SUMMARY
Stem cells undergo a shift in metabolic substrate utilization during specification and/or differentiation, a process that has been termed metabolic reprogramming. Here, we report that during the transition from quiescence to proliferation, skeletal muscle stem cells experience a metabolic switch from fatty acid oxidation to glycolysis. This reprogramming of cellular metabolism decreases intracellular NAD+ levels and the activity of the histone deacetylase SIRT1, leading to elevated H4K16 acetylation and activation of muscle gene transcription. Selective genetic ablation of the SIRT1 deacetylase domain in skeletal muscle results in increased H4K16 acetylation and deregulated activation of the myogenic program in SCs. Moreover, mice with muscle-specific inactivation of the SIRT1 deacetylase domain display reduced myofiber size, impaired muscle regeneration, and derepression of muscle developmental genes. Overall, these findings reveal how metabolic cues can be mechanistically translated into epigenetic modifications that regulate skeletal muscle stem cell biology.
INTRODUCTION
Cellular energy is generated via oxidative-phosphorylation (OXPHOS) in the mitochondria and glycolysis in the cytoplasm. In addition to providing a steady supply of energy, the metabolic state of the cell can influence the epigenome and alter gene expression. This flow of information is afforded by intermediate metabolites that directly or indirectly affect the activity of chromatin-modifying enzymes involved in regulating chromatin dynamics and transcription (Katada et al., 2012; Lu and Thompson, 2012). Cellular substrate and oxygen availability, as well as energy demand, determine which metabolic pathway is employed to generate ATP. Under reduced oxygen tension, ATP is generated via anaerobic glycolysis, while in aerobic conditions ATP is produced mainly via OXPHOS, a process involving the breakdown of substrates to acetyl-CoA, and leading to the production of the reduced form of nicotinamide adenine dinucleotide (NAD+) NADH via the tricarboxylic cycle (TCA). Compared to OXPHOS, glycolysis is an inefficient method to generate ATP. However, it provides a number of important advantages for cells, including the ability to rapidly generate ATP in response to acute changes in energy demand, as well as generating the necessary glycolytic intermediates for the biosynthesis of new macromolecules essential for proliferating cells (Lunt and Vander Heiden, 2011; Ryall, 2013; Shyh-Chang et al., 2013).
The enzymatic activity of sirtuin 1 (SIRT1), a member of the class III deacetylase family (Guarente, 2000; Michan and Sinclair, 2007), is regulated by the free concentration of the intermediate metabolite NAD+ (Imai et al., 2000). While a plethora of non-histone proteins are deacetylated by SIRT1 (Houtkooper et al., 2012; Rodgers et al., 2005), acetylated lysine 16 of histone H4 (H4K16ac) serves as a preferred SIRT1 histone substrate (Vaquero et al., 2004). Even though regulation of sirtuin enzymology is energy-demanding and complex( Sauve and Youn, 2012), it establishes, in principle, a rapid and finely tunable biochemical system through which changes in metabolism can be effectively converted into distinct epigenetic states and gene expression patterns.
Satellite cells (SCs) are skeletal muscle stem cells required for muscle growth and tissue repair (Brack and Rando, 2012; Tajbakhsh, 2009; Yin et al., 2013). Following intense proliferation associated with mouse postnatal muscle growth, SCs enter a quiescent state, representing 3–5% of the total number of adult muscle fiber nuclei( Yin et al., 2013). In response to muscle injury, the niche is remodeled and quiescent SCs enter the cell cycle (become activated). Activated SCs are characterized by the presence of the muscle-specific transcription factor MyoD, and give rise to committed proliferating muscle precursors which, upon expression of the myogenic transcription factor Myogenin, differentiate and fuse to repair damaged muscles (Tajbakhsh, 2009; Yin et al., 2013). Due to changes in requirements placed on SCs during the transition from quiescence to activation, significant differences in the underlying metabolism of these cellular states are likely to occur. Here we describe a metabolic shift from fatty acid (FA) and pyruvate oxidation in quiescent SCs to increased glycolysis and glutaminolysis during SC activation and proliferation. In addition, we document that this process of SC metabolic reprogramming is associated with a decrease in the intracellular NAD +/NADH ratio, reduced SIRT1-mediated deacetylation of H4K16ac, and activation of the myogenic program. SCs derived from mice with muscle-specific inactivation of the SIRT1 deacetylase domain (SIRT1mKO mice)display increased H4K16 ac and deregulated activation of the myogenic program. Finally, SIRT1mKO mice have reduced myofiber size, exhibit impaired muscle regeneration, and reveal a derepression of several muscle developmental genes.
RESULTS
Quiescent Skeletal Muscle Stem Cells Undergo a Switch from Fatty-Acid and Pyruvate Oxidation to Glycolysis during Culture in Growth Permissive Conditions
To analyze transcriptomes of quiescent and proliferating SCs, we used fluorescence-activated cell sorting (FACS) followed by RNA sequencing (RNA-seq). FACS isolation of SCs from two month-old C57BL/6 mouse hindlimb muscles was based on selection of α7-integrin+, Hoechst+, PI− and Lin− (CD11/CD31/CD45/Sca1) cells (modified from Kuang et al., 2007, Figure S1A–D). Freshly isolated (FI) SCs were immediately processed following the completion of sorting, while an aliquot of FACS-isolated SCs were cultured (Cul) on collagen-coated plates in growth media for 48hrs. Quiescent SCs are Pax7+/MyoD− whereas activated SCs are Pax7+/MyoD+( Kuang et al., 2008), therefore we analyzed aliquots of FI or Cul SCs and confirmed them to be Pax7+/MyoD− and Pax7+/MyoD+, respectively (Figure 1A,B).
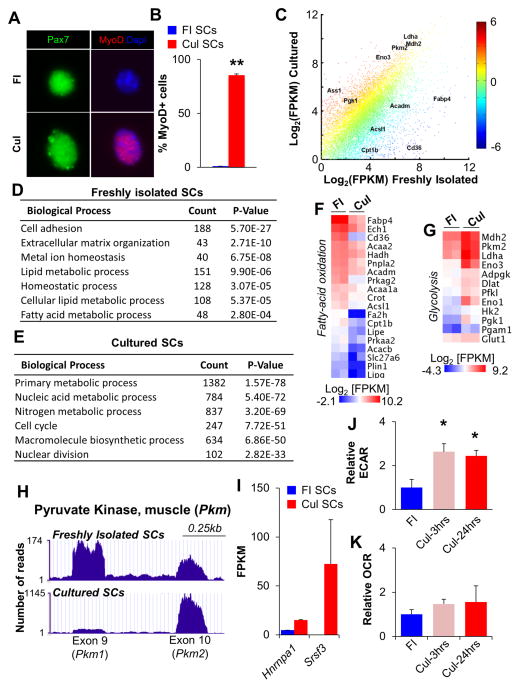
Figure 1. Satellite Cells Undergo a Switch from Oxidative to Glycolytic Metabolism Following Culture in Growth Media.
(A) Immunofluorescent analyses of freshly isolated (FI, quiescent) and cultured (Cul, activated) SCs with Pax7 (green) and MyoD (red) antibodies. Dapi identifies nuclei. (B) Quantification of MyoD+ SCs in FI and Cul cell populations. (C) RNA-seq scatter plot with key metabolic regulators indicated. Each data point represents the mean Log2[FPKM] from two independent biological replicates with color indicating the relative fold change in gene expression. (D,E) Gene ontology analyses of RNA-seq revealed an enrichment of biological processes specific to FI SCs (D) or Cul SCs (E). The number of genes enriched by greater than 1.5-fold is indicated under the ‘Count’ column. (F,G) Heat maps indicating absolute gene expression (Log2[FPKM]) of specific metabolic regulators in FI and Cul SCs. Each gene listed had a mean fold-change of greater than 1.5. (H) RNA-seq traces (UCSC genome browser) for Pkm 1 and 2 isoforms in FI and Cul WT SCs. (I) Expression of the Pkm splice regulators Hnrnpa1 and Srsf3 in FI and Cul SCs. (J,K) Cellular bioenergetics in SCs during culture in growth conditions were evaluated with Seahorse XF96 bioanalyzer. Glycolysis (ECAR) was increased 2.5 fold in Cul-3hrs and Cul-24hrs SCs (J), while basal oxygen consumption (OCR) was not different between FI, Cul-3hrs or Cul-24hrs SCs (K). Data are presented as mean ± SEM. *p < 0.05 and **p < 0.01 (FI SCs versus Cul SCs).
RNA-seq results from FI and Cul SCs identified a large number of differentially regulated genes (Figure 1C and Table S1). Gene ontology (GO) analyses of biological processes highlighted the expected changes following the transition from quiescence to proliferation, with adhesion- and homeostatic-related terms enriched in FI SCs (Figure 1D, Figure S1E and Table S1), and cell-cycle and nuclear division terms enriched in Cul SCs (Figure 1E, Figure S1F and Table S1). Moreover, the two markers of SC quiescence Sprouty 1 (Spry1, Shea et al., 2010)and calcitonin receptor (Calcr, Fukada et al., 2007) were enriched in FI SCs (Table S1). In addition, changes in a number of genes encoding for metabolic regulators were observed in the transition from quiescence to proliferation(Figure 1 C, E). The expression of genes corresponding to proteins that regulate FA metabolic processes and lipid catabolic processes was down-regulated (Figure 1F), whereas that of genes regulating glucose catabolic processes, glutaminolysis, macromolecular biosynthesis (including the pentose phosphate pathway, PPP) and amino-acid transporters was increased in Cul SCs (Figure 1G, Figure S1G,H). Indicating that transcriptional activation of the glycolytic program was not simply the result of in vitro culture conditions, transcription of a similar subset of genes was increased in proliferating SCs derived from regenerating skeletal muscle (Liu et al., 2013). In addition to a global upregulation of genes encoding glycolytic enzymes, we observed that the muscle pyruvate kinase 2 (Pkm2) isoform, known to promote “aerobic-glycolysis” (or the Warburg effect), predominated over its alternative spliced isoform Pkm1 in Cul SCs (Figure 1H). Consistently, expression of Hnrnpa1 and Srsf3, two mediators of Pkm alternative splicing, was also increased (Figure 1I).
While these results are suggestive of a shift in substrate uptake and utilization during the switch from quiescent to proliferating SCs, to directly confirm that glycolysis was elevated in Cul SCs, we employed the Seahorse extracellular flux bioanalyzer to examine both the basal oxygen consumption rate (OCR, an indicator of mitochondrial oxidative activity) and the basal extracellular acidification rate (ECAR, a marker of glycolysis). We observed a 2.5 fold-increase in the ECAR (i.e., increased glycolysis) in both Cul-3hrs and Cul-24hrs SCs, compared to FI SCs (Figure 1J). However, there was no difference in the basal mitochondrial OCR between FI SCs and the two stages of cultured SCs examined (Figure 1K). As a recent study identified an increase in mitochondria following 60hrs of SC activation in vivo (Rodgers et al., 2014), we examined mitochondrial content in FI and Cul SCs. Similarly to that observed in vivo, there was a progressive increase in Mitotracker fluorescence in Cul SCs during 48hrs of culture (Figure S2A) and an enrichment of genes known to regulate the TCA cycle (Figure S2B). These results suggest that Cul SCs preferentially increase their glycolytic rate, despite increasing their mitochondrial content. This observation is reminiscent of mouse epiblast stem cells, which have larger, and more complex mitochondria than mouse embryonic stem cells, and yet exhibit a lower level of oxygen consumption (Zhou et al., 2012).
Altogether, these results indicate that Cul SCs undergo a shift away from FA oxidation, towards glycolysis, glutaminolysis, and activation of the PPP for macromolecule biosynthesis.
SIRT1 Deacetylase Activity and NAD+ Levels are Elevated in Quiescent Compared to Activated Skeletal Muscle Stem Cells
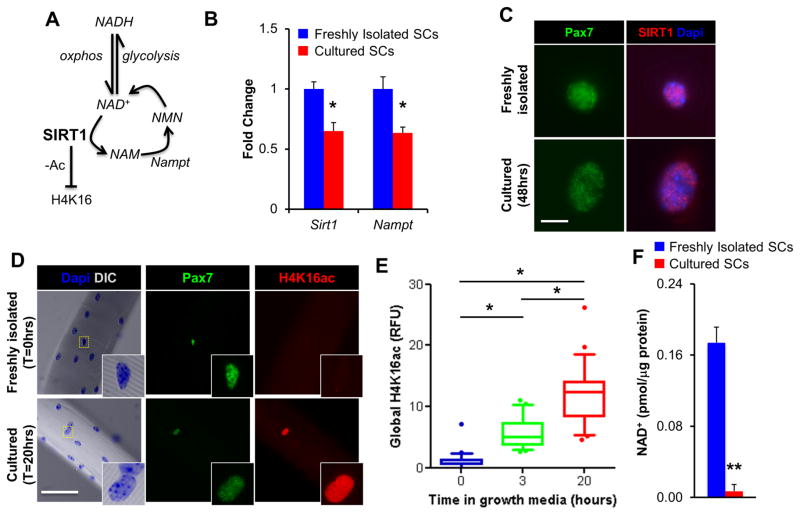
An increased reliance upon oxidative metabolism has been proposed to lead to elevated SIRT1 deacetylase activity, either via changes in absolute SIRT1 levels or changes in the NAD+/NADH ratio (Guarente, 2011; Ryall, 2012, Figure 2A). The shift away from FA oxidation in Cul SCs was associated with a 1.5 fold decrease in the expression of both SIRT1 and the NAD+-generating enzyme Nampt, as measured by qPCR (Figure 2B). However, despite this decline, SIRT1 protein was readily detected and predominantly localized to the nucleus of both FI and Cul SCs (Figure 2C). Global H4K16ac, a substrate for SIRT1-mediated deacetylation (Vaquero et al., 2004), was examined in SCs associated with freshly isolated single extensor digitorum longus (EDL) muscle fibers, and fibers that had been cultured in growth media for 3 or 20 hrs (Figure 2D,E). Single myofiber isolation permits SCs to remain attached to the myofiber and, thus, to maintain their position in the physiological niche between the basal lamina and the sarcolemma (Collins and Zammit, 2009). Myofiber-associated SCs cultured in growth media for 3 or 20hrs displayed a 7 and 16-fold increase in the median level of H4K16ac, compared to SCs on freshly isolated myofibers (Figure 2D,E), respectively. Thus, while SIRT1 protein was observed in the nucleus of Cul SCs, global H4K16ac was strongly elevated during the early stages of culture, suggesting a possible regulation of SIRT1 activity. As SIRT1 deacetylase activity is dependent upon NAD+ supply (Imai et al., 2000), we used FACS-isolated SCs to examine NAD+ levels. While NADH was undetectable in both FI and Cul SCs, the amount of total NAD+ was ten-fold higher in FI than Cul SCs (Figure 2F). Therefore, H4K16 deacetylation, a proxy for SIRT1 deacetylase activity, and NAD+ were elevated in FI SCs, and reduced in Cul SCs.
Figure 2. The Histone Deacetylase Activity of SIRT1 is Reduced Following Satellite Cell Activation.
(A) Scheme depicting how the deacetylase activity of SIRT1 is dependent on both metabolic processes (NAD+↔NADH) and the NAD+ salvage pathway (NAD+↔NAM). H4K16, histone H4 lysine 16; NAD, nicotinamide adenine dinucleotide; NAM, nicotinamide; NMN, nicotinamide mononucleotide nucleosidase, Nampt, nicotinamide phosphoribosyltransferase. (B) Sirt1 and Nampt expression in FI and Cul SCs, as measured by qPCR (n=3). (C) SIRT1 and Pax7 immunofluorescence of FACS-isolated FI and Cul SCs. White scale bar indicates 5μm. (D) Pax7 and H4K16ac immunofluorescence of SCs on freshly isolated single muscle fibers. White scale bar indicates 50μm, inset is magnified by a magnitude of 4. (E) Quantification of relative fluorescence (RFU) in SCs labeled for H4K16ac (n=2 mice, >50 fibers/timepoint). Results are presented as box-and-whisker plots, with a significant difference indicated when the median ± 95% CI does not overlap. (F) Total NAD levels in FI and Cul SCs (48hrs in growth media, n=3). Data is presented as mean ± SEM, *p < 0.05 and **p < 0.01 (FI SCs versus Cul SCs).
Metabolic Reprogramming of Proliferating Muscle Cells to Oxidative Phosphorylation Leads to a SIRT1-Dependent Repression of MyoD
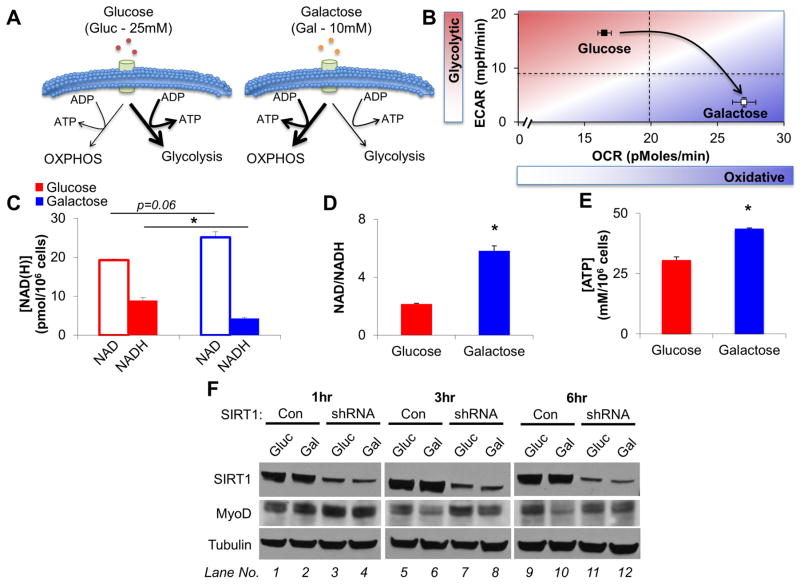
In contrast to glucose, for galactose to be used as an energy substrate it must first be converted to glucose-6-phosphate in an ATP consuming reaction, leading to no net gain in ATP via glycolysis (Gohil et al., 2010). Cells incubated with galactose must therefore rely on OXPHOS to generate ATP (Figure 3A). To test whether metabolic reprogramming impacts upon the NAD+/NADH ratio, H4K16ac, and muscle gene expression, we employed myogenic C2C12 cells. While C2C12 cells do not recapitulate all the features of SCs, they nonetheless retain some shared characteristics. C2C12 cells incubated for 3hrs in growth media containing galactose (10mM) exhibited significantly reduced glycolysis (as indicated via decreased ECAR) and increased oxygen consumption (measured as OCR) compared to cells incubated in growth media containing glucose (25mM, Figure 3B). Culturing C2C12 cells in growth media containing galactose resulted in an increase in the amount of NAD+, with a concomitant decrease in NADH, and a net 2.5 fold increase in the NAD+/NADH ratio of cells (Figure 3C,D). We confirmed that incubation with galactose did not impair ATP generation (and activate starvation pathways), and in fact resulted in elevated levels of ATP (Figure 3E).
Figure 3. A Forced Shift to Oxidative Metabolism Increases NAD+ Levels and Reduces MyoD in Control but not SIRT1 shRNA cells.
(A) Schematic depicting how glucose is employed to preferentially generate ATP via glycolysis. Replacing glucose with galactose forces cells to shift to predominantly utilize OXPHOS for the generation of ATP. (B) C2C12 basal cellular bioenergetics were analyzed on a Seahorse XF96 bioanalyzer during culture in either glucose or galactose based growth media (n=15 replicates/group). (C,D) Culturing C2C12 cells in galactose based growth media increases the amount of NAD+, at the expense of NADH such that there was a three-fold increase in the NAD/NADH ratio (n=3). (E) Replacing glucose with galactose in C2C12 growth media led to an elevation in ATP levels (n=3). (F) Incubating C2C12 cells with galactose instead of glucose for 3 and 6hrs results in a decrease in MyoD in WT (compare lanes 5–6, and 9–10), but not SIRT1 shRNA C2C12 cells (compare lanes 7–8, and 11–12). Data are presented as mean ± SEM. *p < 0.05.
The results presented in the preceding paragraphs indicate that a switch from oxidative phosphorylation to glycolysis accompanying SC activation has the potential to modulate the deacetylase activity of SIRT1 via decreased NAD+ availability. To directly test this possibility, we transduced C2C12 cells with a SIRT1 shRNA retrovirus. The reduced SIRT1 protein levels in these cells was associated with increased MyoD expression, elevated global H4K16ac, and a reduced rate of proliferation (Figure S3A, B). Consistent with a role of SIRT1 in mediating these phenomena, addition of increasing concentrations of the SIRT1 inhibitor nicotinamide (NAM) to proliferating C2C12 cells, similarly led to an increase in global H4K16ac and MyoD expression (Figure S3C). C2C12 cells cultured in galactose (Gal) exhibited a progressive decline in the expression of MyoD protein levels starting at 3hrs and further decreasing at 6hrs (Figure 3F, compare lanes 5–6, 9–10). Importantly, the galactose-induced decline in MyoD protein was abrogated by SIRT1 shRNA (Figure 3F, compare lanes 6 and 8, and 10 and 12), indicating a role for SIRT1 at the nexus between changes in cellular metabolism and expression of the master regulator MyoD.
Ablation of the SIRT1 Deacetylase Domain Leads to Premature Differentiation of Skeletal Muscle Stem Cells
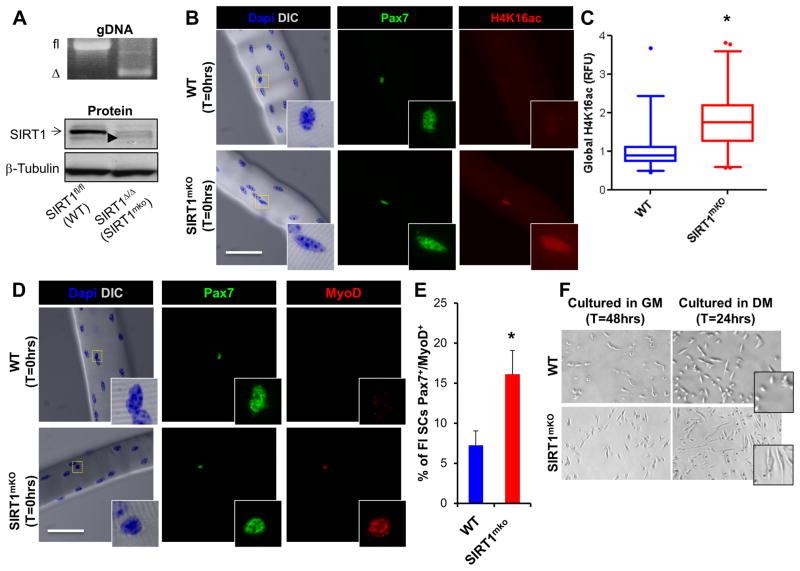
To investigate whether SIRT1 influences SC biology in vivo, we generated a Pax7-specific SIRT1 knockout mouse (SIRT1muscle KO) by crossing mice containing the Cre-recombinase under the control of the Pax7 locus (Pax7-Cre, Keller et al., 2004) with mice containing a modified SIRT1 gene where exon 4 (located within the catalytic deacetylase domain) is flanked by loxP sites (SIRT1 flox mice, Li et al., 2007). The floxed alleles were detected in genomic DNA isolated from skeletal muscle of Cre-negative (WT) mice, while the Δex4 alleles were detected in the presence of Cre-recombinase in SIRT1mKO mice (Figure 4A, top). A band corresponding to the SIRT1 protein (Figure 4A bottom, arrow) was lost and, concomitantly, a slightly faster migrating band corresponding to SIRT1Δex4 appeared (Figure 4A bottom, arrowhead), specifically in the skeletal muscles of SIRT1mKO mice, compared with WT. Freshly isolated single EDL muscle fibers were obtained from WT and SIRT1mKO mice, stained with Pax7 to identify SCs and co-stained with H4K16ac-specific antibodies. While WT myofibers had a tight distribution of SCs expressing low-levels of global H4K16ac, SCs on SIRT1mKO myofibers had a wider distribution of H4K16ac levels, such that there was a two-fold increase in the median level of global SC H4K16ac (Figure 4B,C). These results indicate that, in SCs, SIRT1 is required to maintain H4K16 in a deacetylated state. Moreover, the percentage of Pax7+/MyoD+ SCs on SIRT1mKO fibers was increased (Figure 4D,E and Figure S3D). In light of these findings, we asked whether SIRT1mKO SCs may also undergo premature differentiation and, if so, whether this is a cell-autonomous phenomenon. FACS-isolated SCs were cultured for 48hrs in growth conditions, or induced to differentiate for 24hrs. Compared to WT cells, SIRT1mKO SCs mice exhibited an overt spindle-like, elongated morphology, indicative of early differentiation (Figure 4F).
Figure 4. Ablation of the Catalytic Domain of SIRT1 Results in Increased Global H4K16ac and Precocious Activation/Differentiation of SCs.
(A) Skeletal muscle from WT mice demonstrated the presence of the SIRT1 floxed allele (fl, top panel) and detectable levels of SIRT1 protein (arrow, bottom panel), while SIRT1mKO muscle contained the SIRT1Δex4 allele (Δ, top panel) and ablation of SIRT1 protein. A small level of SIRT1Δex4 protein was detectable in the skeletal muscle of SIRT1mKO mice (arrowhead, bottom panel). (B, C) In SIRT1mKO mice, quiescent SCs (identified as Pax7+) exhibited a two-fold increase in global H4K16ac, compared to WT mice, as determined via relative fluorescence (RFU) in SCs labelled for H4K16ac (n=2 mice, >50 fibers/timepoint). White scale bar indicates 50μm, inset is magnified by a magnitude of four. Results are presented as box-and-whisker plots (5–95 percentiles), with a significant difference indicated when the median ± 95% CI does not overlap. (D, E) MyoD and Pax7 staining (n=2 mice, >50 fibers/timepoint) of fiber-associated SIRT1mKO SCs. Data is presented as mean ± SEM, *p < 0.05. (F) DIC images of SCs isolated from WT and SIRT1mKO mice during proliferation in growth media (GM, 48hrs), or early differentiation in differentiation media (24hrs DM). Note the overt spindle-like, elongated morphology of SCs from SIRT1mKO mice indicating premature differentiation.
SIRT1 Regulates H4K16 Acetylation and Expression of Selected Genes
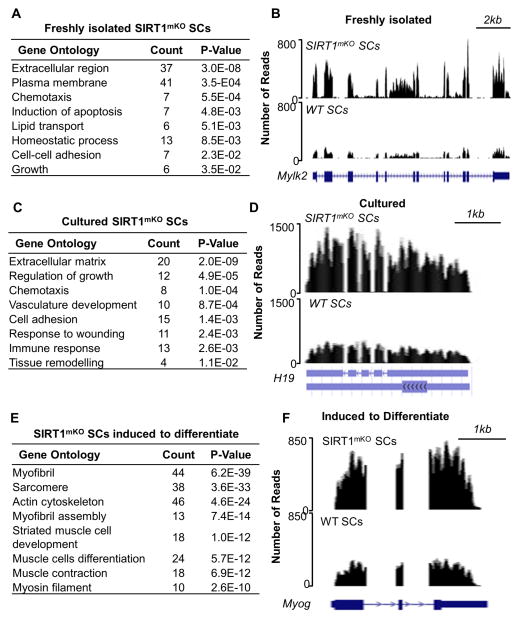
To thoroughly investigate the global impact of inactivating the SIRT1 deacetylase activity on the transcriptome, we performed RNA-seq on SCs derived from WT or SIRT1mKO mice at several stages of myogenesis, including quiescence (FI), activation (Cul) and early differentiation (Diff). In FI SIRT1mKO SCs, 361 genes were down-regulated compared to FI WT SCs (Table S2). On the other hand, transcripts corresponding to 287 genes were up-regulated (Figure 5A and Table S2). Among the up-regulated genes in FI SIRT1mKO SCs were those for muscle-specific myosin light chain kinase 2 (Mylk2, Figure 5B, Table S2), myosin light chain phosphorylatable (Mylpf), the skeletal muscle proteoglycan biglycan (Bgn), and the skeletal muscle-specific ryanodine receptor 1 (Ryr1, Table S2). Expression of these genes is physiologically observed in differentiated skeletal muscle cells. Other up-regulated transcripts corresponded to the BMP inhibitors chordin (Chrd) and follistatin (Fst), known to play functional roles in SC proliferation and differentiation (Jeong et al., 2013; Ono et al., 2011), and transcription factors eyes-absent 1 (Eya1-required for somitic myogenesis, Grifone et al., 2007) and Foxo3 (Table S2, Dentice et al., 2010). Overall, these findings indicate a propensity of FI SIRT1mKO SCs to activate genes physiologically expressed in activated/differentiating SCs. In Cul SIRT1mKO SCs, 322 genes were down-regulated, while 253 transcripts were up-regulated (Figure 5C, Table S2). Bgn, a transcript up-regulated in FI SIRT1mKO SCs, continued to be increased, and transcripts for the decorin (Dcn) gene, whose expression is increased during muscle regeneration of mdx mice (Abe et al., 2009) were augmented (Table S2). Transcripts corresponding to the H19 long noncoding (lnc) RNA were also increased (Figure 5D). Processing of H19 lnc RNA generates miR-675-3p and -5p, two microRNAs induced and required for skeletal muscle differentiation (Dey et al., 2014). In SIRT1mKO SCs induced to differentiate, 177 genes were down-regulated (Table S2) and 757 gene transcripts were increased (Figure 5E and Table S2). The up-regulated transcripts included actins, myosins, and troponins (Table S2). Transcription factors influencing different aspects of myogenesis, such as Myog (Figure 5F), Mef2a-d, Eya4, Nfatc, Nfix, and Smad3 were also increased (Table S2). The H19 lncRNA, up-regulated in Cul (Figure 5D), was also increased in SIRT1mKO SCs induced to differentiate (Figure S5C). Of particular interest, transcripts typically expressed in either embryonic muscle and/or upregulated during muscle regeneration including the embryonic myosin heavy chain isoform (Myh3), filamin C (Flnc), and brain expressed 1 (Bex1) were increased in SIRT1mKO SCs (Goetsch et al., 2005; Koo et al., 2007; Weydert et al., 1987, Table S2, see below Figure 7G).
Figure 5. Ablation of the SIRT1 Catalytic Domain Leads to Modifications in Gene Expression in both Freshly Isolated and Cultured Satellite Cells.
(A) Gene ontology (GO) of genes upregulated in FI SIRT1mKO SCs. (B) RNA-seq profiles (UCSC genome browser) of the Mylk2 gene in FI WT SCs (bottom) and FI SIRT1mKO SCs (top). (C) GO of genes upregulated in Cul SIRT1mKO SCs. (D) RNA-seq profiles of the H19 gene in Cul WT SCs (bottom) and Cul SIRT1mKO SCs (top). (E) GO of genes upregulated in SIRT1mKO SCs induced to differentiate (24hrs DM). (F) RNA-seq profiles of the Myog gene in WT (bottom) and SIRT1mKO SCs (top) induced to differentiate. RNA-seq experiments were done with either three (FI SCs) or two (Cul and Differentiating SCs) biological replicates.
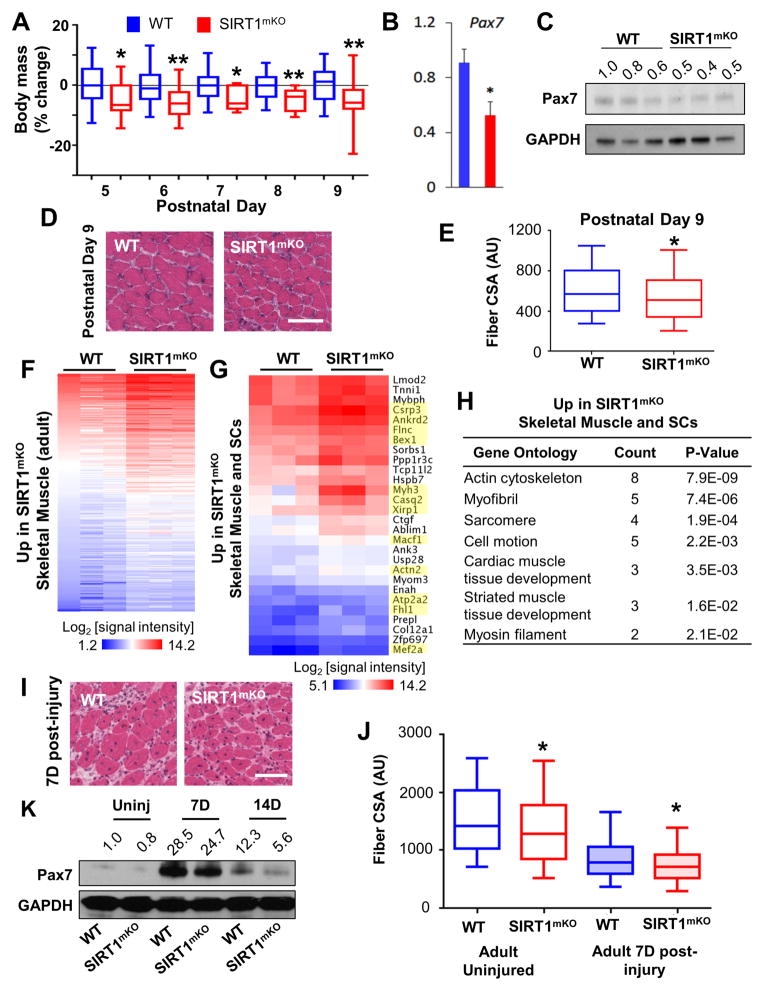
Figure 7. Loss of SIRT1 Deacetylase Activity Leads to Developmental and Regenerative Defects in Skeletal Muscle.
(A) Body mass was evaluated starting at postnatal (P) days 5 through 9 in WT and SIRT1mKO mice (WT n=27; SIRT1mKO =24). Significance was determined using a Student’s t-test (two-tailed distribution without assuming equal variance). (B,C) Pax7 mRNA (left) and protein expression (right) were evaluated in the skeletal muscles of P9 WT and SIRT1mKO mice. (D) H&E staining of skeletal muscles in P9 WT and SIRT1mKO mice. White scale bar indicates 50μM. (E) Quantification of skeletal muscle fiber CSA from P9 mice (>1,000 fibers analyzed/muscle). Results are presented as a box-and-whisker plot. (F) Heat map of the microarray results (Log2[signal intensity]) for genes upregulated in SIRT1mKO skeletal muscle. Each gene listed has a mean fold change of >1.5 and p < 0.05 (n=3 samples/group). (G) Heat map of the microarray results (Log2[signal intensity]) for genes upregulated in both SIRT1mKO SCs induced to differentiate and SIRT1mKO skeletal muscle, with developmentally-regulated muscle genes highlighted in yellow. (H) Gene Ontology for genes upregulated in both SIRT1mKO SCs induced to differentiate and SIRT1mKO skeletal muscle. (I) H&E skeletal muscle staining of two-months old (adult) WT and SIRT1mKO mice seven days after CTX injection. (J) Quantification of skeletal muscle fiber CSA from adult WT and SIRT1mKO mice regenerating muscles (seven days after CTX, >1,000 fibers analyzed/muscle). Results are presented as a box-and-whisker plot. (K) Pax7 protein levels in uninjured, and 7 and 14 days regenerating muscles of WT and SIRT1mKO mice. Data is presented as mean ± SEM, *p < 0.05; **p < 0.01.
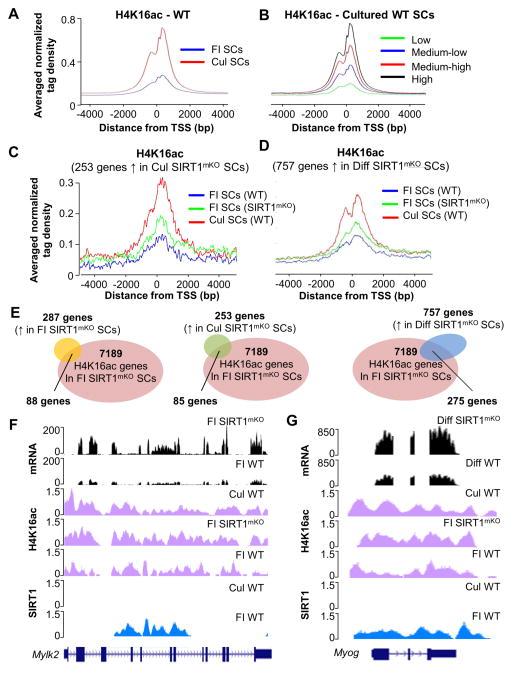
Of all histone lysines, acetylation of H4K16 profoundly alters chromatin structure by disrupting formation of the 30nm chromatin fibers and preventing cross-fiber formation (Shogren-Knaak et al., 2006). Accordingly, subtle enrichment of H4K16ac significantly and positively impacts transcription (Taylor et al., 2013). To correlate transcription and H4K16ac enrichment, ~1x106 SCs derived from 7–8 WT or SIRT1mKO mice were employed for ChIP-seq after immunoprecipitation with an H4K16ac antibody. H4K16 acetylation was observed to predominantly occur at and around the transcriptional start site (TSS) and was higher in Cul SCs compared to FI SCs (Figure 6A). Moreover, there was a positive correlation between H4K16ac and gene expression, such that the genes with the highest expression were also the most H4K16 acetylated in both Cul SCs (Figure 6B) and FI SCs (Figure S4A). We addressed the genome-wide role of SIRT1 on H4K16ac and gene expression by correlating H4K16ac ChIP-seq and RNA-seq datasets from WT and SIRT1mKO SCs. Global H4K16ac at the 253 up-regulated genes in Cul SIRT1mKO SCs was already increased in FI SIRT1mKO SCs (Figure 6C). Similarly, H4K16ac at the 757 genes whose transcription was increased in SIRT1mKO SCs induced to differentiate was also increased in FI SIRT1mKO SCs, being positioned at an intermediate H4K16ac level between that observed in FI and Cul WT SCs (Figure 6D). Of the 287 genes up-regulated in FI SIRT1mKO SCs, one third (88/287, 30%, Table S3) displayed significantly increased H4K16ac (Figure 6E). However, the vast majority of H4K16ac was found to occur at genes whose transcription was not concomitantly increased (Figure 6E). In Cul SIRT1mKO SCs, 85 of the 253 up-regulated genes (33%, Table S3) had increased H4K16ac (Figure 6E), and 275 out of the 757 up-regulated genes (36%, Table S3) in SIRT1mKO SCs induced to differentiate, manifested increased H4K16ac when assayed in FI SIRT1mKO SCs (Figure 6E).
Figure 6. Ablation of the SIRT1 Catalytic Domain Alters H4K16 Acetylation and Influences Expression of Selected Genes.
(A) Average signal of normalized tag density for H4K16ac ChIP-seq in FI WT SCs (blue line) and Cul WT SCs (red line). (B) Average signal of normalized tag density for H4K16ac ChIP-seq in Cul WT SCs for genes with low expression (green line), medium-low expression (blue line), medium high (red line) and high expression (black line). (C) Average signal of normalized tag density for H4K16ac ChIP-seq in FI WT SCs (blue line), FI SIRT1mKO SCs (green line) and Cul WT SCs for genes up-regulated in Cul SIRT1mKO SCs. (D) Average signal of normalized tag density for H4K16ac ChIP-seq in FI WT SCs (blue line), FI SIRT1mKO SCs (green line) and Cul WT SCs induced to differentiate for genes up-regulated in SIRT1mKO induced to differentiate. (E) Venn diagram representing genes up-regulated in FI SIRT1mKO SCs (287genes), Cul SIRT1mKO SCs (253 genes), and SIRT1mKO SCs induced to differentiate (757 genes), and genes acquiring H4K16ac in FI SIRT1mKO SCs (7189 genes). (F) ChIP-seq and RNA-seq profiles of the Mylk2 gene. Bottom to top: SIRT1 ChIP-seq profile in FI and Cul WT SCs (blue signals); H4K16ac profile in FI WT and SIRT11mKO SCs, and WT Cul SCs (magenta signals); Mylk2 mRNA expression profile in FI WT and SIRT1mKO SCs (black signals). (G) ChIP-seq and RNA-seq profiles of the Myog gene. Bottom to top: SIRT1 ChIP-seq profile in FI and Cul WT SCs (blue signals); H4K16ac profile in WT and SIRT1mKO FI SCs, and WT Cul SCs (magenta signals); Myog mRNA expression profile in FI WT and SIRT1mKO SCs (black signals).
To directly analyze a link between SIRT1, H4K16ac and gene expression, we performed SIRT1 ChIP-seq in FI and Cul SCs (Table S4). Examples of selected genes are illustrated in genome browser screen views for SIRT1 ChIP-seq, H4K16ac ChIP-seq as well as RNA-seq traces (Figure 6F,G, and Figure S4 and S5). SIRT1 was enriched at the Mylk2, H19, and Myog loci in FI but absent in Cul SCs, its enrichment negatively correlating with H4K16ac and gene transcription (Figure 6F,G). H4K16 acetylation and SIRT1 enrichment at the Mylk2 and Myog were validated using ChIP-qPCR (Figure S6). These findings are in agreement with our previous results indicating a repressive role of SIRT1 for Myog expression (Fulco et al., 2003). Overall, the results described in this paragraph indicate that while SIRT1 regulates the H4K16ac status at several thousand loci, its deletion only permits the expression of a few hundred genes.
Metabolic Reprogramming of Skeletal Muscle Stem Cells Leads to a SIRT1-Dependent Delay in H4K16ac Acquisition and MyoD Expression
To further examine the link between metabolism, H4K16ac and SC activation, we used isolated single fibers from WT and SIRT1mKO mice and incubated them for 20hrs in either glucose- (25mM) or galactose- (10mM) based growth media (Figure S7). Similarly to what observed in C2C12 cells (Figure 3), SCs from WT mice incubated in galactose-supplemented media exhibited a reduction in global H4K16ac and MyoD expression, compared to SCs incubated in glucose media (Figure S7A, C). In contrast SCs from SIRT1mKO mice did not exhibit any appreciable difference in global H4K16ac or MyoD expression in either glucose or galactose media (Figure S7B, D). Overall, these results provide evidence for a direct link between a change in metabolism and SC activation and support a role for SIRT1 in this process, downstream of the change in metabolism.
Ablation of SIRT1 Deacetylase Activity Leads to Developmental and Regenerative Defects in SIRT1mKO Mice
To determine the effects of SIRT1 ablation in skeletal muscle, we investigated phenotypic changes in SIRT1mKO mice during both development and regeneration. SIRT1mKO mice were initially smaller than littermate controls (Figure 7A), and had reduced Pax7 gene and protein expression (Figure 7B,C). In addition, at postnatal day 9 (P9), skeletal muscles from SIRT1mKO mice exhibited smaller fiber cross-sectional areas (CSA, Figure 7D,E). By day P14 the body mass of SIRT1mKO mice became indistinguishable from littermate controls (data not shown). Interestingly, it is approximately at P14 that SCs gradually start reducing their proliferation to enter quiescence at P21 (Chang and Rudnicki, 2014). A microarray assay of RNA derived from the gastrocnemius hindlimb muscles revealed an enrichment of 219 and a reduction of 191 transcripts in the muscles of SIRT1mKO compared to WT mice (Figure 7F and Table S5). Of the upregulated transcripts in SIRT1mKO muscle, 28 (corresponding to 12% of the total transcripts) were also increased in SIRT1mKO SCs induced to differentiate (Figure 7G and Table S5). A GO analysis of the transcripts upregulated in both SIRT1mKO muscle and SIRT1mKO SCs returned biological terms related to skeletal muscle development (‘myofibril’, ‘sarcomere’ and ‘striated muscle development’, Figure 7H). The most highly upregulated transcript in SIRT1mKO muscles corresponded to the embryonic myosin heavy chain isoform Myh3 (5.8-fold increase, Table S5), which is physiologically expressed in fetal muscle and repressed in the adult (Weydert et al., 1987). In addition to Myh3, several other transcripts upregulated in both SIRT1mKO muscles and SIRT1mKO SCs induced to differentiate are known to be preferentially expressed during muscle development and/or in regenerating muscles (highlighted in Figure 7G, Table S5).
In response to injury, quiescent SCs become activated, enter mitosis, giving rise to myogenic progenitor cells, which ultimately differentiate to restore damaged muscles (Chang and Rudnicki, 2014). To evaluate whether muscle regeneration was altered in SIRT1mKO mice, muscle damage was induced by injecting the tibialis anterior (TA) muscle of two month-old WT or SIRT1mKO mice with the myogenic agent cardiotoxin (CTX). Seven days after CTX injection, regenerating muscles of SIRT1mKO mice were composed of myofibers with smaller CSA (Figure 7H,I), and reduced Pax7 protein (Figure 7J, both at 7 and 14 days after CTX injection), compared to littermate controls. Overall, these findings indicate that SIRT1 is required for appropriate postnatal muscle growth and adult muscle regeneration.
DISCUSSION
In this study, SIRT1 was found to lie at the nexus between SC metabolic reprogramming and muscle gene expression. Our findings are consistent with a model wherein the metabolic state influences the gene expression program of SCs via modulation of the metabolite NAD+ and SIRT1 deacetylase activity. Ablation of a SIRT1 domain conferring deacetylase activity resulted in widespread H4K16ac and derepression of several hundred genes in SCs. However, such hyperacetylation did not result in a generalized and immediate transcriptional response of all target genes. For instance, enrichment of H4K16ac at the Myog locus was increased in FI SIRT1mKO SCs but its expression was augmented only in SIRT1mKO SCs induced to differentiate. Local chromatin architecture and additional transcriptional events - including histone dynamics and transcription factor availability - likely determine whether, at specific loci, transcriptional derepression is concomitant with SIRT1 removal or will occur at later stages of cell differentiation. Furthermore, that only approximately 30–40% of the genes derepressed upon SIRT1 ablation acquired H416ac indicates that either additional histone lysines or non-histone substrates are involved in SIRT1-mediated transcriptional repression (Houtkooper et al., 2012). It is interesting to note that SIRT1 does not appear to regulate the expression of metabolic regulators. Thus, while SIRT1 responds to the process of SC metabolic reprogramming by influencing H4K16 acetylation and gene expression, it does not play a direct role in the observed switch towards glycolysis in proliferating SCs. Despite the presence of MyoD+ SCs on SIRT1mKO fibers, the corresponding Myod1 transcripts were not elevated. This observation is consistent with findings reporting that Myod1 transcripts are already present in quiescent SCs, not exhibiting a significant increase during SC activation in vivo (Kanisicak et al., 2009; Liu et al., 2013; Pallafacchina et al., 2010; Rodgers et al., 2014). Analogous to microRNA-mediated Myf5 regulation (Crist et al., 2012), post-transcriptional processing of Myod1 mRNA transcripts may be responsible for the appearance of the MyoD protein in activated SCs. Moreover, while increased Myog transcripts in Cul (24hrs DM) SIRT1mKO SCs correlated with the enrichment of H4K16ac at its locus in quiescent SIRT1mKO SCs, and SIRT1 disengagement in cultured WT SCs, we cannot exclude that additional regulatory mechanisms controlled by SIRT1 also contribute to Myog activation. In this regard, SIRT1 deacetylates the histone acetyltransferases PCAF and p300, and transcription factors MyoD and Foxo3 (Brunet et al., 2004; Fulco et al., 2003; Motta et al., 2004), making it plausible that non-histone substrates relevant to myogenesis are affected in SIRT1mKO SCs. Recently, SIRT1 has been implicated in regulating autophagic flux and SC activation (Tang and Rando, 2014). In this study, authophagy in fiber-associated SCs was detected after 24–36 hrs of culture, during which time SCs had entered the S-phase of the cell cycle, as measured via an EdU assay. Interestingly, using an inducible SC-specific SIRT1-KO mouse (Pax7CreERT2xSIRT1fl/fl) these authors demonstrated that SIRT1 ablation inhibited autophagic flux and delayed entry into the S-phase of the cell cycle. The different timing employed to evaluate SC activation, along with the mode of SIRT1 excision (inducible vs constitutive) and the use of MyoD versus EdU as a measure of SC activation, may account for the observed differences between this study and the current results.
Altogether, our findings suggest a role for metabolism in the regulation of SC biology, beyond simply providing the building blocks and ATP required for new cell growth. Within this context, it has previously been shown that the generation of nucleocytoplasmic acetyl-CoA is dependent upon glycolytic production of citrate (Wellen et al., 2009). In contrast, FA oxidation leads to the production of mitochondrial acetyl-CoA for entry into the TCA cycle (Wellen et al., 2009). Thus any shift towards glycolysis would be expected to lead to both a decrease in SIRT1 deacetylase activity (through reduced NAD+), and an increased supply of acetyl-CoA for histone/protein acetylation, and together likely explain the observed increase in global H4K16ac in active SCs. There is previous evidence that the behavior of adult SCs is influenced by regulatory pathways controlled by oxygen tension and metabolic states (Csete et al., 2001; Liu et al., 2012). A link between metabolism and SC function has been proposed, with recent studies indicating a link between OXPHOS and SC clonogenic capacity. Elevated mitochondrial abundance and OXPHOS activity, induced by calorie restriction (CR), was found to be associated with an increase in the number of cells capable of initiating myogenic colony formation (Cerletti et al., 2012). Regulation of MyoD and Myogenin by increased SIRT1 activity elicited by CR would keep SCs in a stem-like state and favor their clonogenicity. Consistent with this, both the SIRT1 level and the metabolic milieu conducive to its activation were augmented in SCs derived from CR animals (Cerletti et al., 2012). A similar increase in clonogenic capacity was observed in HSC with increased FA oxidation (Ito et al., 2012) while, more recently, glucose and glutamine metabolism has been reported to regulate human HSC lineage specification (Oburoglu et al., 2014).
Finally, our gene expression profiling of whole muscles revealed deregulated transcription in SIRT1mKO mice. Intriguingly, a subset of the upregulated transcripts was also increased in SIRT1mKO SCs induced to differentiate (Figure 7G). Since in adult mice SCs have completed their differentiation process to give rise to mature myofibers, we interpret these findings to indicate that the upregulated transcripts identified in SIRT1mKO muscles derive from the myofiber compartment. The presence of such transcripts in both differentiating SIRT1mKO SCs and SIRT1mKO adult muscles suggests that their deregulation likely originates in SCs and is further maintained in the adult muscle.
In summary, our results provide insights into the developing paradigm linking the process of metabolic reprogramming, transcriptional regulation, and acquisition of defined cell states. In the absence of SIRT1 quiescent SCs lose their respective blueprint of carefully regulated H4K16ac and undergo a progressive deregulation of gene expression during activation and differentiation. We conclude that metabolic modifications affect the activity of SIRT1, which acts as a relay able to interpret the rapid change in metabolism (via NAD+) and induce subsequent changes in H4K16 acetylation status and gene expression.
EXPERIMENTAL PROCEDURES
Full details are provided in the Supplemental Experimental Procedures.
Fluorescence-Activated Cell Sorting
SCs were sorted based on our previously described technique (Juan et al., 2011), which utilizes a modified version of the method described by Kuang et al. (2007). Briefly, hindlimb muscles from mice were digested with 0.2% collagenase for 60 mins, SCs were liberated by treating the resultant slurry with collagenase/dispase. All cells were incubated with primary antibodies raised against α7-integrin, and PE labelled CD11, CD31, CD45, and Sca1, and Hoechst 33342 (10μg/ml) and PI (30mins). SCs were isolated via FACS, gating on positive α7-integrin and Hoechst staining, and negative PI and PE-CD/11/CD31/CD45/Sca1 staining.
Single Muscle Fiber Isolation and Culture
Single EDL fibers were isolated and cultured as previously described (Juan et al. 2011). Briefly, single fibers were liberated in a collagenase solution and then immediately fixed in 4% PFA, or cultured in growth media for the specified time (see Supplemental Experimental Procedures for growth media conditions). Measurements of relative fluorescent units (RFU) were completed via ImageJ (NIH, Bethesda MD) and represent grey intensity multiplied by nuclear size (see Supplemental Experimental Procedures for details).
Chromatin Immunoprecipitation
For ChIP-seq, ~1x106 SCs (isolated via FACS from the hindlimbs of 7–8 mice) were immediately cross-linked (for FI SCs) or cultured for 48 hrs (Cul SCs) and subsequently cross-linked in 1% formaldehyde and processed according to published protocols (Metivier et al., 2003; Mousavi et al., 2012). Briefly, cells were lysed in RIPA buffer (1x PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and centrifuged at 2000 rpm for 5 minutes. The chromatin fraction was sheared by sonication (4x30sec) in 1.5 ml siliconized Eppendorf tubes. The resulting sheared chromatin samples were cleared for 1 hour, immunoprecipitated overnight and washed in buffer I (20 mM TrisHCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), buffer II (20 mM TrisHCl pH 8.0, 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), buffer III (10 mM TrisHCl pH 8.0, 250 mM LiCl, 1% NP-40; 1% sodium deoxycholate, 1 mM EDTA) and Tris-EDTA (pH 8.0). All washes were performed at 4°C for 5 min. Finally, cross-linking was reversed in elution buffer (100 mM sodium bicarbonate (NaHCO3), 1% SDS) at 65°C overnight.
Cellular Bioenergetics
Oxygen consumption and extracellular acidification rates of both SCs and C2C12 cells were measured on an extracellular flux bioanalyzer (XF96, Seahorse Biosciences, North Billerica, MA), and seeded at a density of 50,000 and 12,000 cells per well, respectively (see Supplemental Experimental Procedures for details).
Mice
The initial characterization of the metabolic profile of SCs was conducted in WT mice (C57BL/6 background). SIRT1mKO mice were generated via breeding of Pax7cre/+ knock-in mice (Keller et al., 2004) with mice containing the floxed exon 4 SIRT1 allele (Li et al., 2007). Skeletal muscle injury was induced via a single intramuscular injection of the snake venom cardiotoxin to the TA muscle. Briefly, cardiotoxin (20μg/mL) was injected into the TA muscle until the maximal holding capacity of the muscle was reached (30–50μL). In all cases using SIRT1mKO mice, littermates were used as controls. All experiments were performed according to the National Institutes of Health Animal Care and Use regulations.
Statistical Analyses
All data are presented as mean ± SEM, excluding RFU measurements of H4K16ac and MyoD and muscle fiber CSA which are expressed as box-and-whisker plots. Statistical analyses were performed using a Student’s t-test to calculate differences between two groups, or a two-way ANOVA followed by a Tukey’s post-hoc analysis for multiple comparisons. For data presented as box-and-whisker plots, data were determined to be statistically different when the median ± 95% CI did not overlap.
Supplementary Material
Acknowledgments
We thank Yansong Gu (University of Washington, Seattle) for sharing the SIRT1 floxed mice, Kristina Zaal (Light Imaging Section, NIAMS) for help in evaluating Pax7+/MyoD+ immunofluorescence staining, and Jim Simone (Flow Cytometry Section, NIAMS) for help with FACS. JGR was supported, in part, via an Overseas Biomedical Research Fellowship from the National Health & Medical Research Council of Australia (NH&MRC). This work was supported by the Intramural Research Program of the National Institute of Arthritis, and Musculoskeletal and Skin diseases (NIAMS) at the National Institutes of Health.
Footnotes
ACCESSION NUMBER
Accession Number for RNA-seq and ChIP-seq datasets pending
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at.
AUTHOR CONTRIBUTIONS
J.G.R, S.D., and V.S. designed the experiments and wrote the manuscript. J.G.R. performed most of the presented experiments. S.D. performed RNA-seq and ChIP-seq experiments. A.D. performed FACS and RNA-seq experiments. A.J. performed single fiber isolation and immunofluorescence staining. H.Z. conducted bioinformatics analysis of RNA-seq and ChIP-seq data. X.F. performed muscle injury experiments, muscle sectioning, immunofluorescence staining, and helped with animal breeding. D.C performed FACS experiments. M.K. supervised animal breeding and analyzed animal body mass. G.G-C prepared libraries for RNA-seq and ChIP-seq and performed sequencing. M.F. provided technical and intellectual advice.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe S, Hirose D, Kado S, Iwanuma O, Saka H, Yanagisawa N, Ide Y. Increased expression of decorin during the regeneration stage of mdx mouse. Anat Sci Int. 2009;84:305–311. doi: 10.1007/s12565-009-0039-9. [DOI] [PubMed] [Google Scholar]
- Brack AS, Rando TA. Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell. 2012;10:504–514. doi: 10.1016/j.stem.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Cerletti M, Jang YC, Finley LW, Haigis MC, Wagers AJ. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang NC, Rudnicki MA. Satellite cells: the architects of skeletal muscle. Current Topics in Developmental Biology. 2014;107:161–181. doi: 10.1016/B978-0-12-416022-4.00006-8. [DOI] [PubMed] [Google Scholar]
- Collins CA, Zammit PS. Isolation and grafting of single muscle fibres. Methods Mol Biol. 2009;482:319–330. doi: 10.1007/978-1-59745-060-7_20. [DOI] [PubMed] [Google Scholar]
- Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Csete M, Walikonis J, Slawny N, Wei Y, Korsnes S, Doyle JC, Wold B. Oxygen-mediated regulation of skeletal muscle satellite cell proliferation and adipogenesis in culture. Journal of Cellular Physiology. 2001;189:189–196. doi: 10.1002/jcp.10016. [DOI] [PubMed] [Google Scholar]
- Dentice M, Marsili A, Ambrosio R, Guardiola O, Sibilio A, Paik JH, Minchiotti G, DePinho RA, Fenzi G, Larsen PR, et al. The FoxO3/type 2 deiodinase pathway is required for normal mouse myogenesis and muscle regeneration. Journal of Clinical Investigation. 2010;120:4021–4030. doi: 10.1172/JCI43670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes and Development. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Goetsch SC, Martin CM, Embree LJ, Garry DJ. Myogenic progenitor cells express filamin C in developing and regenerating skeletal muscle. Stem Cells Dev. 2005;14:181–187. doi: 10.1089/scd.2005.14.181. [DOI] [PubMed] [Google Scholar]
- Gohil VM, Sheth SA, Nilsson R, Wojtovich AP, Lee JH, Perocchi F, Chen W, Clish CB, Ayata C, Brookes PS, et al. Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat Biotechnol. 2010;28:249–255. doi: 10.1038/nbt.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Giordani J, Niro C, Souil E, Bertin F, Laclef C, Xu PX, Maire P. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Developmental Biology. 2007;302:602–616. doi: 10.1016/j.ydbio.2006.08.059. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- Guarente L. Sirtuins, aging, and metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:81–90. doi: 10.1101/sqb.2011.76.010629. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Conboy MJ, Conboy IM. Pharmacological inhibition of myostatin/TGF-beta receptor/pSmad3 signaling rescues muscle regenerative responses in mouse model of type 1 diabetes. Acta Pharmacol Sin. 2013;34:1052–1060. doi: 10.1038/aps.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Derfoul A, Feng X, Ryall JG, Dell’Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ. Progenitors of skeletal muscle satellite cells express the muscle determination gene, MyoD. Developmental Biology. 2009;332:131–141. doi: 10.1016/j.ydbio.2009.05.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Keller C, Hansen MS, Coffin CM, Capecchi MR. Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev. 2004;18:2608–2613. doi: 10.1101/gad.1243904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo JH, Smiley MA, Lovering RM, Margolis FL. Bex1 knock out mice show altered skeletal muscle regeneration. Biochemical and Biophysical Research Communications. 2007;363:405–410. doi: 10.1016/j.bbrc.2007.08.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Gillespie MA, Rudnicki MA. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell. 2008;2:22–31. doi: 10.1016/j.stem.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Rajendran GK, Liu N, Ware C, Rubin BP, Gu Y. SirT1 modulates the estrogen-insulin-like growth factor-1 signaling for postnatal development of mammary gland in mice. Breast Cancer Res. 2007;9:R1. doi: 10.1186/bcr1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wen Y, Bi P, Lai X, Liu XS, Liu X, Kuang S. Hypoxia promotes satellite cell self-renewal and enhances the efficiency of myoblast transplantation. Development. 2012;139:2857–2865. doi: 10.1242/dev.079665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annual Review of Cell and Developmental Biology. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Molecular Cell. 2012;45:255–262. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PubMed] [Google Scholar]
- Oburoglu L, Tardito S, Fritz V, de Barros SC, Merida P, Craveiro M, Mamede J, Cretenet G, Mongellaz C, An X, et al. Glucose and Glutamine Metabolism Regulate Human Hematopoietic Stem Cell Lineage Specification. Cell Stem Cell. 2014;15:169–184. doi: 10.1016/j.stem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Ono Y, Calhabeu F, Morgan JE, Katagiri T, Amthor H, Zammit PS. BMP signalling permits population expansion by preventing premature myogenic differentiation in muscle satellite cells. Cell Death and Differentiation. 2011;18:222–234. doi: 10.1038/cdd.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina G, Francois S, Regnault B, Czarny B, Dive V, Cumano A, Montarras D, Buckingham M. An adult tissue-specific stem cell in its niche: a gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 2010;4:77–91. doi: 10.1016/j.scr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai CR, et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert) Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Ryall JG. The role of sirtuins in the regulation of metabolic homeostasis in skeletal muscle. Curr Opin Clin Nutr Metab Care. 2012;15:561–566. doi: 10.1097/MCO.0b013e3283590914. [DOI] [PubMed] [Google Scholar]
- Ryall JG. Metabolic reprogramming as a novel regulator of skeletal muscle development and regeneration. FEBS J. 2013;280:4004–4013. doi: 10.1111/febs.12189. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Youn DY. Sirtuins: NAD(+)-dependent deacetylase mechanism and regulation. Current Opinion in Chemical Biology. 2012;16:535–543. doi: 10.1016/j.cbpa.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Shea KL, Xiang W, LaPorta VS, Licht JD, Keller C, Basson MA, Brack AS. Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell. 2010;6:117–129. doi: 10.1016/j.stem.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development. 2013;140:2535–2547. doi: 10.1242/dev.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S. Skeletal muscle stem cells in developmental versus regenerative myogenesis. J Intern Med. 2009;266:372–389. doi: 10.1111/j.1365-2796.2009.02158.x. [DOI] [PubMed] [Google Scholar]
- Tang AH, Rando TA. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO Journal. 2014 doi: 10.15252/embj.201488278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GC, Eskeland R, Hekimoglu-Balkan B, Pradeepa MM, Bickmore WA. H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Research. 2013;23:2053–2065. doi: 10.1101/gr.155028.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Molecular Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weydert A, Barton P, Harris AJ, Pinset C, Buckingham M. Developmental pattern of mouse skeletal myosin heavy chain gene transcripts in vivo and in vitro. Cell. 1987;49:121–129. doi: 10.1016/0092-8674(87)90762-8. [DOI] [PubMed] [Google Scholar]
- Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C, et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO Journal. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.