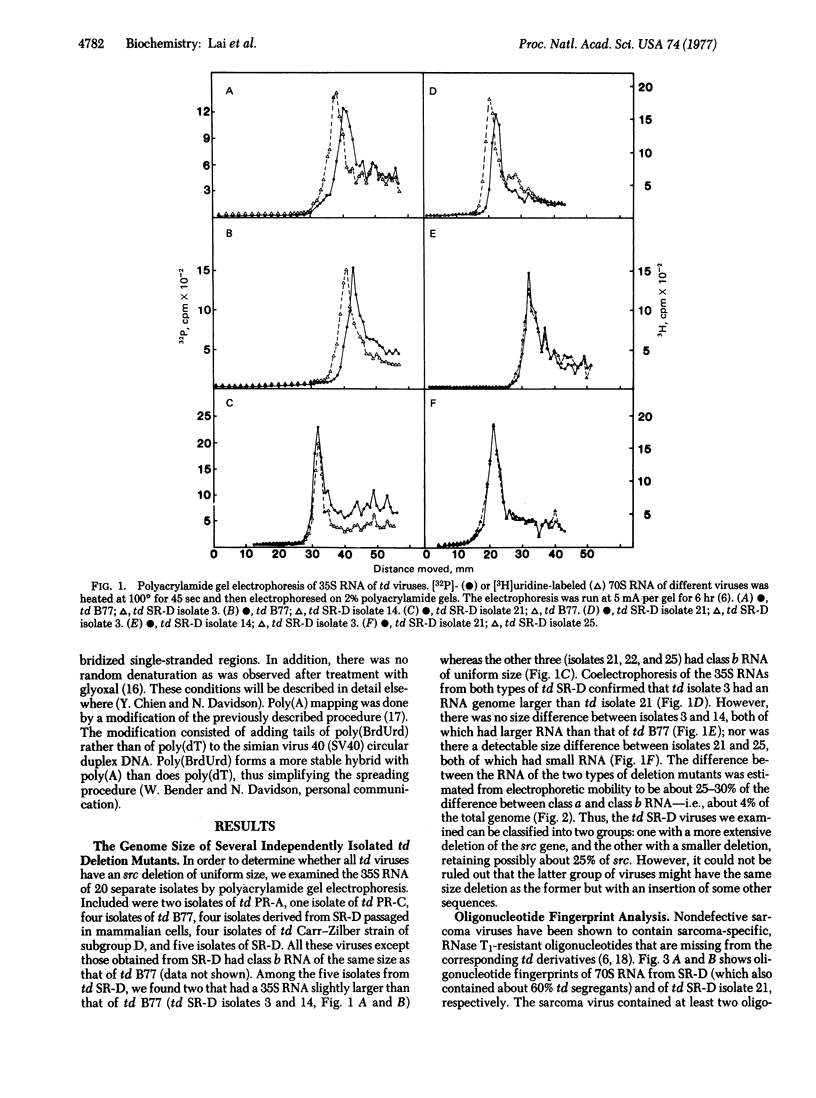
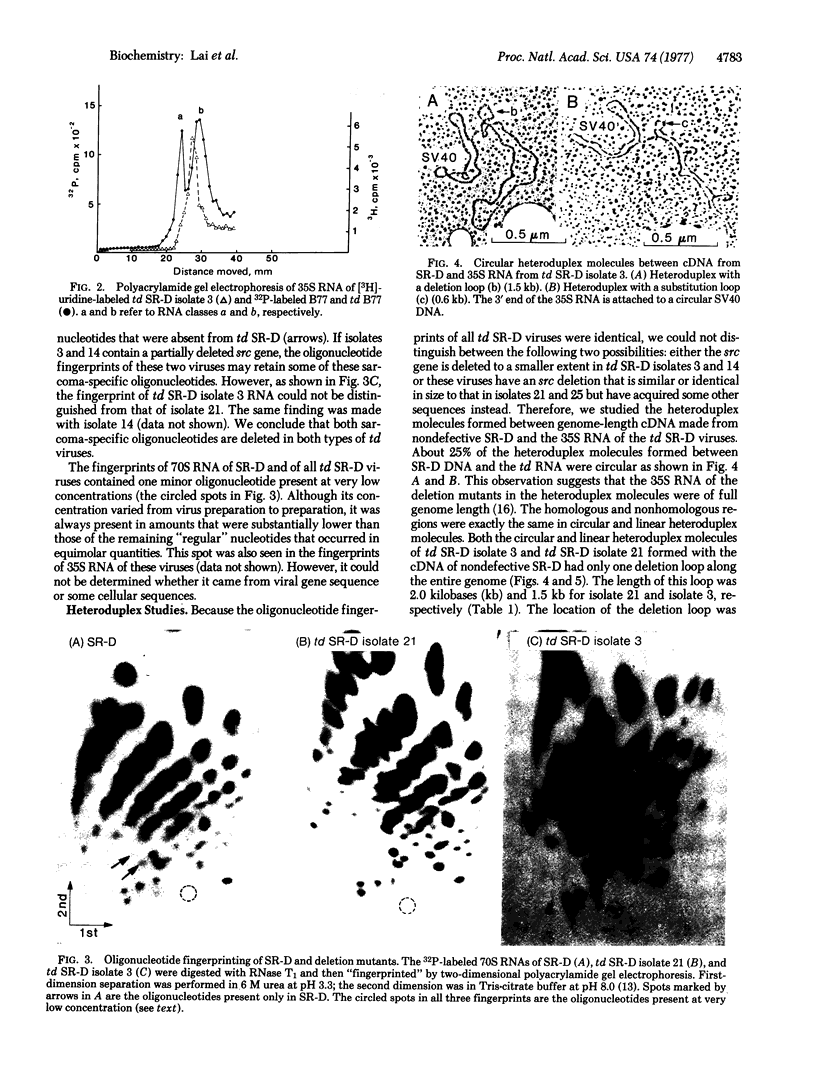
The genome size of 20 transformation-defective (td) viruses derived from different strains of Rous sarcoma viruses [Prague (subgroups A and C), Schmidt-Ruppin (subgroups A and D) (SR-D), Bratislava 77, and Carr-Zilber subgroup D)] was examined by polyacrylamide gel electrophoresis. All of the td viruses except td SR-D have 35S RNA of the same size—i.e., class b RNA. Two of five td SR-D viruses examined have a slightly larger RNA, corresponding to a td deletion that is about 25% smaller than that of class b RNA. However, the RNase T1-oligonucleotide fingerprints of all the td SR-D viruses are identical, lacking two sarcoma-specific oligonucleotides. The fingerprints of these viruses also showed a minor oligonucleotide present at very low concentration. A study of heteroduplex molecules formed between genome-length cDNA made from wild-type SR-D and 35S RNA of td SR-D showed a deletion loop of 2.0 and 1.5 kilobases, respectively, at the map position of the src gene for these two classes of td SR-D viruses, confirming the results of polyacrylamide gel electrophoresis. In addition, some heteroduplex molecules with a substitution loop of 0.6-0.7 kilobase at the same site as the deletion loop were observed in all five of the td SR-D viruses. We conclude that some of the td SR-D viruses have a partially deleted src gene and that all of the td SR-D viruses have incorporated heterologous sequences of distinct length in some RNA molecules at the position of the src gene. The nature and origin of these heterologous sequences are discussed.
Keywords: deletion mutant, 35S RNA, polyacrylamide gel electrophoresis, oligonucleotide mapping, heteroduplex
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender W., Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976 Apr;7(4):595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- Bernstein A., MacCormick R., Martin G. S. Transformation-defective mutants of avian sarcoma viruses: the genetic relationship between conditional and nonconditional mutants. Virology. 1976 Mar;70(1):206–209. doi: 10.1016/0042-6822(76)90254-3. [DOI] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P. H., Vogt P. K. Differences between the ribonucleic acids of transforming and nontransforming avian tumor viruses. Proc Natl Acad Sci U S A. 1970 Dec;67(4):1673–1680. doi: 10.1073/pnas.67.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P. H., Vogt P. K. Gel electrophoresis of avian leukosis and sarcoma viral RNA in formamide: comparison with other viral and cellular RNA species. J Virol. 1973 Sep;12(3):594–599. doi: 10.1128/jvi.12.3.594-599.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P. H., Vogt P. K. RNA species obtained from clonal lines of avian sarcoma and from avian leukosis virus. Virology. 1973 Jul;54(1):207–219. doi: 10.1016/0042-6822(73)90130-x. [DOI] [PubMed] [Google Scholar]
- Graf T., Bauer H., Gelderblom H., Bolognesi D. P. Studies on the reproductive and cell-converting abilities of avian sarcoma viruses. Virology. 1971 Feb;43(2):427–441. doi: 10.1016/0042-6822(71)90315-1. [DOI] [PubMed] [Google Scholar]
- Junghans R. P., Duesberg P. H., Knight C. A. In vitro synthesis of full-length DNA transcripts of Rous sarcoma virus RNA by viral DNA polymerase. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4895–4899. doi: 10.1073/pnas.72.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans R. P., Hu S., Knight C. A., Davidson N. Heteroduplex analysis of avian RNA tumor viruses. Proc Natl Acad Sci U S A. 1977 Feb;74(2):477–481. doi: 10.1073/pnas.74.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S., Hanafusa H. Genetic recombination with avian tumor virus. Virology. 1972 Jul;49(1):37–44. doi: 10.1016/s0042-6822(72)80005-9. [DOI] [PubMed] [Google Scholar]
- Lai M. M., Duesberg P. H., Horst J., Vogt P. K. Avian tumor virus RNA: a comparison of three sarcoma viruses and their transformation-defective derivatives by oligonucleotide fingerprinting and DNA-RNA hybridization. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2266–2270. doi: 10.1073/pnas.70.8.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. F., Wimmer E. "Fingerprinting" high molecular weight RNA by two-dimensional gel electrophoresis: application to poliovirus RNA. Nucleic Acids Res. 1976 Jul;3(7):1647–1658. doi: 10.1093/nar/3.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. S., Duesberg P. H. The a subunit in the RNA of transforming avian tumor viruses. I. Occurrence in different virus strains. II. Spontaneous loss resulting in nontransforming variants. Virology. 1972 Feb;47(2):494–497. doi: 10.1016/0042-6822(72)90287-5. [DOI] [PubMed] [Google Scholar]
- Neiman P. E., Wright S. E., McMillin C., MacDonnell D. Nucleotide sequence relationships of avian RNA tumor viruses: measurement of the deletion in a transformation-defective mutant of Rous sarcoma virus. J Virol. 1974 Apr;13(4):837–846. doi: 10.1128/jvi.13.4.837-846.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif H. J., Saedler H. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol Gen Genet. 1975;137(1):17–28. doi: 10.1007/BF00332538. [DOI] [PubMed] [Google Scholar]
- Starlinger P., Saedler H. IS-elements in microorganisms. Curr Top Microbiol Immunol. 1976;75:111–152. doi: 10.1007/978-3-642-66530-1_4. [DOI] [PubMed] [Google Scholar]
- Stone M. P., Smith R. E., Joklik W. K. 35S a and b RNA subunits of avian RNA tumor virus strains cloned and passaged in chick and duck cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):859–868. doi: 10.1101/sqb.1974.039.01.100. [DOI] [PubMed] [Google Scholar]
- Vogt P. K., Ishizaki R. Reciprocal patterns of genetic resistance to avian tumor viruses in two lines of chickens. Virology. 1965 Aug;26(4):664–672. doi: 10.1016/0042-6822(65)90329-6. [DOI] [PubMed] [Google Scholar]
- Vogt P. K. Spontaneous segregation of nontransforming viruses from cloned sarcoma viruses. Virology. 1971 Dec;46(3):939–946. doi: 10.1016/0042-6822(71)90092-4. [DOI] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P. H., Kawai S., Hanafusa H. Location of envelope-specific and sarcoma-specific oligonucleotides on RNA of Schmidt-Ruppin Rous sarcoma virus. Proc Natl Acad Sci U S A. 1976 Feb;73(2):447–451. doi: 10.1073/pnas.73.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P., Beemon K., Vogt P. K. Mapping RNase T1-resistant oligonucleotides of avian tumor virus RNAs: sarcoma-specific oligonucleotides are near the poly(A) end and oligonucleotides common to sarcoma and transformation-defective viruses are at the poly(A) end. J Virol. 1975 Oct;16(4):1051–1070. doi: 10.1128/jvi.16.4.1051-1070.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P., Mellon P., Vogt P. K. Distribution of envelope-specific and sarcoma-specific nucleotide sequences from different parents in the RNAs of avian tumor virus recombinants. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1073–1077. doi: 10.1073/pnas.73.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]





