Summary
RNA-mediated gene silencing in human cells requires the accurate generation of ∼22-nucleotide microRNAs (miRNAs) from double-stranded RNA substrates by the endonuclease Dicer. Although the phylogenetically conserved RNA-binding proteins TRBP and PACT are known to contribute to this process, their mode of Dicer binding and their genome-wide effects on miRNA processing have not been determined. We solved the crystal structure of a human Dicer–TRBP interaction complex comprising two domains of previously unknown structure. Interface residues conserved between TRBP and PACT show that the proteins bind to Dicer in a similar manner and by mutual exclusion. Based on the structure, a catalytically active Dicer that cannot bind TRBP or PACT was designed and introduced into Dicer-deficient mammalian cells, revealing selective defects in guide strand selection. These results demonstrate the role of Dicer-associated RNA binding proteins in maintenance of gene silencing fidelity.
Introduction
MicroRNAs (miRNAs) regulate a diverse array of eukaryotic cellular processes including differentiation, proliferation, and apoptosis (He and Hannon, 2004). In mammals, the RNA-induced silencing complex (RISC) performs miRNA-mediated gene silencing by targeting an Argonaute protein (e.g. Ago2) to repress mRNAs bearing sequence complementarity to a bound ∼22 nucleotide (nt) miRNA guide (Liu et al., 2004). Critical to this process are the production and loading into Argonaute of mature guide miRNAs, which are functions of the endoribonuclease Dicer and its double-stranded RNA-binding protein (dsRBP) partners (Gregory et al., 2005). During formation of RNA-induced silencing complexes (RISCs), Dicer cleaves pre-miRNA hairpin substrates to generate roughly symmetric miRNA duplexes of a specified length. Each product duplex binds to Argonaute in an orientation that defines the guide strand, while the opposing passenger strand is ejected and degraded, a process known as strand selection (Khvorova et al., 2003; Matranga et al., 2005). In humans, the Dicer-associated dsRBP paralogs TRBP and PACT have been shown to influence both cleavage and strand selection activities, although their functions have not been fully defined (Lee et al., 2006). In addition, TRBP and PACT have been implicated in mediating miRNA isoform (isomiR) processing by Dicer to generate related miRNAs of differing lengths and altered targeting specificity (Fukunaga et al., 2012; Lee et al., 2013). TRBP has also been shown to stabilize Dicer (Paroo et al., 2009), and dsRBP homologs in flies are responsible for sorting of small RNAs between distinct pathways (Hartig and Förstemann, 2011; Okamura et al., 2011).
Dicer partner dsRBPs TRBP and PACT may be somewhat functionally redundant in miRNA biogenesis due to their conserved sequence and domain organization: they both consist of three double-stranded RNA-binding domains (dsRBDs), with the first two binding to double-stranded RNA (dsRNA) and the third participating in protein-protein interactions. Accordingly, previous studies of potential defects resulting from the absence of just one of the two proteins may have been hindered due to functional compensation by the remaining dsRBP. In this work, we used crystallography to determine the previously unknown structures of mutually interacting domains in human Dicer and TRBP, revealing that Dicer employs a conserved interface to bind both TRBP and PACT. The structure elucidated the molecular details of a distinct partner-binding interface on Dicer, enabling cellular experiments in which Dicer binding to both proteins was abolished. These experiments revealed unforeseen effects on miRNA length determination and guide strand selection for a subset of cellular miRNAs that associate with Ago2, implicating Dicer partner proteins in maintenance of miRNA production and subsequent gene silencing.
Results
Structure of the Dicer–TRBP interface
To determine how Dicer associates with its dsRBP partners and how these complexes influence miRNA function in mammalian cells, we determined the crystal structure of a human Dicer-TRBP interaction domain complex at 3.2 Å resolution (Table 1). This complex includes the partner-binding domain of the Dicer N-terminal helicase (DicerPBD) bound to the third dsRBD of TRBP (TRBP3), the regions shown experimentally to be both necessary and sufficient for Dicer–TRBP association in vivo (Daniels et al., 2009) (Figure 1A). DicerPBD is a four-helix bundle of 97 amino acid residues and the stable globular domain TRBP3 contains a 69-residue α/β sandwich typical of the dsRBD fold extended at its N-terminus by a partially disordered 31-residue linker containing one helix (Figure 1B). The Dicer–TRBP interface comprises a hydrophobic core surrounded by complementary electrostatic interactions over an area of 1006 Å2 (Figure 1C). Both TRBP3 and full-length TRBP exhibit similar low nanomolar affinities for full-length Dicer as determined by isothermal titration calorimetry, indicating that TRBP3 is responsible for Dicer association (Figure S1A).
Table 1.
Crystallographic statistics.
| Native | Se (peak) | |
|---|---|---|
| Crystal properties | ||
| Space group | F4132 | F4132 |
| Unit cell | ||
| a, b, c (Å) | 292, 292, 292 | 294, 294, 294 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Data collection | ||
| Wavelength (Å) | 1.11111 | 0.979663 |
| Resolution range (Å) | 49.35-3.2 (3.28-3.2) | 169.5 - 3.7 (3.79-3.70) |
| Total reflections | 239356 | 1006047 |
| Unique reflections | 18127 | 21915 |
| Completeness (%) | 100.00 (99.80) | 99.8 (99.3) |
| Redundancy | 13.2 (13.6) | 45.9 (39.5) |
| Rmeas (%) | 18.9 (194.4) | 20.4 (142.3) |
| I/sigma (I) | 18.55 (1.70) | 26.96 (3.67) |
| Wilson B-factor | 79.26 | 103.67 |
| Refinement | ||
| R-factor | 0.2280 | |
| R-free | 0.2593 | |
| Number of atoms | 3192 | |
| Protein residues | 410 | |
| RMS (bonds) | 0.013 | |
| RMS (angles) | 1.59 | |
| Ramachandran favored (%) | 97 | |
| Ramachandran outliers (%) | 0.25 | |
| Clashscore | 22.46 | |
| Average B-factor | 89.5 | |
Figure 1.
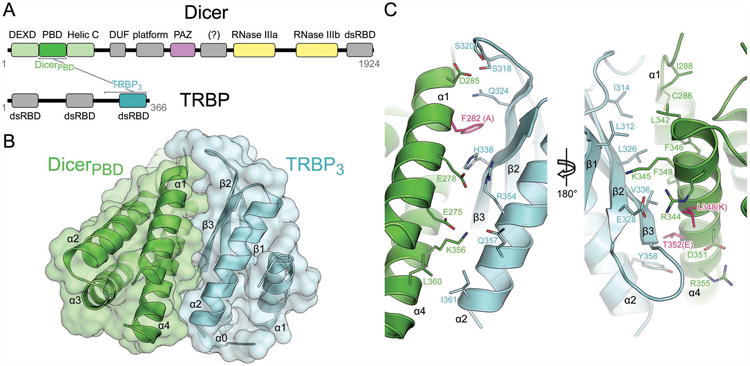
Structure of the Dicer–TRBP interface. (A) Cartoon representation of the primary sequence of Dicer and TRBP with brackets indicating the interacting domains. (B) Overlaid backbone cartoon and surface representations of the Dicer partner-binding domain (PBD) and the third dsRBD of TRBP. (C) Front and back views with interfacial residues shown. Dicer residues mutated to abrogate TRBP and PACT binding are shown in pink with resulting residues indicated in parentheses. See also Figure S1.
The TRBP3 (residues 258–366) core contains an αβ sandwich (residues 289–363), embodying the αβββα fold typical of a dsRBD. An N-terminal extension of 31 residues beyond the canonical dsRBD domain is necessary for TRBP3 stability and is part of a protease-resistant fragment (Figure S1B). This extension contributes to electron density readily observed in the initial experimental map obtained via single wavelength anomalous dispersion, yet the amino acid identity of the sequence remains ambiguous due to poor electron density and was modeled as a separate chain of alanines (Figure S1C). This N-terminal feature abuts a cleft between helices α1 and α2 of the dsRBD core, and its C-terminus is ∼11 Å away from the N-terminus of one crystallographic copy of the TRBP3 dsRBD core and ∼8 Å away from the N-terminus of the other (Figure S1D). Thus, this N-terminal feature may be docking to the dsRBD core in cis, in trans (via domain swapping), or in some heterogeneous combination of the two in the crystal. This potential homodimer interface is unlikely to be biologically relevant due to the weak dissociation constant of 54 μM reported for dimerization (Yamashita et al., 2010). Notably, mRNA decay factor Staufen also contains a degenerate dsRBD that lacks RNA binding activity and mediates homodimerization through an N-terminal helical extension via domain swapped docking onto a similar cleft between the two helices of its dsRBD core (Gleghorn et al., 2013), suggesting a shared structural motif. Indeed the dsRBD core of TRBP3 finds its top DALI match in this fifth dsRBD of Staufen, with a backbone RMSD of 1.9 Å over the 69 core residues (Figure S1E) (Holm and Park, 2000).
The DicerPBD (residues 269–401) model contains neither the disordered loop between α1 and α2 (residues 290–293) nor the disordered C-terminal region (residues 392–401). The C-terminal helix (α5) and the preceding loop (residues 370–391) form a crystallographic homodimer by packing against helices α2 and α3 of a DicerPBD protomer from the neighboring asymmetric unit (Figure S1F). The conformation of residues 370–391 observed in the crystal is almost certainly artifactual based on its divergence from the expected helicase architecture (Figure S1G) and thus is omitted from figures.
Implications for Dicer–PACT binding
To assess whether TRBP paralog PACT might bind to Dicer in a similar manner, we used the extensive sequence conservation between the third dsRBD of PACT (PACT3) and TRBP3 (Figure S2A, B) to generate a homology model based on the Dicer–TRBP interface structure (Figure 2A). Conserved interfacial PACT residues (Figure 2B) provide complementarity to the same DicerPBD surface that binds TRBP3, suggesting that PACT binds to Dicer in the same location as TRBP and that Dicer interactions with the two partner proteins are mutually exclusive. To test this premise, Dicer was mutated in three positions (Dicermut: F282A, L348K, T352E) to disrupt the conserved TRBP- and PACT-binding interface (Figures 1C, 2A, 2C). Observations that TRBP stabilizes Dicer (Chendrimada et al., 2005; Paroo et al., 2009) may stem from inherent instability of the hydrophobic interface region of Dicer that is exposed in the absence of TRBP and PACT. To promote Dicer stability in the absence of partner proteins, Dicermut was designed to enhance polarity of the TRBP/PACT binding site (Figures 1C, 2A, 2C). Dicermut exhibits in vitro behavior and catalytic activity indistinguishable from wild-type (WT) Dicer (Figure S2C). As expected, binding experiments showed that Dicermut has diminished affinity for both TRBP and PACT, supporting the conclusion that these two partner proteins bind the same site on Dicer (Figure 2D).
Figure 2.
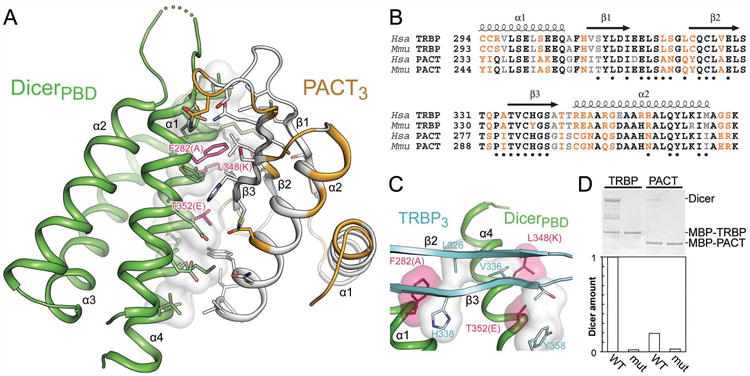
A single surface of Dicer binds TRBP or PACT. (A) A homology model of PACT3 based on the TRBP3 structure reveals conservation of interfacial residues (white, identical or similar; orange, dissimilar). Mutated Dicer residues are shown as in Figure 1C. (B) Sequence alignment comparing TRBP and PACT in human and mouse. Black, identical residues; grey, similar residues; orange, dissimilar residues; dots, TRBP residues located within 5 Å of Dicer in the crystal structure. (C) A view of interfacial contacts between TRBP and Dicer, showing Dicer residues targeted for mutation as in 1C. (D) MBP-tagged TRBP or PACT was used to pull down WT Dicer or Dicermut, demonstrating the latter protein's lack of affinity for dsRBP partner proteins. See also Figure S2.
Role of Dicer partner proteins in miRNA biogenesis
We wondered how the recruitment of TRBP or PACT to Dicer might influence global mammalian miRNA processing and RISC loading in mammalian cells. To investigate the role of this interaction, we took advantage of high conservation between the human and mouse Dicer–TRBP and Dicer–PACT interfaces to introduce the point mutations of human Dicermut into the mouse Dicer gene (Figure S2D, E). This experimental design offers the benefit of displacing both of the potentially functionally redundant proteins TRBP and PACT from Dicer without removing them from the other cellular pathways in which they participate (Daniels and Gatignol, 2012). Deep sequencing was performed for three biological replicates on Ago2-coimmunoprecipitated small RNAs from WT mouse embryonic fibroblast (MEF) cells and on those from a Dcr-/- MEF line (Betancur and Tomari, 2011; Yang et al., 2010a; Yi et al., 2006) transfected for 24 h with either an empty vector or a vector encoding WT Dicer or Dicermut. Under these rescue conditions Dicer was confirmed to be present at levels comparable to the endogenous levels in MEF cells, and undetectable in the Dcr-/- MEF cells instead transfected with empty vector (Figure S2F). Ago2 is known to be destabilized when Dicer is down-regulated (Martinez and Gregory, 2013), and we observed a depletion of Ago2 and other Ago isoforms in the Dcr-/- condition and recovery of these levels upon rescue with WT Dicer or Dicermut (Figure S2G). TRBP and PACT levels were not markedly affected by varying Dicer levels, or by the form of Dicer used for rescue (Figure S2G). In the absence of Dicer, we observed a predominant pool of Ago2-associated RNAs shorter than 15 nt, much smaller than the 20–25 nt species typically resulting from Dicer cleavage (Figure S3A). After rescue of dicing activity via transfection with WT Dicer or Dicermut, pools of canonically sized 20–24 nt Ago2-associated miRNAs are regenerated, but to differing degrees: canonical miRNAs represent 12% of all reads after WT Dicer rescue and 33% of all reads after Dicermut rescue (Figure S3B). We observed striking changes in the abundance of several miRNAs between the two rescue conditions, with eight miRNAs constituting 60% of all canonically sized reads in the Dicermut rescue condition compared to 33% during WT Dicer rescue. We conclude that the unexpected yet reproducible increase in loading of Ago2 with canonically sized miRNAs primarily results from changes in regulation of a class particular miRNAs (including let7f-2, 10b, 99a, 99b, 100, and 125a) that are responsive to levels of free cytoplasmic TRBP as previously reported (Table S1) (DeVito et al., 2012; Melo et al., 2009). Since we observe no change in the total levels of TRBP under differing experimental conditions (Figure S2G), we suspect that the TRBP population liberated from a complex with Dicer under the Dicermut rescue condition is responsible for this varying regulation of certain miRNAs. Accordingly, we are unable to address potential contributions of TRBP and PACT to the efficiency of Ago2 loading. Subsequent analysis was performed using ≥15 nt Ago2-associated miRNAs produced by Dicer.
We wondered whether TRBP and PACT participate in fine-tuning of small RNA processing, performing quality control that influences a subset of substrates. We assessed whether these Dicer partner proteins contribute to two key aspects of miRNA processing: miRNA strand selection and length determination. Strand selection (scored as log〈5p arm coverage/3p arm coverage〉) behavior for pre-miRNA duplexes in a WT MEF reference condition was correlated to rescue conditions with either WT Dicer or Dicermut (Table S2), revealing a reproducible and marked defect in the global fidelity of strand selection when Dicer is unable to recruit a dsRBP partner (Figures 3A and S3C, D). Although most miRNAs exhibit indistinguishable strand selection behavior between the two rescue conditions and the WT MEF reference, in the presence of Dicermut 14 of the 108 miRNAs analyzed show a pronounced change in the proportion of 5′ versus 3′ miRNA strands associated with Ago2 (Table S3), including an instance where the strand preference is markedly inverted (miR-30e) (Figure S3E). Thus Dicer partner proteins contribute to correct strand selection for a subset of miRNA duplexes.
Figure 3.
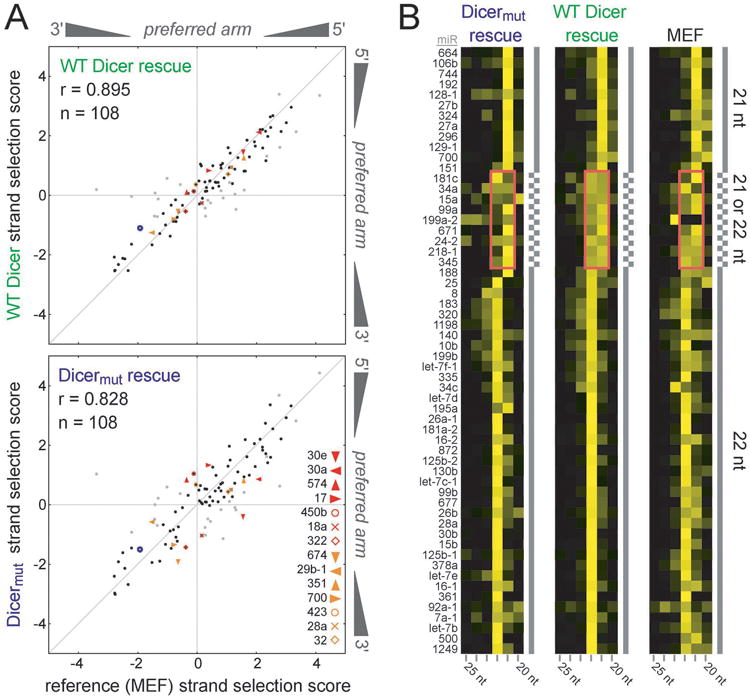
Roles of Dicer partner proteins TRBP and PACT in miRNA biogenesis. (A) Correlation of strand selection behavior (scored as log〈5′ arm coverage/3′ arm coverage〉 among Ago2-associated miRNA) between WT MEF (X axis) and rescue conditions (Y axis) with either WT Dicer or Dicermut demonstrates the importance of TRBP and PACT in maintaining fidelity of strand selection. In the Dicermut condition, an increased deviation from the diagonal is observed due to impaired strand selection fidelity. Labels denote the 14 miRNA duplexes most dramatically affected (to >1 standard deviation) by the loss of TRBP and PACT recruitment to Dicer. 23 candidate duplexes varying in strand selection behavior by more than one standard deviation between MEF and WT rescue conditions (shown in grey) are not considered based on the possibility that they are behaving aberrantly due to the Dcr-/- MEF context. The blue open circle represents miR-132. (B) Cluster analysis of miRNA lengths observed in WT MEF cells or Dicer knockout cells rescued with either WT Dicer or Dicermut, revealing an increased propensity for formation of 22 nt products instead of 21 nt products when Dicer can recruit TRBP and PACT for certain sensitive miRNAs (pink boxes). Ordering is based on results obtained in the WT Dicer rescue condition. See also Figure S3.
Examination of the size of Ago2-associated RNAs revealed that Dicer's recruitment of dsRBP partners also has an effect on Dicer product length for a subset of miRNAs (Figure 3B). This is apparent for miRNAs whose length varies between 21 and 22 nt, the two most abundant functional miRNA lengths. Our analysis did not discriminate between strands, but this should not influence our interpretation since the nine miRNAs implicated here do not overlap with the 14 cases that are sensitive to TRBP/PACT regarding their strand selection. This observation supports the idea that formation of a stable Dicer–dsRBP complex can promote generation of a product with an additional nucleotide (an isomiR) in the case of miRNAs that Dicer alone cleaves with imperfect precision.
Because thermodynamic asymmetry of miRNA duplexes has been implicated in the process of strand selection, we examined our panel of miRNA duplexes for a correlation between duplex terminal base pairing stability and propensity for strand selection to be sensitive to TRBP/PACT recruitment to Dicer (Figure S3A). We observed no such correlation, and similarly detected no correlation between the thermodynamic asymmetry of miRNA duplexes with their strand selection score under any single experimental condition (Figure S3A). These observations underscore the difficulty of predicting miRNA strand selection behavior based on thermodynamic properties alone (Malefyt et al., 2014).
We next considered the role of Argonaute protein binding specificity for the 5′ nucleotide of miRNAs (Frank et al., 2010) in defining which miRNA duplexes exhibited strand selection behavior linked to the recruitment of TRBP/PACT by Dicer. Since Ago2 binds guide strands bearing 5′-terminal U or A nucleotides preferentially versus strands bearing C or G (Frank et al., 2010), differing nucleotides at these termini could be the primary factor determining which strand is selected as the guide in some cases. Notably, a change in the position of Dicer-mediated pre-miRNA cleavage induced by TRBP (Fukunaga et al., 2012; Lee et al., 2013) can result in a change in the 5′ nucleotide identity of the product miRNA duplex. In the case of miR-30e, for which strand preference was inverted in the absence of Dicer-dsRBP interaction (Figures 3A and S3E), a TRBP-induced change in Dicer cleavage position could trigger a change in 5′-terminal nucleotide identity. The longer potential miR-30e duplex bears a 5′-U on the 5p arm and a Dicer-generated 5′-C terminus on the 3p arm, and the 5′-U-containing strand is the selected guide that binds to Ago2 (Figure 4A). In contrast, a shorter potential isomiR duplex results in both ends of the mature duplex bearing 5′-U, eliminating the nucleotide identity distinction that might otherwise drive Ago2-mediated strand selection preference. Thus the inversion of miR-30e strand selection observed in the absence of TRBP/PACT recruitment to Dicer could be triggered by a change in Dicer cleavage position linked to the presence or absence of a dsRBP partner.
Figure 4.
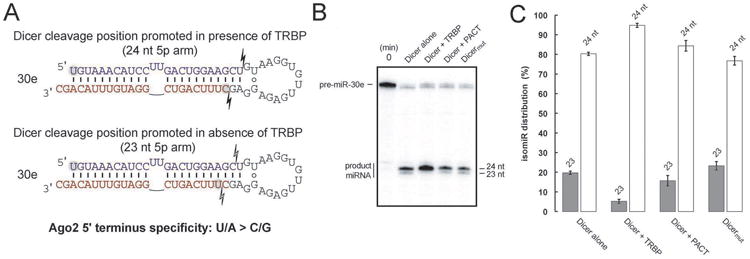
Impact of TRBP on Dicer cleavage position of miR-30e. (A) In vitro dicing of pre-miR-30e shows a dependence on TRBP for consistent cleavage of a 24 nt product. (B) Quantification of isomiR distributions resulting from dicing assays of pre-miR-30e. Error bars represent standard deviation based on triplicate experiments. (C) Schematic showing the effect of altered dicing on pre-miR-30e and ramifications for downstream Ago2 loading. See also Figure S4.
To test the roles of TRBP and/or PACT in the formation of mir-30e isomiRs, we assayed for Dicer cleavage position in vitro, comparing WT Dicer with Dicermut both alone and in the presence of TRBP or PACT. Since ejected passenger strands are not detected via sequencing, interpretation of those data will be complicated for cases in which cleavage position variability is linked to changes in strand selection preference; therefore we examined Dicer cleavage position directly. We observed similar cleavage behavior of pre-miR-30e for Dicermut and for WT Dicer alone or in the presence of PACT, but a distinct change in isomiR production in the presence of TRBP (Figure 4B, C). This enhanced formation of a 1-nt longer isomiR in the presence of TRBP is consistent with the change in Ago2-loaded mir-30e strands observed in the cellular sequencing data, providing a plausible explanation for the inversion of strand selection when Dicer is incapable of recruiting TRBP. The ability of TRBP to influence isomiR formation during dicing while PACT cannot agrees with previous studies distinguishing the activity of these Dicer binding partners. The strand selection behavior of miR-30a was also sensitive to TRBP recruitment by Dicer (Figure 3A), and due to a pre-miR structure nearly identical to pre-miR-30e, a similar mechanism by which isomiR formation influences Ago2-mediated strand selection is likely. Indeed, a recent report has shown a TRBP-based dependency on Dicer cleavage position of pre-miR-30a analogous to our findings for pre-miR-30e (Kim et al., 2014). Among four other pre-miRs that were candidates for this mechanism and displayed a dsRBP sensitivity for their strand selection, we found that miR-423 and miR-32 bear a similar sensitivity to TRBP for isomiR formation during dicing (Figure S4B).
Overall structure of TRBP
To probe the mechanistic link between these effects on miRNA biogenesis and the behavior of Dicer partner proteins during RISC loading, we used NMR to investigate the inter-domain interactions in TRBP. 1H-15N HSQC spectra were collected using the full-length protein or using constructs representing its constituent globular domains (Figures 5 and S5). No pronounced chemical shift perturbations are induced in the presence of neighboring domains, demonstrating that the three domains of TRBP do not associate with each other in the absence of protein or RNA interacting partners, consistent with a previous study that examined the first two domains and not the third (Benoit et al., 2013). Hence TRBP is expected to sample a conformation wherein the maximum distance between its three globular domains is limited only by the number of residues tethering the domains.
Figure 5.
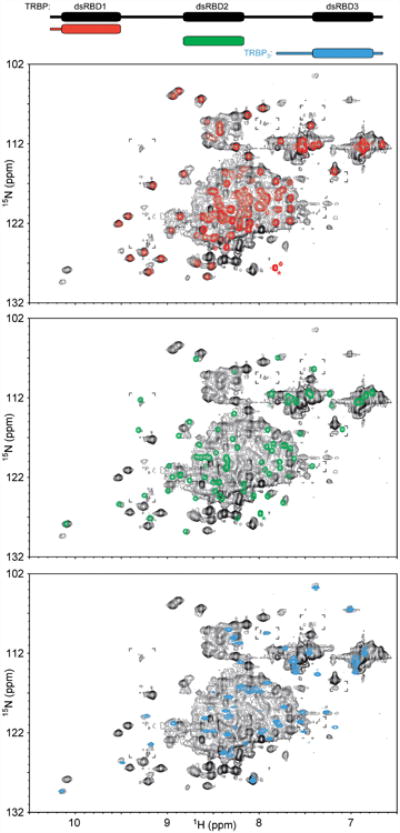
The absence of inter-domain interactions in TRBP. 1H-15N HSQC spectra of individual TRBP dsRBD constructs (dsRBD1, red, top; dsRBD2, green, middle; TRBP3 which includes dsRBD3, blue, bottom) show no pronounced chemical shift perturbations when overlaid with a spectrum of full-length TRBP containing all three dsRBDs (black), indicating the absence of inter-domain interactions. Asterisks mark peaks assigned via their diagnostic intensity to the C-termini of the dsRBD1 or dsRBD2 constructs, expected to be shifted in the spectrum of the full-length TRBP due to change in chemical environment. Brackets mark regions wherein the full-length TRBP spectrum is displayed at a slightly lower threshold to show low-intensity peaks. See also Figure S5.
Discussion
Many organisms encode Dicer-associated RNA-binding proteins that are thought to influence substrate recognition and processing during miRNA maturation. To understand how human Dicer interacts with its partner protein TRBP, we determined the crystal structure of the interacting portions of Dicer and TRBP, revealing previously unknown structures of two domains contributing to the Dicer–TRBP interface. To help understand these domains in their functional contexts, they were compared to related protein structures. Alignment of DicerPBD to the structurally similar lobe of the dsRNA helicase RIG-I (Figure S6A) (Luo et al., 2012) showed that TRBP3 does not block the expected helicase-RNA interaction. Furthermore, alignment of TRBP3 to the RNA-bound form of the second RNA-binding domain of TRBP (Yang et al., 2010b) revealed that TRBP3 binds Dicer using a surface distinct from that used for dsRNA recognition (Figure S6B). Amino acid substitutions on the Dicer-distal face of TRBP3 appear to be responsible for the domain's distinct loss of dsRNA binding affinity relative to that of canonical dsRBDs (Figure S6B).
PACT, a human paralog of TRBP, also interacts with Dicer, but it has been unknown whether PACT and TRBP bind Dicer simultaneously, by mutual exclusion, or in tandem (Kok et al., 2007). Conservation between TRBP and PACT within the Dicer–dsRBP interface suggested that the proteins bind in a mutually exclusive manner (Figure 2A), as confirmed using Dicer mutations that abolish binding to both dsRBP partners (Figure 2D). This experiment ruled out simultaneous direct binding to Dicer, although it remains possible that TRBP and PACT associate with each other while only one is bound to Dicer (Kok et al., 2007; Laraki et al., 2008).
TRBP has been suggested to extend the half-life of Dicer in cells through its physical association with Dicer and its phosphorylation state (Paroo et al., 2009). The stability of the Dicer–TRBP complex is reflected in our observation that DicerPBD cannot be successfully expressed and purified unless TRBP3 is present. This effect is likely due to the large hydrophobic patch on Dicer that is exposed to solvent in the absence of a dsRBP partner. Phosphorylation of TRBP occurs at four sites, two of which are located in the linker peptide preceding the folded structure of TRBP3 and distal to the Dicer interface (Figures S1D and S2A). This observation suggests that TRBP phosphorylation does not directly alter the affinity of TRBP for Dicer and may instead influence Dicer stability as a downstream consequence of altered TRBP stability.
A persistent question in the field has been the role of TRBP and PACT in guide strand selection (Betancur and Tomari, 2011; Noland and Doudna, 2013; Tomari, 2004), the process in which one arm of a miRNA duplex is discarded (the passenger strand or miR*) while the other (the guide strand or miR) is loaded onto Argonaute to provide targeting specificity. Failure of this crucial step in miRNA biogenesis could result in spurious gene silencing and/or the loss of intended silencing. Early studies implicated dsRBPs as robust players in discrimination of potential guide strands (Tomari, 2004), but subsequent studies suggested that they may not be necessary for this step (Betancur and Tomari, 2011; Noland and Doudna, 2013). Comparing the miRNA populations bound to Ago2 in the context of a Dicer knockout MEF line rescued via transfection of either WT Dicer or the dsRBP binding–incompetent Dicermut revealed subtle but reproducible defects in global strand selection fidelity. For 13% of the miRNA duplex pairs analyzed, strand selection diverged from typical behavior due to the absence of TRBP and PACT from the miRNA biogenesis pathway. Notably, the miRNAs most sensitive to such dsRBP-influenced strand selection have been linked to processes including cellular differentiation, apoptosis, and cancer metastasis (Table S3).
Inspection of miRNA length and its dependence on the recruitment of partner dsRBPs to Dicer reveals an influence in cases where Dicer product length typically varies between 21–22 nt, the two most abundant lengths for miRNA (Figure 3B). The populations of these variable miRNAs tend to shift towards 22 nt when Dicer is able to bind TRBP and PACT, suggesting that these proteins can contribute to miRNA length determination. This is consistent with evidence that TRBP can change the position of Dicer cleavage and promote formation of an RNA product with an additional nucleotide (Fukunaga et al., 2012; Lee et al., 2013).
We found no trend in sequence or predicted pre-miRNA structure to distinguish the top 14 miRNAs identified as sensitive to TRBP/PACT recruitment to Dicer (Fig. 3A) from those miRNAs least sensitive to dsRBP recruitment regarding strand selection behavior. Furthermore, 5 of 14 sensitive miRNAs obeyed the “loop counting rule” (5′ end of 3p arm is two base pairs away from a disruption of the helix), in good agreement with the ∼33% of endogenous mammalian miRNAs for which this rule was previously reported to apply (Gu et al., 2012). We also observed no correlation between strand selection behavior and thermodynamic asymmetry (Figure S4A, B). However, in some cases TRBP-induced changes in 5′ nucleotide identity following Dicer processing likely triggers a switch in strand selection based on Ago2 RNA binding preferences. In vitro cleavage assays support this mechanism for miR-30e and related miRNAs, as well as for miR-423 and miR-32, all of which showed altered strand selection behavior in cells containing the Dicermut protein that cannot recruit TRBP.
A previous report identified one mammalian miRNA duplex, miR-132, for which a 21-nt rather than 22-nt product is generated preferentially in the absence of TRBP (Fukunaga et al., 2012). Examination of our sequencing results reveals a similar TRBP dependency in miR-132, with an increase in 21-nt product generated when Dicer cannot recruit TRBP/PACT (Figure S4C). However, strand selection of the miR-132 duplex is not affected by dsRBP recruitment to Dicer (Figure 3A). This may be because the 5′-terminal nucleotide of the guide strand remains either A or U depending on the altered cleavage position, both of which are favorably bound by Ago2 and would not be expected to influence strand selection based on Ago2 specificity as in the case of miR-30e (Figure 4).
The domain organization of TRBP and its mode of Dicer association were used together with recent structural studies of Dicer (Lau et al., 2012) to generate a model of the Dicer–TRBP complex that provides insight into the spatial organization and associated processing mechanisms of the miRNA biogenesis machinery (Figure 6A). As detailed above, TRBP3 was positioned relative to the Dicer helicase domain based on DicerPBD homology to RIG-I (Luo et al., 2012), while the first and second dsRBDs of TRBP are shown with their linker connectivities and orientations of the N- and C-termini of each domain based on published structures (Yamashita et al., 2010). HSQC experiments found no evidence for inter-domain interactions in TRBP (Figures 5 and S5), suggesting that the span of the protein is defined by the lengths of the inter-domain linkers. The 90 residues of TRBP inter-domain linkers is expected to span up to 270 Å, a distance exceeding the 150 Å measured across the longest axis of Dicer and providing access to all RNA-interacting functional groups in Dicer. Additionally, since Ago2 binds to the RNase IIIa domain of Dicer, TRBP may be capable of binding to Dicer products to enhance the rate and influence orientation of loading into Ago2 (Sasaki and Shimizu, 2007).
Figure 6.
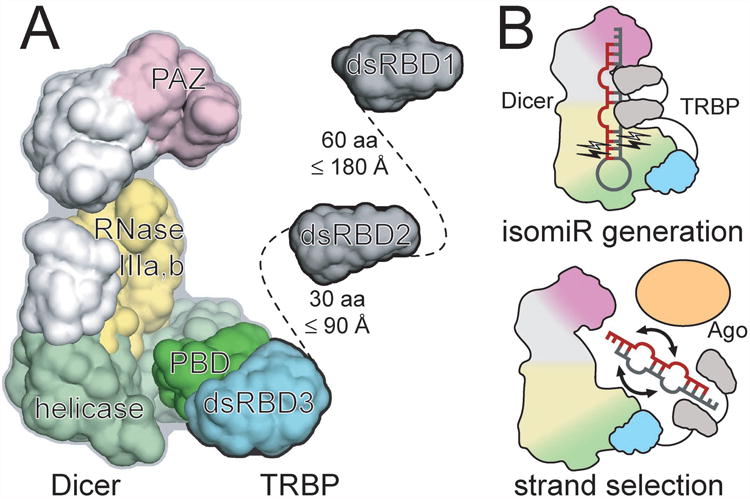
Mechanisms of Dicer partner proteins in the context of the miRNA biogenesis machinery. (A) The human Dicer architecture (as determined by electron microscopy) is colored according to functional domains (PAZ, pink; RNase IIIa/b, yellow; helicase, green), with the Dicer–TRBP interface structure determined in the present crystallographic work shown in dark green (DicerPBD) and cyan (TRBP3). NMR results suggest that the two N-terminal RNA-binding domains of an extended TRBP can readily access an RNA bound near the paired RNase III active sites of Dicer. (B) Models for how Dicer partner proteins contribute to isomiR formation (top) and strand selection fidelity during transfer of a Dicer product duplex to Argonaute (bottom). See also Figure S6.
Since these dsRBDs bind preferentially to canonical A-form dsRNA (Masliah et al., 2013), their binding could compress a pre-miRNA duplex containing mismatched or bulged nucleotides towards an A-form geometry that positions an additional base pair within the region of Dicer that determines miRNA length (Figures 6A and 6B, top) (Fukunaga et al., 2012). In addition, the ability of dsRBDs to slide along dsRNA ligands (Koh et al., 2013) could help ensure preferential binding at the more thermodynamically stable (thus likely closer to A-form) end of a miRNA duplex produced by Dicer, thereby occluding that end and increasing the likelihood that the Ago2 MID domain might bind the 5′ terminus at the other, less stable end (Figure 6B, bottom). This would result in behavior consistent with previously observed determinants of strand selection wherein the 5′ terminus at the less thermodynamically stable end of a duplex is more likely to serve as the guide in RISC (Noland and Doudna, 2013). Additionally, the MID domain of Ago2 is known to exhibit specificity in its binding of the different 5′ terminal nucleotides (U/A > C/G) (Frank et al., 2010). This property of Ago2 can result in changes in strand selection behavior downstream of TRBP-induced shifting of Dicer cleavage position and 5′-terminal nucleotide identity of product miRNA duplexes.
The fact that only certain miRNA duplexes exhibit Dicer partner dsRBP-dependent strand selection behavior is consistent with a pathway involving contributions from Dicer, Argonaute, and Dicer-binding proteins to varying degrees depending on substrate characteristics, an idea supported by previous work (Betancur and Tomari, 2011; Noland and Doudna, 2013). The critical yet nuanced role of Dicer partner proteins in miRNA biogenesis parallels the delicately balanced regulation of cellular processes imparted by miRNAs themselves (Flynt and Lai, 2008). Together, the crystal structure of the human Dicer–TRBP interface and the global analysis of Argonaute-loaded miRNAs resulting from abolition of this interaction in cells show that the Dicer–TRBP and Dicer–PACT complexes contribute to proper miRNA length and strand selection in a subset of mammalian miRNAs.
Experimental Procedures
Protein Expression & Purification
For clarity, the 366 residue isoform TRBP2 will be referred to as “TRBP” throughout, and residue numbers will refer to the full-length protein. Dicer, TRBP, or PACT samples were purified as previously described (Lee et al., 2013). The Dicer–TRBP interface complex was isolated via coexpression of untagged DicerPBD (residues 269–401) and His6-MBP-tagged TRBP3 (residues 258–366) followed by copurification (protocol as above) with tag removal. Additional TRBP constructs comprised residues 1–105 (dsRBD1), 154–234 (dsRBD2), 228–366 (dsRBD3 and the preceding linker), and 98–366 (deletion of dsRBD1). 15N-labeled TRBP construct samples were prepared by using M9 minimal media containing 15N-labeled ammonium chloride and purifying as usual. Selenomethionine (SeMet) derivatized protein was prepared by supplementing M9 minimal media (Sambrook et al., 1989) via the pathway-inhibition protocol (Ent et al., 1999).
Trypsin Digest
A 1 g/L sample of a TRBP construct containing residues 228–366 (dsRBD3 and the preceding linker) was incubated with either 0.1% or 1% (by mass) trypsin at room temperature for 60 min and quenched with SDS-PAGE loading dye (Figure S1B). Subsequent mass spectrometry indicated a stable fragment: residues 258–366, leading to generation of the construct used for subsequent experiments (Figure S2A, B).
Crystallography, Data Processing, & Model Building
Native crystals were grown at 16°C via hanging drop vapor diffusion and typically appeared within two weeks. Drops were prepared by mixing 2 μL of 7.3 g/L TRBP-Dicer interface complex in crystallization buffer (10 mM sodium acetate pH 5, 50 mM sodium chloride, 1 mM TCEP, and 5% (w/v) glycerol) with 1 μL reservoir solution (260 mM potassium sulfate, 1 mM TCEP, and 18% (w/v) PEG 3350). SeMet derivatized crystals were prepared similarly, but by mixing 1 μL 9.4 g/L protein in crystallization buffer with 1 μL water and 0.5 μL reservoir solution (250 mM potassium sulfate, 1 mM TCEP, and 17% (w/v) PEG 3350). Crystals were cryo-protected via soaking for 15 s in modified reservoir solution supplemented with 6% (w/v) glycerol, mounted on nylon loops, and plunged into liquid nitrogen. Data were collected at the Advanced Light Source (Lawrence Berkeley National Laboratory), beamline 8.3.1.
Data were processed using XDS (Kabsch, 2010), with heavy atom search performed using SHELX (Sheldrick, 2008). Initial map generation and solvent flattening was performed using SOLVE/RESOLVE and CCP4 (Winn et al., 2011). Initial modeling of helices was performed using ARP/wARP, followed by manual building of the model using Coot. Crystals belonged to the cubic space group F4132 with two copies of the Dicer–TRBP interface heterodimer per asymmetric unit. The model was subjected to iterative rounds of refinement against the isomorphous native map with consideration of real space XYZ coordinates, group B-factors, secondary structure-based hydrogen bond restraints, backbone non-crystallographic symmetry restraints, and translation/libration/screw (TLS) parameters.
Isothemal Titration Calorimetry
Protein samples were dialysed against 300 mM KCl, 5% glycerol, 1 mM TCEP, and 20 mM HEPES NaOH pH 7.5 for > 24 h. Using an ITC-200 Auto (GE Healthcare), WT TRBP (residues 1–366) or TRBP3 (residues 258–366) at 100 or 180 μM was injected into 10 μM Dicer using one 0.5 μL injection followed by nineteen 2 μL injections at 25°C. Experiments were performed in duplicate. Data were analyzed via modified Origin software (GE Healthcare), fitting to a one-site binding model following baseline correction. Control experiments with TRBP injected into buffer were performed to account for heats possibly associated with dimer dissociation of TRBP (Yamashita et al., 2010), but no such heat could be observed.
Mutagenesis
To abrogate binding by TRBP or PACT, the following Dicer mutations were made: F282A, L348K, and T352E. This was performed for the human enzyme via site directed mutagenesis using the following primers: F282A-f, GCTGATGGAATTAGAAGAAGCACTTAATGCTATCAATGATTGTAATATATCTCTG; F282A-r, CAGAGATATATTACAATCATTGATAGCATTAAGTGCTTCTTCTAATTCCATCAGC; L348K+T352E-f, GGAAATTTTTAAAGTTTACAGACGAATTCCTAAGGAAAATACATGC; L348K+T352E-r, CCTTAGGAATTCGTCTGTAAACTTTAAAAATTTCCTGTGCAG.
Mutations at equivalent positions of mouse Dicer were made using the following primers: F270A-f, GATGGAGTTAGAAGCAGCACTTGATGCTATCAATGATTGTAATGTAGCTGTAC; F270A-r, GTACAGCTACATTACAATCATTGATAGCATCAAGTGCTGCTTCTAACTCCATC; L336K+T340E-f, GGAAGTTCCTAAAGTTTACAGACGAATTGTTAAGGAAAATACACGC; L336K+T340E-r, CCTTAACAATTCGTCTGTAAACTTTAGGAACTTCCTGTGTAGCTCC.
Dicing Assays
5′-32P-labeled RNA substrates were annealed in 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM DTT, 1% glycerol, and 1.5 mM Igepal CA-630 by heating at 95°C for 5 min followed by snap cooling on ice for 5 min. Single turnover dicing assays were performed in 5 μL mixtures, which consisted of 100 nM Dicer (or previously purified Dicer-dsRBP complex (Lee et al., 2013)) and < 1 nM 5′ 32P-labeled substrate. Reactions were incubated for 15 min at 37°C with 10 nM pre-let-7 hairpin RNA in 20 mM Tris-HCl pH 7.5, 25 mM NaCl, 1 mM DTT, 1% glycerol, 1.5 mM MgCl2, and 0.01% Igepal CA-630. Reactions were quenched using EDTA and the denaturing PAGE gels were visualized by phosphorimaging. All RNA substrates were gel purified using 12% acrylamide denaturing PAGE prior to use. RNA transcripts contained a 3′-terminal sequence for cloning [GGGAAUUCUAAUACGACUCACUAUAGGGAGA], a 5′-terminal HDV ribozyme sequence [GGGUCGGCAUGGCAUCUCCACCUCCUCGCGGUCCGACCUGGGCAUCCGAGGAAAC UCGGAUGGCUAAGGGAGAGCCAACGAGUAGUGGGAUCCGGG], and a variable internal sequence containing a hammerhead ribozyme [CUGAUGAGUCCGUGAGGACGAAACGGUACCCGGUACCGUC] (HH) and a pre-miR (underlined) that was released after self-cleavage of the adjacent ribozymes:
| 30e | GGAUGUUUACA[HH]UGUAAACAUCCUUGACUGGAAGCUGUAAGGUGUUGAGAGGAGCUUUCAGUCGGAUGUUUACAGC |
| 574 | ACACACACUCA[HH]UGAGUGUGUGUGUGUGAGUGUGUGUCGCUCCAAGUCCACGCUCAUGCACACACCCACA |
| 17 | UAAGCACUUUG[HH]CAAAGUGCUUACAGUGCAGGUAGUGAUGUGUGCAUCUACUGCAGUGAGGGCACUUGUAG |
| 423 | UCUGCCCCUCA[HH]UGAGGGGCAGAGAGCGAGACUUUUCUAUUUUCCAAAAGCUCGGUCUGAGGCCCCUCAGU |
| 32 | AAUGUGCAAUA[HH]UAUUGCACAUUACUAAGUUGCAUGUUGUCACGGCCUCAAUGCAAUUUAGUGUGUGUGAUAUU |
| let-7 | CUACUACCUCA[HH]UGAGGUAGUAGGUUGUAUAGUUUUAGGGUCACACCCACCACUGGGAGAUAACUAUACAAUCUACUGUCUUUC |
Pulldown
13 pmol His6-MBP-tagged TRBP or PACT was allowed to bind to equimolar amounts of WT Dicer or Dicermut in 200 μL binding buffer (300 mM NaCl, 30 mM imidazole, 5% glycerol, 20 mM HEPES NaOH 7.5, and 1 mM TCEP) for 20 min at RT. This mixture was bound to equilibrated nickel affinity resin for 15 min at RT and then washed twice with 500 μL binding buffer. Beads were boiled in 1× loading dye to elute bound protein and run on SDS-PAGE. Protein bands were quantified using Image Lab (BioRad) and bound Dicer quantities were plotted.
NMR Spectroscopy
NMR data were collected at 298 K at the NMR Facility at the University of California, Berkeley on Bruker Avance II 900 MHz spectrometer equipped with a Bruker cryogenic probe. HSQC spectra were collected using 1 mM protein at 25°C in a buffer of 20 mM HEPES NaOH pH 7.5, 100 mM potassium chloride, 5% (w/v) glycerol, 1 mM TCEP, and 5% D2O. Data were processed using NMRpipe and analyzed using NMRviewJ.
Transfection, Library Preparation, and Sequencing
Plasmids expressed mouse Dicer with an N-terminal 3×FLAG tag, driven by a TK promoter. 10 cm2 plates of WT or Dcr-/- (Yang et al., 2010a) mouse embryonic fibroblast (MEF) cells were transfected with 10 μg of plasmid using 30 μL of Lipofectamine 2000 (Life Technologies). Fresh media (Glutamax DMEM (Gibco) supplemented with 10% fetal bovine serum and 1% pennicillin/streptomycin) was introduced after 5 h. Cells were harvested after 24 h via scraping followed by washing with PBS. For lysis, cells were resuspended in a hypotonic buffer (10 mM HEPES NaOH pH 8, 1.5 mM MgCl2, 10 mM KCl, 1 mM TCEP, and 1 mM PMSF) to induce swelling, followed by eight passes through a 27G½ needle. Nuclei and cellular debris were subsequently pelleted by centrifugation. Total protein was quantified using Bradford reagent and subsequent steps used equivalent amounts of total protein. Western blots were performed using DM1A (Abcam) for tubulin control, 610418 (BD Biosciences) for Hsp90 control, J101 (a gift from Chrysi Kanellopoulou, NIH) against Dicer, 2D4 (Wako) against Ago2, 2A8 (Millipore) against pan-Ago (Ago1/2/3/4), 1B9-1A7 (Abcam) against PACT, and ab42018 (Abcam) against TRBP. For immunoprecipitation, 3 μL (4 μg) anti-mouse AGO2 antibody (2D4, Wako) was bound to 50 μL of Dynabeads G (Life Technologies) slurry in 100 μL citrate-phosphate buffer (25 mM and 50 mM, respectively; pH 5) for 90 min at RT with agitation. 200 μL cleared lysate (supplemented with 4 M sodium chloride to a final concentration of 200 mM) was bound to the antibody-coated beads with agitation for 4 h at 4°C. Beads were washed with 200 μL PBS and the RNA was isolated via extraction with acid phenol followed by ethanol precipitation. 75 ng of isolated RNA was used in library preparation with the miRvana kit (Life Technologies) modified with a custom 5′ adapter with eight degenerate nucleotides (N), 5′ GUUCAGAGUUCUACAGUCCGACGAUCNNNNNNNNGUAC 3′, to incorporate a distinct primer I.D. for each ligation event, eliminating signal noise from PCR amplification (Jabara et al., 2011). After library amplification, samples were gel purified and ∼150 bp bands were excised, eluted, and ethanol precipitated. Samples were run on an Illumina HiSeq2500 in high output mode and single-end sequenced.
Sequencing Analysis
Raw sequencing data was pre-processed before alignment by filtering for read quality, collapsing PCR replicates (based on primer I.D.) and removing adapter regions, using the Fastx Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). After pre-processing, ∼1.5 million reads remained in each sample. Pre-processed sequences were aligned to a reference pre-miRNA transcriptome (Kozomara and Griffiths-Jones, 2011) with Bowtie2 in global alignment mode, allowing for up to 100 degenerate mappings (Bowtie2 settings–very-sensitive –k 100) (Langmead and Salzberg, 2012). 60-70% of reads were aligned to the pre-miRNA transcriptome under these settings. This is true even for the predominantly <15 nt reads observed under the Dcr-/- condition, although such sequences derive from pre-miRNAs cleaved or degraded by Dicer-independent processes. Ambiguous mappings were resolved by using eXpress (eXpress settings –output-align-prob, -B 20, and all other settings as default) to produce an estimated probability of correct alignment, such that the sum of these probabilities for all alignments for a given read sequence was set to 1 (Roberts and Pachter, 2013). This step re-scales each mapping of a multi-mapped read based on its probability of alignment, and ensures quantitative treatment of alignments. This step is especially important for this experiment because the short read lengths of miRNAs are expected to produce many ambiguously mapped reads. Downstream analysis of alignments was performed using custom-written Python scripts using the open-source libraries PySam and BioPy. Read counts at each step of analysis are presented in Table S4. Our analysis of strand selection behavior was as follows: for each mouse pre-miR annotated in miRbase with at least one read at each arm, we plotted log〈5′ arm coverage/3′ arm coverage〉 – defined as the “strand selection score” – in a scatter plot with the WT MEF condition as reference and compared this to values obtained under the rescue conditions using WT Dicer or Dicermut. To determine the miRNAs most drastically affected by the presence or absence of Dicer partner dsRBPs, we averaged the values from the equivalent replicate datasets, sorted the miRNAs by strand selection score, and removed from consideration (due to potentially atypical behavior) miRNA duplexes where magnitude of strand selection score differed by more than a standard deviation (0.49) between WT MEF reference and WT Dicer rescue conditions. Top remaining cases differing by more than one standard deviation in magnitude (0.60) between WT MEF reference and the Dicermut rescue condition are reported in Table S3. Cluster analysis was performed on the miRNAs meeting the above criteria for strand selection scoring. Data for both potential strands were considered in tandem for each miRNA. Ordering of miRNAs was determined based on the WT Dicer rescue clustering, and this ordering was applied to the other two samples for comparison.
Supplementary Material
Figure S1, related to main figure 1. TRBP and Dicer: structure and interactions. (A) Thermograms resulting from titration of WT TRBP (left) or TRBP3 (right) into Dicer. Duplicate runs yielded the following values for WT TRBP: Kd = 2.8 ± 1.7 nM, ΔH = -15.9 ± 0.48 kcal/mol, n = 0.84 ± 0.013; TRBP3: Kd = 4.3 ± 2.5 nM, ΔH = -23.2 ± 1.3 kcal/mol, n = 1.35 ± 0.025. Stoichiometric deviations from n=1 likely result from protein samples that are partially misfolded, aggregated, or otherwise incompetent for binding. (B) Trypsin digestion of human TRBP residues 228–366 (dsRBD3 plus the 70 residues between it and dsRBD2) reveals the stable TRBP3 core (residues 258–366) used for crystallography. This corresponds to a globular domain ∼50% larger than a typical dsRBD. Protein marker ladder is labeled with kDa values. (C) An omit map contoured at 1.0 σ showing unbiased density for the partially helical portion of TRBP3 (α0) N-terminal of the dsRBD fold. (D) The canonical dsRBD fold consists of α1, β1, β2, β3, and α2. Orientation of adjacent TRBP3 protomers in the crystal reveals the possibilty of domain-swapping involving the 36 amino acid extension N-terminal to the dsRBD fold. Disorder prevents assignment of the fold as cis (∼11 Å distance) or trans (∼8 Å distance). It is plausible that the domain-swapped trans conformation is adopted above the 54 μM Kd observed for TRBP dimerization, while the cis conformation is adopted when the protein is a monomer. A red asterisk marks the approximate location of two phosphorylation sites (S283, S286) found within a linker region disordered in the crystal. (E) Staufen (PDB ID: 4DKK)(Gleghorn et al., 2013) contains a non-canonical dsRBD with a 1.9 Å backbone root-mean-square deviation to the non-canonical dsRBD found in TRBP3. Staufen contains an N-terminal extension involved in domain swapping. (F) Crystal contacts between neighboring copies of DicerPBD, mediated by helix α5 and the preceding loop. (G) Homodimeric contacts between Dicer protomers are not likely to be biologically relevant. For the dimer to form, helix α5 must adopt a conformation inconsistent with the fold of its natural helicase context, as demonstrated by alignment to the structure of homologous helicase RIG-I (PDB ID: 4AY2)(Luo et al., 2012).
Figure S2, related to main figure 2. Sequences of TRBP and PACT, and mutation of Dicer. (A) Domain structure and conservation of TRBP and PACT. The dsRBDs are marked by red, green, or blue for the first, second, or third domains, respectively. The structured extension N-terminal to dsRBD3 is shown in grey. Sequence alignment of human TRBP and PACT. Secondary structure elements from independently reported TRBP domain structures (PDB ID: dsRBD 1, 3LLH; dsRBD 2, 2CPN)(Yamashita et al., 2010) or from the present crystal structure (dsRBD3) are shown above the sequences. Labels for potential α0 helices outside the canonical dsRBD region are based on a prediction from the Jpred server. The globular TRBP3 domain (grey, blue regions) is defined based on mass spectrometric analysis following trypsin digestion. Notably, trypsin cleaved after arginine 257, less efficiently after arginine 259, and not detectably after arginine 269, providing precision in the domain assignment. Red asterisks mark known TRBP phosphorylation sites. (B) Scale representation of the domain structure in TRBP and PACT. Inset numbers represent the estimated length of flexible linkers. Based on conservation and secondary structure prediction, PACT is expected to feature a structured region in the linker between the second and third domains analogous to the one observed in TRBP. (C) Cleavage assay demonstrating indistinguishable pre-let-7 dicing activity of Dicermut and WT Dicer. (D) DicerPBD alignment between human and mouse; secondary structure observed in the crystal structure is indicated above the aligned sequences. Interfacial TRBP or Dicer residues found within 5 Å of the binding partner are indicated with a black dot. Alignment of the human DicerPBD domain residues used in crystallization vs. the equivalent mouse residues. Pink letters indicate the three Dicer mutations used to abrogate binding to TRBP or PACT. (E) Alignment of TRBP3 and the equivalent region in PACT between human and mouse; secondary structure observed for TRBP in the crystal structure is indicated above the aligned sequences. Cartoons marking likely α0 helices outside the canonical dsRBD region are based on predictions from the Jpred server. (F) Western blot demonstrating comparable levels of Dicer in transfected (rescue) conditions and in WT MEF cells. This also confirms the absence of Dicer in the Dcr-/- MEF line. (G) Western blots for Ago proteins (the pan-Ago antibody detects Ago1/Ago2/Ago3/Ago4), TRBP, and PACT under various experimental conditions. Ago levels are dependent on the presence of Dicer, while TRBP and PACT levels are generally independent of Dicer/Ago levels.
Figure S3, related to main figure 3. RNA sequencing data. (A) Histogram plots of normalized read coverage versus length for Ago2-coimmunoprecipitated RNA in WT MEF cells, Dcr -/- MEF, Dcr -/- MEF rescued with WT Dicer, and Dcr-/- MEF rescued with Dicermut. Two distinct populations are observed: shorter reads (mean ≈ 8 nt) are associated with Dicer-independent Ago2 loading while longer reads (mean ≈ 22 nt) are associated with canonical, Dicer-dependent miRNA processing. (B) Quantification of read density for Dicer-independent (< 15 nt RNA) and Dicer-dependent (≥ 15 nt RNA) loading of Ago2 demonstrates that canonical loading of Argonaute is not assisted by the presence of TRBP and PACT. Each data point represents an average value ±SD from biological triplicates. Rescue conditions are not able to recover fully to the typical ratio between the two pools of miRNA length during the 24 h following transfection, likely due to the extremely long (∼1 week) half-life of Ago2 (Nabanita et al., 2013). (C) Strand selection data showing error bars from three biological replicates. Correlation of strand selection behavior (scored as log〈5′ arm coverage/3′ arm coverage〉) between WT MEF (X axis) and rescue conditions (Y axis) with either WT Dicer or Dicermut demonstrates the importance of TRBP and PACT in maintaining fidelity of strand selection. In the Dicermut condition, an increased deviation from the diagonal is observed due to impaired ;strand selection fidelity. Labels denote the miRNA duplexes most dramatically affected. Duplexes varying by more than one standard deviation between MEF and WT rescue conditions are not considered based on the likelihood that they are behaving aberrantly due to use of the Dicer-KO cell line. (D) Paired correlations between biological replicate samples for three conditions demonstrates reproducibility of strand selection observations. Strand selection score is defined as: log〈5′ arm coverage/3′ arm coverage〉. Similar behavior is observed between replicates. These plots include more points than the plots comparing between conditions (Fig. 3A) because those cases require that qualifying counts be present in all of the conditions being compared. (E) Deep sequencing coverage for the 5′ and 3′ arms of four miRNA duplexes where Dicer protein partners TRBP and PACT contribute most to the fidelity of strand selection. Biological triplicate results for WT MEF, WT Dicer rescue, and Dicermut rescue are shown in black, green, and blue, respectively. Inset plots show quantified strand selection scores (defined as log〈5′ arm coverage/3′ arm coverage〉) by sample. Each data point represents an average value ±SD from replicates. WT Dicer rescue (green) traces approximate strand selection behavior of WT MEF cells (black), while Dicermut rescue (blue) traces deviate from the traditional strand selection behavior.
Figure S4, related to main figure 4. RNA sequencing and biochemical data. (A) Assessment of thermodynamic asymmetry in miRNA duplexes as correlated to the strand selection score (log〈5p arm coverage/3p arm coverage〉). The top row of plots compares each observed strand selection score to the difference in calculated thermodynamic stability between the two termini of a miRNA duplex. A positive dddG value corresponds to greater stability at the 5′ end of the 5p arm. Using a Pearson R test for correlation, none of these correlations are significant; correlation in all cases is between -0.3 and +0.3. (B) In vitro dicing assay for miR length. Lanes show uncleaved pre-miR, cleavage with Dicer alone, Dicer plus TRBP, Dicer plus PACT, or Dicermut alone. For pre-miR-423 and pre-miR-32, the cleavage position is influenced by the presence of TRBP (asterisk) but is insensitive to PACT, while the other two pre-miRNAs assayed are insensitive to dsRBPs. Secondary structures of pre-miR substrates are shown, with filled or open icons respectively representing the typical or variant Dicer cleavage positions. Grey circles represent potential resulting 5′ termini of the miRNA duplex. (C) Distribution of length variants in the predominant, 3p arm of miR-132 under different experimental conditions. Values are by percentage of the total reads for 20, 21, and 22 nt RNAs. Error bars represent the standard deviation between three replicates. 21 nt isomiRs become more abundant when Dicer cannot recruit TRBP/PACT.
Figure S5, related to main figure 5. 1H-15N HSQC spectra of TRBP domains. (A) Legend for the different domains used. Black, wild type; red, dsRBD1; green, dsRBD2; blue, dsRBD3 with the preceding 36 residues (TRBP3); teal, dsRBD3 with the preceding 67 residues; brown, dsRBD2 and dsRBD3 with the linker residues N-terminal to each domain. (B) Spectrum of the full length (wild type) TRBP with all domains contained in a single polypeptide (1 mM). (C) Spectrum of dsRBD1 (1 mM). (D) Spectrum of dsRBD2 (1 mM). (E) Spectrum of dsRBD3 with the preceding 36 residues (TRBP3, 138 μM). (F) Overlaid spectra of dsRBD2 (green, 1 mM), TRBP3 (blue, 138 μM), and the construct containing both domains plus the additional linker residues (brown, 116 μM). This demonstrates that dsRBD2 peaks are not perturbed in the context of dsRBD3 and the intervening linker. It has previously been shown that dsRBD2 does not interact with dsRBD1 in the absence of binding partners(Benoit et al., 2013). (G) Overlaid spectra of wild type TRBP (black, 1 mM), TRBP3 (blue, 1 mM), and dsRBD3 with the preceding 67 residues (teal, 1 mM). Asterisks mark prominent peaks (not accounted for in the context of Figure 4) corresponding to residues found in the 31 residue linker N-terminal to TRBP3.
Figure S6, related to main figure 6. Comparison of domains in the Dicer:TRBP interface structure. (A) Structural similarity between the DicerPBD and the equivalent portion of the helicase RIG-I (PDB ID: 4AY2)(Luo et al., 2012). The RIG-I structure (grey) contains a dsRNA duplex (red). In the likely case that Dicer's helicase adopts a tertiary structure similar to that of RIG-I, TRBP will rest on the outside of the helicase “clamp” that binds dsRNA. In the bottom view, the N-terminus extends “up” from α0, in the same direction that the Dicer architecture extends with its paired RNase III domains and its PAZ domain. (B) The dsRBD architecture is conserved between the second and third domains of TRBP, but different faces of the domain are utilized in binding. TRBP3 (cyan) binds to DicerPBD using a face distinct from that used by TRBP2 (green, PDB ID: 3ADL)(Yang et al., 2010) to bind dsRNA. TRBP2 residues involved in RNA recognition are shown as sticks, as are the TRBP3 residues in equivalent positions (except in the case of glycine or a shortened loop). TRBP3 bears dissimilar features in positions corresponding to TRBP2 residues Q165, H188, and R215, contributing to the domain's loss of dsRNA affinity.
Table S1, related to main figure 3. Relative abundance of the miRNAs most different between the Dcr-/-rescue conditions using WT Dicer or Dicermut. The “%” value refers to the share of reads for a given miRNA for a particular replicate, and the standard deviation is reported for that value between three replicates. The “Total%” value demonstrates that the majority of the Ago2-bound miRNAs in the Dicermut condition correspond to one of these eight miRNAs.
Table S2, related to main figure 3. The spreadsheet lists strand selection scores (log〈5p arm coverage/3p arm coverage〉) for the 108 microRNAs that could be compared across MEF, WT Dicer rescue, and Dicermut rescue conditions. Scores are reported for three biological replicates and as an averaged value with standard deviation.
Table S3, related to main figure 3. MicroRNAs whose strand selection behavior is dramatically altered in the absence of Dicer partner dsRBPs. Human miR equivalent are noted in parentheses when homolog naming differs from mouse miR. The “Δs.s.” values denote the change in strand selection score (defined as log〈5′ arm coverage/3′ arm coverage〉) between the WT MEF reference sample and the Dicermut sample.
Table S4, related to main figure 3. Sequencing reads throughout data processing and analysis. Note that total input RNA was normalized before library preparation, so the “input” values do not reflect the amount of RNA obtained via Ago2 immunoprecipitation under the various experimental conditions.
Acknowledgments
We thank Doudna Lab members for helpful discussion, Lior Pachter for essential insights, Jamie Cate for critically reading the manuscript, Jeff Pelton for assistance with NMR, David King for providing mass spectrometry, the Lawrence Berkeley National Laboratory beam line staff, Ian MacRae for sharing of Dicer architecture coordinates, Eric Lai and Alexander Tarakhovsky for contributing the Dcr-/- MEF cell line, Chrysi Kanellopoulou for the kind gift of Dicer antibodies, Ann Fischer for assistance with cell culture, Minyong Chung at the Vincent J. Coates Genomics Sequencing Laboratory for providing sequencing, Kendall Condon and Megan Hochstrasser for technical assistance, Mary Matyskiela for providing reagents, and Amy Lee for providing reagents and discussion. This work was supported by NIH grants GM073794 and GM096689.
Footnotes
Accession Numbers: The molecular coordinates have been deposited to the protein data bank (PDB: 4WYQ). Sequencing data have been uploaded in .fastq format (Bioproject ID: PRJNA267577).
Author Contributions: R.C.W., A.T., and J.A.D. designed the experiments. R.C.W., C.L.N., and C.P.S. prepared samples. R.C.W., C.L.N., C.P.S., and M.A.K. conducted the experiments. R.C.W. and A.T. analyzed the data. R.C.W. and J.A.D. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Benoit M, Imbert L, Palencia A, Pérard J, Ebel C, Boisbouvier J, Plevin M. The RNA-binding region of human TRBP interacts with microRNA precursors through two independent domains. Nucleic Acids Research. 2013;41:4241–52. doi: 10.1093/nar/gkt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur JG, Tomari Y. Dicer is dispensable for asymmetric RISC loading in mammals. RNA. 2011;18:24–30. doi: 10.1261/rna.029785.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels SM, Melendez-Peña CE, Scarborough RJ, Daher A, Christensen HS, Far El M, Purcell DF, Lainé S, Gatignol A. Characterization of the TRBP domain required for Dicer interaction and function in RNA interference. BMC Molecular Biology. 2009;10:38. doi: 10.1186/1471-2199-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S, Gatignol A. The multiple functions of TRBP, at the hub of cell responses to viruses, stress, and cancer. Microbiology And Molecular Biology Reviews : MMBR. 2012;76:652–66. doi: 10.1128/MMBR.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVito C, Riggi N, Cornaz S, Suvà M, Baumer K, Provero P, Stamenkovic I. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell. 2012;21:807–21. doi: 10.1016/j.ccr.2012.04.023. [DOI] [PubMed] [Google Scholar]
- Ent F, Lockhart A, Kendrick-Jones J, Löwe J. Crystal structure of the N-terminal domain of MukB: a protein involved in chromosome partitioning. Structure (London, England : 1993) 1999;7:1181–7. doi: 10.1016/s0969-2126(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Flynt A, Lai E. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nature Reviews Genetics. 2008;9:831–42. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Han BW, Hung J, Xu J, Weng Z, Zamore PD. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleghorn M, Gong C, Kielkopf C, Maquat L. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nature Structural & Molecular Biology. 2013;20:515–24. doi: 10.1038/nsmb.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R, Chendrimada T, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis P, Kay M. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–11. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Förstemann K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Research. 2011;39:3836–3851. doi: 10.1093/nar/gkq1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon G. MicroRNAs: small RNAs with a big role in gene regulation. Nature Reviews Genetics. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics (Oxford, England) 2000;16:566–7. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- Jabara C, Jones C, Roach J, Anderson J, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2011;108:20166–71. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallographica Section D, Biological Crystallography. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim Y, Yeo J, Lee J, Jung Lee N, Lee JH, Cho J, Seo D, Kim J, Kim NV, et al. Deletion of Human tarbp2 Reveals Cellular MicroRNA Targets and Cell-Cycle Function of TRBP. Cell Reports. 2014;0:1–15. doi: 10.1016/j.celrep.2014.09.039. [DOI] [PubMed] [Google Scholar]
- Koh H, Kidwell M, Ragunathan K, Doudna J, Myong S. ATP-independent diffusion of double-stranded RNA binding proteins. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2013;110:151–6. doi: 10.1073/pnas.1212917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok KH, Ng MJ, Ching Y, Jin D. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laraki G, Clerzius G, Daher A, Melendez-Peña C, Daniels S, Gatignol A. Interactions between the double-stranded RNA-binding proteins TRBP and PACT define the Medipal domain that mediates protein-protein interactions. RNA Biology. 2008;5:92–103. doi: 10.4161/rna.5.2.6069. [DOI] [PubMed] [Google Scholar]
- Lau P, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ. The molecular architecture of human Dicer. Nature Structural & Molecular Biology. 2012;19:436–440. doi: 10.1038/nsmb.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Zhou K, Smith A, Noland C, Doudna J. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Research. 2013;41:6568–76. doi: 10.1093/nar/gkt361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park S, Kim Y, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell M, Rivas F, Marsden C, Thomson J, Song J, Hammond S, Joshua-Tor L, Hannon G. Argonaute2 is the catalytic engine of mammalian RNAi. Science (New York, NY) 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Luo D, Kohlway A, Vela A, Pyle A. Visualizing the determinants of viral RNA recognition by innate immune sensor RIG-I. Structure (London, England : 1993) 2012;20:1983–8. doi: 10.1016/j.str.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malefyt A, Wu M, Vocelle D, Kappes S, Lindeman S, Chan C, Walton S. Improved asymmetry prediction for short interfering RNAs. The FEBS Journal. 2014;281:320–30. doi: 10.1111/febs.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N, Gregory R. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA (New York, NY) 2013;19:605–12. doi: 10.1261/rna.036434.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah G, Barraud P, Allain FH. RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cellular And Molecular Life Sciences : CMLS. 2013;70:1875–95. doi: 10.1007/s00018-012-1119-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nature Genetics. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Noland C, Doudna J. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA (New York, NY) 2013;19:639–48. doi: 10.1261/rna.037424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Robine N, Liu Y, Liu Q, Lai EC. R2D2 organizes small regulatory RNA pathways in Drosophila. Molecular And Cellular Biology. 2011;31:884–896. doi: 10.1128/MCB.01141-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the Human MicroRNA-Generating Complex Mediates MAPK/Erk Signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nature Methods. 2013;10:71–3. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning 1989 [Google Scholar]
- Sasaki T, Shimizu N. Evolutionary conservation of a unique amino acid sequence in human DICER protein essential for binding to Argonaute family proteins. Gene. 2007;396:312–320. doi: 10.1016/j.gene.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallographica Section A, Foundations Of Crystallography. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Tomari Y. A Protein Sensor for siRNA Asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D, Biological Crystallography. 2011;67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Nagata T, Kawazoe M, Takemoto C, Kigawa T, Güntert P, Kobayashi N, Terada T, Shirouzu M, Wakiyama M, et al. Structures of the first and second double-stranded RNA-binding domains of human TAR RNA-binding protein. Protein Science. 2010;20:118–130. doi: 10.1002/pro.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O′Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2010a;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Chen H, Yang J, Machida S, Chua N, Yuan YA. Structure of Arabidopsis HYPONASTIC LEAVES1 and Its Molecular Implications for miRNA Processing. Structure/Folding And Design. 2010b;18:594–605. doi: 10.1016/j.str.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, O′Carroll D, Pasolli H, Zhang Z, Dietrich F, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nature Genetics. 2006;38:356–62. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to main figure 1. TRBP and Dicer: structure and interactions. (A) Thermograms resulting from titration of WT TRBP (left) or TRBP3 (right) into Dicer. Duplicate runs yielded the following values for WT TRBP: Kd = 2.8 ± 1.7 nM, ΔH = -15.9 ± 0.48 kcal/mol, n = 0.84 ± 0.013; TRBP3: Kd = 4.3 ± 2.5 nM, ΔH = -23.2 ± 1.3 kcal/mol, n = 1.35 ± 0.025. Stoichiometric deviations from n=1 likely result from protein samples that are partially misfolded, aggregated, or otherwise incompetent for binding. (B) Trypsin digestion of human TRBP residues 228–366 (dsRBD3 plus the 70 residues between it and dsRBD2) reveals the stable TRBP3 core (residues 258–366) used for crystallography. This corresponds to a globular domain ∼50% larger than a typical dsRBD. Protein marker ladder is labeled with kDa values. (C) An omit map contoured at 1.0 σ showing unbiased density for the partially helical portion of TRBP3 (α0) N-terminal of the dsRBD fold. (D) The canonical dsRBD fold consists of α1, β1, β2, β3, and α2. Orientation of adjacent TRBP3 protomers in the crystal reveals the possibilty of domain-swapping involving the 36 amino acid extension N-terminal to the dsRBD fold. Disorder prevents assignment of the fold as cis (∼11 Å distance) or trans (∼8 Å distance). It is plausible that the domain-swapped trans conformation is adopted above the 54 μM Kd observed for TRBP dimerization, while the cis conformation is adopted when the protein is a monomer. A red asterisk marks the approximate location of two phosphorylation sites (S283, S286) found within a linker region disordered in the crystal. (E) Staufen (PDB ID: 4DKK)(Gleghorn et al., 2013) contains a non-canonical dsRBD with a 1.9 Å backbone root-mean-square deviation to the non-canonical dsRBD found in TRBP3. Staufen contains an N-terminal extension involved in domain swapping. (F) Crystal contacts between neighboring copies of DicerPBD, mediated by helix α5 and the preceding loop. (G) Homodimeric contacts between Dicer protomers are not likely to be biologically relevant. For the dimer to form, helix α5 must adopt a conformation inconsistent with the fold of its natural helicase context, as demonstrated by alignment to the structure of homologous helicase RIG-I (PDB ID: 4AY2)(Luo et al., 2012).
Figure S2, related to main figure 2. Sequences of TRBP and PACT, and mutation of Dicer. (A) Domain structure and conservation of TRBP and PACT. The dsRBDs are marked by red, green, or blue for the first, second, or third domains, respectively. The structured extension N-terminal to dsRBD3 is shown in grey. Sequence alignment of human TRBP and PACT. Secondary structure elements from independently reported TRBP domain structures (PDB ID: dsRBD 1, 3LLH; dsRBD 2, 2CPN)(Yamashita et al., 2010) or from the present crystal structure (dsRBD3) are shown above the sequences. Labels for potential α0 helices outside the canonical dsRBD region are based on a prediction from the Jpred server. The globular TRBP3 domain (grey, blue regions) is defined based on mass spectrometric analysis following trypsin digestion. Notably, trypsin cleaved after arginine 257, less efficiently after arginine 259, and not detectably after arginine 269, providing precision in the domain assignment. Red asterisks mark known TRBP phosphorylation sites. (B) Scale representation of the domain structure in TRBP and PACT. Inset numbers represent the estimated length of flexible linkers. Based on conservation and secondary structure prediction, PACT is expected to feature a structured region in the linker between the second and third domains analogous to the one observed in TRBP. (C) Cleavage assay demonstrating indistinguishable pre-let-7 dicing activity of Dicermut and WT Dicer. (D) DicerPBD alignment between human and mouse; secondary structure observed in the crystal structure is indicated above the aligned sequences. Interfacial TRBP or Dicer residues found within 5 Å of the binding partner are indicated with a black dot. Alignment of the human DicerPBD domain residues used in crystallization vs. the equivalent mouse residues. Pink letters indicate the three Dicer mutations used to abrogate binding to TRBP or PACT. (E) Alignment of TRBP3 and the equivalent region in PACT between human and mouse; secondary structure observed for TRBP in the crystal structure is indicated above the aligned sequences. Cartoons marking likely α0 helices outside the canonical dsRBD region are based on predictions from the Jpred server. (F) Western blot demonstrating comparable levels of Dicer in transfected (rescue) conditions and in WT MEF cells. This also confirms the absence of Dicer in the Dcr-/- MEF line. (G) Western blots for Ago proteins (the pan-Ago antibody detects Ago1/Ago2/Ago3/Ago4), TRBP, and PACT under various experimental conditions. Ago levels are dependent on the presence of Dicer, while TRBP and PACT levels are generally independent of Dicer/Ago levels.
Figure S3, related to main figure 3. RNA sequencing data. (A) Histogram plots of normalized read coverage versus length for Ago2-coimmunoprecipitated RNA in WT MEF cells, Dcr -/- MEF, Dcr -/- MEF rescued with WT Dicer, and Dcr-/- MEF rescued with Dicermut. Two distinct populations are observed: shorter reads (mean ≈ 8 nt) are associated with Dicer-independent Ago2 loading while longer reads (mean ≈ 22 nt) are associated with canonical, Dicer-dependent miRNA processing. (B) Quantification of read density for Dicer-independent (< 15 nt RNA) and Dicer-dependent (≥ 15 nt RNA) loading of Ago2 demonstrates that canonical loading of Argonaute is not assisted by the presence of TRBP and PACT. Each data point represents an average value ±SD from biological triplicates. Rescue conditions are not able to recover fully to the typical ratio between the two pools of miRNA length during the 24 h following transfection, likely due to the extremely long (∼1 week) half-life of Ago2 (Nabanita et al., 2013). (C) Strand selection data showing error bars from three biological replicates. Correlation of strand selection behavior (scored as log〈5′ arm coverage/3′ arm coverage〉) between WT MEF (X axis) and rescue conditions (Y axis) with either WT Dicer or Dicermut demonstrates the importance of TRBP and PACT in maintaining fidelity of strand selection. In the Dicermut condition, an increased deviation from the diagonal is observed due to impaired ;strand selection fidelity. Labels denote the miRNA duplexes most dramatically affected. Duplexes varying by more than one standard deviation between MEF and WT rescue conditions are not considered based on the likelihood that they are behaving aberrantly due to use of the Dicer-KO cell line. (D) Paired correlations between biological replicate samples for three conditions demonstrates reproducibility of strand selection observations. Strand selection score is defined as: log〈5′ arm coverage/3′ arm coverage〉. Similar behavior is observed between replicates. These plots include more points than the plots comparing between conditions (Fig. 3A) because those cases require that qualifying counts be present in all of the conditions being compared. (E) Deep sequencing coverage for the 5′ and 3′ arms of four miRNA duplexes where Dicer protein partners TRBP and PACT contribute most to the fidelity of strand selection. Biological triplicate results for WT MEF, WT Dicer rescue, and Dicermut rescue are shown in black, green, and blue, respectively. Inset plots show quantified strand selection scores (defined as log〈5′ arm coverage/3′ arm coverage〉) by sample. Each data point represents an average value ±SD from replicates. WT Dicer rescue (green) traces approximate strand selection behavior of WT MEF cells (black), while Dicermut rescue (blue) traces deviate from the traditional strand selection behavior.
Figure S4, related to main figure 4. RNA sequencing and biochemical data. (A) Assessment of thermodynamic asymmetry in miRNA duplexes as correlated to the strand selection score (log〈5p arm coverage/3p arm coverage〉). The top row of plots compares each observed strand selection score to the difference in calculated thermodynamic stability between the two termini of a miRNA duplex. A positive dddG value corresponds to greater stability at the 5′ end of the 5p arm. Using a Pearson R test for correlation, none of these correlations are significant; correlation in all cases is between -0.3 and +0.3. (B) In vitro dicing assay for miR length. Lanes show uncleaved pre-miR, cleavage with Dicer alone, Dicer plus TRBP, Dicer plus PACT, or Dicermut alone. For pre-miR-423 and pre-miR-32, the cleavage position is influenced by the presence of TRBP (asterisk) but is insensitive to PACT, while the other two pre-miRNAs assayed are insensitive to dsRBPs. Secondary structures of pre-miR substrates are shown, with filled or open icons respectively representing the typical or variant Dicer cleavage positions. Grey circles represent potential resulting 5′ termini of the miRNA duplex. (C) Distribution of length variants in the predominant, 3p arm of miR-132 under different experimental conditions. Values are by percentage of the total reads for 20, 21, and 22 nt RNAs. Error bars represent the standard deviation between three replicates. 21 nt isomiRs become more abundant when Dicer cannot recruit TRBP/PACT.
Figure S5, related to main figure 5. 1H-15N HSQC spectra of TRBP domains. (A) Legend for the different domains used. Black, wild type; red, dsRBD1; green, dsRBD2; blue, dsRBD3 with the preceding 36 residues (TRBP3); teal, dsRBD3 with the preceding 67 residues; brown, dsRBD2 and dsRBD3 with the linker residues N-terminal to each domain. (B) Spectrum of the full length (wild type) TRBP with all domains contained in a single polypeptide (1 mM). (C) Spectrum of dsRBD1 (1 mM). (D) Spectrum of dsRBD2 (1 mM). (E) Spectrum of dsRBD3 with the preceding 36 residues (TRBP3, 138 μM). (F) Overlaid spectra of dsRBD2 (green, 1 mM), TRBP3 (blue, 138 μM), and the construct containing both domains plus the additional linker residues (brown, 116 μM). This demonstrates that dsRBD2 peaks are not perturbed in the context of dsRBD3 and the intervening linker. It has previously been shown that dsRBD2 does not interact with dsRBD1 in the absence of binding partners(Benoit et al., 2013). (G) Overlaid spectra of wild type TRBP (black, 1 mM), TRBP3 (blue, 1 mM), and dsRBD3 with the preceding 67 residues (teal, 1 mM). Asterisks mark prominent peaks (not accounted for in the context of Figure 4) corresponding to residues found in the 31 residue linker N-terminal to TRBP3.
Figure S6, related to main figure 6. Comparison of domains in the Dicer:TRBP interface structure. (A) Structural similarity between the DicerPBD and the equivalent portion of the helicase RIG-I (PDB ID: 4AY2)(Luo et al., 2012). The RIG-I structure (grey) contains a dsRNA duplex (red). In the likely case that Dicer's helicase adopts a tertiary structure similar to that of RIG-I, TRBP will rest on the outside of the helicase “clamp” that binds dsRNA. In the bottom view, the N-terminus extends “up” from α0, in the same direction that the Dicer architecture extends with its paired RNase III domains and its PAZ domain. (B) The dsRBD architecture is conserved between the second and third domains of TRBP, but different faces of the domain are utilized in binding. TRBP3 (cyan) binds to DicerPBD using a face distinct from that used by TRBP2 (green, PDB ID: 3ADL)(Yang et al., 2010) to bind dsRNA. TRBP2 residues involved in RNA recognition are shown as sticks, as are the TRBP3 residues in equivalent positions (except in the case of glycine or a shortened loop). TRBP3 bears dissimilar features in positions corresponding to TRBP2 residues Q165, H188, and R215, contributing to the domain's loss of dsRNA affinity.
Table S1, related to main figure 3. Relative abundance of the miRNAs most different between the Dcr-/-rescue conditions using WT Dicer or Dicermut. The “%” value refers to the share of reads for a given miRNA for a particular replicate, and the standard deviation is reported for that value between three replicates. The “Total%” value demonstrates that the majority of the Ago2-bound miRNAs in the Dicermut condition correspond to one of these eight miRNAs.
Table S2, related to main figure 3. The spreadsheet lists strand selection scores (log〈5p arm coverage/3p arm coverage〉) for the 108 microRNAs that could be compared across MEF, WT Dicer rescue, and Dicermut rescue conditions. Scores are reported for three biological replicates and as an averaged value with standard deviation.
Table S3, related to main figure 3. MicroRNAs whose strand selection behavior is dramatically altered in the absence of Dicer partner dsRBPs. Human miR equivalent are noted in parentheses when homolog naming differs from mouse miR. The “Δs.s.” values denote the change in strand selection score (defined as log〈5′ arm coverage/3′ arm coverage〉) between the WT MEF reference sample and the Dicermut sample.
Table S4, related to main figure 3. Sequencing reads throughout data processing and analysis. Note that total input RNA was normalized before library preparation, so the “input” values do not reflect the amount of RNA obtained via Ago2 immunoprecipitation under the various experimental conditions.


