Summary
Polycomb repressive complex-2 (PRC2) is a histone methyltransferase required for epigenetic silencing during development and cancer. Early works suggested binding specificity of PRC2 to certain long non-coding RNAs for recruitment to chromatin. More recent studies provided evidence both in favor and against this idea. Here, we bridge the two existing models of PRC2-RNA interaction. RepA RNA is a good binding partner for PRC2, while multiple non-relevant RNAs, including bacterial mRNAs, also bind PRC2; with Kd's depend to some extent on the experimental conditions. Human and mouse PRC2 have broadly similar RNA-binding properties in vitro. Examination of evidence supporting an existing model for site-specific recruitment of PRC2 by a well-defined RNA motif in cells reveals that results are PRC2-independent. We conclude that promiscuous and specific RNA-binding activities of PRC2 in vitro are not mutually exclusive, and that binding specificity in vivo remains to be demonstrated.
Introduction
Polycomb repressive complex-2 (PRC2) is required for epigenetic silencing of transcription during embryonic development and cancer. It is a histone methyltransferase that mono-, di- and tri-methylates lysine 27 of histone H3, providing an epigenetic mark of repressed chromatin. The functional significance of this posttranslational modification has been demonstrated by a point mutation in lysine 27 of histone H3 that leads to homeotic transformations, like those seen in PRC2 deficiency (Pengelly et al., 2013). In Drosophila, PRC2 is recruited to chromatin through polycomb response elements (reviewed in (Schwartz and Pirrotta, 2007)). Despite the discovery of functionally similar elements in vertebrates ((Cuddapah et al., 2012; Sing et al., 2009; Woo et al., 2010)), the understanding of PRC2-specific recruiters to chromatin is far from complete and may rely on an ensemble of factors, including DNA elements, bridging proteins and RNA (reviewed in (Di Croce and Helin, 2013; Margueron and Reinberg, 2011; Simon and Kingston, 2013)).
Evidence indicates that long non-coding RNAs (lncRNAs) can recruit PRC2 to loci designated for silencing. RepA ncRNA recruits PRC2 during X-chromosome inactivation (Zhao et al., 2008). An RNA sequence that was predicted to form two hairpins is tandemly repeated 8 to 9 times within the A repeat region of Xist and RepA. The full A repeat sequence recruits PRC2 (Zhao et al., 2008), but what elements within the A repeat attract PRC2 are currently unknown. In vitro evidence suggests that a single repeat monomer can bind PRC2, albeit weakly (Zhao et al., 2008), but more complex RNA structures (Duszczyk et al., 2011; Maenner et al., 2010) involving multiple repeat units have greater affinity (Cifuentes-Rojas et al., 2014). Inspired by the potential of the single repeat to form a two-hairpin motif (Wutz et al., 2002), one study proposed that a two-hairpin motif of approximately 20 to 30 bases long was enriched within a subclass of non-coding RNAs that associate with PRC2 (Kanhere et al., 2010), particularly where there was an absence of tandem repeats. Their luciferase reporter system suggested that the two-hairpin motif may be responsible for the recruitment of PRC2 for epigenetic repression in vivo. Furthermore, to our knowledge this work provides the sole evidence that minimal point mutations in a short and well-defined PRC2-binding RNA motif can disrupt repression in vivo.
More recently, a quantitative binding study showed that PRC2 binds RepA RNA selectively, with high specificity compared to non-relevant RNA transcripts (Cifuentes-Rojas et al., 2014). These findings are in good agreement with the proposed role of RepA in the recruitment of PRC2 (Zhao et al., 2008). The study further indicated that, whereas RNA targets PRC2 in cis, RNA inhibits the histone methyltransferase activity of PRC2 until the complex comes in contact with JARID2 (Cifuentes-Rojas et al., 2014), thereby ascribing multiple physiological functions to the interaction between RNA and PRC2. It has also been shown that PRC2 associates with hundreds to thousands of RNAs in various cell types (Kaneko et al., 2013; Kanhere et al., 2010; Khalil et al., 2009; Zhao et al., 2010) and recent studies showed that PRC2 binds RNA promiscuously in vitro and in vivo (Davidovich et al., 2013; Kaneko et al., 2013). In vitro, the observed binding affinities of PRC2 for RNA differed in two reports (Cifuentes-Rojas et al., 2014; Davidovich et al., 2013). Moreover, one study suggested that the A repeat sequence element is neither sufficient nor essential for the recruitment of PRC2 in vivo (da Rocha et al., 2014), though the recruitment of PRC2 in the absence of the Repeat A sequence is significantly attenuated (Jeon and Lee, 2011).
Together, these studies have presented seemingly contradictory evidence for interactions between RNA and PRC2. A foremost question in the current debate is whether PRC2 binds RNA specifically, promiscuously, or nonspecifically. In particular, the term “promiscuous”, when applied to RNA binding, has caused some confusion in the field. “Promiscuous” means binding to many RNAs without the requirement for an obvious or well-defined protein-binding motif and with affinities that are not enormously different. Importantly, promiscuous does not mean nonspecific, as the latter term implies that the binding constants of different RNAs cannot be distinguished from each other. As PRC2 has become a model system for the study of the recruitment of chromatin remodeling factors by lncRNAs, resolving the issue of the specificity of RNA binding by PRC2 is important in order to better interpret results emerging in this active field.
Here we have combined resources from two independent laboratories to undertake a fresh examination of the issue by testing different experimental conditions and protein preparations. Our data argue that PRC2 exhibits both specificity and promiscuity in RNA binding in vitro. We show that human and mouse PRC2 complexes both bind RNA with mid to low nanomolar affinity. RepA RNA has 3- to 8-fold higher affinity than size-matched irrelevant bacterial mRNAs under specific binding conditions. We also find that RNA length increases binding affinities irrespective of sequence and that various longer-length RNAs can bind PRC2. Finally, we examined the model of a two-hairpin binding motif conferring transcription regulation by PRC2 in vivo and observe PRC2-independent effects that therefore leave open the question of structural binding motifs for further investigation.
Results
Controlling for RNA length is essential to assess binding specificity by PRC2
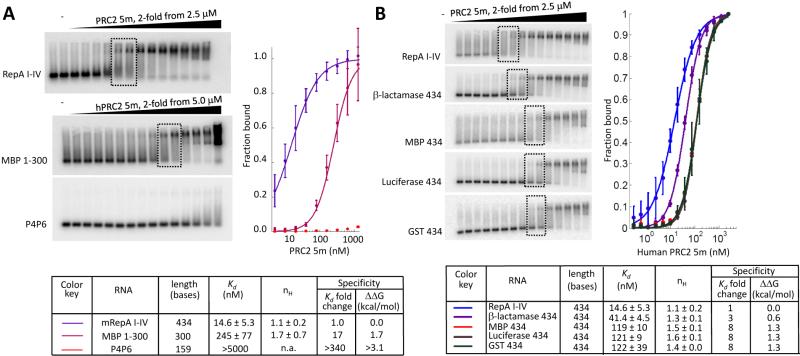
Following the recent finding that mouse PRC2 binds RepA RNA in vitro with higher affinity compared to non-relevant RNAs (Cifuentes-Rojas et al., 2014), we repeated these binding assays using the same RNAs and protocol but using human PRC2 5m purified as previously described (Davidovich et al., 2014; Davidovich et al., 2013). Quantitative EMSAs (Figure 1A) gave dissociation constants of human PRC2 5m to RepA I-V RNA (14.6 ± 5.3 nM) and to the two non-relevant RNAs MBP 1-300 (245 ± 77 nM) and P4P6 (Kd > 5000 nM) RNAs, which were used by (Cifuentes-Rojas et al., 2014) as negative controls. Qualitatively, these observations with human PRC2 are in good agreement with the mouse PRC2 study (Cifuentes-Rojas et al., 2014), showing higher affinity of RepA to PRC2 compared to the two non-relevant, yet shorter, RNAs.
Figure 1. Adequate control for RNA length is necessary to assess RNA binding specificity.
EMSA performed as previously described (Cifuentes-Rojas et al., 2014), using human PRC2 5m. (A) Large variation in affinity is observed when non-specific RNAs are shorter than the canonical RNA for which they are controlling. Same RNAs as used in (Cifuentes-Rojas et al., 2014): RepA I-IV, 434 bases long RNA, including all tandem repeats from the repeat A region within mouse RepA RNA; MBP 1-300, 300 bases from the 5’ end of MBP mRNA from E. coli ; P4P6, 159 bases long RNA from Tetrahymena group I self-splicing intron. (B) Experiment repeated with RepA and size-matched RNAs, serving as an adequate control for RepA length, including 434 bases from the 5’ ends of protein coding mRNA sequences from bacterial β-lactamase, E. coli maltose binding protein (MBP), Firefly luciferase and glutathione S-transferase (GST) from Schistosoma japonicum (blood fluke). Dissociation constants and Hill coefficients are indicated. Dashed boxes indicate the protein concentrations required for shifting half of the radiolabeled RNA, namely Kd, as identified by densitometry. Error bars within binding curves and standard deviations for binding constants represent three independent experiments, performed on different days. Kd stands for dissociation constant and nH for Hill coefficient. Binding specificity represented as fold-change between dissociation constants that were observed for RepA and non-specific RNAs, as previously defined (Johansson et al., 1998). Same values used to calculate the corresponding differences in Gibbs free energy (ΔΔG). See also Figure S1 and Table S1.
We previously showed that PRC2 binds RNA promiscuously in a length-dependent manner (Davidovich et al., 2013), as expected for a protein binding to a nucleic acid partner with multiple binding sites (Broderick et al., 2011; Epstein, 1979; Kowalczykowski et al., 1986). MBP 1-300 (300 bases) and P4P6 (159 bases) are both shorter than mouse RepA I-IV (434 bases). We therefore repeated this experiment under the same conditions (Cifuentes-Rojas et al., 2014), but with a panel of non-relevant RNAs all of the same length as mouse RepA I-IV RNA (Figure 1B). Mouse RepA I-IV indeed showed 3- to 8-fold higher affinity for PRC2, compared to the other RepA size-matched non-relevant RNAs that we tested. Yet, PRC2 did bind all the size-matched non-relevant RNAs as well, with dissociation constants spanning from 40 nM to 120 nM (Figure 1B). These differences in affinity represent ΔΔGs of around 1 kcal/mol (Figure 1B).The Hill coefficient ranged from 0.9 to 1.6 (Figures 1-2), with RepA typically at the lower end.
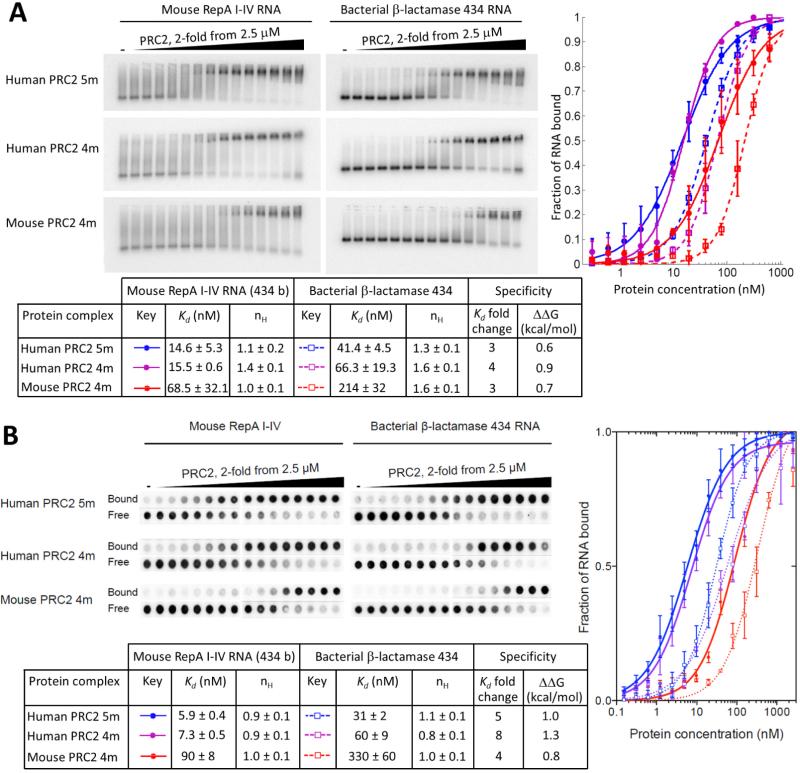
Figure 2. Complete binding curves of mouse RepA I-IV in the presence of different concentrations of human PRC2 5m, human PRC2 4m and mouse PRC2 4m, in comparison to a non-relevant RNA control of the same length.
(A) Binding and EMSA conditions are as previously described (Cifuentes-Rojas et al., 2014). To control for RepA I-IV length, a 434 bases long RNA comprising the 5’ end of the coding sequence of the non-relevant bacterial beta-lactamase mRNA (β-lactamase 434) was used. Error bars within binding curves and standard deviations within the table represent two to three independent experiments, performed on different days. (B) Filter Binding assay performed as previously described (Cifuentes-Rojas et al., 2014) for the same RNAs and PRC2s. Specificity defined as in Figure 1. Error bars within binding curves and standard deviations within the table represent at least three independent experiments, performed on different days. See also Figure S1 and Table S1.
These experiments demonstrate elevated affinity of PRC2 to RepA, under binding conditions that were previously used (Cifuentes-Rojas et al., 2014). At the same time, they also reinforce the previous observation that PRC2 can bind various RNAs above a certain length with affinities within an order of magnitude of each other, which has been called “promiscuous binding” (Davidovich et al., 2013).
Direct comparison of human vs. mouse PRC2 for binding RepA and non-relevant RNA
Although PRC2 subunits are relatively conserved between mammals, mouse and human PRC2 protein subunits might in principle have different RNA-binding properties, and could potentially account for affinity differences previously reported (Cifuentes-Rojas et al., 2014; Davidovich et al., 2013). To exclude this concern (Cifuentes-Rojas et al., 2014), we tested various binding conditions for quantitative EMSA using mouse RepA I-IV and three different PRC2 complexes (Figure S1): human PRC2 5m (EZH2, SUZ12, EED, RBBP4 and AEBP2), human PRC2 4m (same as 5m, except no AEBP2) and mouse PRC2 4m (Figure 2, left EMSAs). Dissociation constants spanned a range of four-fold for all PRC2 complexes binding to RepA I-IV RNA (14.6 ± 5.3 nM to 68.5 ± 32.1 nM). All three PRC2 complexes bound the sized-matched non-relevant β-lactamase 434 RNA, bearing 434 bases from the coding sequence of bacterial beta-lactamase mRNA, about 3- to 4-fold more weakly (41.4 ± 4.5 nM to 214 ± 32 nM, Figure 2A). These results were consistent with those obtained by filter binding assays, as previously used (Cifuentes-Rojas et al., 2014) (Figure 2B). Collectively, this panel of experiments (Figure 2) indicates that human and mouse PRC2 bind both RepA RNA and size-matched non-relevant RNAs, with RepA RNA having 3- to 4-fold higher affinity. These results further strengthen the idea that specific and promiscuous binding properties of PRC2 are not mutually exclusive.
Effects of experimental conditions and RNA length on observed binding specificity
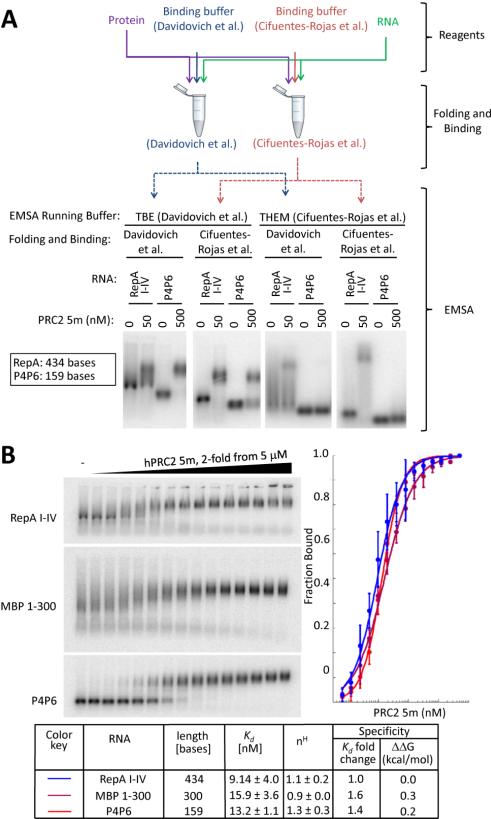
Binding affinity in vitro can be altered by experimental conditions, as certain conditions can reduce or increase the stability of some RNPs (Alves and Cunha, 2012; Hellman and Fried, 2007; Ryder et al., 2008). To test potential effects of the experimental conditions on complex stability, we performed a comparative binding experiment with human PRC2 5m. RNA folding and RNA-protein binding were carried out using conditions previously published (Cifuentes-Rojas et al., 2014; Davidovich et al., 2013). We performed this test with mouse RepA I-IV RNA (434 bases) and the shorter non-relevant RNA P4P6 (159 bases). Each sample was split in half and each portion was loaded on a different gel for EMSA. The gels were then run in TBE (Davidovich et al., 2013) or THEM (Cifuentes-Rojas et al., 2014) buffer (Figure 3A). Remarkably, the RepA-PRC2 complex was readily evident regardless of experimental conditions, while PRC2 binding to P4P6 RNA was only observed when TBE was the electrophoresis buffer, not when THEM was used. These results are in good agreement with both previous works (Cifuentes-Rojas et al., 2014; Davidovich et al., 2013) and with our findings within this study, showing that various RepA-size-matched mRNAs bind PRC2 under both these conditions (Figures 1 and 2).
Figure 3. Apparent RNA binding affinity depends on experimental conditions.
(A) RepA I-IV and P4P6 RNAs, as previously used (Cifuentes-Rojas et al., 2014), were subjected to a qualitative binding experiment, where the RNA folding protocol and PRC2-RNA incubation were performed according to either (Davidovich et al., 2013) or (Cifuentes-Rojas et al., 2014). Each sample was split into two and each portion loaded on a non-denaturing gel, buffered either with TBE or THEM. Agarose gel density and running conditions were as published within these references. (B) Same RNAs and PRC2, as in Figure 1, were subjected to quantitative binding experiments using binding buffer including 50 mM Tris HCl, pH 7.5 (@ RT), 100 mM KCl, 2.5 mM MgCl2, 0.1 mM ZnCl2, 2 mM 2-mercaptoethanol, 0.1 mg/ml BSA, 0.1 mg/ml yeast tRNA (Sigma R5636), 0.05 % v/v NP40 and 5 % v/v glycerol. Samples were loaded on a 0.7% (w/v) agarose gel, buffered with TBE, and EMSA carried out as previously described (Davidovich et al., 2013). Error bars within binding curves and standard deviations within the table represent three independent experiments, performed on different days. See also Figures S2 and S3 and Table S1.
We then performed the same binding experiment as within (Cifuentes-Rojas et al., 2014) and herein (Figure 1), using another binding buffer including 50 mM Tris HCl, pH 7.5 at RT, 100 mM KCl, 2.5 mM MgCl2, 0.1 mM ZnCl2, 2 mM 2-mercaptoethanol, 0.1 mg/ml BSA, 0.1 mg/ml yeast tRNA Sigma R5636, 0.05 % v/v NP40 and 5 % v/v glycerol. EMSA was carried out with TBE, as previously described (Davidovich et al., 2013). In agreement with our mix-and-match experiment (Figure 3A), PRC2 associated with all three RNAs with similar affinities (Figure 3B), regardless of RNA length. Collectively, binding experiments thus far (Figure 1, 2 and 3) indicate that apparent binding specificity and affinity of PRC2 to RNA can be altered by the experimental protocol to a certain degree. Differences in availability of magnesium ions in the two running buffers could play a role. Under some conditions (Figure 3B), the affinity differences were smaller than previously observed with RNAs of different sizes (Cifuentes-Rojas et al., 2014). Such binding conditions can potentially be useful in order to reproducibly emphasize small variations in affinity (Figure 1B).
Examination of recruitment of PRC2 by a two-hairpin motif
The possible existence of an in vivo RNA motif for PRC2 recognition has been of great interest to the field. Two-hairpin motifs have been shown to be capable of binding PRC2 in qualitative binding assays in vitro (Kanhere et al., 2010; Zhao et al., 2010; Zhao et al., 2008), potentially providing a basis for structural recognition. In vivo tethering experiments in which the addition of such motifs repressed gene expression from luciferase reporters indicated that the motifs could function in a repressive mechanism (Kanhere et al., 2010). To examine PRC2's binding properties to these RNAs, we transcribed five RNAs bearing binding sequences previously identified, each fused to the luciferase reporter sequence used within the original work (Kanhere et al., 2010). Specifically, two constructs included wild-type two-hairpin motifs originating from RepA RNA (RepA) or an independent short ncRNA (Short RNA) (Figure S2A). Two other constructs included mutations that were designed to disrupt these motifs (RepA mut and Short RNA mut, respectively) (Kanhere et al., 2010). A fifth construct included the R and U5 regions within the HIV LTR (LTR), originally used to estimate the basal expression level in a luciferase reporter assay (Kanhere et al., 2010). Quantitative EMSAs were performed with PRC2 5m as previously described (Davidovich et al., 2013). Notably, all RNAs bound PRC2 in vitro with similar Kd's, whether in the presence or absence of the two-hairpin motif (Figure S2B, S2C and S2D). This is consistent with similar affinities observed for other RNAs tested using these conditions (Davidovich et al., 2013).
We next asked whether these inserted elements repress transcription of a luciferase reporter, and indeed they do (Fig. S3A), as previously reported (Kanhere et al., 2010). Expression was higher in cells transfected with plasmids without the two-hairpin motif and lower in its presence (Figure S3A). In light of our finding that the in vitro affinities of all RNAs were similar (Fig. S2), we tested whether the repressive effect was dependent on PRC2 by measuring luciferase activity after knockdown of the essential PRC2 subunit, SUZ12 (Figure S3A and S3B). Although substantial depletion of SUZ12 and of the H3K27me3 mark were observed after knockdown (Figure S3B), expression levels from all vectors were unchanged (Figure S3A). Thus, although there are reproducible differences in expression levels between these plasmids, the differences do not appear to be PRC2-dependent.
In search of an alternative explanation for the differences in luciferase expression among the different plasmids, we next sequenced the U3 promoters in the five plasmids that were used in the original work (Kanhere et al., 2010). We found a promoter mutation (U3 Δ115-116) only in the two plasmids carrying the wild-type sequences of the two-hairpin motifs (RepA and Short RNA, Figure S3E), the two vectors that had lower luciferase expression levels. When we corrected this promoter mutation, the expression level of the corrected reporter (RepA corrected) increased back to the basal level (Figure S3C, left bar plot). This observation indicates that the expression difference observed between the RepA and RepA mut reporters (Kanhere et al., 2010) resulted from the U3 Δ115-116 promoter mutation, rather than differences in efficiency of recruitment of PRC2 by the transcribed RNA. We corrected also the two mutations that we identified within the U3 promoter of the Short RNA construct (Figure S3F), but not present in Short RNA mut. Correction of each mutation resulted in partially increased expression, rising to about one-half the basal level (Figure S3D). Because the reporters were designed with the two-hairpin motif within the same transcript as the luciferase coding sequence, it could be that structure in the leader RNA affects the initiation or elongation of transcription, the initiation or efficiency of translation, or RNA stability, any of which is presumptively PRC2-independent. Additional tests (Fig. S3C, right bar graph) showed that the promoter mutation U3 del115-116 gave a two-fold expression decrease even in the absence of any leader RNA. These results demonstrate that the two-hairpin form of Repeat A is not sufficient to achieve silencing in the reporter-based system.
Discussion
Here we have shown empirically that specific and promiscuous RNA binding by PRC2 are not necessarily mutually exclusive, and we elucidated biochemical conditions under which binding behavior can, to some extent, be modulated experimentally. It is well established that the apparent dissociation constant of a given protein to RNA can be affected by binding conditions in vitro (Alves and Cunha, 2012; Hellman and Fried, 2007; Ryder et al., 2008). We show here that either by EMSA or Filter Binding Assay, PRC2 binds canonical and non-canonical RNA targets with reasonably high affinities (Kd's in the range of 15 – 200 nM) (Figure 1B and 2). Longer RNAs in general bind better, though length alone does not determine the affinity of binding. An example is the difference in affinity of PRC2 for the identically-sized RepA (Kd = 81 nM) and its antisense RNA, Tsix (Kd = 320 nM) (Cifuentes-Rojas et al., 2014). Our observations are consistent with previous evidence indicating higher affinity of PRC2 to RepA RNA compared to non-relevant RNAs (Cifuentes-Rojas et al., 2014), with the apparent specificity depending on the binding conditions. They also show that PRC2 can bind noncanonical RNAs. We further tested the binding of multiple PRC2 complexes from both human and mouse to RepA and noncanonical RNAs and found that the observed binding specificity is very similar across these species, regardless of whether EMSA or Filter Binding is used as the experimental assay. Thus, in vitro, PRC2 engages with multiple binding partners (“promiscuous”), at the same time that is has up to 8-fold specificity for RepA, of the Xist locus.
The Hill coefficient that was measured for different complexes and RNAs ranged from 0.9 to 1.6, and depends on the protein, RNA, binding conditions, and assays used (Figures 1-3), with RepA typically at the lower end. A value greater than 1.0 indicates positive cooperativity, commonly involving multiple proteins that cooperate in order to increase the binding enthalpy through protein-protein interactions. While PRC2 is a large complex that binds long RNAs, there have been some complications in the establishment of an EMSA-based assay that is sufficiently sensitive to detect multiple binding events. An experimentally measured Hill coefficient is affected mainly by the number of proteins that bind cooperatively to a single binding site, but also by the number of binding sites on the RNA (Senear and Brenowitz, 1991). Therefore, whether binding of PRC2 to RNA demonstrates cooperative protein-protein interactions remains to be determined. We have previously used a series of titration experiments to show multiple binding events of PRC2 on a single RNA, with 3 to 4 PRC2 complexes per 200 bases long RNA (Davidovich et al., 2014). One should not expect that binding stoichiometry will always scale linearly with RNA length, in the case of multiple binding sites. Yet, it should be safe to assume that longer RNAs, of approximately 400 bases, would offer PRC2 multiple binding sites as well. It is possible that some variation in the binding stoichiometry of PRC2:RNA would exist between different RNAs, especially in light of the variations in Hill coefficients, as we observed herein.
Under binding conditions that indicate the greatest differences between the canonical target RepA and non-relevant size-matched RNAs (Figure 1B), differences in affinity correspond to around 1 kcal/mol differences in free energy (ΔΔG) (Figure 1B). This energy difference is on the order of the enthalpy that can be contributed by a single hydrogen bond (Pauling, 1960). Previous work characterizing specific protein-RNA interactions commonly saw larger ΔΔG values with minor sequence changes. For instance, an AU bulge deletion within the telomerase RNA from yeast significantly reduced its affinity to its binding protein Ku, with Kd increased by >270 fold, representing ΔΔG >2.9 kcal/mol (Dalby et al., 2013). Bacteriophage MS2 coat protein undergoes >1000-fold reduction in affinity (ΔΔG >3.0 kcal/mol) to its target RNA upon a point mutation in a bulged adenosine within its specific structural binding motif (Romaniuk et al., 1987). The N-terminal RNP domain of U1A protein showed reduction in affinity of >100 fold, or ΔΔG = 2.5 kcal/mol, after a single base modification, adenosine to purine riboside, within its binding site on U1 snRNA (Nolan et al., 1999). The small ΔΔG for PRC2-RepA interactions (relative to PRC2-MBP, for example) may reflect the fact that PRC2 is not a single-purpose RNA-binding protein. Indeed, PRC2 has the ability to interact with various transcripts in vivo and in vitro. Furthermore, from a biophysical perspective, an energetic difference of 1 kcal/mol may seem minor, considering the size of these relatively long RNAs (434 bases) and their sequence diversity. However, in physiological circumstances, small differences in affinity and energetics as measured in vitro can have profound effects in vivo. Experiments performed in cellular models will be necessary to determine the functional significance of both large and small differences identified here under various biochemical conditions. Indeed, specific binding RNAs, such as RepA/Xist, may be folded differently in vivo than in an in vitro-transcribed system. If so, the Kd's and ΔΔG's calculated here would need re-interpretation. Another possibility is that binding specificity is achieved in vivo through competition with other RNPs that specifically bind selected nascent RNA transcripts (Herzog et al., 2014).
The model of RNA-binding specificity by PRC2 would be well supported if RNA characteristics that are determinants for PRC2 binding could be identified experimentally, with the evaluation of specificity guided by the available literature. Relevant to this, a previous publication (Kanhere et al., 2010) proposed that a two-hairpin motif may be responsible for specific recognition between PRC2 and short promoter-associated RNAs. This work has been influential and, to our knowledge, provides the only evidence for a two-hairpin model in cells. Importantly, we reproduced the reporter-based observations obtained within the original study (Kanhere et al., 2010). However, we also found that the expression differences between reporter plasmids are PRC2-independent (Figure S3A and S3B) and some expression variations could be attributed to promoter mutations (Figures S3C through S3F). It is important to note that the two-hairpin motif contains only one of 8-9 repeat units within RepA and may therefore be less efficient at recruiting PRC2 to achieve silencing, perhaps explaining why effects could not be measured above background. Our work in no way challenges the remaining conclusions of the original study, nor does it rule out the possibility that specific RNA motifs could, in principle, recruit PRC2 for epigenetic repression in vivo. Our findings therefore leave open the search for RNA motifs that bind PRC2.
Experimental Procedures
Protein expression and purification
PRC2 complexes (Figure S1) were expressed in insect cells and purified as previously described for the human (Davidovich et al., 2014) and mouse (Cifuentes-Rojas et al., 2014) complexes.
In vitro RNA-binding assays
RepA I-IV, MBP 1-300 and P4P6 RNAs were transcribed and labeled as previously described (Cifuentes-Rojas et al., 2014). Other RNAs were transcribed from DNA templates that were PCR amplified using plasmids and primers as described within Table S1. Electrophoretic mobility shift assays (EMSAs) and filter binding assays were performed as previously described (Cifuentes-Rojas et al., 2014; Davidovich et al., 2013), unless otherwise indicated.
Tissue culture and reporter plasmids
Reporter constructs were kindly donated by Richard Jenner, University College London, UK. Knockdown of SUZ12 in HEK293T/17 cells was performed as previously described (Davidovich et al., 2013). Reporter plasmids were transfected 48 h later using Lipofectamine 2000 (Life Technologies catalog number 11668019), using the manufacturer's protocol. Firefly and Renilla luciferase activities were measured 24 h after plasmids were transfected, using the Dual-Glo luciferase assay system (Promega catalog number E2920), following the manufacturer's instructions. Immunoblotting protocol and antibodies were as previously described (Davidovich et al., 2013). Sequences of primers used for sequencing and PCR mutagenesis are given in Table S1.
Supplementary Material
Acknowledgements
We thank Dr. Richard Jenner for providing luciferase reporter plasmids and for open discussions to resolve the differences in our results. C.C-R. is supported by NIH-F32-GM101828 and J.T. L. by NIH-R01-GM090278. T.R.C. and J.T.L. are investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
C.D., C.C-R., J.T.L. and T.R.C designed the experiments. C.D., X.W., C.C-R., K.J.G and A.R.G performed the experiments. C.D., C.C-R., J.T.L. and T.R.C wrote the manuscript.
References
- Alves C, Cunha C. In: Electrophoretic Mobility Shift Assay: Analyzing Protein - Nucleic Acid Interactions. Magdeldin M, editor. InTech; 2012. [Google Scholar]
- Broderick JA, Salomon WE, Ryder SP, Aronin N, Zamore PD. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. Rna. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory Interactions between RNA and Polycomb Repressive Complex 2. Molecular cell. 2014 doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Roh TY, Cui K, Jose CC, Fuller MT, Zhao K, Chen X. A novel human polycomb binding site acts as a functional polycomb response element in Drosophila. PloS one. 2012;7:e36365. doi: 10.1371/journal.pone.0036365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, et al. Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Molecular cell. 2014;53:301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Dalby AB, Goodrich KJ, Pfingsten JS, Cech TR. RNA recognition by the DNA end-binding Ku heterodimer. Rna. 2013;19:841–851. doi: 10.1261/rna.038703.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Goodrich KJ, Gooding AR, Cech TR. A dimeric state for PRC2. Nucleic acids research. 2014 doi: 10.1093/nar/gku540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nature structural & molecular biology. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nature structural & molecular biology. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Duszczyk MM, Wutz A, Rybin V, Sattler M. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. Rna. 2011;17:1973–1982. doi: 10.1261/rna.2747411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein IR. Kinetics of nucleic acid-large ligand interactions: exact Monte Carlo treatment and limiting cases of reversible binding. Biopolymers. 1979;18:2037–2050. doi: 10.1002/bip.1979.360180815. [DOI] [PubMed] [Google Scholar]
- Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nature protocols. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog VA, Lempradl A, Trupke J, Okulski H, Altmutter C, Ruge F, Boidol B, Kubicek S, Schmauss G, Aumayr K, et al. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nature genetics. 2014 doi: 10.1038/ng.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson HE, Dertinger D, LeCuyer KA, Behlen LS, Greef CH, Uhlenbeck OC. A thermodynamic analysis of the sequence-specific binding of RNA by bacteriophage MS2 coat protein. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9244–9249. doi: 10.1073/pnas.95.16.9244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nature structural & molecular biology. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Molecular cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC, Paul LS, Lonberg N, Newport JW, McSwiggen JA, von Hippel PH. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986;25:1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS biology. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan SJ, Shiels JC, Tuite JB, Cecere KL, Baranger AM. Recognition of an essential adenine at a protein-RNA interface: Comparison of the contributions of hydrogen bonds and a stacking interaction. Journal of the American Chemical Society. 1999;121:8951–8952. [Google Scholar]
- Pauling L. The nature of the chemical bond and the structure of molecules and crystals; an introduction to modern structural chemistry. 3d edn Cornell University Press; Ithaca, N.Y.: 1960. [Google Scholar]
- Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- Romaniuk PJ, Lowary P, Wu HN, Stormo G, Uhlenbeck OC. RNA binding site of R17 coat protein. Biochemistry. 1987;26:1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- Ryder SP, Recht MI, Williamson JR. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods in molecular biology. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature reviews. Genetics. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Senear DF, Brenowitz M. Determination of binding constants for cooperative site-specific protein-DNA interactions using the gel mobility-shift assay. The Journal of biological chemistry. 1991;266:13661–13671. [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Molecular cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nature genetics. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.