Abstract
Background
Chinese cabbage is an important leaf vegetable that experienced long-term cultivation and artificial selection. Dof (DNA-binding One Zinc Finger) transcription factors, with a highly conserved Dof domain, are members of a major plant-specific transcription factor family that play important roles in many plant biological processes. The Dof family transcription factors, one of the most important families of transcriptional regulators in higher plants, are involved in massive aspects of plant growth, development, and response to abiotic stresses. Our study will supply resources for understanding how Dof transcription factors respond to abiotic stress and the interaction network of these genes in tolerance mechanism.
Results
In this study, we performed a comprehensive analysis of Dof family factors in Chinese cabbage. In total, 76 genes encoding BraDof family transcription factor were identified from Chinese cabbage, and those BraDof factors were divided into nine classes. Fifteen motifs were found based on Dof amino acid sequence alignments. Chromosome locations and gene duplications of BraDof family genes were also analyzed. Ten duplicate events of BraDof genes were discovered in Chinese cabbage chromosomes. The uneven distribution of BraDof genes in Brassica chromosomes may cause the expansion of BraDof genes. In the Dof family, 37 and 7 orthologous genes were identified between Chinese cabbage and Arabidopsis and between Chinese cabbage and Oryza sativa, respectively. The interaction networks of Dof factors in Chinese cabbage were also constructed. Expression profiles of nine selected genes from different nine classes subjected to four abiotic stresses (cold, heat, salt and drought) were further investigated by quantitative real-time PCR to obtain a better understanding of the functions and regulation mechanisms of BraDof family transcription factors in two Chinese cabbage varieties, ‘Lubaisanhao’ and ‘Qingdao 87-114’.
Conclusions
Dof-family transcription factors were analyzed in genome of Chinese cabbage. Chromosomal locations showed that duplication might result in expansion. Response to abiotic stresses was elucidated in Chinese cabbage varieties. The results provide novel insights into the stress responses of BraDof genes and promote a better understanding of the construction and function of Dofs in Chinese cabbage.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1242-9) contains supplementary material, which is available to authorized users.
Keywords: Dof, Transcription factor, Chromosomal location, Gene duplication, qRT-PCR, Chinese cabbage
Background
Chinese cabbage (Brassica rapa L. ssp. pekinensis), an economically important vegetable in Asia, belongs to the family Cruciferae, species Brassica [1,2]. ‘Lubaisanhao’ and ‘Qingdao 87-114’ are two important Chinese cabbage varieties that share several identical traits, such as high yield, good quality, and disease resistance. In green-house condition, ‘Lubaisanhao’ grow better than ‘Qingdao 87-114’ in lower temperature condition, however, ‘Qingdao 87-114’ showed better resistance to high temperature in summer. ‘Lubaisanhao’ can be planted in relatively low temperature environments, while ‘Qingdao 87-114’ can grow in Nanjing District, southern China with heat tolerance. ‘Lubaisanhao’ suit cooler and wetter condition during seedling stage, however, ‘Qingdao 87-114’ can fit hotter temperature environment [3].
Plants grow in complicated environments and survive many stresses, such as cold, heat, drought, and soil salinization. These abiotic stresses cause drops in crop production and declines in quality [4,5]. To resist damage, plants must evolve to regulate plant-specific signals by gene expression for specific physiological and metabolic pathways [6,7]. Transcription factors are a special type of regulators with highly conserved specific DBDs (DNA-binding domains) involved in stress resistance [5,8].
Dof (DNA-binding One Zinc Finger) transcription factors, a family of plant-specific transcription factors, play important roles in many fundamental processes in higher plants, such as photosynthesis, stress responses, seed germination, flower induction, and light-mediated circadian rhythms [9-12]. Dof transcription factors generally contain 200–400 amino acids with a highly conserved 52-amino acid Dof domain [10,13]. The Dof domain is located on the N-terminus, which has been identified as a DNA-binding domain, and features the structure of a Cys2/Cys2 Zn2+ finger recognizing a cis-regulatory element comprising the common core sequence (AT)/AAAG [10,13-17]. The DNA-binding domain is a critical region regarded as a bi-functional domain that combines with DNA and interacts with other proteins [10,14,18]. A transcriptional regulation domain at the C-terminal region may perform diverse functions because of its interactions with different regulatory proteins and activation of gene expression [10,14,19,20].
Many studies have demonstrated that Dof transcription factors are involved in plant growth and development, seed germination, photosynthesis, and biotic/abiotic stress responses in many species, but little is known in Chinese cabbage. Till now, the abiotic stress responses of BraDof genes in Chinese cabbage are unknown. The objective of this study was to establish an extensive picture of the BraDof family transcription factors in Chinese cabbage.
In this study, we identified 76 BraDof members in Chinese cabbage based on genome sequences and divided them into nine classes. The proportions of these nine classes of BraDof family genes, gene duplications, and chromosomal locations were also investigated for further study. Expression profiles under four stress treatments (cold, heat, salt, and drought) were evaluated to determine the responses of BraDof genes to abiotic stresses in two varieties of Chinese cabbage. The results will provide novel insights into the stress responses of BraDof genes and promote a better understanding of the construction and function of Dofs in Chinese cabbage.
Results
Identification and analysis of BraDof transcription factors
To identify and analyze BraDof factors, we identified the BraDof members in Chinese cabbage based on genome sequences. A total of 76 genes encoding BraDof family transcription factors were identified from genome sequence analysis in Chinese cabbage (Additional file 1: Tables S1 to S11). The BraDof family factors were divided into nine classes on the basis of the predicted Dof domains as follows: Classes A, B1, B2, C1, C2.1, C2.2, C3, D1, and D2. Class D1 showed the largest number of BraDof factors (18.42%), whereas Class C2.2 showed the least number of BraDof family factors (5.26%) (Figure 1). These different 9 classes (A, B1, B2, C1, C2.1, C2.2, C3, D1, and D2) represent each sub-family of Dof family transcription factors, and the 9 classes we classified based on the predicted Dof domains analogize by the phylogenetic tree by using 46 A. thaliana Dof genes as references [9,10,21] . The proportion of the nine classes is not even. Two sets of classes shared the same proportions: Classes A and B2 showed individual proportions of 10.53% (eight members) and Classes C1 and D2 showed individual proportions of 7.89% (six members) (Figure 1).
Figure 1.
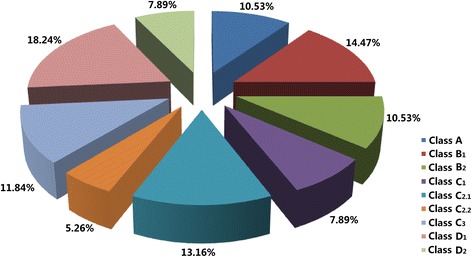
Proportions of various BraDof classes in Chinese cabbage. The nine different colors indicate the nine BraDof classes of Chinese cabbage.
Characterization of the deduced amino acid sequences of BraDofs
Most BraDofs share the similar physical and chemical characteristics, but there were several exceptions. ExPASy, Sequence Manipulation Suite, and RPSP were used to analyze the physical and chemical characteristics of BraDofs from Chinese cabbage; the data are summarized in Additional file 1: Table S12. The theoretical pI of Class B1 was about 9, and only one BraDof factor (BraDof028) showed a theoretical pI of 8.77. The results in theoretical pI value were also observed in Class B2, however, this class had a pI of 10.14. Classes A and C2.1 showed theoretical pI values of 7–10. Class C2.2 yielded the lowest theoretical pI of 5. All other classes had complex theoretical pI ranging in value from 5 to 10. The percentage of aliphatic amino acids in all of the BraDofs was slightly higher than the percentage of aromatic amino acids. The percentage of positive amino acids was twice as large as the percentage of negative amino acids. The proportions of aliphatic and positive amino acids in most of the BraDofs were > 10%, contrasting with the proportions of aromatic and negative amino acids, which were < 10%.
Theoretical predictions of protein solubility are important in determining the structure and stability of a protein. In total, the percentage of insoluble recombinant proteins of most of the BraDofs was > 97%. The percentage of insoluble recombinant proteins in all of the BraDofs of Classes B1, C1, and D2 was > 97%. By contrast, the percentage of insoluble recombinant proteins of some BraDofs of Classes A, B2, and C2.1 was < 97%. Class C2.2 showed a percentage of insoluble recombinant proteins relatively lower than those of other classes (<90%). The percentages of insoluble recombinant proteins in Classes C3 and D1 ranged from 50 to > 97%.
Motif location and phylogenetic relationship analysis of BraDofs
Most of the 76 BraDof family proteins in Chinese cabbage classified into nine classes have 15 motifs. A total of 76 BraDof family proteins were analyzed by multiple sequence alignment of their Dof domains. All 76 BraDof family proteins had a very highly conserved zf-Dof domain. MEME Suite was used to identify the motifs of the amino acid sequences of the BraDof proteins, and 15 motifs were identified (Figures 2 and 3). All of the BraDofs contained motifs 1 and 2, which suggests that all BraDofs have a highly conserved domain. Moreover, each class of BraDofs had several special motifs at their C-terminal regions. One BraDof factor of Class A did not have motif 6, and one BraDof factor of Class D2.1 did not have motifs 13 and 14. Similar motifs on the C-terminal regions determine the phylogenetic relationship of BraDofs.
Figure 2.
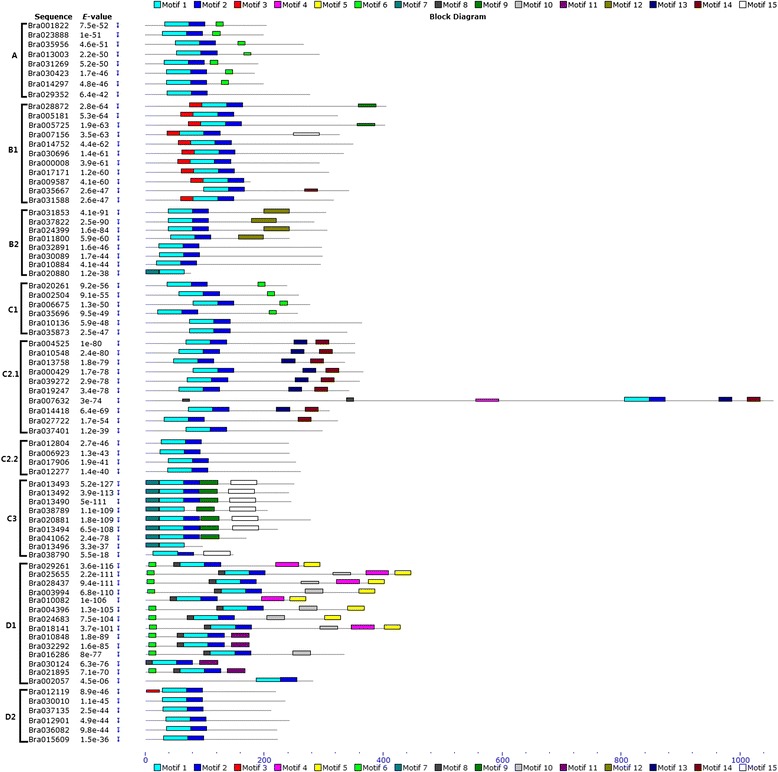
Common motifs of BraDof family proteins in Chinese cabbage. Dof domains are represented by striped boxes.
Figure 3.
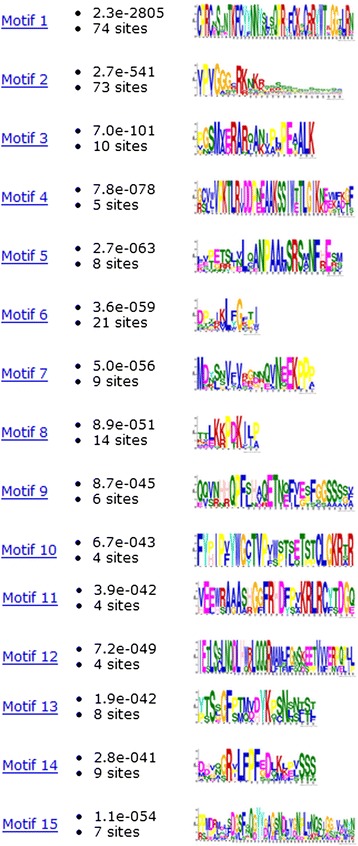
Sequence logos of Dof domains in Chinese cabbage. The overall height of the stack indicates the level of sequence conservation. Heights of residues within a stack indicate the relative frequency of each residue at that position.
To explain the phylogenetic relationships of the BraDof family transcription factors, a neighbor-joining tree was constructed for phylogenetic analysis to classify the 76 BraDof transcription factors into nine classes, based on the full-length amino acid sequences of BraDofs from Chinese cabbage and AtDofs from Arabidopsis [21] (Figure 4). These AtDofs’ amino acid sequences are respectively most similar to BraDofs’ from the same class.
Figure 4.
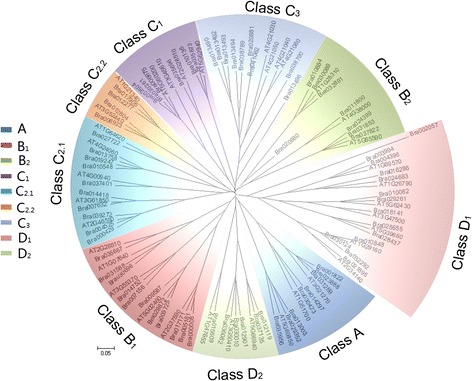
Unrooted phylogenetic tree of BraDofs in Chinese cabbage. The amino acid sequences of the DBD were aligned using Clustal W, and the phylogenetic tree was constructed using MEGA 5.
The evolution of BraDofs transcription factors among plant species
To research the evolution of BraDofs in plant species, comparisons of Dof transcription factors in different plant species were constructed. Compared with other species, Chinese cabbage has a relatively big Dof family, with the 76 Dof members. Except soy and Chinese cabbage, the numbers of Dof members in other higher plants are almost ranged from 30 to 50. Only one or two members were identified in each algae, and there were all belonged to Class D1. Moreover, Class D1 is also the biggest class in many plants (Figure 5). A total of 76 BraDof family factors were identified in Chinese cabbage, and only 47 AtDof factors was found in Arabidopsis. The genome sizes of Chinese cabbage and Arabidopsis were 485 Mb and 125 Mb, respectively. The Dof factors density decreased from 0.376/Mb to 0.157/Mb in Arabidopsis and Chinese cabbage. The results were consistent with the report by Mun et al. [22].
Figure 5.
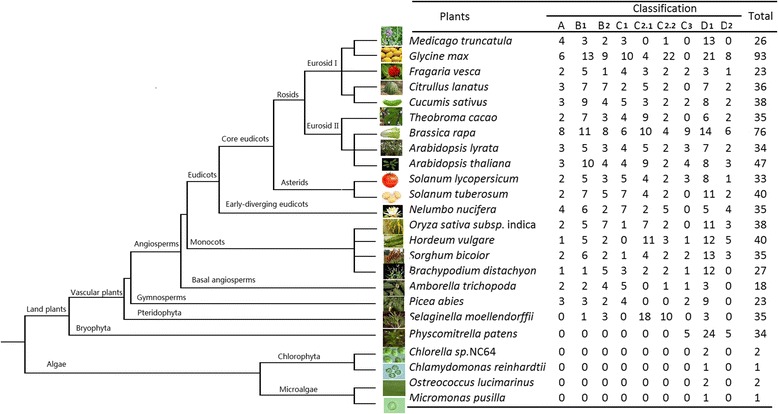
Comparisons of Dof transcription factors in different species.
Location and duplication of BraDof genes in Chinese cabbage chromosomes
To determine the genomic distribution of BraDof genes, the DNA sequence of each BraDof gene was used to search the Chinese cabbage genome database using BLASTN. Although each of the 10 Chinese cabbage chromosomes contained several BraDof genes, their distributions seemed uneven, and a single BraDof gene was located on Scaffold 000435 (Figure 6). Fourteen BraDof genes were located on chromosome 01; the largest number of BraDof genes was observed in this chromosome. Chromosomes 05 and 10 had only 4 BraDof genes each, showing the fewest BraDof genes among all of the chromosomes studied. Chromosomes A04 and A08 had 8 BraDof genes. No BraDof genes were found on the short arms of chromosomes A03 and A07, but 11 and 6 BraDofs were respectively found on their long arms. Several gene clusters, such as BraDof 002–006 on A01 and BraDof 025 and BraDof 026 on A03, were also found on the chromosomes of Chinese cabbage. Interestingly, all of these genes belonged to Class C3.
Figure 6.
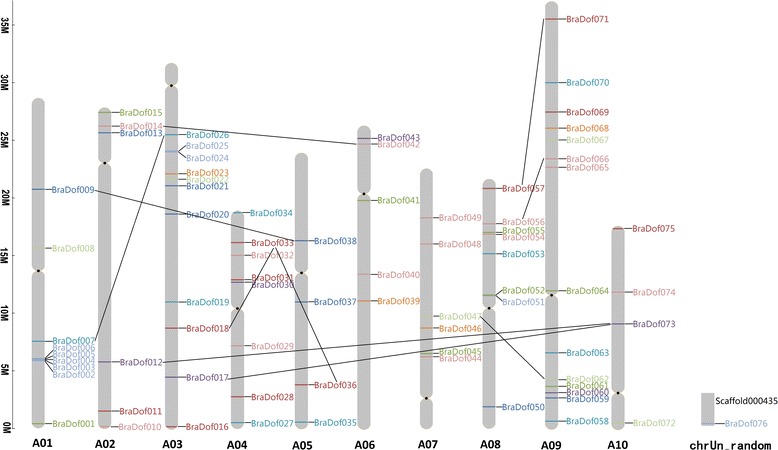
Chromosomal locations and predicted clusters of BraDof genes in Chinese cabbage.
Potential duplicated genes were marked on the 10 chromosomes and Scaffold 000435 according to the synonymous substitution rate and synteny data (Additional file 1: Tables S13 and S14). Ten duplication events were predicted on these chromosomes, and these duplication events occurred in six classes of genes, including Classes A, B1, C1, C2.1, D1, and D2 (Figure 6). None of the potential duplications were intra chromosomal duplication events, and all of the duplications occurred on two different chromosomes. Furthermore, the duplicate genes belonged to the same classes. Two special genes duplicating with two genes were discovered: BraDof033, which was classified in Class B1, duplicated with BraDof018 and BraDof036, and BraDof073, which belonged to Class C1, duplicated with BraDof012 and BraDof017.
Now, the whole genome A. thaliana and Chinese cabbage were analyzed very deeply and thoroughly, and found that Chinese cabbage has undergone triplication events since its divergence from A. thaliana [23]. Moreover, A. thaliana and O. sativa are two model plants, which were belonged to dicotyledon and monocotyledon, respectively. Therefore, we constructed two comparative syntenic maps of Chinese cabbage associated with A. thaliana and O. sativa, respectively (Figure 7). Thirty-seven and seven pairs of orthologous Dof genes were found between BraDofs and AtDofs and between BraDofs and OsDofs, respectively. And there were nine and thirty paralogs located in genome of O. sativa and Chinese cabbage, respectively. In addition, thirteen and ten pairs of corthologous Dof genes were investigated in the Chinese cabbage genome and O. sativa genome (Figure 7).
Figure 7.
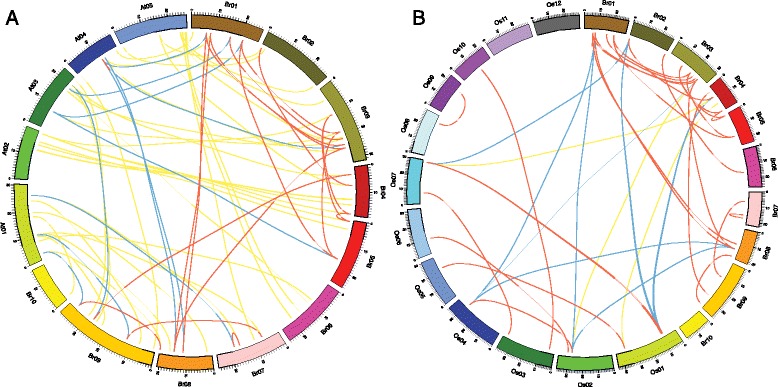
Gene duplication and synteny analysis of Dof genes between Chinese cabbage and two other model plant species. Ten Chinese cabbage (A01 to A10), five Arabidopsis chromosome (Chr1 to Chr5) and twelve rice (Os1 to Os12) maps were based on the orthologous and paralogous pair positions, and demonstrate highly conserved synteny. The red lines represent the orthologous Dof genes between Chinese cabbage and two other model plant species. (A) Gene duplication and synteny analysis of Dof genes between Chinese cabbage and Arabidopsis. The green and blue lines represent the paralogous Dof genes in Chinese cabbage and Arabidopsis, respectively. Colored lines connecting two chromosomal regions denote syntenic regions of genome. (B) Gene duplication and synteny analysis of Dof genes between Chinese cabbage and O. sativa. The green and blue lines represent the paralogous Dof genes in Chinese cabbage and O. sativa, respectively. Colored lines connecting two chromosomal regions denote syntenic regions of genome.
The interaction network of BraDof genes
To further research the BraDof genes how to interact with other genes, an interaction network associated with AtDofs Arabidopsis orthologs was built according to BraDof genes from Chinese cabbage (Figure 8). The red, purple and green lines stand for positive correlation (Pearson correlation coefficient >0) with seventy-one pairs of interacting genes, negative correlation (Pearson correlation coefficient <0) with sixteen pairs of interacting genes and unclear correlation (Pearson correlation coefficient not calculated) with sixteen pairs of interacting genes. The interaction network of BraDofs showed a very complicated correlation with other genes in Chinese cabbage, which may indicate BraDof genes involve in many fundamental mechanisms by regulating many downstream factors or being regulated by many upstream genes.
Figure 8.
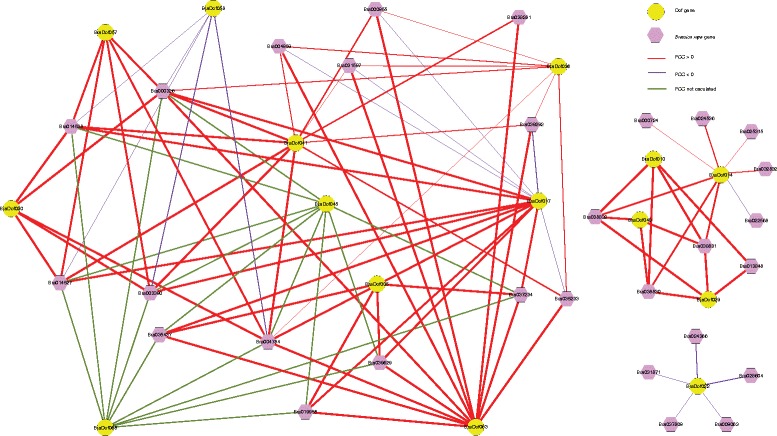
The interaction networks of Dofs in Chinese cabbage according to the orthologs in Arabidopsis . The PCC represents the Pearson Correlation Coefficient.
Expression analysis of BraDof genes under stress treatments in two Chinese cabbage varieties
To further investigate the functions of BraDof genes correlated with their expression, 9 BraDof genes were subjected to qRT-PCR to examine the expression profiles of BraDof genes under cold, heat, salt, and drought treatments in ‘Lubaisanhao’ and ‘Qingdao 87-114’. The 9 BraDof genes were selected from the different 9 subgroups on behalf of these 9 subgroups to do qRT-PCR for expression analysis. The 9 genes were representative and can explain the expression profiles of genes from 9 different classes. Each of these 9 genes having a highly conserved Dof domain should be more similar to their homologous genes, and the efficiency of the primers based on their 3’ terminal sequences are better.
While most of the BraDof genes were up regulated under the four stress treatments (Figures 9 and 10), some notable exceptions were observed. The BraDof072 gene was down regulated by cold, heat, and drought stress treatments in ‘Lubaisanhao’ and by salt stress treatment in ‘Qingdao87-114’. In addition, BraDof048 was down regulated in ‘Lubaisanhao’ by heat treatment, and BraDof003 and BraDof074 were down regulated in ‘Qingdao87-114’ by salt treatment. BraDof023 in ‘Qingdao87-114’ was initially down regulated by heat and salt stress treatments and then up regulated after 4 h. The gene of BraDof074 showed complicated expression. The gene was down regulated in ‘Qingdao87-114’ by salt treatment. However, in ‘Lubaisanhao’, the gene was first down regulated by cold stress treatment and then up regulated later on. Under heat conditions, opposite findings were observed (Figure 9).
Figure 9.
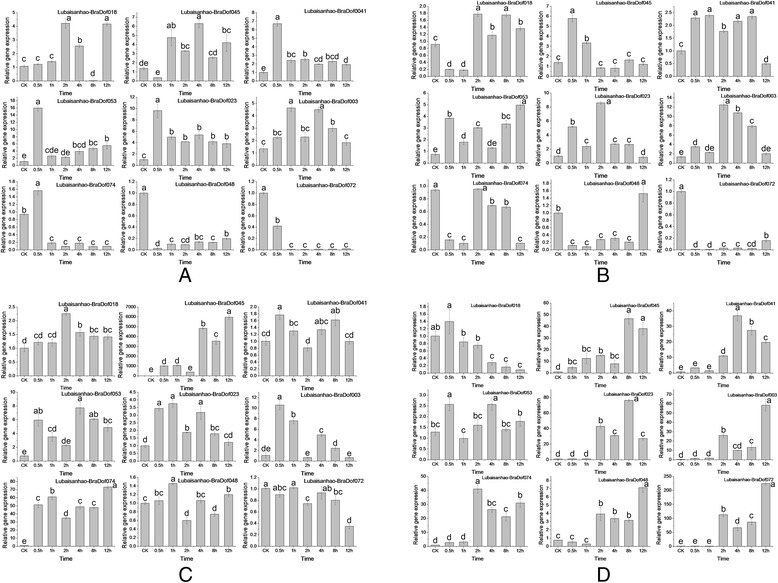
Expression patterns of BraDof genes in the Chinese cabbage variety ‘Lubaisanhao’ under heat, cold, salt, and drought treatments. (A). Expression patterns at 38°C; (B). Expression patterns at 4°C; (C). Expression patterns under salt treatment; (D). Expression patterns under drought treatment. Different letters indicate significant differences between different time treatments stages in the same cultivar (P < 0.05).
Figure 10.
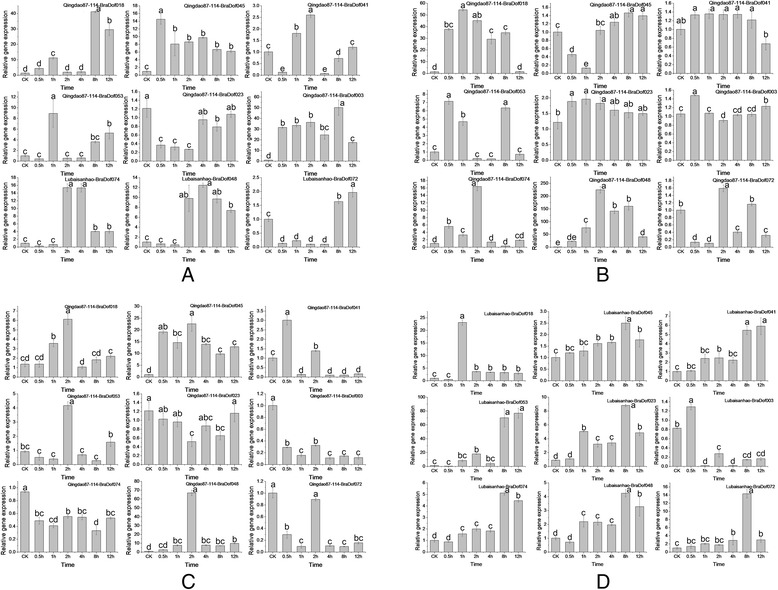
Expression patterns of BraDof genes in the Chinese cabbage variety ‘Qingdao87-114’ under heat, cold, salt, and drought treatments. (A). Expression patterns at 38°C; (B). Expression patterns at 4°C; (C). Expression patterns under salt treatment; (D). Expression patterns under drought treatment. Different letters indicate significant differences between different time treatments stages in the same cultivar (P < 0.05).
Discussion
Overall Dof gene families in maize, Arabidopsis, barley, wheat and rice
The gene encoding Dof protein, named ZmDof1, was first isolated in maize and found to be involved in C4 photosynthesis [24]. A total of 30 Dof family genes in rice, 36 genes in Arabidopsis, 24 genes in barley, 31 genes in wheat, and 54 genes in maize have been identified and analyzed since then [21,25-27]. In higher plants, Dof family transcription factors play important roles in many biological and physiological processes regulating growth, development, defense mechanisms, and flowering in plants. In Arabidopsis, AtDAG1, a phloem-specific transcription factor, is involved in light-quality responses during photomorphogenesis; AtCOG1 and OBF3 function in the same manner [16,28-30]. Another Dof factor in Arabidopsis, cycling DOF factor 1, principally negatively mediates the expression of CONSTANS to control the photoperiodic pathway of flowering time; CDF2 and CDF3 also show this function [13,31]. Overexpression of OsDof12 in transgenic rice promotes early flowering [32], and the OsDof3 gene has been identified to participate in the gibberellin-regulated expression of target genes in rice [33,34]. Several barley Dof family transcription factors, including BPBF (HvDof24), SAD (HvDof23), HvDof 17, and HvDof19, were involved in the regulation of hormonal balance between gibberellin and abscisic acid during seed germination [35-37]. Similar to other zinc fingers, the Dof domain, as a bi-functional domain, regulates both DNA-binding and protein-protein interactions with other regulatory proteins including basic leucine zipper (bZIP), myeloblastosis (MYB) transcription factors and nuclear high mobility group (HMG) proteins [10,13,19,33,35,38-41].
Genome evolution of the Dof family transcription factors in plant
Dof transcription factors were widely identified in many higher plants [10,28,30,35,37]. With further study, some Dof genes were also found in low plants, such as, algaes and moss [25]. A comparison of Dof genes from algaes to higher plants in our study showed that higher plants have far more Dof genes than algaes, which indicating a huge expansion of Dof family transcription factor members occurred in the evolution process of low plants to land plants. Furthermore, only class D1 members exist in algaes inferred that the other Dof classes’ members may originate from class D1 members.
The genome of Chinese cabbage has undergone triplication events since its divergence from A. thaliana [23]. The genome size of Chinese cabbage (485 Mb) is more than three times larger than that of Arabidopsis (125 Mb), while the number of BraDof transcription factors (76 members) is only less than twice of Arabidopsis (47 members). This outcome may result from alternative actions during the triplicate evolution. The number of Dof members varies among land plants [20], potato contains a great amount of family members of Dof transcription factors (93 members), notably more than that of lower plants, such as in algae [10,14,42], only one Dof factor was found in the green alga C. reinhardtii and no identifiable Dof factor in the red alga Cyanidioschyzon merolae. This result demonstrated that the origin of the Dof transcription factors pre-dates the divergence of the green algae and the ancestors of terrestrial plants [25,42]. Dof transcription factors are associated with the evolution of plants [42,43].
Gene duplication of BraDof genes in Chinese cabbage
It is well known that gene or genome duplication events are the primary sources of genetic novelty [43-45]. Gene duplication was the main effect in gene family expansion, the more Dof genes emerged in higher plants indicated the domain duplication was occurred in eukaryotic evolution [43]. Novel Dof genes arise through divergence of duplicated genes after either single gene duplication, segmental duplication, or whole-genome duplication from aglae to angiosperm [23,46]. The triplication event between Chinese cabbage and A. thaliana has revealed that A. thaliana as a main ancestral constituting the current diploid Chinese cabbage. The distribution of the Dof family genes in the whole Chinese cabbage genome likely resulted from segment and whole-genome duplication [23,43]. A diagram of the chromosomal position of BraDof genes was constructed to determine the distribution of BraDof genes in Chinese cabbage. Although each of the 10 Chinese cabbage chromosomes contained several BraDof genes and a single BraDof gene, BraDof076, was located on Scaffold 000435, the distributions of the genes were uneven. Genome duplication has occurred throughout plant evolution [47]. Ten pairs of potential duplicate genes were found in Chinese cabbage chromosomes. Interestingly, each pair of duplicate genes belonged to the same class. Two special genes had duplicate relationships with two other genes: BraDof033 duplicated with BraDof018 and BraDof036 and BraDof073 duplicated with BraDof012 and BraDof017. Several BraDof genes were clustered on chromosomes 01 and 03 of Chinese cabbage, and these clustered genes were all classified in Class C3. These results indicate that the expansion of BraDof genes in Chinese cabbage chromosomes is related to the network and duplication events of BraDof genes of Chinese cabbage. The uneven distribution ofBraDof gene locations on the chromosomes maybe related to the genome duplication of Chinese cabbage.
Moreover, an amount of BraDof-AtDof and BraDof-OsDof gene pairs were identified in several pairs of duplicated genomic patterns. The number of orthologous events of BraDof-AtDof genes is more than the number of BraDof-OsDof genes. Furthermore, pairwise orthologs were found between Arabidopsis and Chinese cabbage (Figure 7). During the evolution of Chinese cabbage from Arabidopsis, triplication events had occurred by whole-genome analysis [23,44]. For this reason, this result may indicate BraDof genes in Chinese cabbage share the similar structure and function with AtDof genes in Arabidopsis. Higher plants have developed complex response mechanisms of gene regulation network and pathway to stress resistance mechanisms, which include biochemical, physiological, cellular and molecular processes [7,48]. When plants are exposed to various abiotic stresses, thousands of transcription factors, including Dof family genes, are activated to adjust the physiological and biochemical pathways [49,50].
The interaction of Dof transcription factors with other factors
Dof proteins have been shown to interact with specific basic leucine zipper (bZIP) proteins, nuclear high mobility group (HMG) proteins, and other Dof proteins [38-40]. Most of the BraDof positively or negatively interact with a wide range of Protein kinase C (PKC) (Figure 8). Furthermore, according to NCBI blast, BraDof 022 was potentially involved in negatively interacting with five other bZIPs (Figure 8), which is similar with previous studies. Some other BraDofs associated with some tolerant factors, such as TPr (translocated promoter region) protein, TCP (named after the first three characterized members: teosinte-branched 1 (tb1) from Zea mays [51], CYCLOIDEA (CYC) from Antirrhinum majus [52] and PROLIFERATION CELL FACTOR 1 and 2 (PCF1 and PCF2) from Oryza sativa [53]) proteins, and so on. Protein kinase C isozymes transduce a massive number of extracellular signals causing generation of the lipid second messenger diacylglycerol (DAG), therefore regulating diverse cellular behaviors, such as survival, growth and proliferation, migration, and apoptosis; consequently, their dysregulation is associated with a plethora of pathophysiologies [54,55]. Moreover, nucleoporin Tpr is a component of the nuclear pore complex (NPC) that localizes exclusively to intranuclear filaments. Tpr functions as a scaffolding element in the nuclear phase of the NPC and plays a role in mitotic spindle checkpoint signaling [56,57]. These genes positively or negatively regulated by BraDofs were involved in abiotic stress tolerance more or less. The BraDofs mediate other factors might be a complicated pathway to respond abiotic stress in Chinese cabbage.
Dof transcription factors response abiotic stress in Chinese cabbage
The Dof family of transcription factors is a large group of factors that are found mainly in plants. In plant, Dof family transcription factors play very important roles in development as well as in hormonal regulation and stress responses. However, little is known about this family in Chinese cabbage [10,14,25,37]. In particular, the abiotic stress response mechanisms of BraDof genes in Chinese cabbage are unknown [58]. Understanding BraDofs functions in Chinese cabbage will help to develop characterize of Chinese cabbage cultivars such as salt and drought tolerance. The expression profiles analysis of large numbers of BraDof genes provides a powerful tool for identifying groups of the BraDof genes and discovering novel regulators involved in the signal transport of abiotic stress responses in Chinese cabbage. The mechanism of the BraDofs’ responses to the four abiotic stresses (cold, heat, high salinity, and drought) also needs further investigation.
The expression profiles of nine selected BraDof genes from nine classes under four stress treatments (salt, drought, heat, and cold) were investigated using qRT-PCR in the Chinese cabbage varieties ‘Qingdao87-114’ and ‘Lubaisanhao’. Most genes showed up-regulated expression profiles under these four stresses in both varieties (Figures 9 and 10). Moreover, up-regulated expressions of these genes were always observed about 2 h after stress treatments, which indicate that BraDof gene expression increases immediately under application of abiotic stresses in Chinese cabbage.
As revealed by qRT-PCR expression profiles analysis, all BraDof genes respond to different abiotic stresses, high salinity, drought, and extreme temperatures with different timing in leaves, indicating that these genes might participate in abiotic stress responses. Corrales and his colleagues demostrated that transgenic Arabidpsis hosting the SlCDF3 gene (a tomato Dof factor) exhibited higher expression levels of abitioc stress-responsive genes under non-stress conditions, like COP15, RD29A and ERD10, which suggested that SlCDFs might function as up-stream regulators in drought and salt stress response pathways [58]. Most of the BraDof genes were up-regulated quickly (before 2 h) by the four stress treatments, although notable exceptions may be observed. Some genes were down-regulated, and the up-regulated expression was not simply up regulated. The results indicated that the defense actions of BraDof genes respond quickly to induce other BraDof family genes, and they also might cause the down regulation of gene expression in activation of plant resistance. Our study demonstrates that BraDof transcription factors play a crucial role in plant resistance to abiotic stresses via a complicated reaction mechanism. With the accumulation or reduction of BraDof genes expression levels, the interacting factors may be regulated to accumulating or reducing for abiotic tolerance. The results provide a better understanding of the functions of BraDof genes in Chinese cabbage.
Conclusions
We performed a comprehensive analysis of Dof family factors in Chinese cabbage. A total of 76 genes encoding BraDof family transcription factor were identified from Chinese cabbage, and those BraDof factors were divided into nine classes: Classes A, B1, B2, C1, C2.1, C2.2, C3, D1 and D2. The proportion of each class of BraDof factors was calculated based on phylogenetic relationships. Ten duplicate events of BraDof genes were discovered in Chinese cabbage chromosomes. The uneven distribution of BraDof genes in Brassica chromosomes may cause the expansion of BraDof genes. Most of the selected BraDof genes were induced by the four abiotic stresses (cold, heat, salt and drought) treatments. In addition, the comparative study of the Dof family factors between Chinese cabbage and other plant species provided valuable information for further function studies of the Dof factors in Chinese cabbage. Our study will support recourses for understanding how BraDof transcription factors respond to abiotic stresses and the interaction of these genes for abiotic stress response mechanism.
Methods
Database searches and analysis for BraDof family members in Chinese cabbage
The nucleotide and protein sequences of BraDof factors from B. rapa L. ssp. pekinensis were collected from the BRAD (http://brassicadb.org/brad/) database [23]. We used the BLAST program available on the NCBI (National Center for Biotechnology Information) database to search for potential Dof domains (http://www.ncbi.nlm.nih.gov/). The sequences of all Dof transcription factors in other species were downloaded from the plant TFDB database (http://planttfdb.cbi.edu.cn/) [59].
BraDof domain, motif identification, and phylogenetic analysis
MEME Suite was used to identify all motifs in the BraDof protein sequences [60]. Analysis was performed using the following parameters: number of repetitions, if any; optimum width of the motif, 6–50; and maximum number of motifs, 15. Identified BraDof domains were aligned using ClustalW with default parameters. A phylogenetic tree was constructed using MEGA 5.0, comparing 47 Arabidopsis thaliana Dof domains using the neighbor-joining method with a bootstrap value of 1,000 [61,62].
Putative protein characteristic prediction and duplication analysis of BraDofs
Several physiological and biochemical indices were also analyzed. The theoretical pI was calculated by ExPASy (http://web.expasy.org/compute_pi/) [63]. Sequence Manipulation Suite (http://www.bio-soft.net/sms/) was used to analyze protein statistics, and the solubility of recombinant proteins was predicted by the RPSP program (http://biotech.ou.edu) [64]. The synteny of the BraDof genes was calculated by MicroSyn software according to Cai’s means (http://fcsb.njau.edu.cn/microsyn/) [65].
Identification of orthologous and paralogous genes
OrthoMCL was used to search for orthologous and paralogous genes in Chinese cabbage, Arabidopsis, and rice using the entire Dof protein sequences [66]. We used an E-value of 1e−5 for BLASTP and OrthoMCL analyses, gathered the orthologous and paralogous relationships, and displayed them using the Circos software (http://circos.ca/) [67]. An interaction network of BraDof genes from Chinese cabbage was constructed to understand the genome-wide regulation network. An interaction work of Arabidopsis Dof proteins was constructed using the Arabidopsis Interactions Viewer (http://bar.utoronto.ca/interactions/cgi-bin/arabidopsis_interactions_viewer.cgi). By replacing Arabidopsis proteins with corresponding orthologous and co-orthologous Chinese cabbage proteins, an interaction network of BraDof proteins from Chinese cabbage was constructed and displayed using the Cytoscape software (http://www.cytoscape.org/).
Plant materials, growth conditions and stress treatments
Seeds of ‘Lubaisanhao’ and ‘Qingdao87-114’ were obtained from the State Key Laboratory of Crop Genetics and Germplasm Enhancement in Nanjing Agricultural University. The seeds were germinated at 25°C for 2 d and grown in a growth chamber with a photoperiod of 14 h of light and 10 h of darkness at 28°C. Young leaves were harvested for RNA isolation. For temperature stress treatments, whole four-leaf seedlings were exposed to 4°C or 38°C. For salt stress treatments, four-leaf seedlings were incubated with 100 mM NaCl. For drought stress treatments, four-leaf seedlings were incubated with 15% PEG 6000. Leaves were harvested 0, 0.5, 1, 2, 4, 8, and 12 h after application of the stress treatments, immediately frozen in liquid nitrogen, and then stored at −70°C for RNA isolation.
Total RNA isolation and cDNA synthesis
Total RNA was isolated from plant materials using a total RNA kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. A total of 1 μg of RNA of each sample was used to synthesize first strand cDNA using M-MLV reverse transcriptase according to the manufacturer’s instructions (TaKaRa, Dalian, China). cDNA was diluted 20-fold for quantitative real-time PCR (qRT-PCR) analysis.
qRT-PCR analysis
To analyze BraDof gene transcription levels under abiotic stress, we performed qRT-PCR on an Applied Biosystems 7500 real-time PCR system using a SYBR Green RT-PCR kit (Novland, Shanghai, China). The PCR thermal cycle conditions were as follows: denaturation at 95°C for 30 s, 40 cycles of 95°C for 10 s, and 58°C for 20 s. At the end of each cycle, fluorescence intensities were measured for qRT-PCR. A melting curve (61 cycles at 65°C for 10 s) was generated to check for specific amplification. The method of calculating relative gene expression was performed as described by Pfaffl using the 2−ΔΔCq method [68]. Briefly, the expression profiles were performed with modifications as follows: Ratio = 2−ΔΔCq; ΔΔCq = ΔCq (sample) − ΔCq (control); ΔCq = Cq(target) − Cq(reference). The experiments were repeated three times for different abiotic stress treatments. Standard errors of means among replicates were also calculated. A statistical method (t test) for significant differences was analyzed using the SPSS statistics software (version 17.0) [69].
The primers used for relative quantification were designed from the coding sequences of BraDof transcription factors in Chinese cabbage; all of the primers used in this experiment are provided in Table 1. All primers used for relative quantification were synthesized by Genscript Nanjing Inc. (Nanjing, China).
Table 1.
Primer sequences used for qRT-PCR amplification of actin and 9 Dof genes
| Name | Oligonucleotides sequences |
|---|---|
| Actin-Forward primer | AGTGGGCGTACTACTGGTATTGTG |
| Actin-Reverse primer | AGAGAATCAGTGAGGTCCCTACCC |
| Bra030423F1-Forward primer | CCCGACCAGAAACCTAAC |
| Bra030423R1-Reverse primer | CGTAACTAATCCCGACATCC |
| Bra014752F1-Forward primer | ATCTTCAAATGCGTTGTATC |
| Bra014752R1-Reverse primer | ACTCCGTTCCCATCCTTA |
| Bra030089F1-Forward primer | CTATTTGGGTCTTCGGTAT |
| Bra030089R1-Reverse primer | ATTGACAACACTCGCATT |
| Bra035873F1-Forward primer | TGGGAGTGAACAATAACAA |
| Bra035873R1-Reverse primer | CCAATCAAGCGATAACAG |
| Bra010548F1-Forward primer | TGCCAGTTTCTTCGTCTT |
| Bra010548R1-Reverse primer | TCCTCCACTCAGCATCCC |
| Bra012804F1-Forward primer | ATGGGTTGCCTCCGTTTC |
| Bra012804R1-Reverse primer | GCTCCAGCTTCCGAACAA |
| Bra013492F1-Forward primer | CAAGGTTATTACGGTGCT |
| Bra013492R1-Reverse primer | TGCCGACATTATGGTTCA |
| Bra002057F1-Forward primer | CTGGTGGAAAGATGAGGT |
| Bra002057R1-Reverse primer | CATTGATTCGCAGAGGAC |
| Bra012119F1-Forward primer | ACAACGGTGATGTTACGGC |
| Bra012119R1-Reverse primer | CAAGTCCAAACCCTTCCAG |
Acknowledgements
The research was supported by National Natural Science Foundation of China (31272175), New Century Excellent Talents in University (NCET-11-0670), Jiangsu Natural Science Foundation (BK20130027), Priority Academic Program Development of Jiangsu Higher Education Institutions, and Jiangsu Shuangchuang Project. Thanks to Hua-Wei Tan for his assistance of bioinformatics analysis. We apologize that we cannot cite all relevant articles. We thank some content of the figure from a network in which we cannot cite the relevant reference. We thank the anonymous reviewers for the inspiring comments on the manuscript.
Additional file
C2C2-Dof-like genes in Chinese cabbage chromosome 1. Table S2. C2C2-Dof-like genes in Chinese cabbage chromosome 2. Table S3. C2C2-Dof-like genes in Chinese cabbage chromosome 3. Table S4. C2C2-Dof-like genes in Chinese cabbage chromosome 4. Table S5. C2C2-Dof-like genes in Chinese cabbage chromosome 5. Table S6. C2C2-Dof-like genes in Chinese cabbage chromosome 6. Table S7. C2C2-Dof-like genes in Chinese cabbage chromosome. Table S8. C2C2-Dof-like genes in Chinese cabbage chromosome 8. Table S9. C2C2-Dof-like genes in Chinese cabbage chromosome 9. Table S10. C2C2-Dof-like genes in Chinese cabbage chromosome 10. Table S11. C2C2-Dof-like genes in Chinese cabbage scaffold. Table S12. Theoreticalvalues of Dofs in Chinese cabbage. Table S13. K s values of potentially duplicated genes in Chinese cabbage. Table S14. Number of homologies of potentially duplicated genes in Chinese cabbage.
Footnotes
Jing Ma and Meng-Yao Li contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: ASX JM. Performed the experiments: JM MYL FW JT ASX. Analyzed the data: JM MYL ASX. Contributed reagents/materials/analysis tools: ASX. Wrote the paper: JM. Revised the paper: ASX JM. All authors read and approved the final manuscript.
Contributor Information
Jing Ma, Email: 2014204025@njau.edu.cn.
Meng-Yao Li, Email: 2012204021@njau.edu.cn.
Feng Wang, Email: wangf@njau.edu.cn.
Jun Tang, Email: t.jun525@163.com.
Ai-Sheng Xiong, Email: xiongaisheng@njau.edu.cn.
References
- 1.Qi J, Yu S, Zhang F, Shen X, Zhao X, Yu Y, et al. Reference gene selection for real-time quantitative polymerase chain reaction of mRNA transcript levels in Chinese cabbage (Brassica rapa L. ssp. pekinensis) Plant Mol Biol Rep. 2010;28(4):597–604. doi: 10.1007/s11105-010-0185-1. [DOI] [Google Scholar]
- 2.Zhang FL, Takahata Y, Watanabe M, Xu JB. Agrobacterium-mediated transformation of cotyledonary explants of Chinese cabbage (Brassica campestris L. ssp. pekinensis) Plant Cell Rep. 2000;19(6):569–75. doi: 10.1007/s002990050775. [DOI] [PubMed] [Google Scholar]
- 3.Wu H. The breeding technology of Chinese cabbage. Beijing: Jindun publishing house; 2014. [Google Scholar]
- 4.Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol. 1999;17(3):287–91. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- 5.Singh K. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5(5):430–6. doi: 10.1016/S1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 6.Rushton PJ, Somssich IE. Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol. 1998;1(4):311–5. doi: 10.1016/1369-5266(88)80052-9. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 8.Riechmann JL. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290(5499):2105–10. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 9.Noguero M, Atif RM, Ochatt S, Thompson RD. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sc: Int J Exp Plant Biol. 2013;209:32–45. doi: 10.1016/j.plantsci.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7(12):555–60. doi: 10.1016/S1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- 11.Paolis A, Sabatini S, Pascalis L, Costantino P, Capone I. A rolB regulatory factor belongs to a new class of single zinc finger plant proteins. Plant J. 1996;10(2):215–23. doi: 10.1046/j.1365-313X.1996.10020215.x. [DOI] [PubMed] [Google Scholar]
- 12.Kisu Y, Esaka M, Suzuki M. Putative zinc-binding fomain of plant transcription factor, AOBP, is related to DNA-binding domains of steroid hormone receptors and GATA1. Proc Jpn Acad Ser B Phys Biol Sci. 1995;71(9):288–92. doi: 10.2183/pjab.71.288. [DOI] [Google Scholar]
- 13.Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309(5732):293–7. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- 14.Yanagisawa S. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004;45(4):386–91. doi: 10.1093/pcp/pch055. [DOI] [PubMed] [Google Scholar]
- 15.Yanagisawa S. A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Res. 1995;23(17):3403–10. doi: 10.1093/nar/23.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J: Cell Mol Biol. 1999;17(2):209–14. doi: 10.1046/j.1365-313X.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- 17.Umemura Y, Ishiduka T, Yamamoto R, Esaka M. The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J: Cell Mol Biol. 2004;37(5):741–9. doi: 10.1111/j.1365-313X.2003.01997.x. [DOI] [PubMed] [Google Scholar]
- 18.Cavalar M, Moller C, Offermann S, Krohn NM, Grasser KD, Peterhansel C. The interaction of DOF transcription factors with nucleosomes depends on the positioning of the binding site and is facilitated by maize HMGB5. Biochemistry. 2003;42(7):2149–57. doi: 10.1021/bi026761r. [DOI] [PubMed] [Google Scholar]
- 19.Yanagisawa S, Sheen J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell. 1998;10(1):75–89. doi: 10.1105/tpc.10.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, Tuskan GA, Cheng MZ. Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 2006;142(3):820–30. doi: 10.1104/pp.106.083642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol. 2003;3:17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mun JH, Kwon SJ, Yang TJ, Seol YJ, Jin M, Kim JA, et al. Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 2009;10(10):R111. doi: 10.1186/gb-2009-10-10-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43(10):1035–9. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 24.Yanagisawa S, Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J Biol Chem. 1993;268(21):16028–36. [PubMed] [Google Scholar]
- 25.Moreno-Risueno MA, Martinez M, Vicente-Carbajosa J, Carbonero P. The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol Genet Genomics: MGG. 2007;277(4):379–90. doi: 10.1007/s00438-006-0186-9. [DOI] [PubMed] [Google Scholar]
- 26.Shaw LM, McIntyre CL, Gresshoff PM, Xue GP. Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct Integr Genomics. 2009;9(4):485–98. doi: 10.1007/s10142-009-0130-2. [DOI] [PubMed] [Google Scholar]
- 27.Libault M, Joshi T, Benedito VA, Xu D, Udvardi MK, Stacey G. Legume transcription factor genes: what makes legumes so special? Plant Physiol. 2009;151(3):991–1001. doi: 10.1104/pp.109.144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward JM, Cufr CA, Denzel MA, Neff MM. The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell. 2005;17(2):475–85. doi: 10.1105/tpc.104.027722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papi M, Sabatini S, Altamura MM, Hennig L, Schafer E, Costantino P, et al. Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol. 2002;128(2):411–7. doi: 10.1104/pp.010488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park DH, Lim PO, Kim JS, Cho DS, Hong SH, Nam HG. The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J: Cell Mol Biol. 2003;34(2):161–71. doi: 10.1046/j.1365-313X.2003.01710.x. [DOI] [PubMed] [Google Scholar]
- 31.Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 2006;11(11):550–8. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Iwamoto M, Higo K, Takano M. Circadian clock- and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant Cell Environ. 2009;32(5):592–603. doi: 10.1111/j.1365-3040.2009.01954.x. [DOI] [PubMed] [Google Scholar]
- 33.Washio K. Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochim Biophys Acta. 2001;1520(1):54–62. doi: 10.1016/S0167-4781(01)00251-2. [DOI] [PubMed] [Google Scholar]
- 34.Washio K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 2003;133(2):850–63. doi: 10.1104/pp.103.027334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Isabel-La Moneda I, Carbonero P. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J: Cell Mol Biol. 2002;29(4):453–64. doi: 10.1046/j.0960-7412.2001.01230.x. [DOI] [PubMed] [Google Scholar]
- 36.Diaz I, Martinez M, Isabel-LaMoneda I, Rubio-Somoza I, Carbonero P. The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J: Cell Mol Biol. 2005;42(5):652–62. doi: 10.1111/j.1365-313X.2005.02402.x. [DOI] [PubMed] [Google Scholar]
- 37.Moreno-Risueno MA, Diaz I, Carrillo L, Fuentes R, Carbonero P. The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. Plant J: Cell Mol Biol. 2007;51(3):352–65. doi: 10.1111/j.1365-313X.2007.03146.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang B, Chen W, Foley RC, Buttner M, Singh KB. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell. 1995;7(12):2241–52. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci. 1997;94(14):7685–90. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanagisawa S. Dof DNA-binding domains of plant transcription factors contribute to multiple protein-protein interactions. Eur J Biochem/FEBS. 1997;250(2):403–10. doi: 10.1111/j.1432-1033.1997.0403a.x. [DOI] [PubMed] [Google Scholar]
- 41.Krohn NM, Yanagisawa S, Grasser KD. Specificity of the stimulatory interaction between chromosomal HMGB proteins and the transcription factor Dof2 and its negative regulation by protein kinase CK2-mediated phosphorylation. J Biol Chem. 2002;277(36):32438–44. doi: 10.1074/jbc.M203814200. [DOI] [PubMed] [Google Scholar]
- 42.Shigyo M, Tabei N, Yoneyama T, Yanagisawa S. Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol. 2007;48(1):179–85. doi: 10.1093/pcp/pcl044. [DOI] [PubMed] [Google Scholar]
- 43.Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–43. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 44.Moore RC, Purugganan MD. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 2005;8(2):122–8. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Semon M, Wolfe KH. Consequences of genome duplication. Curr Opin Genet Dev. 2007;17(6):505–12. doi: 10.1016/j.gde.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Kellogg EA. What happens to genes in duplicated genomes. Proc Natl Acad Sci U S A. 2003;100(8):4369–71. doi: 10.1073/pnas.0831050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4(1):10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhuang J, Zhang J, Hou X-L, Wang F, Xiong A-S. Transcriptomic, Proteomic, Metabolomic and Functional Genomic Approaches for the Study of Abiotic Stress in Vegetable Crops. Crit Rev Plant Sci. 2014;33(2–3):225–37. doi: 10.1080/07352689.2014.870420. [DOI] [Google Scholar]
- 49.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6(5):410–7. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- 50.Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149(1):88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386(6624):485–8. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 52.Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383(6603):794–9. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- 53.Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell. 1997;9(9):1607–19. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyde R, Corkins ME, Somers GA, Hart AC. PKC-1 acts with the ERK MAPK signaling pathway to regulate Caenorhabditis elegans mechanosensory response. Genes Brain Behav. 2011;10(3):286–98. doi: 10.1111/j.1601-183X.2010.00667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu-Zhang AX, Newton AC. Protein kinase C pharmacology: refining the toolbox. Biochem J. 2013;452(2):195–209. doi: 10.1042/BJ20130220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajanala K, Nandicoori VK. Localization of nucleoporin Tpr to the nuclear pore complex is essential for Tpr mediated regulation of the export of unspliced RNA. PLoS One. 2012;7(1):e29921. doi: 10.1371/journal.pone.0029921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.David-Watine B. Silencing nuclear pore protein Tpr elicits a senescent-like phenotype in cancer cells. PLoS One. 2011;6(7):e22423. doi: 10.1371/journal.pone.0022423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corrales A-R, Nebauer SG, Carrillo L, Fernández-Nohales P, Marqués J, Renau-Morata B, et al. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J Exp Bot. 2014;65(4):995–1012. doi: 10.1093/jxb/ert451. [DOI] [PubMed] [Google Scholar]
- 59.Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014;42(Database issue):D1182–7. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–8. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilkinson DL, Harrison RG. Predicting the solubility of recombinant proteins in Escherichia coli. Nat Biotechnol. 1991;9(5):443–8. doi: 10.1038/nbt0591-443. [DOI] [PubMed] [Google Scholar]
- 65.Cai B, Yang X, Tuskan GA, Cheng ZM. MicroSyn: a user friendly tool for detection of microsynteny in a gene family. BMC Bioinformatics. 2011;12:79. doi: 10.1186/1471-2105-12-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stern L. A visual approach to SPSS for Windows: a guide to SPSS 17.0, 2nd edn. Boston: Allyn & Bacon; 2010. [Google Scholar]


