Abstract
Background
Bifidobacteria is one of the major gut commensal groups found in infants. Their colonization is commonly associated with beneficial effects to the host through mechanisms like niche occupation and nutrient competition against pathogenic bacteria. Iron is an essential element necessary for most microorganisms, including bifidobacteria and efficient competition for this micronutrient is linked to proliferation and persistence. For this research we hypothesized that bifidobacteria in the gut of iron deficient infants can efficiently sequester iron. The aim of the present study was to isolate bifidobacteria in fecal samples of iron deficient Kenyan infants and to characterize siderophore production and iron internalization capacity.
Results
Fifty-six bifidobacterial strains were isolated by streaking twenty-eight stool samples from Kenyan infants, in enrichment media. To target strains with high iron sequestration mechanisms, a strong iron chelator 2,2-dipyridyl was supplemented to the agar media. Bifidobacterial isolates were first identified to species level by 16S rRNA sequencing, yielding B. bifidum (19 isolates), B. longum (15), B. breve (11), B. kashiwanohense (7), B. pseudolongum (3) and B. pseudocatenulatum (1). While most isolated bifidobacterial species are commonly encountered in the infantile gut, B. kashiwanohense was not frequently reported in infant feces. Thirty strains from culture collections and 56 isolates were characterized for their siderophore production, tested by the CAS assay. Siderophore activity ranged from 3 to 89% siderophore units, with 35 strains (41%) exhibiting high siderophore activity, and 31 (36%) and 20 (23%) showing intermediate or low activity. The amount of internalized iron of 60 bifidobacteria strains selected for their siderophore activity, was in a broad range from 8 to118 μM Fe. Four strains, B. pseudolongum PV8-2, B. kashiwanohense PV20-2, B. bifidum PV28-2a and B. longum PV5-1 isolated from infant stool samples were selected for both high siderophore activity and iron internalization.
Conclusions
A broad diversity of bifidobacteria were isolated in infant stools using iron limited conditions, with some strains exhibiting high iron sequestration properties. The ability of bifidobacteria to efficiently utilize iron sequestration mechanism such as siderophore production and iron internalization may confer an ecological advantage and be the basis for enhanced competition against enteropathogens.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-014-0334-z) contains supplementary material, which is available to authorized users.
Keywords: Bifidobacteria, Iron, Siderophore, Nutrient competition, CAS assay
Background
Bifidobacteria represent one of the most predominant groups of commensal bacteria residing in the human and animal gastrointestinal tract [1]. Bifidobacterium spp. are among the first anaerobes able to reach high levels within the first week of life and generally represent more than 40% of infant gut microbiota [2,3]. Different Bifidobacterium species are associated with a broad range of beneficial effects on host’s health. Their colonization in the gut is associated with modulation of the intestinal microbiota composition and activity, immune-modulation and attenuation of inflammatory symptoms [4,5]. Additionally, mechanisms involved in host protection include nutrient competition, competitive exclusion, direct antagonism by inhibitory substances and host-mediated effects such as improved barrier function and altered immune response [6,7]. Numerous studies have demonstrated the efficacy against enteric pathogens of selected bifidobacterial strains but the mechanistic basis supporting the observed benefits is still often lacking [8]. Strain dependency is one of the features that have been revealed while seeking for new probiotic strains [9], thus the importance of selection of suitable candidates through biochemical assays targeting potential probiotic functions.
Iron is an essential micronutrient for most intestinal commensal and pathogenic bacteria, except for Lactobacillus [10]. Iron is involved in many essential metabolic processes like cell proliferation, electron transport, and is a cofactor for different enzymes [11]. For instance, iron plays an important role in most pathogenic bacteria, as iron sequestration mechanism have been associated to cell replication and persistence and thus being involved in pathogenesis [12]. However, iron is restricted in most environments including the mammalian intestine [13]. The limited availability of iron in the host provides one form of non-specific immune-defence that bacteria, for instance, bacterial pathogens need to overcome in order to grow and cause infection [14]. Therefore microorganisms have evolved different acquisition systems like import of iron from different sources and production of siderophores to utilize, accumulate and sequester iron [15]. Various physiological mechanisms such as low/high affinity iron transport systems are employed by bacteria to tightly control iron uptake, intracellular concentration and storage, preventing toxic effects [16]. Iron starvation signals the up-regulation of acquisition systems and as a response many gram positive bacteria synthesize low molecular weight iron chelating ligands, known as siderophores, to compete for iron [17]. In contrast, iron excess activates the expression of efflux pumps and ferritin like proteins involved in iron accumulation [18,19].
Withholding iron has been identified as a major competitive and defense mechanisms in many gram positive and gram negative bacteria, because of its general limited availability in different environments, such as the intestine [20,21]. The concept of restricting iron to pathogens has been coined as nutritional immunity [22,23] and is usually associated with efficient iron sequestration systems. It has been hypothesized that one of the beneficial actions of health promoting bifidobacteria is to sequester iron, thus making it less available to pathogens, hence representing another form of nutritional immunity [24]. The ability of bifidobacteria to outcompete by producing siderophores and/or concentrating iron from the environment may therefore provide a competitive advantage against competing microorganism by limiting the iron available in complex ecosystems such as the gut microbiota. Thus, bifidobacteria, may cause iron starvation of competing microorganism; for instance, enteropathogens. However to our knowledge, the ability to produce siderophores in culture supernatant and to internalize iron of diverse bifidobacteria species has never been investigated. We hypothesized that bifidobacteria present in the gut of iron deficient infants efficiently sequester iron. Therefore our aim was to isolate Bifidobacterium strains from stool samples from iron deficient Kenyan infants and to investigate their ability to produce siderophores as well as to sequester iron in comparison to culture collection strains.
Results
Isolation and genotypic identification
The bifidobacteria gene copy numbers (CN) was, assessed in the twenty-eight stool samples of Kenyan infants by qPCR targeting the xylulose-5-phosphate/fructose-6-phosphatephosphoketolase gene (xfp) gene. The mean bifidobacteria CN was log 9.8 SD +/− 0.4 copies per gram feces, in the range from log 6.4 to log 10.4 copies per gram feces sample. Twenty-eight baseline stool samples from Kenyan infants were streaked in BRS, MRS-cys, TPY and BSM agar supplemented with a strong iron chelator 2,2-dipiridyl. A total of 208 strains were isolated and analyzed by 16S rRNA gene sequencing for identification. 16S rRNA partial sequences with a length of 1100 to 1300 bp after quality trimming were aligned for genotypic identification using BLAST, yielding 41% Enterococcus spp. (94 isolates), 29% Bifidobacterium spp. (56), 28% Lactobacillus spp. (54), 1% Weisella spp. (2), 0.5% Streptoccocus spp. (1) and 0.5% Leuconostoc spp. (1). Bifidobacterium 16S rRNA partial sequences were assigned to six phylogenetic taxa: 34% B. bifidum (19 isolates), 27% B. longum (15), 19% B. breve (11), 13% B. kashiwanohense (7), 5% B. pseudolongum (3), 2% B. pseudocatenulatum (1) (Table 1). B. bifidum was most frequently isolated in BSM and Beerens agar, while B. longum and B. breve were obtained from TPY plates and B. kashiwanohense from TPY and BSM plates. Bifidobacterium isolated on BSM agar represented five species (5), compared to four species isolated on Beerens, three on TPY and only one on MRS-cys (1) agar. Three different bifidobacteria species were isolated from each stool samples 3, 5, 8 and 11 (Table 2), while either one or two bifidobacterial species were obtained from the other samples. In addition, B. kashiwanohense was only found in three fecal samples 11, 20 and 25 (Table 2).
Table 1.
Occurrence and isolate numbers of isolated Bifidobacterium species in the infant stool samples (n = 28)
| Distribution of bifidobacterial species in infant fecal samples | No. isolates obtained | |
|---|---|---|
| n = 28 | n = 56 | |
| B. bifidum | 13 | 19 |
| B. longum | 8 | 15 |
| B. breve | 8 | 11 |
| B. kashiwanohense | 3 | 7 |
| B. pseudolongum | 1 | 3 |
| B. pseudocatenulatum | 1 | 1 |
Table 2.
Bifidobacterial population and species isolated from Kenyan infant stool samples (n = 28)
| Feces sample | Hb (g/dl) | Total bacteria | Bifidobacteria population | Isolated species from different iron restricted media |
|---|---|---|---|---|
| 1 | 10.1 | 11.6 | 9.7 | B. breve BSM1-2 |
| 2 | 8.7 | 10.4 | 9.4 | B. bifidum BSM2-1, B. bifidum BSM2-3, B. breve TPY2-1, B. breve TPY2-3, B. breve TPY2-4 |
| 3 | 10.2 | 10.9 | 9.5 | B. breve BSM3-1, B. longum TPY3-1, B. longum TPY3-2, B. pseudocatenulatum BRS3-2, B. bifidum BRS-300 |
| 4 | 9.9 | 11.8 | 6.4 | B. longum TPY4-1 |
| 5 | 9.6 | 11.3 | 8.9 | B. longum PV5-1, B. bifidum BRS5-3, B. breve TPY5-1, B. breve TPY5-2 |
| 6 | 9.2 | 10.3 | 9.0 | B. bifidum TPY6-2, B. bifidum MRSc6-292, B. bifidum MRSc6-312 |
| 7 | 8.6 | 10.8 | 9.5 | No Bifidobacterium isolates obtained |
| 8 | 8.1 | 11.7 | 10.0 | B. pseudolongum PV8-2, B. pseudolongum BSM8-1, B. pseudolongum BSM8-3, B. breve BSM8-4, B. bifidum BRS8-1, B. bifidum BRS8-2, B. bifidum BRS8-3, B. longum TPY8-1, B. longum TPY8-2 |
| 9 | 9.9 | 11.1 | 9.5 | No Bifidobacterium isolates obtained |
| 10 | 9.8 | 11.2 | 9.9 | B. breve TPY10-1, B. breve TPY10-2 |
| 11 | 9.5 | 12.1 | 10.1 | B. kashiwanohense BSM11-1, B. kashiwanohense BSM11-4, B. longum BSM11-5, B. kashiwanohense TPY11-1, B. kashiwanohense BRS11-1, B. kashiwanohense TPY11-2 |
| 12 | 11.3 | 12.2 | 9.7 | B. bifidum BSM12-2, B. longum TPY12-1 |
| 13 | 10.3 | 9.9 | 9.2 | B. longum TPY13, B. longum TPY13-2, B. longum TPY13-3 |
| 14 | 10.4 | 10.6 | 9.4 | No bifidobacteria isolate obtained |
| 15 | 10.4 | 10.4 | 9.1 | B. bifidum BSMd15, B. bifidum BSM15-2, |
| 16 | 10.5 | 10.6 | 9.3 | B. bifidum BRS16-1, B. breve TPY16-2 |
| 17 | 10.5 | 10.7 | 9.4 | No Bifidobacterium isolates obtained |
| 18 | 10.8 | 11.1 | 9.1 | B. bifidum BSM18 |
| 19 | 10.8 | 10.5 | 8.9 | No Bifidobacterium isolates obtained |
| 20 | 11.1 | 10.7 | 9.0 | B. kashiwanohense PV20-2 |
| 21 | 11.3 | 10.7 | 9.0 | B. bifidum BRS21 |
| 22 | 11.4 | 10.1 | 8.9 | No Bifidobacterium isolates obtained |
| 23 | 11.8 | 11.7 | 10.4 | No Bifidobacterium isolates obtained |
| 24 | 11.8 | 11.7 | 10.2 | No Bifidobacterium isolates obtained |
| 25 | NDA | 10.4 | 9.3 | B. kashiwanohense TPY25-1 |
| 26 | NDA | 11.1 | 9.0 | B. bifidum BRS26-2 , B. longum TPY26-1 |
| 27 | NDA | 10.1 | 9.0 | B. breve TPY27, B. bifidum BRS27-3 |
| 28 | NDA | 12.0 | 9.7 | B. bifidum PV28-2a , B. bifidum BSM28-1, B. bifidum BSMd28-2 |
Bifidobacterial population is expressed as bifidobacterial log copies per gram feces. SD for bifidobacteria population among samples was +/− 0.35 and for total bacteria SD +/− 0.56. NDA: No data available. Hb: Hemoglobin level (g/dL).
Characterization based on siderophore production
Eighty-six bifidobacterial strains comprising 56 isolates and 30 culture collection strains were characterized for their ability to produce siderophores using the CAS assay (Additional file 1: Figure S1). Siderophore activity (SA) was found to be widely distributed and strain dependent among bifidobacterial strains. Percentage of SA ranged from 3% to 89% siderophore units. To group strains according to SA values, the following ranges were established according to OD630nm measurements and color change after mixing with the CAS dye. Three groups were defined: strains with high SA in the range from 90-60% siderophore units and a color change to orange; strains with intermediate SA in the range from 59-30% and a color change to pink, and strains with intermediate SA in the range from 29-0% and no visible color change. Thirty-four strains (41%) belonged to the first group of high SA, 32 strains (36%) showed intermediate SA, and 20 strains (23%) showed low SA. No correlation was found between SA and hemoglobin levels of the infant stool donor where the strains were isolated (Additional file 2: Table S1). From the thirty-four strains showing high SA, 10 strains belonged to public culture collections and 24 strains belonged to the isolates. Compare to B. bifidum MRSc6-292, the strain with the lowest SA (3 +/−1%), eight B. breve, five B. bifidum, three B. pseudolongum, four B. kashiwanohense and three B. thermoacidophilum strains showed significantly higher SA (p < 0.0001). B. breve showed the highest SA value among species, with 76 +/− 21% (n = 12) (Figure 1a).
Figure 1.
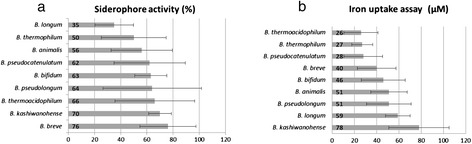
Siderophore activity (a) and iron internalization within bifidobacterial species (b) .
Strains showing similar genotypic identification in the 16S rRNA sequence, isolated from same stool sample and plate and that also showed similar SA value; as well as, strains showing low siderophore activity, were excluded from the iron internalization (II) characterization. Therefore 43 isolates and 19 culture collection strains were available for II characterization.
DNA finger printing
Phylogenetic diversity of sixty-two Bifidobacterium strains included in the II characterization was evaluated by DNA-finger printing using RAPD and REP-PCR (Additional file 3: Figure S2). DNA finger prints showed that strains retained for the II characterization were phylogenetically different, except for B. breve TPY 10–2 and B. pseudolongum BSM 8–1 as they showed identical DNA finger prints with B. breve TPY 10–1 and B. pseudolongum BSM 8–2, respectively.
Characterization based on iron internalization
Sixty bifidobacterial strains comprising 41 isolates and 19 culture collection strains were characterized by the iron uptake assay determined by using graphite furnace atomic absorption spectrophotometer. Iron internalization (II) was found to be strain dependent and B. kashiwanohense was the bifidobacterial species with the highest overall iron internalization of 78 μM +/− 27 (n = 7) (Figure 1b). Iron concentrations tested in bifidobacterial pellets ranged from 8 μM to 118 μM (Figure 2). Eight B. longum strains, six B. kashiwanohense, three B. bifidum, one B. pseudolongum and one B. breve showed significantly higher II values (p < 0.0001) than B. thermoacidophilum LMG21397 exhibiting the lowest II of 8 μM.
Figure 2.
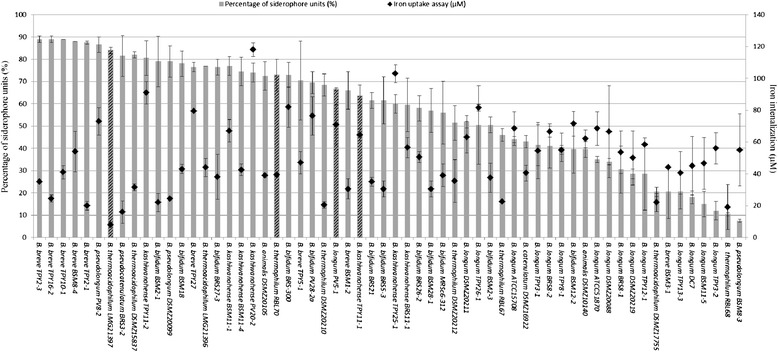
Characterization of bifidobacterial strains for siderophore activity and iron uptake measured by the CAS assay and iron internalization test, respectively. Columns show siderophore activity (%) (n = 2) and squares show the results of iron internalization (μM) (n = 2). Striped columns indicate selected strains after strain characterization. Error bars correspond to standard deviations calculated for two independent replicates for iron uptake and siderophore activity respectively.
Discussion
In the present study, 56 bifidobacterial strains with high iron sequestration properties were isolated from fecal samples of iron deficient Kenyan infants, most of them anemic, using an iron depleted culture dependent approach. We assumed for our research that in breast-fed infants little iron enters the colon and thus low iron availability is expected in the gut. Indeed, iron content in breast milk was reported to be in the range from 0.4 mg/L to 0.5 mg/L [25-27]. Thus, a high competition of strains for iron would be present in the gut of breast fed infants, creating selective pressure for strains with high iron sequestration properties. After the genotypic identification the iron sequestration properties of the isolates were measured and compared to those of culture collection strains.
Using qPCR targeting the xfp gene, high populations of bifidobacteria were found in fecal samples of Kenyan infants in agreement with previous studies where population was found to be in the range of 109-1010 gene copy numbers per gram feces in breast fed infants [28,29]. To perform the enrichment of bifidobacterial diversity with high iron sequestration mechanisms a culture dependent approach was applied, combining the low iron environment characterized from Kenyan infants, with the use of 2,2-dipiridyl which is a strong chelator for iron. The most frequently isolated bifidobacterial species were B. bifidum, B. longum, B. breve, B. pseudolongum and B. pseudocatenulatum which are species frequently found in the infant gut [5,30]. The application of strong iron limitation during the isolation process may have limited the growth of some bifidobacterial species that could not grow under these stringent conditions. On the other hand the identification of strains from six phylogenetic taxa present in infant stool suggest that bifidobacteria is able to efficiently overcome iron limitation as one of the mechanisms active in the gut.
Using culture independent methods Turroni et al. [3] showed that a broad diversity of species were present in the gut of European infants. Major bifidobacterial groups identified were B. breve, B. bifidum, B. longum, B. pseudocatenulatum and B. pseudolongum in agreement with our study. As shown in Table 1, the most frequently isolated species in the present study were B. breve, B. bifidum and B. longum which are bifidobacterial species highly encountered in breast fed infants. In addition, using culture dependent studies, Turroni et al. [31] and Matsuki et al. [32] showed that the most frequently isolated species in fecal samples from European infants were B. longum, B. adolescentis and B. breve, B. longum, respectively. This is in contrast to our study where B. animalis or B. catenulatum were not detected while B. kashiwanohense was isolated in three fecal samples, possibly reflecting species distribution between African and European infants, a bias resulting from the stringent iron limitation, or a non-identified species in previous studies. B. kashiwanohense has been reported for the first time from feces of a healthy Japanese infant in 2011 and is known to be close to B. pseudocatenulatum and B. catenulatum, that are species commonly found in the infantile gut [33]. In addition, B. kashiwanohense was only found in three samples which can be associated with an interindividual variability in species distribution which was previously shown for premature and term infants [5,34,35].
Iron is an essential micronutrient for most microorganisms and directly impacts the metabolism of microorganisms. Functions regulated by iron in bacteria are those involved in iron utilization or acquisition and two transport systems have been described in iron uptake pathways [11]. Bacteria secrete iron scavenging compounds, siderophores, to fulfill their iron requirement for growth. A high fraction (41%) of bifidobacterial strains were in the high SA group, with B. breve exhibiting the highest siderophore activity. In addition, the major fraction of the strains exhibiting high SA belonged to the isolates from infants stool samples, which highlights the importance of testing new isolates for this property. Previous studies demonstrated that bifidobacterial strains can inhibit competing organisms in an iron concentration dependant manner [9]. However, it was also recently reported that the growth of some bifidobacterial strains was not affected by the presence of iron chelators such as 2,2-dipiridyl and 8-hydroxyquinoline [36,37]. Although different attempts have been done to identify iron sequestration mechanisms no study demonstrated the production of iron scavenging compounds in culture supernatant. Production of iron scavenging compounds is a common strategy to compete in environments where iron is limited and activation of this mechanism could represent an advantage for bifidobacteria in the presence of competing bacteria of the gut microbiota.
Bifidobacterial strains were characterized towards iron internalization which is a potential mechanism to scavenge iron and to provide competition. Interestingly, B. kashiwanohense showed the highest II activity suggesting that B. kashiwanohense has efficient sequestration systems that may play a role in competing for iron in the intestine [16]. Thus, the efficient iron uptake systems of bifidobacteria, such as production of iron scavenging compounds and iron internalization, may be important from the point of view of nutritional immunity [36,37].
Despite high SA in B. breve species and II in B. kashiwanohense species were identified, standard deviation within each species group was very high, reflecting a remarkably strain specificity towards SA and II. Different SA and II properties between species and strains enable the selection of strains with the highest iron sequestration activities with possible competitive advantage when competing in the iron limited intestine. No correlation was found between siderophore activity and iron internalization (Spearman’s correlation: −0.22) implicating that these two properties are independently controlled, in agreement with previously reported studies where the two properties were not related [12,38]. In our study sixteen out of twenty strains that showed high SA and II are strains isolated from iron deficient infant stools. Four strains exhibited both high SA and II characteristics: B. pseudolongum PV8-2, B. kashiwanohense PV20-2, B. bifidum PV28-2a, B. longum PV5-1 (Figure 2). The mechanisms involved in iron acquisition have been shown to play an important role during enteric infection [39-41]. It has been recognized that most of the dominant colonizers of an environment have efficient iron scavenging systems and can inhibit the growth of other competing organisms by depriving them from iron [12]. Herein, selected strains could be further tested for the utilization of iron sequestration mechanisms for host protection in complex gut ecosystems where mechanisms involved in iron competition have been addressed to play an important role [39].
Conclusions
A wide diversity of bifidobacterial strains with high iron sequestration properties were obtained from fecal samples of iron deficient Kenyan infants in a culture dependent approach targeting low iron environments. Bifidobacterial species from public culture collections and newly isolated strains were characterized for siderophore production in culture supernatant and iron internalization which enabled the selection of four strains isolated from infant stools with high iron sequestration mechanisms. The implication of an efficient utilization of iron sequestration mechanisms by selected bifidobacteria strains to inhibit enteric pathogens could be further tested in complex gut ecosystems and may uncover one of the mechanisms of bifidobacteria for host protection against infection.
Methods
Culture collection strains
Twenty-one Bifidobacterium strains belonging to different culture collections (DSMZ, ATCC, LMG) and nine belonging to our own culture collection were used for strain characterization and are listed in Table 3. Additionally, 56 bifidobacterial strains typed to species level were isolated from 28 stool samples from Kenyan infants (Table 2).
Table 3.
Strains from culture collections used for characterization of siderophore activity and iron internalization
| Strain | Culture collection | Number |
|---|---|---|
| Bifidobacterium adolescentis | DSMZ | 20083 |
| Bifidobacterium animalis | DSMZ | 10140 |
| Bifidobacterium animalis | DSMZ | 20105 |
| Bifidobacterium bifidum | DSMZ | 20456 |
| Bifidobacterium boum | DSMZ | 20432 |
| Bifidobacterium breve | DSMZ | 20213 |
| Bifidobacterium catenulatum | DSMZ | 16992 |
| Bifidobacterium lactis | LFB culture collection | DC1 |
| Bifidobacterium lactis | LFB culture collection | DC5 |
| Bifidobacterium longum | ATCC | 15708 |
| Bifidobacterium longum | ATCC | 51870 |
| Bifidobacterium longum | LFB culture collection | DC7 |
| Bifidobacterium longum | LFB culture collection | NCC2705 |
| Bifidobacterium longum | DSMZ | 20088 |
| Bifidobacterium longum | DSMZ | 20211 |
| Bifidobacterium longum | DSMZ | 20219 |
| Bifidobacterium pseudocatenulatum | DSMZ | 20438 |
| Bifidobacterium pseudolongum | DSMZ | 20099 |
| Bifidobacterium thermoacidophilum | DSMZ | 15837 |
| Bifidobacterium thermoacidophilum | DSMZ | 17755 |
| Bifidobacterium thermoacidophilum | LMG | 21395 |
| Bifidobacterium thermoacidophilum | LMG | 21396 |
| Bifidobacterium thermoacidophilum | LMG | 21397 |
| Bifidobacterium thermophilum | LFB culture collection | RBL67 |
| Bifidobacterium thermophilum | LFB culture collection | RBL68 |
| Bifidobacterium thermophilum | LFB culture collection | RBL70 |
| Bifidobacterium thermophilum | DSMZ | 20210 |
| Bifidobacterium thermophilum | DSMZ | 20212 |
| Bifidobacterium thermophilum | LFB culture collection | F9K9 |
| Bifidobacterium spp. | LFB culture collection | BB46 |
DSMZ: German Collection of Microorganisms, ATCC: American Type Culture Collection, LMG: Belgian co-ordinated Collections of Micro-organisms, LFB: Laboratory of Food Biotechnology ETH Zurich culture collection.
Cultivation conditions
Working cultures were obtained by inoculating strain stocks (1% inoculation rate) maintained at −80°C in 10 mL de Man, Rogosa and Sharpe broth (Biolife, Italy) (MRS), pH 6.0, supplemented with 0.5 g/L L-cysteine (cys) (Sigma-Aldrich, Switzerland). Incubation was carried out in an anaerobic chamber (Coy Laboratory Products Inc., Inc.) with an atmosphere of 85% N2, 10% CO2 and 5% H2 (PanGas AG, Switzerland) at 37°C for 48 h. Peptone water (Oxoid, Switzerland) supplemented with 0.05% cys, pH 6.5 (peptone-cys) was used as a re-suspension solution. In order to achieve iron limited conditions a chemically semi-defined low iron medium (CSDLIM, tested with 1.5 μM iron) was used, containing: 5.64 g/L 5X M9 minimal salts, 5.0 g/L proteose-peptone (Becton Dickinson, Switzerland), 0.2 g/L cys, 20 mL/L of 20% glucose, 2 mL/L of 1 M MgSO4 and 0.2 mL/L of 1 M CaCl2 2H2O (Sigma-Aldrich, Switzerland). The M9 minimal salts, proteose-peptone and cys were dissolved in 980 mL of double distilled water and autoclaved. Glucose, magnesium and calcium were solubilized in double distilled water to the desired concentration (see above), filter-sterilized (0.22 μm pore size, Millipore, Switzerland), and added to the autoclaved portion. For measuring the ability to internalize iron an iron uptake solution (IUS), with 0.05 μM Fe, pH 6.5 was used, containing: 0.4 g/L KCl; 8 g/L NaCl; 0.14 g/L CaCl2 2H2O; 2 g/L glucose; 8.2 g/L sodium acetate (Sigma-Aldrich, Switzerland); 0.2 g/L cys was used.
Fecal samples for Bifidobacterium strains isolation
Twenty-eight baseline stool samples from Kenyan infants (fifteen male, thirteen female) were used for Bifidobacterium strain isolation [42]. Stools were obtained from eighteen anemic and six iron deficient infants (hemoglobin level <11 g/dL) and from four infants for whom no hemoglobin data was available. All infants were 6 +/−0.25 months old, vaginally delivered, breast fed, unrelated, and they did not receive any antibiotic/probiotic treatment in the previous three months. Stool samples of approx. 1 g of fresh fecal material were taken up with a sterile spatula and collected in cryo-vials containing 20% glycerol (Sigma-Aldrich, Switzerland) and 0.05% (final volume) cys (Sigma-Aldrich, Switzerland). The samples were frozen at −20°C until processing for isolation. This study was approved by the Ethics and Research Committees of the Kenyatta National Hospital/ University of Nairobi (KNH-ERC/A/337), the University of KwaZulu-Natal (BF121/08) and the Swiss Federal Institute of Technology Zurich (EK 2009-N-53). Caregivers signed an informed consent. This study is registered at clinicaltrials.gov as NCT01111864.
Characterization of fecal samples
DNA extraction and quantitative PCR (qPCR)
DNA was extracted from 250 +/− 50 mg fecal sample by a FastDNA SPIN kit for soil (MP Biomedicals, France) and quantification was carried out with a Nanodrop Spectrophotometer (ND-1000, Witec AG, Switzerland). Total bacteria and bifidobacteria populations were detected by qPCR with KAPA™ SYBR® FAST qPCR kit (Biolabo Scientifics Instruments SA, Switzerland) and a 7500 Fast Real-Time PCR System (Applied Biosystems, Switzerland). Total bacteria were quantified using plasmid pLME21 containing the 16S rRNA gene from Escherichia coli JM109 which was amplified with the following primers, Eub338 F (5′-ACTCCTACGGGAGGCAGCAG-3′) and Eub518R (5′-ATTACCGCGGCTGCTGG-3′) [22]. Bifidobacterium spp. was quantified by targeting the xylulose-5-phosphate/fructose-6-phosphatephosphoketolase gene (xfp) with the following primers, xfp-fw (5′-ATC TTCGGACCBGAYGAGAC-3′) and xfp-rv (5′-CGATVACGTGVACGAAGGAC-3′) [43,44]. Samples were analyzed twice in a total volume of 25 μL using KAPA™SYBR®FAST qPCR Master Mix (24 μL) containing 1 μL template DNA diluted 1:100 and 1:10 for total bacteria and bifidobacteria, respectively. Sample mean gene copy numbers (CN) per gram of feces (n = 2) were obtained from standard curves generated for each qPCR run using serial dilutions of control standard amplicons. PCR cycles consisted of initial activation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and elongation at 60°C for 30 s.
Strain isolation
Stool samples were streaked on Beerens agar (BRS) [45]; de Man, Rogosa and Sharpe agar (Biolife, Italy) (MRS), supplemented with 0.5 g/L L-cysteine HCl (cys) (Sigma-Aldrich, Switzerland), pH 6.0; Trypticase-phytone-yeast (TPY) [46] agar and Bifidus selective agar (BSM) (Fluka, Switzerland) [47].To enhance the isolation of bifidobacteria with high iron sequestration properties a strong iron chelator, 2,2-dipyridyl (Sigma-Aldrich, Switzerland) at 50 μM [48] was supplemented to the enrichment agar. Incubation was carried out in the anaerobic chamber at 37°C for 48 h. Based on different morphologies three colonies were picked per enrichment agar, streaked for purity and cultured in MRS-cys broth. Cultures were grown to an OD600nm (Biowave CO8000, Biochrom, England) of 1 +/− 0.5. Purity was verified microscopically and cultures were stored at −80°C in a final concentration of 20% (vol/vol) glycerol and 0.05% cys. Pellets obtained from liquid cultures were stored at −20°C for DNA extraction.
Genotypic characterization
DNA was extracted from cell pellets using a Wizard Genomic DNA purification kit (Promega AG, Switzerland) according to the manufacturer’s instructions and strains were typed to species level using 16S rRNA sanger sequencing as described by Jost et al., 2013 [49]. Briefly, PCR amplification of 16S rRNA genes was performed using a 4:1 mixture of forward primers 8f (5′-AGAGTTTGATCMTGGCTC AG-3′, universal) and 8f-bif (5′-AGGGTTCGATTCTGGCTCAG-3′, Bifidobacterium-specific) and a universal bacterial reverse primer 1391R (5′-GACGGGCGGTGTGTRCA-3′) (Microsynth AG, Switzerland). PCR reaction mixture of 50 μL contained 25 μL of 2X MasterMix (Fermentas GmbH, Switzerland), 0.2 μM of each primer (−mixture) and 1 μl of template DNA diluted to 1 ng/μL. Thermocycling (BiometraTProfessional Thermocycler; Biolabo Scientific Instruments SA, Switzerland) was performed with an initial denaturation step at 94°C for 300 s, a second denaturation step was carried out by thirty cycles at 94°C for 30 s, annealing at 57°C for 60 s and extension at 72°C for 30 s with a final extension at 72°C for 420 s. Specificity and amplicon size were verified by electrophoresis in 1.5% (w/v) agarose gels, and reactions were purified using an illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare Europe GmbH, Switzerland) according to the manufacturer’s instructions. Cycle sequencing PCR was carried out in 20 μL reaction volumes with 5% (v/v) BigDye v3.1 (Applied Biosystems Europe BV), 4 μL of 5X sequencing buffer (Applied Biosystems, Switzerland), 1 μM of reverse primer 1391R and 1 μL of purified PCR template. Thermocycling (labcycler; SensoQuest GmbH, Switzerland) was performed with an initial denaturation step at 96°C for 300 s, followed by thirty-five cycles of denaturation at 96°C for 10 s, annealing at 55°C for 20 s and extension at 60°C for 240 s. Reactions were purified by dextran gel bead filtration (Sephadex; GE Healthcare, Switzerland) before loading 10 μL for capillary electrophoresis (ABI 3130xl DNA Analyzer; Applied Biosystems, Switzerland). Sequencing trace chromatograms were quality-trimmed and checked for miscalled bases using CLC Genomics Workbench 64 v6.0.5 (CLC bio, Denmark). DNA extraction and Sanger sequencing were carried out twice from two independent cultures. 16S rRNA partial sequences were aligned with the Basic Local Alignment Search Tool algorithm (BLAST) to the GenBank database and genotypic assignments were based on the nearest neighbor (<97% sequence similarity).
Siderophore production
Eighty-six bifidobacteria strains from isolation and culture collections and were tested for siderophore production in culture supernatant by the Chroma Azurol S (CAS) assay [15]. Cells were grown twice in 10 mL MRS-cys broth under anaerobic conditions, centrifuged 10,000 g for 10 min at 24°C, supernatant discarded and re-suspended in peptone-cys water. CSDLIM medium (15 mL) was inoculated with 2% v/v of bacterial re-suspension. Strains were cultured anaerobically to an OD600nm of 1.5 +/− 0.3 corresponding to log 6 +/− 0.3 CFU/mL. Siderophore activity was measured in culture supernatant and calculated as percentage of siderophore units according to Payne et al. [48]. Briefly, 2 mL of culture were centrifuged at 10,000 g for 10 min at 4°C. 0.5 mL of cultured supernatant was mixed with 0.5 mL of CAS dye and 0.01 mL of shuttle solution and absorbance (As) was measured at 630 nm (OD630nm) after 6 hours. 0.5 mL of uncultured media plus 0.5 mL of CAS dye and 0.01 mL of shuttle solution were mixed to obtain the absorbance reference (Ar) and uncultured CSDLIM media served as blank for absorbance measurements. A formula was used to obtain percentage of siderophore units: % of siderophore units = [(Ar – As)/Ar] x 100. L. casei DSMZ 20011 was used as negative control since according to Pandey [10] siderophores are not synthesized by Lactobacillus spp.. E. coli K12 was used as positive control due to their ability to produce siderophores in low iron environments [50]. From the starting 86 bifidobacterial strains tested isolates showing similar (97-100%) genotypic identification in the 16 s rRNA, isolated from same stool sample and plate, and that also show similar SA value; as well as, strains showing low siderophore activity, were excluded for the iron uptake (II) assay.
DNA finger printing
Phylogenetic diversity of sixty-two Bifidobacterium strains included in the II, was evaluated by DNA-finger printing using RAPD and REP-PCR. RAPD reactions were performed according to Jost et al. [51]. RAPD amplification reactions consisted of 25 μL that contained 12.5 μL of 2X Master Mix (Fermentas, Switzerland), 1 μM of primer (100 μM) (Mycrosynth, Switzerland) and 1 μL of DNA template diluted to approximately 50 ng/μL. Four separate RAPD reactions were performed for each species, using primers OPA-02 (5′-TGCCGAGCTG-3′), OPL-07 (5′-GGGAACGTGT-3′), OPL-16 (5′-GGGAACGTGT-3′) and OPA-13 (5′- CAGCACCCAC-3′). For reactions using primers OPA-02, OPL-07, OPL-13 and OPL-16, thermocycling was performed with an initial denaturation step at 95°C for 3 min, followed by 45 cycles of denaturation at 94°C for 1 min, annealing at 30°C for 1 min and extension at 72°C for 2 min. Due to the limited number of bands identified in B. kashiwanohense and B. pseudolongum species, REP-PCR was performed as described previously [52]. REP-PCR was performed using the primer REP1R-I (5′-NNNNCGNCGNCATCNGGC-3′) and REP2-I (5′-NCGNCTTATCNGGCCTAC-3′) (Mycrosynth, Switzerland). REP-PCR reactions were carried out with an initial denaturation step at 95°C for 3 min, followed by 30 cycles at 90°C for 30 s, 40°C for 1 min and 72°C for 1.5 min, and final extension at 72°C for 8 min. RAPD and REP-PCR products were visualized by UV light on ethidium bromide stained agarose (1.5%) after gel electrophoresis in 1 × TAE-buffer (pH 8.0), using 1-kb ladder (Fermentas GmbH, Switzerland) as reference.
Characterization based on iron internalization
Sixty strains, 41 isolates and 19 strains belonging to culture collections, were characterized for iron uptake characterization. For iron internalization the method described by Kot et al. [53] was used with some modifications. Briefly, Bifidobacterium strains were grown twice under anaerobic conditions in MRS-cys broth for 24 h at 37°C. Cultures were centrifuged 10,000 g (Biofuge primo, Heraeus, Switzerland) at 24°C, 10 min and supernatant discarded. A cell suspension was prepared with OD600nm adjusted to 1 with peptone-cys water. This suspension was used to inoculate 25 mL of CSDLIM medium at 2% v/v and cultured to an OD600nm of 1 +/− 0.25. Culture was centrifuged and supernatant discarded, and cells were resuspended in 10 mL volume of the iron uptake solution (IUS) to yield a concentration of log 8+/− 0.3 CFU/mL. Two tubes of same volume containing the standardized culture were prepared to determine the amount of iron internalization. The first tube without added iron served as the reference for the original amount of iron present in the pellet. The second tube was supplemented with 35 μM FeSO4 freshly prepared and was used to measure the amount of internalized iron. Both tubes were incubated for 60 min at 37°C under anaerobic conditions, then placed immediately on ice, centrifuged 15 min at 10,000 g (Biofuge primo, Heraeus), and washed twice with 2 mL sterile double distilled water. All samples were processed in duplicate. Iron concentration in the pellet (μM) was measured by graphite furnace atomic absorption spectrophotometer (AA-240Z, Varian Inc., Australia) according to manufacturer’s instructions. Briefly, pellet was mixed with, 0.5 mL of nitric acid 65% added with 0.5 mL of ultrapure water (Super-Q® plus, Millipore, Switzerland), sonicated for 30 min at room temperature, and centrifuged 5 min, 10,000 g. Supernatants were diluted with ultrapure water to adequate iron concentration. Standard curves were generated by serial dilution of a commercial iron standard solution (Titrisol, Merck Chemicals, Germany).
Statistical analysis
Data from two independent trials were analyzed by comparing the means with the nonparametric, multiple comparisons with control, Dunnett’s method. For statistical analysis of siderophore activity, B. bifidum MRSc6-292 served as control strain with the lowest siderophore activity and was compared to the rest of the strains. Control sample for statistical analysis of iron internalization was B. thermoacidophilum LMG21397 with the lowest iron internalization value. Correlation between the two set of data was assessed by Spearman’s correlation.
Nucleotide accession numbers
The main nucleotide sequences determined in this study have been assigned GenBank Accession numbers: [GenBank: KJ412975-KJ412985].
Acknowledgments
Dr. Ted Jost for his support in the 16S rRNA sequencing, Aurora Papa for her support in the siderophore characterization.
Additional files
Characterization of eighty-six bifidobacterial strains towards siderophore activity measured by the CAS assay and columns show siderophore activity (%) (n = 2). Characterization of eighty-six bifidobacterial strains towards siderophore activity measured by the CAS assay and columns show siderophore activity (%) (n = 2). Error bars correspond to standard deviations calculated for two independent replicates for siderophore activity. Light gray columns correspond to strains belonging to public culture collections (DSMZ, ATCC, LMG). Dark gray columns correspond to isolates from stool samples of Kenyan infants.
Identity and siderophore activity of bifidobacterial strains isolated from stool of Kenyan infant of different iron status.
RAPD and REP fingerprints of Bifidobacterium spp. isolated from stool of breast fed Kenyan infants included in the characterization iron internalization using a) OPA-02 primer, b) OPL-07 primer, c) OPL-13 primer, d) OPL-16 primer for RAPD-PCR and REP1R-I primer, REP2-I primer for REP-PCR. RAPD and REP fingerprints of Bifidobacterium spp. isolated from stool of breast fed Kenyan infants included in the characterization of SA and II using a) OPA-02 primer, b) OPL-07 primer, c) OPL-13 primer, d) OPL-16 primer for RAPD-PCR and REP1R-I primer, REP2-I primer for REP-PCR.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
VGP carried out the isolation, typing and characterization of isolates and drafted the manuscript. JT collected the fecal samples. ZC carried out the AAS measurements. ZMB read and approved the final manuscript. LC participated in the design of the study, interpretation of data and reading and approving the final manuscript. CC participated in the design of the study, interpretation of data and reading and approving the final manuscript. All authors read and approved the final manuscript.
Contributor Information
Pamela Vazquez-Gutierrez, Email: pamela.vazquez@hest.ethz.ch.
Christophe Lacroix, Email: christophe.lacroix@hest.ethz.ch.
Tanja Jaeggi, Email: tanja.jaeggi@hest.ethz.ch.
Christophe Zeder, Email: christophe.zeder@hest.ethz.ch.
Michael Bruce Zimmerman, Email: michael.zimmermann@hest.ethz.ch.
Christophe Chassard, Email: christophe.chassard@hest.ethz.ch.
References
- 1.Zoetendal EG, Vaughan EE, de Vos WM. A microbial world within us. Mol Microbiol. 2006;59(6):1639–50. doi: 10.1111/j.1365-2958.2006.05056.x. [DOI] [PubMed] [Google Scholar]
- 2.Jost T, Lacroix C, Braegger CP, Chassard C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One. 2012;7(8):e44595. [DOI] [PMC free article] [PubMed]
- 3.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS One. 2012;7(5):e36957. [DOI] [PMC free article] [PubMed]
- 4.Broekaert IJ, Walker WA. Probiotics and chronic disease. J Clin Gastroenterol. 2006;40(3):270–4. doi: 10.1097/00004836-200603000-00021. [DOI] [PubMed] [Google Scholar]
- 5.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Toole PW, Cooney JC. Probiotic bacteria influence the composition and function of the intestinal microbiota. Interdiscip Perspect Infect Dis. 2008;2008:175285. [DOI] [PMC free article] [PubMed]
- 7.Corr SC, Hill C, Gahan CG. Understanding the mechanisms by which probiotics inhibit gastrointestinal pathogens. Adv Food Nutr Res. 2009;56:1–15. doi: 10.1016/S1043-4526(08)00601-3. [DOI] [PubMed] [Google Scholar]
- 8.Payne AN, Zihler A, Chassard C, Lacroix C. Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol. 2012;30(1):17–25. doi: 10.1016/j.tibtech.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 9.O’Sullivan DJ. Screening of intestinal microflora for effective probiotic bacteria. J Agric Food Chem. 2001;49(4):1751–60. doi: 10.1021/jf0012244. [DOI] [PubMed] [Google Scholar]
- 10.Pandey A, Bringel F, Meyer JM. Iron requirement and search for Siderophores in Lactic-Acid Bacteria. Appl Microbiol Biot. 1994;40(5):735–9. doi: 10.1007/BF00173337. [DOI] [Google Scholar]
- 11.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27(2–3):215–37. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 12.Nairz M, Schroll A, Sonnweber T, Weiss G. The struggle for iron - a metal at the host-pathogen interface. Cell Microbiol. 2010;12(12):1691–702. doi: 10.1111/j.1462-5822.2010.01529.x. [DOI] [PubMed] [Google Scholar]
- 13.Johnson EE, Wessling-Resnick M. Iron metabolism and the innate immune response to infection. Microbes Infect. 2012;14(3):207–16. doi: 10.1016/j.micinf.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol. 2006;2(3):132–8. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 15.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160(1):47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 16.Bezkorovainy A, Solberg L. Ferrous iron uptake by Bifidobacterium breve. Biol Trace Elem Res. 1989;20(3):251–67. doi: 10.1007/BF02917440. [DOI] [PubMed] [Google Scholar]
- 17.Patel AK, Deshattiwar MK, Chaudhari BL, Chincholkar SB. Production, purification and chemical characterization of the catecholate siderophore from potent probiotic strains of Bacillus spp. Bioresour Technol. 2009;100(1):368–73. doi: 10.1016/j.biortech.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Bezkorovainy A, Kot E, Miller-Catchpole R, Haloftis G, Furmanov S. Iron metabolism in bifidobacteria. Int Dairy J. 1996;6(10):905–19. doi: 10.1016/S0958-6946(96)00003-9. [DOI] [Google Scholar]
- 19.Bezkorovainy A, Kot E. Interaction of bifidobacteria with ferric iron. Int Dairy J. 1998;8(5–6):507–12. doi: 10.1016/S0958-6946(98)00078-8. [DOI] [Google Scholar]
- 20.Collins HL. Withholding iron as a cellular defence mechanism - friend or foe? Eur J Immunol. 2008;38(7):1803–6. doi: 10.1002/eji.200838505. [DOI] [PubMed] [Google Scholar]
- 21.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Baumler AJ. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5(5):476–86. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz-Ochoa VE, Jellbauer S, Klaus S, Raffatellu M. Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis. Front Cell Infect Microbiol. 2014;4:2. [DOI] [PMC free article] [PubMed]
- 23.Weinberg ED. Iron and infection. Microbiol Rev. 1978;42(1):45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey JR, Probert CS, Cogan TA. Identification and characterisation of an iron-responsive candidate probiotic. PLoS One. 2011;6(10):e26507. [DOI] [PMC free article] [PubMed]
- 25.Hannan MA, Faraji B, Tanguma J, Longoria N, Rodriguez RC. Maternal milk concentration of zinc, iron, selenium, and iodine and its relationship to dietary intakes. Biol Trace Elem Res. 2009;127(1):6–15. doi: 10.1007/s12011-008-8221-9. [DOI] [PubMed] [Google Scholar]
- 26.Nakamori M, Ninh NX, Isomura H, Yoshiike N, Hien VTT, Nhug BT, Nguyen NV, Nakano T, Khan NC, Yamamoto S. Nutritional status of lactating mothers and their breast milk concentration of iron, zinc and copper in Rural Vietnam. J Nutr Sci Vitaminol. 2009;55(4):338–45. doi: 10.3177/jnsv.55.338. [DOI] [PubMed] [Google Scholar]
- 27.Ejezie F, Nwagha U, Ikekpeazu E, Ozoemena O, Onwusi E. Assessment of iron content of breast milk in preterm and term mothers in enugu urban. Ann Med Health Sci Res. 2011;1(1):85–90. [PMC free article] [PubMed] [Google Scholar]
- 28.Petry N, Egli I, Chassard C, Lacroix C, Hurrell R. Inulin modifies the bifidobacteria population, fecal lactate concentration, and fecal pH but does not influence iron absorption in women with low iron status. Am J Clin Nutr. 2012;96(2):325–31. doi: 10.3945/ajcn.112.035717. [DOI] [PubMed] [Google Scholar]
- 29.Adlerberth I. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:13–29. doi: 10.1159/000146245. [DOI] [PubMed] [Google Scholar]
- 30.Arboleya S, Binetti A, Salazar N, Fernandez N, Solis G, Hernandez-Barranco A, Margolles A, de Los Reyes-Gavilan CG, Gueimonde M. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol. 2012;79(3):763–72. doi: 10.1111/j.1574-6941.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 31.Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, Cagnasso P, Bizzarri B, de’Angelis GL, Shanahan F, van Sinderen D, Ventura M. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75(6):1534–45. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl Environ Microbiol. 1999;65(10):4506–12. doi: 10.1128/aem.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita H, Nakano A, Onoda H, Toh H, Oshima K, Takami H, Murakami M, Fukuda S, Takizawa T, Kuwahara T, Ohno H, Tanabe S, Hattori M. Bifidobacterium kashiwanohense sp. nov., isolated from healthy infant faeces. Int J Syst Evol Microbiol. 2011;61(Pt 11):2610–5. doi: 10.1099/ijs.0.024521-0. [DOI] [PubMed] [Google Scholar]
- 34.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. [DOI] [PMC free article] [PubMed]
- 35.Magne F, Abely M, Boyer F, Morville P, Pochart P, Suau A. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol. 2006;57(1):128–38. doi: 10.1111/j.1574-6941.2006.00097.x. [DOI] [PubMed] [Google Scholar]
- 36.Cronin M, Zomer A, Fitzgerald GF, van Sinderen D. Identification of iron-regulated genes of Bifidobacterium breve UCC2003 as a basis for controlled gene expression. Bioeng Bugs. 2012;3(3):157–67. doi: 10.4161/bbug.18985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haen CD. Probiotic formulations. United States: Bracco Imagining.P.A; 2011. p. 8. [Google Scholar]
- 38.Anatoly Bezkorovainy NT. Aspects of iron metabolism in Bifidobacterium bifidum var. pennsylvanicus. Int J Biochem. 1983;15(3):361–6. doi: 10.1016/0020-711X(83)90105-2. [DOI] [PubMed] [Google Scholar]
- 39.Raffatellu M, Baumler AJ. Salmonella’s iron armor for battling the host and its microbiota. Gut Microbes. 2010;1(1):70–2. doi: 10.4161/gmic.1.1.10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos RL, Almeida AP, Xavier MN, Paixao TA, Wilson RP, Dandekar S, Raffatellu M, Baumler AJ. Enteric pathology and Salmonella-induced cell death in healthy and SIV-infected rhesus macaques. Vet Pathol. 2011;48(5):933–41. doi: 10.1177/0300985810386468. [DOI] [PubMed] [Google Scholar]
- 41.Ganz T. Iron in innate immunity: starve the invaders. Curr Opin Immunol. 2009;21(1):63–7. doi: 10.1016/j.coi.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaeggi T, Moretti D, Kvalsvig J, Holding PA, Tjalsma H, Kortman GA, et al. Iron status and systemic inflammation, but not gut inflammation, strongly predict gender-specific concentrations of serum hepcidin in infants in rural Kenya. PLoS One. 2013;8(2):e57513. [DOI] [PMC free article] [PubMed]
- 43.Meile L, Ludwig W, Rueger U, Gut C, Kaufmann P, Dasen G, Wenger S, Teuber M. Bifidobacterium lactis sp. nov, a moderately oxygen tolerant species isolated from fermented milk. Syst Appl Microbiol. 1997;20(1):57–64. doi: 10.1016/S0723-2020(97)80048-3. [DOI] [Google Scholar]
- 44.Cleusix V, Lacroix C, Dasen G, Leo M, Le Blay G. Comparative study of a new quantitative real-time PCR targeting the xylulose-5-phosphate/fructose-6-phosphate phosphoketolase bifidobacterial gene (xfp) in faecal samples with two fluorescence in situ hybridization methods. J Appl Microbiol. 2010;108(1):181–93. doi: 10.1111/j.1365-2672.2009.04408.x. [DOI] [PubMed] [Google Scholar]
- 45.Beerens H. Detection of bifidobacteria by using propionic acid as a selective agent. Appl Environ Microbiol. 1991;57(8):2418–9. doi: 10.1128/aem.57.8.2418-2419.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergey DH, Krieg NR, Holt JG. Bergey’s manual of systematic bacteriology. Baltimore: Williams & Wilkins; 1984. [Google Scholar]
- 47.Simpson PJ, Fitzgerald GF, Stanton C, Ross RP. The evaluation of a mupirocin-based selective medium for the enumeration of bifidobacteria from probiotic animal feed. J Microbiol Methods. 2004;57(1):9–16. doi: 10.1016/j.mimet.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 48.Payne SM. Detection, isolation, and characterization of siderophores. Methods Enzymol. 1994;235:329–44. doi: 10.1016/0076-6879(94)35151-1. [DOI] [PubMed] [Google Scholar]
- 49.Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. 2013;110:1253–62. doi: 10.1017/S0007114513000597. [DOI] [PubMed] [Google Scholar]
- 50.Coderre PE, Earhart CF. The entD Gene of the Escherichia coli K12 Enterobactin Gene Cluster. J Gen Microbiol. 1989;135(11):3043–55. doi: 10.1099/00221287-135-11-3043. [DOI] [PubMed] [Google Scholar]
- 51.Jost T, Lacroix C, Braegger C, Chassard C. Stability of the maternal gut microbiota during late pregnancy and early lactation. Curr Microbiol. 2014;68(4):419–27. doi: 10.1007/s00284-013-0491-6. [DOI] [PubMed] [Google Scholar]
- 52.Stephenson DP, Moore RJ, Allison GE. Comparison and utilization of repetitive-element PCR techniques for typing Lactobacillus isolates from the chicken gastrointestinal tract. Appl Environ Microbiol. 2009;75(21):6764–76. doi: 10.1128/AEM.01150-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kot E, Bezkorovainy A. Uptake of iron by Bifidobacterium thermophilum depends on the metal content of its growth-medium. J Dairy Sci. 1991;74(9):2920–6. doi: 10.3168/jds.S0022-0302(91)78475-0. [DOI] [Google Scholar]


