Abstract
Glioblastoma multiforme (GBM) is a highly aggressive brain tumor, with dismal survival outcomes. Recently, cancer stem cells (CSCs) have been demonstrated to play a role in therapeutic resistance and are considered to be the most likely cause of cancer relapse. The identification of CSCs is an important step toward finding new and effective ways to treat GBM. Tenascin-C (TNC) protein has been identified as a potential marker for CSCs in gliomas based on previous work. Here, we have investigated the expression of TNC in tissue microarrays including 17 GBMs, 18 WHO grade III astrocytomas, 15 WHO grade II astrocytomas, 4 WHO grade I astrocytomas, and 7 normal brain tissue samples by immunohistochemical staining. TNC expression was found to be highly associated with the grade of astrocytoma. It has a high expression level in most of the grade III astrocytomas and GBMs analyzed and a very low expression in most grade II astrocytomas, whereas it is undetectable in grade I astrocytomas and normal brain tissues. Double-immunofluorescence staining for TNC and CD133 in GBM tissues revealed that there was a high overlap between theses two positive populations. The results were further confirmed by flow cytometry analysis of TNC and CD133 in GBM-derived stem-like neurospheres in vitro. A limiting dilution assay demonstrated that the sphere formation ability of CD133+/TNC+ and CD133–/TNC+ cell populations is much higher than that of the CD133+/TNC– and CD133–/TNC– populations. These results suggest that TNC is not only a potential prognostic marker for GBM but also a potential marker for glioma CSCs, where the TNC+ population is identified as a CSC population overlapping with part of the CD133– cell population.
Keywords: GBM, cancer stem cell, tenascin-C, marker, tissue microarrays
Introduction
Glioma is the most common primary central nervous system tumor. Astrocytic tumor, the most common type of glioma, is classified into four grades, according to different grades of malignancy (I–IV), by the Word Health Organization (WHO).1 Glioblastoma multiforme (GBM), a WHO grade IV astrocytic tumor, is the most devastating primary human brain tumor with a high morbidity and mortality among patients.2 Even when treated with a combination of surgery, chemotherapy, and radiotherapy, patients with GBM still have a very poor long-term outcome, with a median survival of 14.6 months.3,4 The poor survival is primarily due to the recurrence of tumor, which is resistant to standard therapies.5 Thus, new strategies to treat GBM are urgently needed.
Accumulating evidence has demonstrated that a small population of stem-like cells, termed cancer stem cells (CSC), in tumors are responsible for tumor initiation and ongoing growth.6 These cells contribute to therapeutic resistance and are considered to be the most likely cause of cancer relapse.5,7,8 CSCs have been identified and validated in various solid tumors such as breast, colon, pancreas, and liver.9−12 They have also been shown to exist in GBM,13−16 and recent work has shown the possibility for treatment of brain tumors by targeting CSCs.17−20 The identification of new tumor stem cell markers for GBM is essential for diagnosis and effective treatment of malignant brain tumors.
There have been efforts to identify CSC markers in GBM.21−23 CD133 has been identified and widely used as a marker to characterize CSC population in GBMs.18,21,24 However, CD133 expression is observed in mature luminal ductal epithelial cells in various organs of adults, indicating that CD133 may not be a suitable organ-specific CSC marker.25 Other evidence now suggests that CD133 may serve as a CSC marker in only a subgroup of GBM.26 Moreover, CD133– cells also have the capability to generate GBM tumors in immunodeficient nude mice or rats.27,28 These issues underscore the need for additional markers to identify CSCs in GBM. Recently, CD90 was identified as a marker for GBM CSCs in primary human high-grade gliomas using tissue microarrays.23 The expression level of CD90 was highly correlated with WHO grades and was significantly higher than that in low-grade gliomas. Interestingly, CD90 and CD133 markers were coexpressed in GBM, where 100% of the CD133+ cells were also positive for CD90, but only part of the CD90+ cells were positive for CD133, indicating that the CD133+ stem-like cells are a subpopulation of CD90+ cells in GBMs in vivo.23 Other suggested CSC surface markers that have been identified, such as CD15 and NG2, are still under investigation.29,30
Tenascin-C (TNC) is a large extracellular matrix (ECM) glycoprotein, characterized by a six-armed quaternary structure and a modular construction.31 It is composed of four subunits: a cysteine-rich amino terminal domain, a sequence of epidermal growth factor (EGF)-like repeats, a number of fibronectin type III repeats, and a carboxy-terminal domain homologous to fibrinogen. Its deregulated high expression is causally linked to many diseases, including heart failure, thrombosis, atherosclerosis, and cancer.32 TNC was originally discovered as a glioma–mesenchymal extracellular matrix antigen that is ubiquitously expressed in glioblastomas but not in normal brains.33 The distribution pattern of TNC in GBM showed that it plays a role in angiogenesis and tumor cell proliferation.34 TNC contributes to the generation of a stem cell niche, which is important to the development of neural stem cells.35,36 Recent work also demonstrated that TNC is a promising target of GBM therapy by interference RNA intervention (iRNAi).37 Thus, TNC may play an important role in GBM.
In previous work using mass spectrometry analysis, the expression level of TNC in a GBM-derived stem neurosphere line was shown to be dramatically higher than that in three traditional adherent GBM cell lines, indicating that TNC may be a potential marker for CSCs in GBMs.38 TNC has also been identified as a marker in neuroblastoma.39 In the current work, we showed the potential of TNC as a marker for glioma CSCs using tissue microarray analysis, where a significant number of samples from different grades of glioma could be studied. The stem-like cell characterization of TNC positive cells was then further demonstrated using in vitro cell sorting and a limiting dilution assay to monitor sphere formation to understand the role of TNC as a potential CSC marker. It is shown that TNC is not only a potential prognostic marker for GBM but also a potential marker for glioma CSCs.
Materials and Methods
Tissue Samples
Tissue microarray (TMA) samples were obtained from US Biomax (Rockville, MD; cat. nos. GL722, T174, and GL807). In total, 61 samples were analyzed in these experiments, including 17 GBM samples (age: 35 ± 18 years; 7 females and 10 males), 18 WHO grade III astrocytomas (age: 46 ± 11 years; 7 females and 11 males), 15 WHO grade II astrocytomas (age: 41 ± 12 years; 6 females and 9 males), 4 WHO grade I astrocytomas (age: 41 ± 8 years; 1 female and 3 males), and 7 normal brain tissue samples (age: 38 ± 8 years; 6 females and 1 male). The brain tissue samples originated from different donors, and each sample had at least two replicates. The glioma tissue sections were from the tumor areas and do not include the adjacent normal tissues.
Protein Extraction from Tissue Sections
Paraformaldehyde-fixed GBM sections were obtained from University Hospital at the University of Michigan with approval from the Internal Review Board. They were thawed for 20–30 s at room temperature and rinsed with PBS before protein extraction. The FASP protein extraction kit was used to extract protein from tissue sections (Protein Discovery, Inc., Knoxville, TN). Lysis buffer containing 4% SDS, 0.1 M DTT, and protease inhibitors was added onto the tissue section slides (US Biomax, Inc., Rockville, MD) and incubated for 5 min. The collected tissue cells were boiled using a 100 °C water bath for 5 min. The samples were centrifuged at 15 000g for 10 min, and the supernatant was stored at −80 °C for western blotting analysis.
Immunohistochemical Analysis of Tissue Microarrays
Immunohistochemical staining was performed using tissue microarray samples. The paraffin-embedded tissue arrays with 1.5 mm core diameter and 5 μm thickness were dewaxed in xylene for 10 min twice and rehydrated through a series of alcohol solutions (200 proof, Sigma-Aldrich, St. Louis, MO) (100% ethanol twice, 90% ethanol, and 70% ethanol, 5 min each) to water. Then, the slides were boiled for 15 min in citrate buffer (Teknova, Hollister, CA) at pH 6.0 for antigen retrieval. After returning to room temperature, endogenous peroxidase activity was blocked with 3% H2O2 in methanol for 10 min. The TMAs were then rinsed with water and PBS and subsequently blocked with 2% BSA and incubated with rabbit anti-human Tenascin-C monoclonal antibody (1:100 dilution, Abcam, Cambridge, MA) overnight at 4 °C followed by incubation with a goat anti-rabbit IgG conjugated to horseradish peroxidase (1:250 Abcam, Cambridge, MA). Immunodetection was performed using DAB solution (Vector Laboratories, Burlingame, CA). Hematoxylin counterstain was used to visualize nuclei. The TNC expression level in each tissue section was assessed in non-necrotic areas of three separate microscopic fields of view under a magnification of 200× and was represented by the mean of the percentage of TNC+ cells. The results were confirmed by a pathologist.
Double-Immunofluorescence Staining of TNC and CD133
Double-immunoflourescence staining was performed using tissue microarrays. According to the different properties of each individual antibody, a simultaneous procedure was used for the staining of TNC and CD133. Briefly, the TMAs were dewaxed in xylene for 10 min twice and rehydrated through a series of alcohol solutions to water, followed by boiling for 15 min in 100 mM citrate buffer at pH 6.0 for antigen retrieval. After incubation with 1% BSA in PBS for 1 h at room temperature to block nonspecific binding, a mixture of rabbit anti-human TNC (1:80 dilution, Abcam, Cambridge, MA; cat. no. ab108930) monoclonal antibody and mouse anti-human CD133 (1:100 dilution, Millipore, Temecula, CA; cat. no. MAB 4399) monoclonal antibody was incubated with the slides overnight at 4 °C. DyLight 488 anti-rabbit IgG (H + L) and DyLight 549 anti-mouse IgG (H + L) secondary antibodies (1:150 dilution, Vector Laboratories, Burlingame, CA) were used for immunofluorescence detection, and 4,6-diamidino-2-phenylindole (DAPI) counterstain was used to visualize nuclei. Between each step, three washes were applied with PBST for 10 min each. Finally, TMA slides were dehydrated in alcohol and coverslipped using a CC/Mount permanent mounting medium (Sigma, St. Louis, MO).
Cell Culture
HSR-GBM1 neurosphere cells were derived from a primary GBM patient and have been propagated for hundreds of passages as neurospheres in vitro to enrich the cancer stem-like cell population.18,40 HSR-GBM1 neurosphere cells were used for a coupled cell sorting and limiting dilution assay of sphere formation. As described before, NeuroCult proliferation medium (Stem Cell Technologies, Vancouver, Canada) was supplemented with 10 ng/mL EGF (PeproTech, Rocky Hill, NJ), 10 ng/mL FGFb (PeproTech), and 2 μg/mL heparin (Sigma).14,18 Differentiation of the neurospheres was achieved by plating 0.9–1 × 105 cells/cm2 on a polyornithine (15 μg/mL) coated culture plate and maintaining them in the NeuroCult differentiation medium (Stem Cell Technologies) as described previously.14
Western Blotting Analysis
An equal amount of protein from different samples was separated by 4–15% SDS-PAGE and transferred to poly(vinylidene difluoride) membranes (PVDF, Bio-Rad, Hercules, CA). The membranes were blocked for 1 h by 2% milk (Bio-Rad) in PBST (0.1% Tween-20 in PBS) and then incubated with the following antibodies overnight at 4 °C: anti-TNC (Abcam), anti-CD133 (Millipore), and anti-β-actin (Abcam). After being washed three times with PBST, the membranes were incubated with peroxidase-conjugated secondary antibody IgG (H + L) for 1 h, washed another three times with PBST, and then detected by Supersignal West Pico chemiluminescent substrate (Thermo Scientific).
Cell Sorting and Limiting Dilution Assay of Sphere Formation
Fluorescence-activated cell sorting experiments were performed at the Flow Cytometry Core at the University of Michigan in triplicate. To execute the cell sorting experiments, HSR-GBM1 neurospheres were first dissociated into single cells. TNC (Abcam, Cambridge, MA; cat. no. ab108930) monoclonal antibody, DyLight 488 anti-rabbit IgG (H + L) (Vector Laboratories, Burlingame, CA), and CD133-PE (Miltenyi Biotec, Auburn, CA; cat. no. 130-080-801) antibody for FACS were applied to cell staining according to the manufacturer’s instructions. Cells were gated against isotype controls (mouse IgG1-PE, cat. no. 130-092-212, for CD133 and rabbit IgG monclonal antibody, cat. no. ab 172730, for TNC) and analyzed for CD133+/TNC+, CD133–/TNC+, CD133+/TNC–, and CD133–/TNC– cell populations. Each population of CD133+/TNC+, CD133–/TNC+, CD133+/TNC–, and CD133–/TNC– was sorted into 96-wells for 6 days before examination of sphere formation. The percent of wells that did not form spheres (y axis) was plotted against the number of cells plated per well (x axis). The minimum number of cells required to form spheres was calculated from the x intercept of the graph.
Statistical Analysis
The Student’s t-test was used to analyze the difference in pairwise comparison for TNC expression levels among astrocytoma WHO grade I, grade II, grade III, and grade IV and normal brain tissues. Statistical significance was defined as p < 0.05.
Results
TNC Expression in GBM Tissues
The expression level of TNC in a GBM-derived stem neurosphere line was shown to be dramatically higher than that in three traditional adherent GBM cell lines using mass spectrometry analysis.38 To detect TNC expression in GBM tissues, we extracted protein samples from fresh primary GBM biopsy. Western blotting analysis was applied to analyze GBM tissues and normal brain tissues. The results showed a strong expression of TNC in GBM tissues but not in normal brain tissues, where a dramatically different expression level existed between GBM and normal brain tissues (Figure 1).
Figure 1.
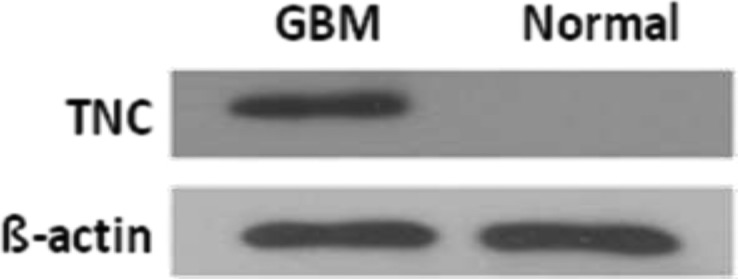
Western blot analysis of TNC expression in GBM (Glioblastoma multiforme) tissues and normal controls. A strong expression is present in GBM patients, whereas in the normal brain tissue, TNC is not detected.
TNC Expression Level Is Highly Correlated with Glioma Tumor Grade
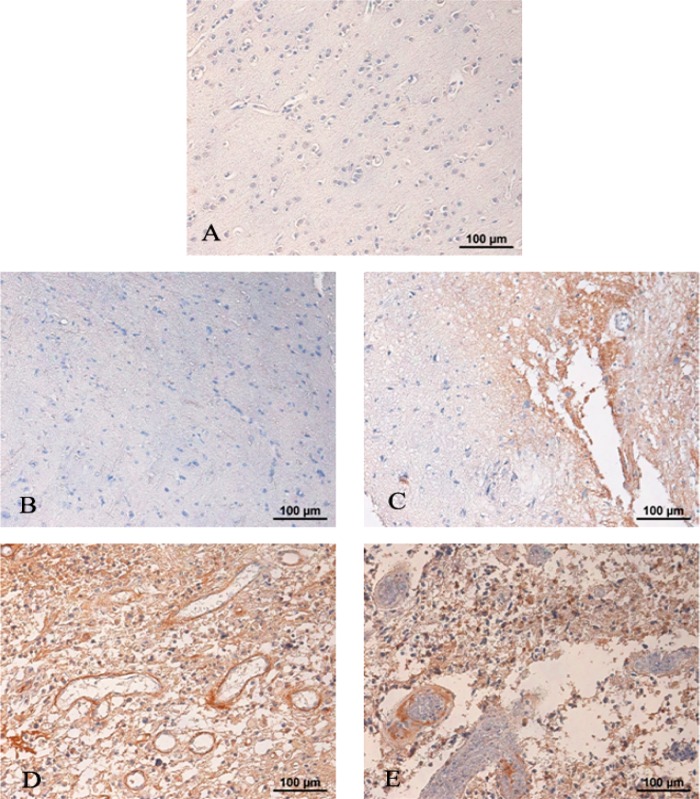
To determine the relevance of aberrant TNC expression in GBM progression, we investigated the expression of TNC using tissue microarrays containing 54 gliomas of different WHO grades and seven normal subjects by immunohistochemical staining. TNC expression was not detectable in normal brains (Figure 2A), WHO grade I astrocytomas (Figure 2B), and part of WHO grade II astrocytomas (Figure 2C). In contrast, it has a dramatically higher expression in all analyzed WHO grade III astrocytomas (Figure 2D) and GBMs (Figure 2E). The results reveal that TNC expression was highly correlated with the grade of astrocytoma, where tumors of higher grades showed higher levels of immunoreactivity.
Figure 2.
Immunohistochemical staining of human TNC in different grades of astrocytomas and normal brains. Paraffin-embedded tissue microarrays (TMA) of glioma were analyzed using anti-TNC monoclonal antibody. A representative figure of each TMA is shown. TNC was not detected in normal brain tissue (A) or astrocytoma WHO grade I (B). In contrast, low or moderate staining for TNC was detected in astrocytoma WHO grade grade II (C), and moderate or strong staining for TNC expression was observed in anaplastic astrocytoma WHO grade III (D) and GBM WHO grade IV (E). Hematoxylin counterstain was used to visualize nuclei. The TNC expression level in each tissue section was assessed in non-necrotic areas of three separate microscopic fields of view under a magnification of 200× and is represented by the mean of the percentage of TNC+ cells. The results were confirmed by a pathologist.
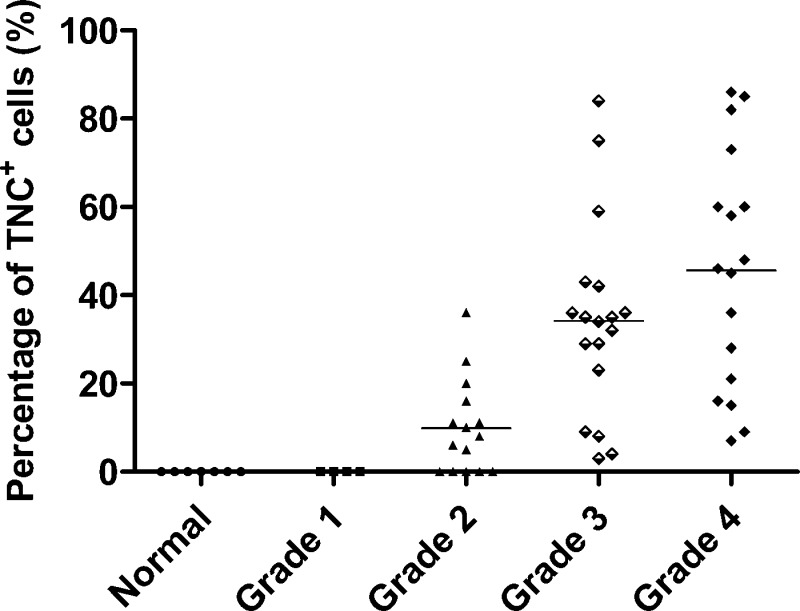
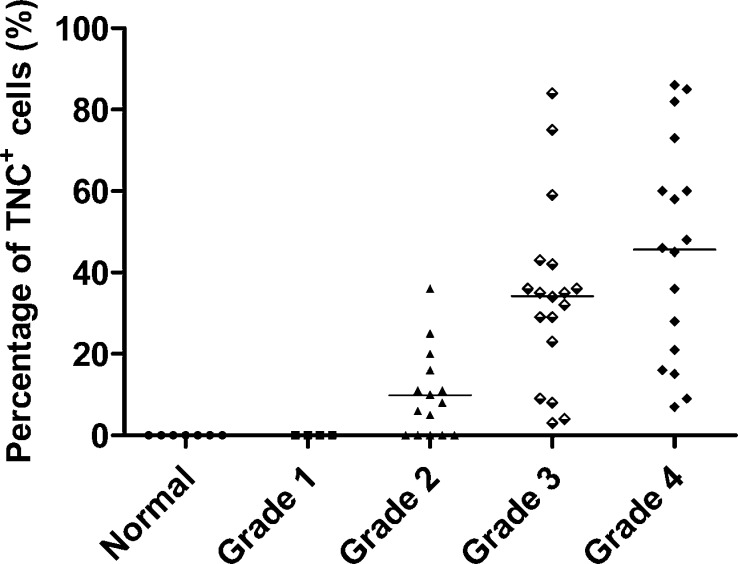
TNC expression was further semiquantitatively analyzed by calculating the percentage of TNC+ cells in the tissue section. The relative expression level of TNC in normal and different glioma grades are shown in Figure 3. The percentage of TNC+ cells was zero in all of the normal brain and WHO grade I astrocytoma tissues. In WHO grade II astrocytoma tissue samples, TNC was not detectable in part of the samples (5 of 15, 33%) and was expressed at weak levels (<16%) in most of the samples (7 of 15, 47%) and at medium levels (>16%, <45%) in a few tissue samples (3 of 15, 20%). In the case of high-grade gliomas, TNC was detectable in all of the samples tested. No significant difference in TNC expression was observed between grade III astrocytomas and GBMs (p > 0.05). However, the TNC expression level in high-grade gliomas (grade III astrocytomas and GBMs) was significantly higher than that in low-grade gliomas (p < 0.001, grade I and grade II astrocytomas). A significant difference of TNC expression level also existed between grade I and grade II astrocytomas. The results showed that TNC was not expressed in normal and grade I astrocytomas patients but that its expression level is highly correlated with late grades of malignancy.
Figure 3.
Scatter plot of TNC expression levels in 54 patients with different grades of astrocytomas and 7 normal subjects. The percentage of TNC+ cells of each tissue sample represents TNC expression level on the y axis. TNC expression levels were found to be highly associated with the grade of astrocytoma. TNC has a high expression level in most of the grade III astrocytomas and GBMs analyzed by immunohistochemical staining, a very low expression in most of the grade II astrocytomas, and undetectable expression in grade I astrocytomas and normal brain tissues.
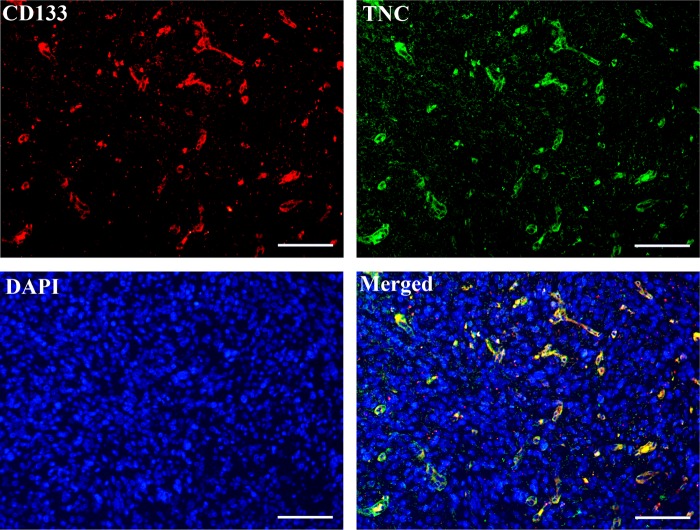
TNC+ Cell Population Has a High Overlap with the CD133+ Cell Population
CD133 has been widely used as a marker to characterize CSC population in GBMs.18,21,24 To determine the relation between the TNC+ and CD133+ cell populations, we performed double-immunofluorescence staining for TNC and CD133 in primary GBM tissue sections. As shown in Figure 4, in most of the GBM tissue cells, CD133 was co-expressed with TNC, where most TNC+ cells were also CD133+ cells, but there are still some TNC+ cells that were not positive for CD133. This indicates that the TNC+ cell population has a high overlap with the CD133+ cell population.
Figure 4.
Double-immunofluorescence staining for TNC and CD133 in a GBM tissue section. TNC expression has a high overlap with CD133 expression, where the coexpression is shown in yellow in the merged image. DAPI counterstain was used to visualize nuclei. Scale bar, 100 μm (200× magnification).
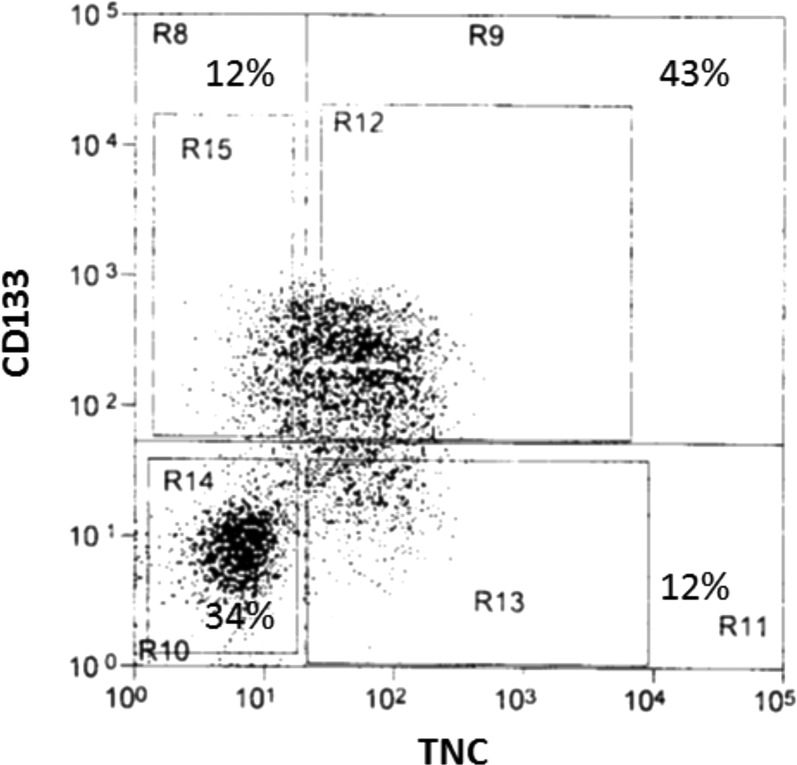
The expression of TNC and CD133 was further investigated in a GBM cell line. Flow cytometry analysis was executed in a GBM-derived stem-like neurosphere line, HSR-GBM1, which has been extensively used in CSC studies.27,40,41 CD133+/TNC+, CD133–/TNC+, CD133+/TNC–, and CD133–/TNC– cell populations were isolated from GBM-derived stem-like neurosphere cells by FACS. The percentage of each subpopulation is shown in Figure 5. The results show that TNC was expressed in about 55% of HSR-GBM1 cells, whereas CD133 was expressed in about 56% of HSR-GBM1 cells. The overlap between the TNC+ and CD133+ cell populations reached 43% (Figure 5).
Figure 5.
Flow cytometry sorting process to isolate CD133+/TNC+, CD133–/TNC+, CD133+/TNC–, and CD133–/TNC– cell populations based on the expression of TNC and CD133. The HSR-GBM1 cell line was used in this experiment. The results show that TNC was expressed in about 55% of HSR-GBM1 cells, whereas CD133 was expressed in about 56% of HSR-GBM1 cells. The overlap between the TNC+ and CD133+ cell populations reached 43%.
TNC Expression Level Decreased along with the Differentiation of GBM Cancer Stem Cells
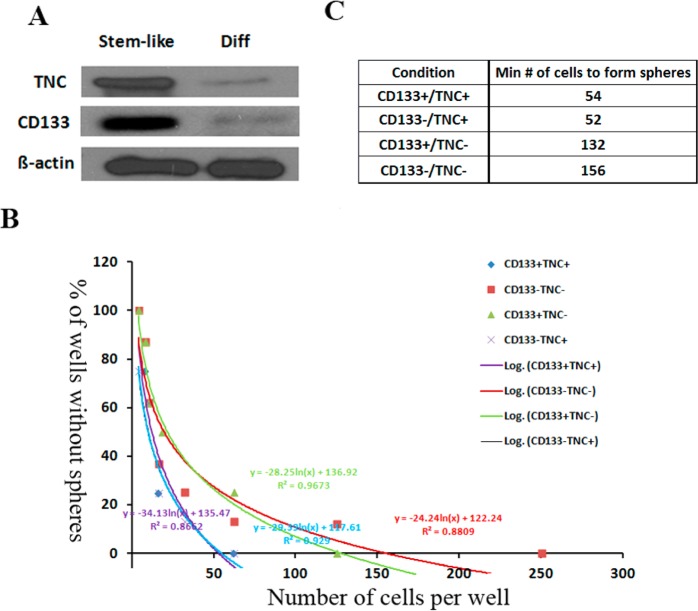
To investigate the TNC expression level changes in the differentiation process of GBM cancer stem cells, undifferentiated HSR-GBM1 stem-like neurospheres were induced to differentiate for 7 to 10 days.40 After collecting the undifferentiated and differentiated cell samples, we measured the changes in the expression level of TNC during the differentiation process of GBM cancer stem cells using western blotting. We found that most stem-like cells were lost after differentiation of HSR-GBM1 cells. The reduced expression of the known GBM stem cell marker CD133 confirmed this differentiation (Figure 6A), which is consistent with previous reports.40,42 The results of western blotting analysis showed that the TNC expression level decreased dramatically in the differentiated cells compared with that in undifferentiated stem-like cells, with a similar differential expression pattern to CD133 (Figure 6A). The results indicate that the expression levels of both TNC and CD133 were decreased along with the differentiation of stem-like cells, where TNC may represent a new marker for CSCs in GBM.
Figure 6.
TNC is a potential maker for GBM stem cells. (A) TNC has the same expression pattern with CD133 along with the differentiation of the stem-like HSR-GBM1 neurospheres, where expression levels of both TNC and CD133 were dramatically reduced in the differentiated cells compared to that in stem-like HSR-GBM1 neurospheres. Stem-like represents the stem-like HSR-GBM1 neurospheres; diff represents the differentiated HSR-GBM1 cells. (B) The ability of CD133+/TNC+, CD133–/TNC+, CD133+/TNC–, and CD133–/TNC– cell populations to form spheres was analyzed. y axis, percent of wells that did not form spheres; x axis, the number of cells plated per well. (C) The minimum number of cells required to form spheres for each population of HSR-GBM1 neurospheres. Compared to the TNC– cell population, the TNC+ cell population (including CD133+/TNC+ and CD133–/TNC+) needs a much lower number of cells to form spheres.
TNC Is a Novel Candidate Marker for GBM Cancer Stem Cells
To determine whether TNC is a novel marker for GBM cancer stem cells, we performed a limiting dilution assay to evaluate the self-renewal ability of the TNC+ cells. GBM-derived stem-like neurosphere cells were fractioned into CD133+/TNC+, CD133–/TNC+, CD133+/TNC–, and CD133–/TNC– cell populations by FACS. The ability of each cell population to form spheres was tested, where the TNC+ cell population has the strongest capacity for sphere formation regardless of CD133 status. On the basis of the data, the percent of wells that do not form spheres (y axis) is plotted against the number of cells plated per well (x axis), where the best line is drawn among the points based on a logarithmic function (Figure 6B). The number of cells required to give rise to colonies is calculated from the x intercept of the graph. The minimum number of cells required for sphere formation for CD133–/TNC+ and CD133+/TNC+ was 52 and 54, respectively. Compared to the TNC+ cell population, the TNC– cell population (CD133+/TNC– and CD133–/TNC–) requires 2–3-fold more cells to form spheres (Figure 6B,C). These results demonstrated that TNC+ cells had a much higher stemness potential than the TNC– cells, which confirmed that TNC is a marker for GBM CSCs. Also, the CD133+/TNC+ cell population required a similar number of cells to form spheres as that of the CD133–/TNC+ cell population. Thus, the TNC+ population as a CSC population contains part of CD133– CSCs.
Discussion
The cancer stem cell hypothesis claims that cancer arises from a tumorigenic subpopulation with self-renewing capacity and stem cell-like properties, designated cancer stem cells. The ability to find cell-surface markers that will allow the prospective identification and isolation of these cells is critical to CSC research. To date, many studies have relied on the enrichment of CSCs based on the cell surface protein CD133.43 However, recent reports have indicated that CD133– GBM cells can form tumors and that the expression of CD133 may be cell cycle regulated.44,45 These issues underscore the need for additional markers to identify CSCs in GBM.
Extracellular matrix (ECM) proteins are key structural components of the perivascular niche and regulate normal stem cell and tumor proliferation and migration.40,46 TNC is an ECM protein known to correlate with prognosis in patients with glioblastoma, probably by stimulation of invasion and tumor angiogenesis.47 TNC also contributes to the regulation of the self-renewal and output of the stem cell population.48 Its functions suggest that TNC plays an important role in GBM tumor initiation. In the current study, it has been shown that TNC is a potential candidate marker for cancer stem cells in glioblastoma.
Tissue microarrays were used to analyze the expression of TNC in glioma tissues of different grades, and TNC expression was found to be highly correlated with the grade of astrocytoma, where tumors of higher grades showed higher levels of immunoreactivity (Figures 2 and 3). TNC was barely detectable in normal and astrocytoma WHO grade I. Western blotting analysis also confirmed these results (Figure 1). These observations suggest that the TNC expression level may be an indicator of the aggressiveness of gliomas, and it appears to be a promising new prognostic marker for gliomas. In the process of differentiation of stem-like cells, TNC expression levels were decreased along with the differentiation of stem-like cells (Figure 6A). A limiting dilution assay further demonstrated the ability of the TNC+ cell population to form spheres (Figure 6B,C). These findings suggest that TNC is a novel marker for CSCs in GBM and overlaps with part of the CD133– cell population.
It should be noted that, in contrast to highly differentiated low-grade gliomas, GBMs are more undifferentiated and therefore are more likely to contain undifferentiated CSCs. Our analysis showed that TNC has a high expression level in most of the grade III astrocytomas and GBMs analyzed by immunohistochemical staining and a very low expression in most grade II astrocytomas, whereas it is undetectable in grade I astrocytomas and normal brain tissues. However, TNC expression level is very heterogeneous in patients, with the expression level varying from only 7% to more than 80%. The average expression level is about 34% in all of the patients of WHO grade III astrocytomas and about 45% in GBM patients (Figure 3). The TNC expression pattern is also consistent with previous reports.49 FACS analysis also demonstrated that TNC has a high expression level in the HSR-GBM1 cell line, with a similar percentage of TNC+ cells (55%) compared to that of CD133+ cells (56%) (Figure 5).
GBM displays significant heterogeneity, so it is unlikely that a single marker will absolutely enrich CSCs. Recently, a number of groups have suggested that different CSCs can coexist in the same tumor.7 Even within a single tumor, different pools of CSCs may be present.50 It is therefore likely that tumors have variability not only in the fraction of TNC+ cells and other CSCs but also in how these populations overlap. In this study, we demonstrated that the TNC+ cell population has a 43% overlap with the CD133+ cell population in the HSR-GBM1 cell line and overlaps with part of the CD133– cell population. Previous investigations have indicated that CD133– cells also have the capability to generate GBM tumors in immunodeficient nude mice or rats.27,28 Therefore, TNC may be a more general marker than CD133. The CD133+ stem-like cells are a subpopulation of CD90+ cells in GBMs.23 It might be interesting to investigate the relationship between the CD90+ and TNC+ cell populations. Unfortunately, the appropriate antibodies are unavailable to perform double staining.
Our findings may provide some potential therapeutic implication for GBM. TNC is also an extracellular matrix protein, which makes it easier to enter into the bloodstream. TNC already has been shown to be significantly elevated in serum in cancer contexts, such as in liver and ovarian cancers.51,52 The magnitude and mechanisms of TNC secretion by astrocytes have also been investigated.53 It would be interesting to investigate the difference in TNC expression in serum associated with GBM.
In conclusion, we have shown that TNC expression is significantly elevated with increasing grades of malignancy and decreases with the differentiation of GBM CSCs. It can be used to distinguish high-grade gliomas from low-grade gliomas and normal brains. Furthermore, it has been shown that TNC is a novel marker for CSCs in GBM and overlaps with part of the CD133– cell population, so TNC itself may be an important therapeutic target for GBMs.
Acknowledgments
We acknowledge support by the National Institutes of Health through grant nos. GM R01 GM49500 (D.M.L), R01CA148621 (X.F.), and R01CA163737 (X.F.). We also would like to acknowledge grant support to Dr. Fan from an Accelerate Brain Cancer Cure Project Award, American Brain Tumor Association Translational Grant, and Voices Against Brain Cancer Research Grant.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Louis D. N.; Ohgaki H.; Wiestler O. D.; Cavenee W. K.; Burger P. C.; Jouvet A.; Scheithauer B. W.; Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley M. R.; Post C.; Ehrlich G. Some speculation on the origin of glioblastoma. Neurosurg. Rev. 2007, 30, 16–20. [DOI] [PubMed] [Google Scholar]
- Stupp R.; Mason W. P.; van den Bent M. J.; Weller M.; Fisher B.; Taphoorn M. J.; Belanger K.; Brandes A. A.; Marosi C.; Bogdahn U.; Curschmann J.; Janzer R. C.; Ludwin S. K.; Gorlia T.; Allgeier A.; Lacombe D.; Cairncross J. G.; Eisenhauer E.; Mirimanoff R. O. Radiotherapy plus concomitant and adjuvant Temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–96. [DOI] [PubMed] [Google Scholar]
- Ohgaki H.; Dessen P.; Jourde B.; Horstmann S.; Nishikawa T.; Di Patre P. L.; Burkhard C.; Schuler D.; Probst-Hensch N. M.; Maiorka P. C.; Baeza N.; Pisani P.; Yonekawa Y.; Yasargil M. G.; Lutolf U. M.; Kleihues P. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004, 64, 6892–9. [DOI] [PubMed] [Google Scholar]
- Bao S.; Wu Q.; McLendon R. E.; Hao Y.; Shi Q.; Hjelmeland A. B.; Dewhirst M. W.; Bigner D. D.; Rich J. N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–60. [DOI] [PubMed] [Google Scholar]
- Dick J. E. Stem cell concepts renew cancer research. Blood 2008, 112, 4793–807. [DOI] [PubMed] [Google Scholar]
- Ortensi B.; Setti M.; Osti D.; Pelicci G. Cancer stem cell contribution to glioblastoma invasiveness. Stem Cell Res. Ther. 2013, 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.; Yuan X.; Zeng Z.; Tunici P.; Ng H.; Abdulkadir I. R.; Lu L.; Irvin D.; Black K. L.; Yu J. S. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 2006, 5, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira L. R.; Jeffrey S. S.; Ribeiro-Silva A. Stem cells in human breast cancer. Histol. Histopathol. 2010, 25, 371–85. [DOI] [PubMed] [Google Scholar]
- Yi S. Y.; Nan K. J. Tumor-initiating stem cells in liver cancer. Cancer Biol. Ther. 2008, 7, 325–30. [DOI] [PubMed] [Google Scholar]
- Bao Q.; Zhao Y.; Renner A.; Niess H.; Seeliger H.; Jauch K. W.; Bruns C. J. Cancer stem cells in pancreatic cancer. Cancers 2010, 2, 1629–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltanian S.; Matin M. M. Cancer stem cells and cancer therapy. Tumour Biol. 2011, 32, 425–40. [DOI] [PubMed] [Google Scholar]
- Ignatova T. N.; Kukekov V. G.; Laywell E. D.; Suslov O. N.; Vrionis F. D.; Steindler D. A. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 2002, 39, 193–206. [DOI] [PubMed] [Google Scholar]
- Galli R.; Binda E.; Orfanelli U.; Cipelletti B.; Gritti A.; De Vitis S.; Fiocco R.; Foroni C.; Dimeco F.; Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004, 64, 7011–21. [DOI] [PubMed] [Google Scholar]
- Singh S. K.; Hawkins C.; Clarke I. D.; Squire J. A.; Bayani J.; Hide T.; Henkelman R. M.; Cusimano M. D.; Dirks P. B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [DOI] [PubMed] [Google Scholar]
- Sanai N.; Alvarez-Buylla A.; Berger M. S. Neural stem cells and the origin of gliomas. N. Engl. J. Med. 2005, 353, 811–22. [DOI] [PubMed] [Google Scholar]
- Piccirillo S. G.; Reynolds B. A.; Zanetti N.; Lamorte G.; Binda E.; Broggi G.; Brem H.; Olivi A.; Dimeco F.; Vescovi A. L. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature 2006, 444, 761–5. [DOI] [PubMed] [Google Scholar]
- Fan X.; Khaki L.; Zhu T. S.; Soules M. E.; Talsma C. E.; Gul N.; Koh C.; Zhang J.; Li Y. M.; Maciaczyk J.; Nikkhah G.; Dimeco F.; Piccirillo S.; Vescovi A. L.; Eberhart C. G. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells 2010, 28, 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.; Matsui W.; Khaki L.; Stearns D.; Chun J.; Li Y. M.; Eberhart C. G. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006, 66, 7445–52. [DOI] [PubMed] [Google Scholar]
- Bao S.; Wu Q.; Li Z.; Sathornsumetee S.; Wang H.; McLendon R. E.; Hjelmeland A. B.; Rich J. N. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008, 68, 6043–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X.; Ma L.; Yi D.; Yoon J. G.; Diercks A.; Foltz G.; Price N. D.; Hood L. E.; Tian Q. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 1591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goidts V.; Bageritz J.; Puccio L.; Nakata S.; Zapatka M.; Barbus S.; Toedt G.; Campos B.; Korshunov A.; Momma S.; Van Schaftingen E.; Reifenberger G.; Herold-Mende C.; Lichter P.; Radlwimmer B. RNAi screening in glioma stem-like cells identifies PFKFB4 as a key molecule important for cancer cell survival. Oncogene 2012, 31, 3235–43. [DOI] [PubMed] [Google Scholar]
- He J.; Liu Y.; Zhu T.; Zhu J.; Dimeco F.; Vescovi A. L.; Heth J. A.; Muraszko K. M.; Fan X.; Lubman D. M. CD90 is identified as a candidate marker for cancer stem cells in primary high-grade gliomas using tissue microarrays. Mol. Cell. Proteomics 2011, 11, M111.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Nguyen D. H.; Dong Q.; Shitaku P.; Chung K.; Liu O. Y.; Tso J. L.; Liu J. Y.; Konkankit V.; Cloughesy T. F.; Mischel P. S.; Lane T. F.; Liau L. M.; Nelson S. F.; Tso C. L. Molecular properties of CD133+ glioblastoma stem cells derived from treatment-refractory recurrent brain tumors. J. Neurooncol. 2009, 94, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmelkov S. V.; Butler J. M.; Hooper A. T.; Hormigo A.; Kushner J.; Milde T.; St Clair R.; Baljevic M.; White I.; Jin D. K.; Chadburn A.; Murphy A. J.; Valenzuela D. M.; Gale N. W.; Thurston G.; Yancopoulos G. D.; D’Angelica M.; Kemeny N.; Lyden D.; Rafii S. CD133 expression is not restricted to stem cells, and both CD133+ and CD133– metastatic colon cancer cells initiate tumors. J. Clin. Invest. 2008, 118, 2111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F.; Zhang S.; Xie R.; Gao B.; Campos B.; Herold-Mende C.; Lei T. The utility and limitations of neurosphere assay, CD133 immunophenotyping and side population assay in glioma stem cell research. Brain Pathol. 2010, 20, 877–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Sakariassen P. O.; Tsinkalovsky O.; Immervoll H.; Boe S. O.; Svendsen A.; Prestegarden L.; Rosland G.; Thorsen F.; Stuhr L.; Molven A.; Bjerkvig R.; Enger P. O. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int. J. Cancer 2008, 122, 761–8. [DOI] [PubMed] [Google Scholar]
- Joo K. M.; Kim S. Y.; Jin X.; Song S. Y.; Kong D. S.; Lee J. I.; Jeon J. W.; Kim M. H.; Kang B. G.; Jung Y.; Jin J.; Hong S. C.; Park W. Y.; Lee D. S.; Kim H.; Nam D. H. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab. Invest. 2008, 88, 808–15. [DOI] [PubMed] [Google Scholar]
- Svendsen A.; Verhoeff J. J.; Immervoll H.; Brogger J. C.; Kmiecik J.; Poli A.; Netland I. A.; Prestegarden L.; Planaguma J.; Torsvik A.; Kjersem A. B.; Sakariassen P. O.; Heggdal J. I.; Van Furth W. R.; Bjerkvig R.; Lund-Johansen M.; Enger P. O.; Felsberg J.; Brons N. H.; Tronstad K. J.; Waha A.; Chekenya M. Expression of the progenitor marker NG2/CSPG4 predicts poor survival and resistance to ionising radiation in glioblastoma. Acta Neuropathol. 2011, 122, 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read T. A.; Fogarty M. P.; Markant S. L.; McLendon R. E.; Wei Z.; Ellison D. W.; Febbo P. G.; Wechsler-Reya R. J. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell 2009, 15, 135–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F. S.; Jones P. L. The tenascin family of ECM glycoproteins: structure, function, and regulation during embryonic development and tissue remodeling. Dev. Dyn. 2000, 218, 235–59. [DOI] [PubMed] [Google Scholar]
- Midwood K. S.; Hussenet T.; Langlois B.; Orend G. Advances in tenascin-C biology. Cell. Mol. Life Sci. 2011, 68, 3175–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon M. A.; Wikstrand C. J.; Furthmayr H.; Matthews T. J.; Bigner D. D. Human glioma–mesenchymal extracellular matrix antigen defined by monoclonal antibody. Cancer Res. 1983, 43, 2796–805. [PubMed] [Google Scholar]
- Behrem S.; Zarkovic K.; Eskinja N.; Jonjic N. Distribution pattern of tenascin-C in glioblastoma: correlation with angiogenesis and tumor cell proliferation. Pathol. Oncol. Res. 2005, 11, 229–35. [DOI] [PubMed] [Google Scholar]
- Jurga M.; Lipkowski A. W.; Lukomska B.; Buzanska L.; Kurzepa K.; Sobanski T.; Habich A.; Coecke S.; Gajkowska B.; Domanska-Janik K. Generation of functional neural artificial tissue from human umbilical cord blood stem cells. Tissue Eng., Part C 2009, 15, 365–72. [DOI] [PubMed] [Google Scholar]
- von Holst A. Tenascin C in stem cell niches: redundant, permissive or instructive?. Cells Tissues Organs 2008, 188, 170–7. [DOI] [PubMed] [Google Scholar]
- Rolle K.; Nowak S.; Wyszko E.; Nowak M.; Zukiel R.; Piestrzeniewicz R.; Gawronska I.; Barciszewska M. Z.; Barciszewski J. Promising human brain tumors therapy with interference RNA intervention (iRNAi). Cancer Biol. Ther. 2010, 9, 396–406. [DOI] [PubMed] [Google Scholar]
- He J.; Liu Y.; Xie X.; Zhu T.; Soules M.; DiMeco F.; Vescovi A. L.; Fan X.; Lubman D. M. Identification of cell surface glycoprotein markers for glioblastoma-derived stem-like cells using a lectin microarray and LC–MS/MS approach. J. Proteome Res. 2010, 9, 2565–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzolo A.; Parodi F.; Marimpietri D.; Raffaghello L.; Cocco C.; Pistorio A.; Mosconi M.; Gambini C.; Cilli M.; Deaglio S.; Malavasi F.; Pistoia V. Oct-4+/Tenascin C+ neuroblastoma cells serve as progenitors of tumor-derived endothelial cells. Cell Res. 2011, 21, 1470–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T. S.; Costello M. A.; Talsma C. E.; Flack C. G.; Crowley J. G.; Hamm L. L.; He X.; Hervey-Jumper S. L.; Heth J. A.; Muraszko K. M.; DiMeco F.; Vescovi A. L.; Fan X. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 2011, 71, 6061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar E. E.; Chaudhry A.; Lin A.; Fan X.; Schreck K.; Matsui W.; Piccirillo S.; Vescovi A. L.; DiMeco F.; Olivi A.; Eberhart C. G. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells 2007, 25, 2524–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.; Liu Y.; Zhu T. S.; Xie X.; Costello M. A.; Talsma C. E.; Flack C. G.; Crowley J. G.; Dimeco F.; Vescovi A. L.; Fan X.; Lubman D. M. Glycoproteomic analysis of glioblastoma stem cell differentiation. J. Proteome Res. 2011, 10, 330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidlingmaier S.; Zhu X.; Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J. Mol. Med. 2008, 86, 1025–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D.; Hau P.; Proescholdt M.; Lohmeier A.; Wischhusen J.; Oefner P. J.; Aigner L.; Brawanski A.; Bogdahn U.; Beier C. P. CD133+ and CD133– glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007, 67, 4010–5. [DOI] [PubMed] [Google Scholar]
- Jaksch M.; Munera J.; Bajpai R.; Terskikh A.; Oshima R. G. Cell cycle-dependent variation of a CD133 epitope in human embryonic stem cell, colon cancer, and melanoma cell lines. Cancer Res. 2008, 68, 7882–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson R. J.; Rich J. N. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat. Rev. Cancer 2007, 7, 733–6. [DOI] [PubMed] [Google Scholar]
- Hau P.; Kunz-Schughart L. A.; Rummele P.; Arslan F.; Dorfelt A.; Koch H.; Lohmeier A.; Hirschmann B.; Muller A.; Bogdahn U.; Bosserhoff A. K. Tenascin-C protein is induced by transforming growth factor-beta1 but does not correlate with time to tumor progression in high-grade gliomas. J. Neurooncol. 2006, 77, 1–7. [DOI] [PubMed] [Google Scholar]
- Garcion E.; Halilagic A.; Faissner A.; ffrench-Constant C. Generation of an environmental niche for neural stem cell development by the extracellular matrix molecule tenascin C. Development 2004, 131, 3423–32. [DOI] [PubMed] [Google Scholar]
- Leins A.; Riva P.; Lindstedt R.; Davidoff M. S.; Mehraein P.; Weis S. Expression of tenascin-C in various human brain tumors and its relevance for survival in patients with astrocytoma. Cancer 2003, 98, 2430–9. [DOI] [PubMed] [Google Scholar]
- Piccirillo S. G.; Combi R.; Cajola L.; Patrizi A.; Redaelli S.; Bentivegna A.; Baronchelli S.; Maira G.; Pollo B.; Mangiola A.; DiMeco F.; Dalpra L.; Vescovi A. L. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene 2009, 28, 1807–11. [DOI] [PubMed] [Google Scholar]
- Didem T.; Faruk T.; Senem K.; Derya D.; Murat S.; Murat G.; Oznur K. Clinical significance of serum tenascin-c levels in epithelial ovarian cancer. Tumour Biol. 2014, 35, 6777–82. [DOI] [PubMed] [Google Scholar]
- Yamauchi M.; Mizuhara Y.; Maezawa Y.; Toda G. Serum tenascin levels in chronic liver disease. Liver 1994, 14, 148–53. [DOI] [PubMed] [Google Scholar]
- Nishio T.; Kawaguchi S.; Iseda T.; Kawasaki T.; Hase T. Secretion of tenascin-C by cultured astrocytes: regulation of cell proliferation and process elongation. Brain Res. 2003, 990, 129–40. [DOI] [PubMed] [Google Scholar]