Abstract
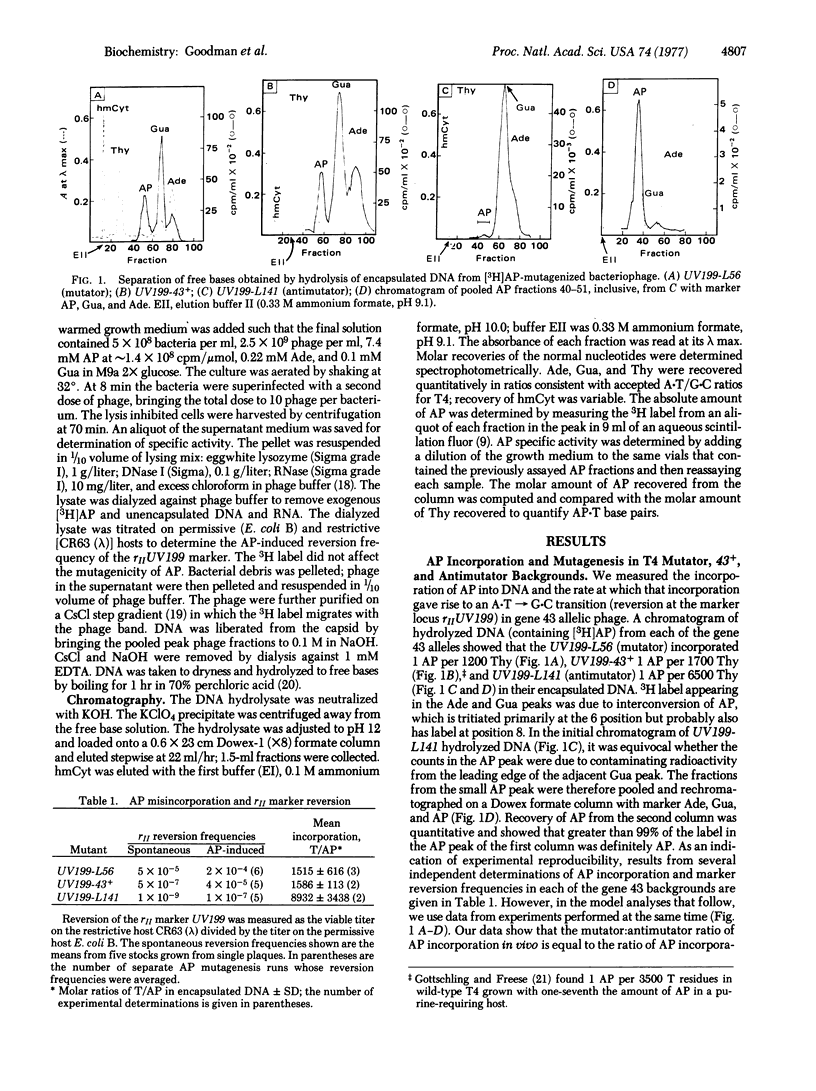
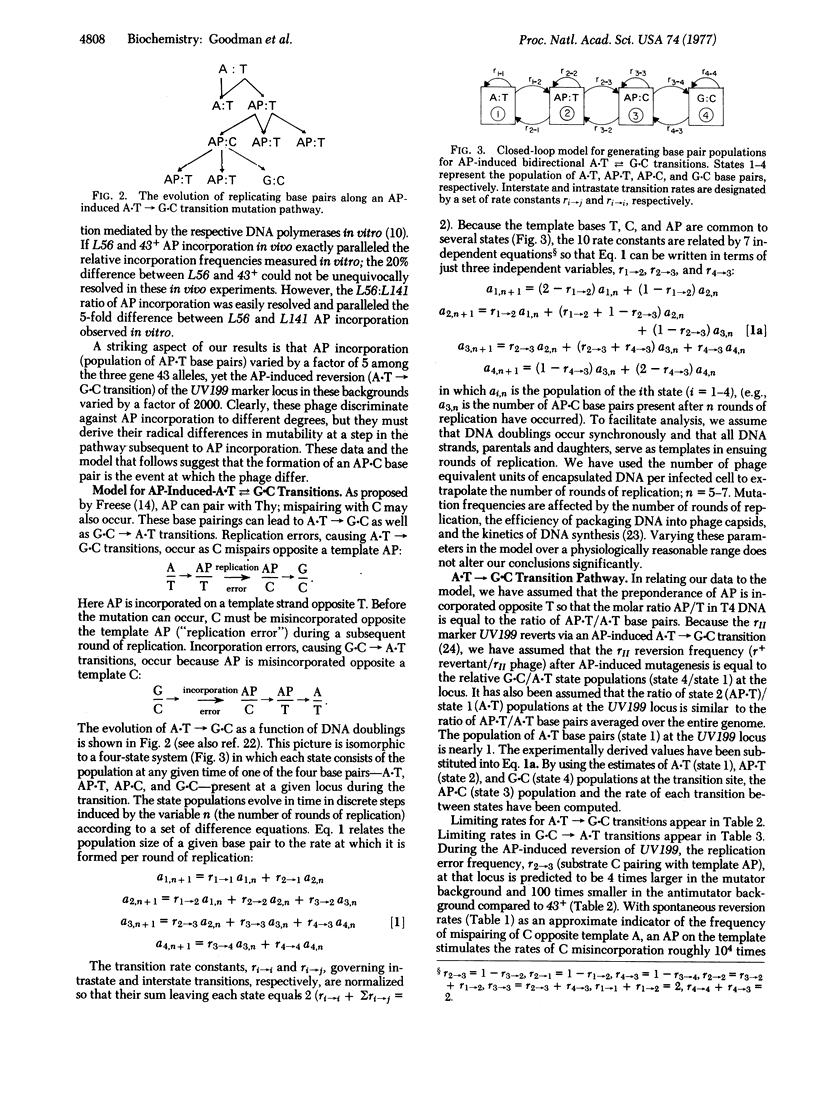
We measured the in vivo incorporation of 2-aminopurine into DNA of T4 bacteriophage allelic for gene 43 (DNA polymerase), mutator (L56), 43+, and antimutator (L141). The magnitude of incorporation (mol/mol of Thy) was 1/1500 in L56, 1/1600 in 43+, and 1/8900 in L141. The incorporation ratio L56:43+:L141 in vivo was equal to that mediated by the purified DNA polymerases of these allelic phages in vitro. A model for 2-aminopurine-induced A-T in equilibrium G-C transitions is discussed. The model is used to predict the magnitudes of replication errors (C mispairing with a template 2-aminopurine) and incorporation errors (2-aminopurine mispairing with a template C) per round of replication and to investigate the asymmetry in 2-aminopurine-induced transitions favoring the A-T leads to G-C pathway over G-C leads to A-T. We suggest that the fidelity of L56 and L141 DNA polymerases exemplifies one-step and two-step editing, respectively.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen E. F., Albrecht I., Drake J. W. Properties of bacteriophage T4 mutants defective in DNA polymerase. Genetics. 1970 Jun;65(2):187–200. doi: 10.1093/genetics/65.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman M. J., Muzyczka N., Goodman M. F., Schnaar R. L. Studies on the biochemical basis of spontaneous mutation. II. The incorporation of a base and its analogue into DNA by wild-type, mutator and antimutator DNA polymerases. J Mol Biol. 1974 Sep 15;88(2):409–421. doi: 10.1016/0022-2836(74)90491-4. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3' leads to 5' exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972 Jan 10;247(1):241–248. [PubMed] [Google Scholar]
- CHAMPE S. P., BENZER S. Reversal of mutant phenotypes by 5-fluorouracil: an approach to nucleotide sequences in messenger-RNA. Proc Natl Acad Sci U S A. 1962 Apr 15;48:532–546. doi: 10.1073/pnas.48.4.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Waard A., Paul A. V., Lehman I. R. The structural gene for deoxyribonucleic acid polymerase in bacteriophages T4 and T5. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1241–1248. doi: 10.1073/pnas.54.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W., Allen E. F. Antimutagenic DNA polymerases of bacteriophage T4. Cold Spring Harb Symp Quant Biol. 1968;33:339–344. doi: 10.1101/sqb.1968.033.01.039. [DOI] [PubMed] [Google Scholar]
- Drake J. W., Allen E. F., Forsberg S. A., Preparata R. M., Greening E. O. Genetic control of mutation rates in bacteriophageT4. Nature. 1969 Mar 22;221(5186):1128–1132. [PubMed] [Google Scholar]
- Freese E. B., Freese E. On the specificity of DNA polymerase. Proc Natl Acad Sci U S A. 1967 Mar;57(3):650–657. doi: 10.1073/pnas.57.3.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese E. B. The mutagenic effect of hydroxyaminopurine derivatives on phage T4. Mutat Res. 1968 Mar-Apr;5(2):299–301. doi: 10.1016/0027-5107(68)90028-6. [DOI] [PubMed] [Google Scholar]
- Hershfield M. S. On the role of deoxyribonucleic acid polymerase in determining mutation rates. Characterization of the defect in the T4 deoxyribonucleic acid polymerase caused by the ts L88 mutation. J Biol Chem. 1973 Feb 25;248(4):1417–1423. [PubMed] [Google Scholar]
- Hopfield J. J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch R. E., McGaw M. K., Drake J. W. Mutator mutations in bacteriophage T4 gene 32 (DNA unwinding protein). J Virol. 1976 Aug;19(2):490–494. doi: 10.1128/jvi.19.2.490-494.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch R. E. The influence of neighboring base pairs upon base-pair substitution mutation rates. Proc Natl Acad Sci U S A. 1971 Apr;68(4):773–776. doi: 10.1073/pnas.68.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzyczka N., Poland R. L., Bessman M. J. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J Biol Chem. 1972 Nov 25;247(22):7116–7122. [PubMed] [Google Scholar]
- Osborn M., Person S., Phillips S., Funk F. A determination of mutagen specificity in bacteria using nonsense mutants of bacteriophage T4. J Mol Biol. 1967 Jun 28;26(3):437–447. doi: 10.1016/0022-2836(67)90314-2. [DOI] [PubMed] [Google Scholar]
- RUDNER R. Mutation as an error in base pairing. Biochem Biophys Res Commun. 1960 Sep;3:275–280. doi: 10.1016/0006-291x(60)90239-4. [DOI] [PubMed] [Google Scholar]
- Speyer J. F., Karam J. D., Lenny A. B. On the role of DNA polymerase in base selection. Cold Spring Harb Symp Quant Biol. 1966;31:693–697. doi: 10.1101/sqb.1966.031.01.088. [DOI] [PubMed] [Google Scholar]
- Speyer J. F., Rosenberg D. The function of T4 DNA polymerase. Cold Spring Harb Symp Quant Biol. 1968;33:345–350. doi: 10.1101/sqb.1968.033.01.040. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Complementary base pairing and the origin of substitution mutations. Nature. 1976 Sep 23;263(5575):285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]


