Abstract
CRISPR RNA-guided nucleases (RGNs) are widely used genome-editing reagents, but methods to delineate their genome-wide off-target cleavage activities have been lacking. Here we describe an approach for global detection of DNA double-stranded breaks (DSBs) introduced by RGNs and potentially other nucleases. This method, called Genome-wide Unbiased Identification of DSBs Enabled by Sequencing (GUIDE-Seq), relies on capture of double-stranded oligodeoxynucleotides into breaks Application of GUIDE-Seq to thirteen RGNs in two human cell lines revealed wide variability in RGN off-target activities and unappreciated characteristics of off-target sequences. The majority of identified sites were not detected by existing computational methods or ChIP-Seq. GUIDE-Seq also identified RGN-independent genomic breakpoint ‘hotspots’. Finally, GUIDE-Seq revealed that truncated guide RNAs exhibit substantially reduced RGN-induced off-target DSBs. Our experiments define the most rigorous framework for genome-wide identification of RGN off-target effects to date and provide a method for evaluating the safety of these nucleases prior to clinical use.
CRISPR-Cas RGNs are robust genome-editing reagents with a broad range of research and potential clinical applications1, 2. However, therapeutic use of RGNs in humans will require a comprehensive knowledge of their off-target effects to minimize the risk of deleterious outcomes. DNA cleavage by S. pyogenes Cas9 nuclease is directed by a programmable ~100 nt guide RNA (gRNA).3 Targeting is mediated by 17-20 nts at the gRNA 5′-end, which are complementary to a “protospacer” DNA site that lies next to a protospacer adjacent motif (PAM) of the form 5′-NGG. Repair of blunt-ended Cas9-induced DNA double-stranded breaks (DSBs) within the protospacer by non-homologous end-joining (NHEJ) can induce variable-length insertion/deletion mutations (indels). Our group and others have previously shown that unintended RGN-induced indels can occur at off-target cleavage sites that differ by as many as five positions within the protospacer or that harbor alternative PAM sequences4-7. In addition, chromosomal translocations can result from joining of on- and off-target RGN-induced cleavage events8-11. For clinical applications, identification of even low frequency alterations will be critically important because ex vivo and in vivo therapeutic strategies using RGNs are expected to require the modification of very large cell populations. The induction of oncogenic transformation in even a rare subset of cell clones (e.g., inactivating mutations of a tumor suppressor gene or formation of a tumorigenic chromosomal translocation) is of particular concern because such an alteration could lead to unfavorable clinical outcomes.
The identification of indels or higher-order rearrangements that can occur anywhere in the genome is a challenge that is not easily addressed and sensitive methods for unbiased, genome-wide identification of RGN-induced off-target mutations in living cells have not yet been described12, 13. Whole genome re-sequencing has been used to attempt to identify RGN off-target alterations in edited single cell clones14, 15 but the exceedingly high projected cost of sequencing very large numbers of genomes makes this method impractical for finding low frequency events in cell populations12. We and others have used focused deep sequencing to identify indel mutations at potential off-target sites identified either by sequence similarity to the on-target site4, 5 or by in vitro selection from partially degenerate binding site libraries6. However, these approaches are biased because they assume that off-target sequences are closely related to the on-target site and, as a result, may miss potential off-target sites in the genome. ChIP-Seq has also been used to identify off-target binding sites for gRNAs complexed with catalytically dead Cas9 (dCas9), but the majority of published work suggests that very few, if any, of these sites represent off-target sites of cleavage by active Cas9 nuclease16-19
Here we describe the development of GUIDE-Seq, which enabled us to generate global specificity landscapes for thirteen different RGNs in living human cells. These profiles revealed that the total number of off-target DSBs varied widely for individual RGNs and suggested that broad conclusions about the specificity of RGNs from S. pyogenes or other species should be based on characterization of large numbers ofgRNAs. Our findings also expanded the range and nature of sequences at which off-target effects can occur and demonstrated that ChIP-Seq of dCas9 and two widely used computational approaches do not identify many of the sites found by GUIDE-Seq. Our method also identified RGN-independent DNA breakpoint hotspots that can participate together with RGN-induced DSBs in higher-order genomic alterations such as translocations. Lastly, we show in direct comparisons that truncating the protospacer complementariy region of gRNAs greatly improved their genome-wide off-target DSB profiles, demonstrating the utility of GUIDE-Seq for evaluating technology advances designed to improve RGN specificities. The experiments outlined here provide the most rigorous strategy described to date for evaluating the specificities of RGNs that may be considered for therapeutic use.
RESULTS
Overview of the GUIDE-Seq method
GUIDE-Seq consists of two stages (Fig. 1a): In Stage I, blunt-ended RGN-induced DSBs in the genomes of living human cells are tagged by integration of a blunt double-stranded oligodeoxynucleotide (dsODN) at these breaks via an end-joining process consistent with NHEJ. In Stage II, dsODN integration sites in genomic DNA are precisely mapped at the nucleotide level using unbiased amplification and next-generation sequencing.
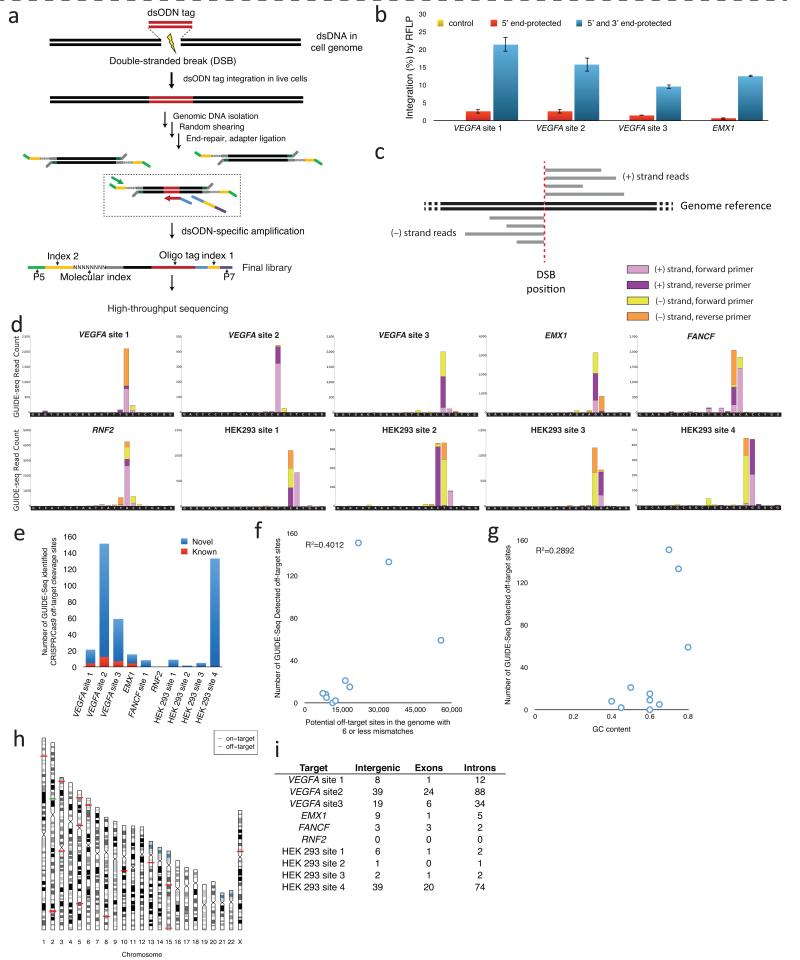
Figure 1. Design, optimization and application of the GUIDE-Seq method.
(a) Schematic overview of the GUIDE-Seq method.
(b) Optimization of dsODN integration into RGN-induced DSBs in human cells. Rates of integration for different modified oligonucleotides as measured by RFLP assay are shown. Control reactions were transfected with only the RGN-encoding plasmids (i.e., without dsODN).
(c) Schematic illustrating how mapping of genomic sequence reads enabled identification of DSB position.
(d) GUIDE-Seq-based identification of RGN-induced DSBs. Start sites of GUIDE-Seq reads mapped back to the genome enable localization of the DSB to within a few base pairs. Mapped reads for the on-target sites of the ten RGNs we assessed by GUIDE-Seq are shown. In all cases, the target site sequence is shown with the 20 bp protospacer sequence to the left and the PAM sequence to the right on the x-axis. Note how in all cases the highest peak falls within 3 to 4 bps of the 5′-edge of the NGG PAM sequence, the expected position of an RGN cleavage event. Equivalent maps for off-target sites are presented in Supplementary Fig. 2.
(e) Numbers of previously known and novel off-target cleavage sites identified by GUIDE-Seq for the ten RGNs analyzed in this study.
(f) Scatterplot of on-target site orthogonality to the human genome (y-axis) versus total number of off-target sites detected by GUIDE-Seq for the ten RGNs of this report. Orthogonality was calculated as the total number of sites in the human genome bearing 1 to 6 mismatches relative to the on-target site.
(g) Scatterplot of on-target site GC content (y-axis) versus total number of off-target sites detected by GUIDE-Seq for the ten RGNs of this report.
(h) Chromosome ideogram of CRISPR/Cas9 on- and off-target sites for the RGN that targets EMX1. Additional ideograms for the remaining RGNs can be found in Supplementary Fig. 3.
(i) Genomic locations of off-target cleavage sites identified by GUIDE-Seq for the ten RGNs examined in this study.
For Stage I, we optimized conditions to integrate a blunt, 5′ phosphorylated 34 bp dsODN into RGN-induced DSBs in human cells. In initial experiments, we failed to observe integration of such dsODNs into RGN-induced DSBs (data not shown). Using dsODNs bearing two phosphothiorate linkages at the 5′ ends of both DNA strands designed to stabilize the oligos in cells20, we observed only modest detectable integration frequencies (Fig. 1b). However, addition of phosphothiorate linkages at the 5′ and 3′ ends of both strands (Online Methods) led to robust integration efficiencies (Fig. 1b). These rates of integration were only two-to three-fold lower than the frequencies of indels induced by RGNs alone at these sites (i.e., in the absence of the dsODN) (data not shown).
For Stage II, we developed a strategy that allowed us to selectively amplify and sequence, in an unbiased fashion, only those fragments bearing an integrated dsODN (Fig. 1a). We accomplished this by first ligating “single-tail” next-generation sequencing adapters to randomly sheared genomic DNA from cells transfected with dsODN and plasmids encoding RGN components. We then performed a series of PCR reactions initiated by one primer that specifically annealed to the dsODN and another that annealed to the sequencing adapter (Fig. 1a and Supplementary Fig. 1). Because the sequencing adapter was only single-tailed, this enabled specific unidirectional amplification of the sequence adjacent to the dsODN, without the bias and background inherent to methods such as linear amplification-mediated (LAM)-PCR21, 22. We refer to this strategy as the Single-Tail Adapter/Tag (STAT)-PCR method. By performing STAT-PCR reactions using primers that annealed to each of the strands in the dsODN, we obtained reads of adjacent genomic sequence on both sides of each integrated tag (Fig. 1c). Incorporation of a random 8 bp molecular barcode during the amplification process (Supplementary Fig. 1) allowed for correction of PCR bias, thereby enabling accurate quantitation of unique sequencing reads obtained from high-throughput sequencing (Supplementary Methods).
Genome-wide off-target cleavage profiles of RGNs in human cells
We performed GUIDE-Seq with Cas9 and ten different gRNAs targeted to various endogenous human genes in either U2OS or HEK293 human cell lines (Supplementary Table 1). By analyzing the dsODN integration sites (Supplementary Methods), we were able to identify the precise genomic locations of DSBs induced by each of the ten RGNs, mapped to the nucleotide level (Fig. 1d and Supplementary Fig. 2). For >80% of these genomic sites, we were able to identify an overlapping target sequence that was either the on-target site or a closely related off-target site (Supplementary Methods). The total number of off-target sites we identified for each RGN varied widely, ranging from zero to >150 (Fig. 1e), demonstrating that unwanted genomic cleavage by any particular RGN can be considerable or minimal on the extremes. Control experiments in which we sequenced across dsODN insertions at on- and off-target sites for five of the RGNs using anchored multiplex PCR (AMP)-based next-generation sequencing (Fig. 3a, Online Methods and Supplementary Methods) revealed that >93% of these sites (123 out of 132) showed detectable evidence of one of more dsODN molecules, consistent with NHEJ-mediated capture into the DSB (data not shown).
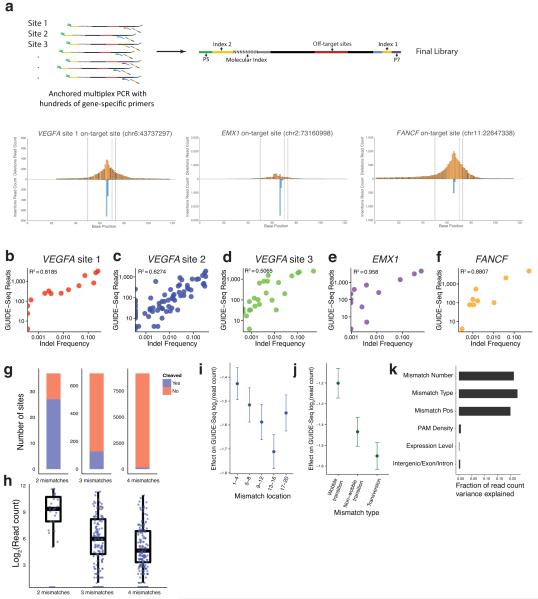
Figure 3. Analysis of RGN-induced off-target sequence characteristics.
(a) Schematic overview of the AMP-based sequencing method used to confirm indel mutations at GUIDE-Seq cleavage sites is shown in the top half of the figure. Histogram plots of mapped indel mutations are shown for three RGN on-target sites. Deletions are shown above the X-axis whereas insertions are shown below. Boundaries of the overall target site (i.e., protospacer and PAM sequence) are shown with dotted lines and the boundary between the protospacer and PAM sequence is shown as a dotted line between the other two. RGN cleavage is predicted to occur 3 to 4 bps from the 5′ edge of the protospacer. Additional on-target and off-target indel histogram plots are shown in Supplementary Fig. 4.
(b)-(f) Scatterplots of indel frequencies (x-axis) and GUIDE-Seq sequencing read counts (y-axis) for cleavage sites identified by GUIDE-Seq for RGNs targeted to: VEGFA site 1, VEGFA site 2, VEGFA site 3, EMX1, and FANCF.
(g) Fraction of potential RGN off-target sites bearing a certain number of mismatches that are cleaved (as detected by GUIDE-Seq).
(h) Plots of GUIDE-Seq read counts (log-scale) for RGN off-target cleavage sites bearing a certain number of mismatches
(i) Effects of mismatch position within the protospacer on GUIDE-Seq read counts for RGN off-target sites. Bases are numbered 1 to 20 with 20 being the base adjacent to the PAM.
(j) Effects of wobble transition, non-wobble transition, and transversion mismatches estimated by linear regression analysis.
(k) Fraction of GUIDE-Seq read count variance explained by individual univariate analyses for the effect of mismatch number, mismatch type, mismatch position, PAM density, expression level, and genomic position (intergenic/exon/intron).
We did not observe any obvious correlation between the total number of off-target sites we observed by GUIDE-Seq and orthogonality of the on-target site relative to the human genome or GC content of the on-target protospacer sequence (Figs. 1f and 1g). Off-target sequences were found dispersed throughout the genome (Fig. 1h and Supplementary Fig. 3) in exons, introns, and non-coding intergenic regions (Fig. 1i). Included among the off-target sequences we identified were all 28 of the bona fide off-target sites previously known for four of the RGNs4, 5 (Figs. 1e, 2a-2j and Supplementary Table 2). GUIDE-Seq also identified a large number of previously unknown off-target sites that map throughout the human genome (Fig. 1e, 1h, 1i, 2a-2j, Supplementary Table 2, and Supplementary Fig. 3).
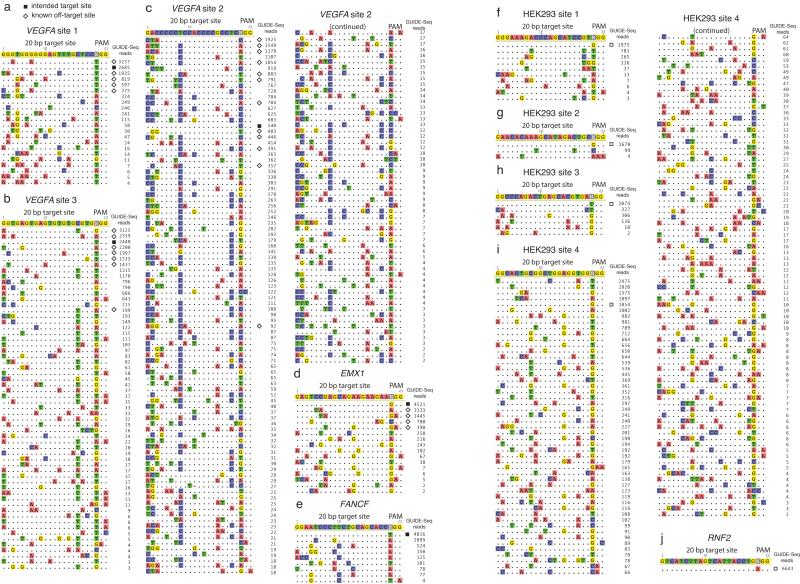
Figure 2. Sequences of off-target sites identified by GUIDE-Seq for ten RGNs.
For each RGN, the intended target sequence is shown in the top line with cleaved sites shown underneath and with mismatches to the on-target site shown and highlighted in color. GUIDE-Seq sequencing read counts are shown to the right of each site. The on-target site is marked with a green square and previously known off-target sites with a red diamond. Data is shown for RGNs targeting the following sites: (a) VEGFA site 1, (b) VEGFA site 2, (c) VEGFA site 3, (d) EMX1, (e) FANCF, (f) HEK293 site 1, (g) HEK293 site 2, (h) HEK293 site 3, (i) HEK293 site4, (j) RNF2. No off-target sites were found for the RGN targeted to the RNF2 site.
We next tested whether the number of sequencing reads for each off-target site identified by GUIDE-Seq (shown in Figs. 2a-2j and hereafter referred to as “GUIDE-Seq read counts”) represented a proxy for the relative frequency of indels that would be induced by an RGN alone (i.e., in the absence of a dsODN). We used AMP sequencing to examine these same sites from cells in which only the nuclease components had been expressed and found that >80% (106 out of 132) harbored variable-length indels characteristic of NHEJ-mediated repair of an RGN cleavage event, further supporting our conclusion that GUIDE-Seq identifies bona fide RGN off-target sites (Fig. 3a and Supplementary Fig. 4). (Many of the sites for which we did not see evidence of indels also had low GUIDE-Seq read counts, suggesting that the inability to detect mutations at these sites may be related to the sensitivity of sequencing and the sampling depth of our experiments). The range of indel mutation frequencies we detected ranged from 0.03% to 60.1%. Notably, we observed positive linear correlations between GUIDE-Seq read counts and indel mutation frequencies for all five RGN off-target sites (Figs. 3b-3f). Thus, we conclude that GUIDE-Seq read counts for a given site represent a quantitative measure of the cleavage efficiency of that sequence by an RGN.
Analysis of RGN-induced off-target sequence characteristics
Visual inspection of the off-target sites we identified by GUIDE-Seq for nine RGNs underscored the diversity of variant sequences at which these nucleases can cleave. These sites harbored as many as six mismatches within the protospacer sequence (consistent with a previous report showing in vitro cleavage of sites bearing up to seven mismatches6), non-canonical PAMs (including previously described NAG and NGA sequences5, 23 but also NAA, NGT, NGC, and NCG sequences), and 1 bp “bulge”-type mismatches24 at the gRNA/protospacer interface (Fig. 2a-2j). Protospacer mismatches tended to occur in the 5′ end of the target site but could also be found at certain 3′ end positions, supporting the concept that there are no simple rules for predicting mismatch effects based on position4. Notably, some off-target sites actually had higher sequencing read counts than their matched on-target sites (Figs. 2a-2c, 2i), consistent with our previous observations that off-target mutation frequencies can in certain cases be higher than those at the intended on-target site4. Many of the previously known off-target sites for four of the RGNs were those with high read counts (Figs. 2a-d), suggesting that earlier analyses4, 5 had primarily identified sites that were most efficiently cleaved.
Quantitative analysis of our GUIDE-Seq data for nine RGNs enabled us to quantify the potential contributions and impacts of different variables such as mismatch number, location, and type on off-target site cleavage. We found that the fraction of total genomic sites bearing a certain number of protospacer mismatches that are cleaved by an RGN decreased with increasing numbers of mismatches (Fig. 3g). In addition, sequence read counts showed an overall downward trend with increasing numbers of mismatches (Fig. 3h). In general, protospacer mismatches positioned closer to the 5′ end of the target site tended to be associated with smaller decreases in GUIDE-Seq read counts than those closer to the 3′ end although mismatches positioned 1 to 4 bp away from the PAM were somewhat better tolerated than those 5 to 8 bps away (Fig. 3i). The nature of the mismatch was also associated with an effect on GUIDE-Seq read counts. Wobble mismatches occurred frequently in the off-target sites and our analysis suggested they are associated with smaller impacts on GUIDE-Seq read counts than other non-Wobble mismatches (Fig. 3j). Consistent with these results, we found that the single factors that explain the greatest degree of variation in off-target cleavage in univariate regression analyses were mismatch number, position and type. By contrast, other factors such as the density of proximal PAM sequences, gene expression level or genomic position (intergenic/intronic/exonic) explained a much smaller proportion of the variance in GUIDE-Seq cleavage read counts (Fig. 3k). A combined linear regression model that considered multiple factors including mismatch position, mismatch type, gene expression level, and density of proximal PAM sequences yielded results consistent with the univariate analyses (Supplementary Fig. 5). This analysis also allowed us to independently estimate that, on average and depending on their position, each additional wobble mismatch decreased off-target cleavage rates by approximately two-to three-fold, while additional non-wobble mismatches decreased cleavage rates by approximately three-fold (Supplementary Fig. 5).
Comparisons with in silico off-target prediction methods
Having established the efficacy of GUIDE-Seq, we next performed direct comparisons of our method with two popular computational programs for predicting off-target mutation sites: the MIT CRISPR Design Tool25 (http://crispr.mit.edu) and the E-CRISP software26 (http://www.e-crisp.org/E-CRISP/). Both of these programs identify potential off-target sites based on “rules” about mismatch number and position. In direct comparisons, we discovered that both programs failed to identify the vast majority of off-target sites found by GUIDE-Seq for the nine RGNs (Figs. 4a and 4b). Many of these sites were missed because the E-CRISP and MIT programs simply did not consider off-targets bearing more than 3 and 4 mismatches, respectively (Figs 4c and 4d). Even among the sequences that were considered, these programs still failed to identify the majority of the bona fide off-target sites (Fig. 4c and 4d), highlighting their currently limited capability to account for the factors that determine whether cleavage will occur. In particular, it is worth noting that sites missed include those with as few as one mismatch (Figs. 4c and 4d), although the ranking scores assigned by the MIT program did have some predictive power among the subset of sites it correctly identified.
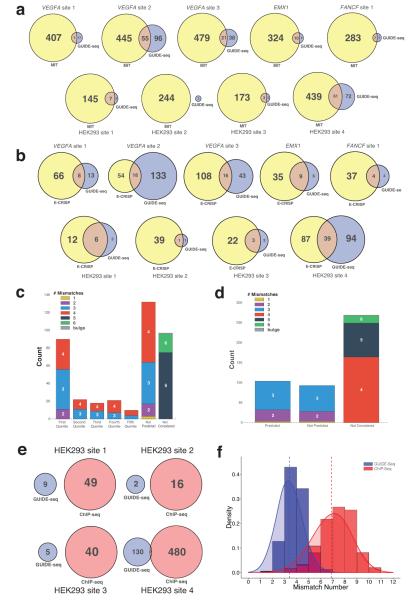
Figure 4. Comparisons of GUIDE-Seq with computational prediction or ChIP-Seq methods for identifying RGN off-target sites.
(a) Venn diagrams illustrating overlap between cleavage sites predicted by the MIT CRISPR Design Tool and GUIDE-Seq for nine RGNs.
(b) Venn diagrams illustrating overlap between off-target sites predicted by the E-CRISP computational prediction program and GUIDE-Seq for nine RGNs.
(c) Histogram showing the numbers of bona fide RGN off-target sites identified by GUIDE-Seq that are predicted, not predicted, and not considered by the MIT CRISPR Design Tool. Sites predicted by the MIT CRISPR Design Tool are divided into quintiles based on the score provided by the program. Each bar has the sites sub-classified based on the number of mismatches relative to the on-target site. Bulge sites are those that have a skipped base position at the gRNA-protospacer DNA interface.
(d) Histogram showing the numbers of bona fide RGN off-target sites identified by GUIDE-Seq that are predicted, not predicted, and not considered by the E-CRISP computational prediction tool. Sites are subdivided as described in (c).
(e) Venn diagrams illustrating overlap between dCas9 binding sites identified by ChIP-Seq and RGN off-target cleavage sites identified by GUIDE-Seq.
(f) Histogram plots of RGN off-target sites identified by GUIDE-Seq and dCas9 binding sites identified by ChIP-Seq classified by the number of mismatches in the sequence relative to the intended on-target site. Kernel density estimation of GUIDE-Seq and ChIP-Seq mismatches is depicted. Dotted lines indicate the mean number of mismatches for each class of sites.
Comparison with off-target binding sites found by ChIP-Seq
We also sought to compare GUIDE-Seq with previously described ChIP-Seq methods for identifying Cas9 binding sites. Four of the RGNs we evaluated by GUIDE-Seq used gRNAs that had been previously characterized in ChIP-Seq experiments with catalytically inactive Cas9 (dCas9)18. Very little overlap exists between Cas9 off-target cleavage sites identified by GUIDE-Seq and dCas9 off-target binding sites identified by ChIP-Seq; among the 149 RGN-induced off-target cleavage sites we identified for the four gRNAs, only three were previously identified by the previously published dCas9 ChIP-Seq experiments using the same gRNAs (Fig. 4e). This lack of overlap is likely because dCas9 off-target binding sites are fundamentally different from Cas9 off-target cleavage sites, a hypothesis supported by our data showing that Cas9 off-target cleavage sites for these four gRNAs identified by GUIDE-Seq harbor on average far fewer mismatches than the binding sites identified by ChIP-Seq (Fig. 4f) and by the results of previous studies showing that very few dCas9 binding sites show evidence of indels in the presence of active Cas916-19. Although GUIDE-Seq failed to identify the seven off-target sites previously identified by ChIP-Seq and reported to be targets of mutagenesis by Cas9, we believe this is because those sites were incorrectly identified in that earlier study18 as bona fide off-target cleavage sites (Supplementary Results and Supplementary Fig. 6) We conclude that very few (if any) dCas9 off-target binding sites discovered by ChIP-Seq actually represent bona fide Cas9 off-target cleavage sites.
RGN-independent DSB hotspots identified by GUIDE-Seq
Our GUIDE-Seq experiments also revealed the existence of a total of 30 unique RGN-independent DSB hotspots in the U2OS and HEK293 cells used for our studies (Supplementary Table 3). We uncovered these when analyzing genomic DNA from control experiments with U2OS and HEK293 cells in which we transfected only the dsODN without RGN-encoding plasmids. In contrast to RGN-induced DSBs that mapped precisely to specific base pair positions, RGN-independent DSB hotspots have dsODN integration patterns that are more broadly dispersed at each locus in which they occur (Supplementary Methods). These 30 breakpoint hotspots were distributed over many chromosomes and appeared to be present at or near centromeric or telomeric regions (Fig. 5a). Only two of these hotspots were common to both cell lines while the majority appeared to be cell line-specific (25 in U2OS and seven in HEK293 cells) (Fig. 5a and Supplementary Table 3).
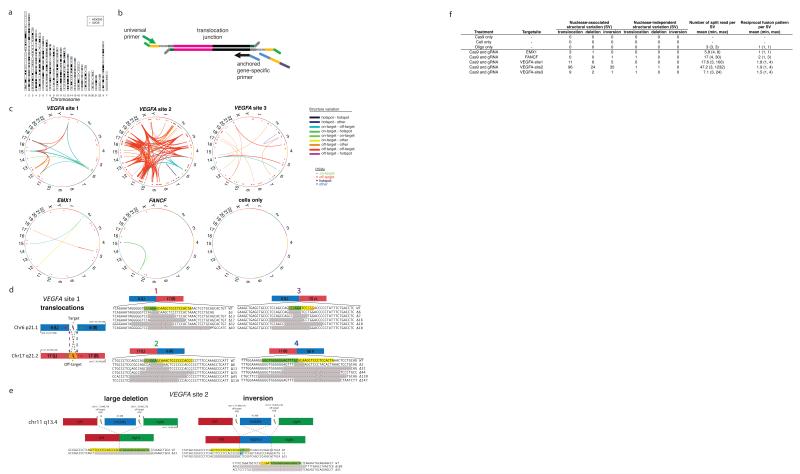
Figure 5. Large-scale structural alterations induced by RGNs.
(a) Chromosome ideogram illustrating the locations of breakpoint hotspots in U2OS and HEK293 cells. Two hotspots overlap at the centromeric regions of chromosomes 1 and 10.
(b) Schematic overview of AMP strategy for detecting translocations. Additional details in Online Methods.
(c) Circos plots of structural variation induced by RGNs. Data for five RGNs and a control of cells are shown. Chromosomes are arranged in a circle with translocations shown as arcs between two chromosomal locations. Deletions or inversions greater than 1 kb in length are shown as straight lines. Sites that are not on-target, off-target, or breakpoint hotspots are classified as “other”.
(d) Example of a translocation detected between the VEGFA site 1 on-target site on chromosome 6 and an off-target site on chromosome 17. All four possible reciprocal translocations were detected using AMP.
(e) Examples of large deletion and inversion between two off-target sites in VEGFA site 2 detected by AMP.
(f) Summary table of different RGN-induced and RGN-independent structural variations observed with five RGNs. Controls with Cas9 only, dsODN oligo only, and cells only are also shown.
Analysis of large-scale genomic rearrangements
In the course of analyzing the results of our AMP-based sequencing experiments designed to identify indels at RGN-induced and RGN-independent DSBs, we also discovered that at least some of these breaks can participate in translocations, inversions and large deletions. The AMP method enabled us to observe these large-scale genomic alterations because, for each DSB site examined, it used nested locus-specific primers anchored at only one fixed end rather than a pair of flanking locus-specific primers (Fig. 5b).
For the five RGNs we examined, AMP sequencing revealed that RGN-induced on-target and off-target DSBs could participate in a variety of translocations (Fig. 5c). In at least one case, we observed all four possible translocation events resulting from a pair of DSBs (Fig. 5d). When two DSBs were present on the same chromosome, we also observed large deletions and inversions (Fig. 5c). We also observed an example of both a large deletion between two RGN-induced breaks as well as an inversion of that same intervening sequence (Fig. 5e). Notably, our results also revealed translocations (and deletions or inversions) between RGN-induced and RGN-independent DSBs (Fig. 5c & 5f), suggesting the need to consider the interplay between these two types of breaks when evaluating the off-target effects of RGNs on cellular genomes. Although our data suggested that the frequencies of these large-scale genomic rearrangements are likely to be very low, precise quantification was not possible with the sequencing depth of our existing dataset. Increasing the number of sequencing reads should increase the sensitivity of detection and enable better quantitation of these important genomic alterations.
GUIDE-Seq profiles of RGNs directed by truncated gRNAs
Previous studies from our group have shown that use of gRNAs bearing truncated complementarity regions of 17 or 18 nts can reduce mutation frequencies at known off-target sites of RGNs directed by full-length gRNAs27. However, because this analysis was limited to a small number of known off-target sites, the genome-wide specificities of these truncated gRNAs (tru-gRNAs) remained undefined in our earlier experiments. We used GUIDE-Seq to obtain genome-wide DSB profiles of RGNs directed by three tru-gRNAs, each of which are shorter versions of three of the full-length gRNAs we had assayed above. In all three cases, the total number of off-target sites identified by GUIDE-Seq decreased substantially with use of a tru-gRNA (Fig. 6a-6d). Mapping of GUIDE-Seq reads enabled us to precisely identify the cleavage locations of on-target (Fig. 6e) and off-target sites (Supplementary Fig. 7). As expected, included in the list of off-target sites were 10 of the 12 previously known off-target sites for RGNs directed by the three tru-gRNAs (Figs. 6f-6h). The sequences of the off-target sites we identified primarily had one or two mismatches in the protospacer but some sites had as many as four (Figs. 6f-6h). In addition, some sites had alternative PAM sequences of the forms NAG, NGA, and NTG (Figs. 6f-6h). These data provide confirmation on a genome-wide scale that truncation of gRNAs can substantially reduce off-target effects of RGNs and show how GUIDE-Seq can be used to assess specificity improvements for the RGN platform.
Figure 6. GUIDE-Seq profiles of RGNs directed by tru-gRNAs.
(a) Numbers of previously known and novel off-target cleavage sites identified for RGNs directed to the to VEGFA site 1, VEGFA site 3, and EMX1 target sites by matched full-length gRNAs and truncated gRNAs. Note that the data for the RGNs directed by full-length gRNAs are the same as those presented in Fig. 1e and is shown again here for ease of comparison.
(b)-(d) Chromosome ideograms showing on- and off-target sites for RGNs directed to the VEGFA site 1, VEGFA site 3, and EMX1 target sites by matched full-length gRNAs and truncated gRNAs. Note that the ideograms for the RGNs directed by full-length gRNAs are the same as those presented in Fig. 1h and Supplementary Fig. 3 and are shown again here for ease of comparison.
(e) GUIDE-Seq-based identification of DSBs induced by RGNs directed by tru-gRNAs. Mapped reads for the on-target sites of the three RGNs directed by tru-gRNAs we assessed by GUIDE-Seq are shown. In all cases, the target site sequence is shown with the 20 bp protospacer sequence to the left and the PAM sequence to the right on the x-axis. As with RGNs directed by full-length gRNAs, note how the highest peak falls within 3 to 4 bps of the 5′-edge of the NGG PAM sequence, the expected position of an RGN cleavage event.
(f)-(h) Sequences of off-target sites identified by GUIDE-Seq for RGNs directed by tru-gRNAs. For each RGN, the intended target sequence is shown in the top line with cleaved sites shown underneath and with mismatches to the on-target site shown and highlighted in color. GUIDE-Seq sequencing read counts are shown to the right of each site. The intended on-target site is marked with a green square, previously known off-target sites of RGNs directed by both a full-length gRNA and a tru-gRNA are marked with a red diamond, and previously known off-target sites found only with RGNs directed by a tru-gRNA are marked with a yellow diamond. Previously known off-target sites were those that were shown to have a mutagenesis frequency of 0.1% or higher in an earlier report27. Data is shown for RGNs directed by tru-gRNAs to the (f) VEGFA site 1, (g) VEGFA site 3, and (h) EMX1 target sites.
DISCUSSION
Our studies show that GUIDE-Seq provides an unbiased, genome-wide, and sensitive method for detecting RGN-induced DSBs. The method is unbiased because it detects DSBs without making assumptions about the nature of the off-target site (e.g., presuming that the off-target site is closely related in sequence to the on-target site). GUIDE-Seq identifies off-target sites genome-wide, including within exons, introns and intergenic regions. Although the current lack of a gold standard method for comprehensively identifying all RGN off-target sites in a cell prevents us from knowing the sensitivity of GUIDE-Seq with certainty, we believe that it very likely has a low false-negative rate for the following reasons: First, all RGN-induced blunt-ended DSBs should take up the blunt-ended dsODN by NHEJ, a hypothesis supported by the strong correlations we observe between GUIDE-Seq read counts (which measure dsODN uptake) and indel frequencies in the presence of the RGN (which measure rates DSB formation and of their mutagenic repair) (Figs. 3b-3f). We note that these correlations include over 130 sites which show a wide range of indel mutagenesis frequencies. Second, using previously identified off-target sites as a benchmark (which is the only way currently to gauge success), GUIDE-Seq was able to detect 38 out of 40 of these sites that show a range of mutagenesis frequencies extending to as low as 0.12%. The method detected all 28 previously known off-target sites for four full-length gRNAs and 10 out of 12 previously known off-target sites for three tru-gRNAs (see Supplementary Discussion for potential explanations of why we did not detect two of the 40 sites).
Although our validation experiments show that GUIDE-Seq can sensitively detect off-target sites that are mutagenized by RGNs with frequencies as low as 0.1%, its detection capabilities might be further improved with deeper sequencing. Strategies that use next-generation sequencing to detect indels are limited by the error rate of the platform (typically ~0.1%). By contrast, GUIDE-Seq uses sequencing to identify dsODN insertion sites rather than indels and is therefore not limited by error rates but by sequencing depth. For example, we believe that the small number of sites detected in our GUIDE-Seq experiments for which we did not find indels in our sequencing validation experiments actually represent sites that likely have indel mutation frequencies below 0.1%. Consistent with this, we note that all but three of these 26 sites had GUIDE-Seq read counts below 100. Taken together, these observations suggest that we may be able to increase the sensitivity of GUIDE-Seq simply by increasing the number of sequencing reads (and by increasing the number of genomes used as template for amplification). For example, use of a sequencing platform that yields 1000-fold more reads would enable detection of sites with mutagenesis frequencies three orders of magnitude lower (i.e., 0.0001%), and we expect further increases to occur with continued improvements in next-generation sequencing technology. Of note, one of the RGNs we assessed did not yield any detectable off-target effects even when we repeated the GUIDE-Seq experiment a second time (data not shown). This finding raises the intriguing possibility that some gRNAs may induce very few, or perhaps no, undesired mutations (at least at the current detection limit of these GUIDE-Seq experiments).
In direct comparisons, we found that two existing computational programs failed to identify the majority of bona fide off-target sites found by GUIDE-Seq. This is not entirely surprising given that parameters used by these programs were based on more restrictive assumptions about the nature of off-target sites that do not account for greater numbers of protospacer mismatches (up to six) and new alternative PAM sequences identified by our GUIDE-Seq experiments. It is possible that better predictive programs might be developed in the future but doing so will require experimentally determined genome-wide off-target sites for a larger number of RGNs. Until such programs can be developed, identification of off-target sites will be most effectively addressed by experimental methods such as GUIDE-Seq.
Our experimental results elaborate a clear distinction between off-target binding sites of dCas9 and off-target cleavage sites of Cas9. Our results strongly suggest that the binding of off-target sites by dCas9 being captured with ChIP-Seq represents a different biological process than cleavage of off-target sites by Cas9 nuclease, consistent with the results of a recent study showing that engagement of the 5′-end of the gRNA with the protospacer is needed for efficient cleavage19. Although ChIP-Seq assays will undoubtedly play a role in characterizing the genome-wide binding of dCas9 fusion proteins, the method is clearly not effective for determining genome-wide off-target cleavage sites of catalytically active RGNs.
GUIDE-Seq has several advantages over other previously described genome-wide methods for identifying DSB sites in cells. The BLESS (breaks labeling, enrichment on streptavidin and next-generation sequencing) oligonucleotide tagging method is performed in situ on fixed, permeabilized cells28. In addition to being prone to artifacts associated with cell fixation, BLESS will only capture breaks that exist at a single moment. By contrast, GUIDE-Seq is performed on living cells and captures DSBs that occur over a more extended period of time (days), thereby making it a more sensitive and comprehensive assay. Capture of integration-deficient lentivirus (IDLV) DNA into regions near DSBs and identification of these loci by LAM-PCR has been used to identify a small number of off-target sites for engineered zinc finger nucleases (ZFNs)22 and transcription activator-like effector nucleases (TALENs)29 in human cells. However, IDLV integration events are generally low in number and widely dispersed over distances as far as 500 bps away from the actual off-target DSB22, 29, making it challenging both to precisely map the location of the cleavage event and to infer the sequence of the actual off-target site. In addition, the LAM-PCR process used in previous IDLV capture experiments suffers from sequence bias and/or low efficiency of useful sequencing reads. Collectively, these limitations may also explain the apparent inability to detect lower frequency ZFN off-target cleavage sites in previous studies30. By contrast, dsODNs are integrated very efficiently and precisely into DSBs with GUIDE-Seq, enabling mapping of breaks with single nucleotide resolution and simple, straightforward identification of the nuclease off-target cleavage sites. Furthermore, in contrast to LAM-PCR, our STAT-PCR method allows for efficient, unbiased amplification and sequencing of genomic DNA fragments in which the dsODN has integrated. We note that STAT-PCR may have more general utility beyond its use in GUIDE-Seq (e.g., to map the integration sites of viruses on a genome-wide scale).
GUIDE-Seq also identified breakpoint hotspots that occur in cells even in the absence of RGNs. We believe that these DSBs are not just an artifact of GUIDE-Seq because our AMP-based sequencing experiments verified not only capture of dsODNs but also the formation of indels (data not shown) and larger-scale genomic rearrangement involving these sites. Of note, the majority of hotspots we found appear to be unique to each of the two cell lines examined in our study, but two appear to be common to both. It will be interesting in future studies to define the parameters that govern why some sites are breakpoint hotspots in one cell type but not another. Also, because our results show that these breakpoint hotspots can participate in translocations, the existence of cell-type-specific breakpoint hotspots might help to explain why certain genomic rearrangements only occur in specific cell types but not others. To our knowledge, GUIDE-Seq is the first method to be described that can identify breakpoint hotspots in living human cells without the need to add drugs that inhibit DNA replication (e.g., aphidicolin)28. Therefore, we expect that it will provide a useful tool for identifying and studying these breaks.
Our work establishes an important qualitative approach approach for identifying translocations induced by RGNs. AMP-based targeted sequencing of RGN-induced and RGN-independent DSBs discovered by GUIDE-Seq can find large-scale genomic rearrangements (translocations, deletions, and inversions) involving both classes of sites, highlighting the importance of examining all of these loci. In addition, presumably not all RGN-induced or RGN-independent DSBs will participate in large-scale alterations and understanding why some sites do and other sites do not contribute to these rearrangements will be an important area for further research.
GUIDE-Seq will also provide an important way to evaluate alterations to the RGN platform on a genome-wide scale. In this report, we used GUIDE-Seq to show that the use of truncated gRNAs can reduce genome-wide off-target effects. We envision that GUIDE-Seq might also be used to assess the specificities of alternative Cas9 nucleases from other bacteria or archaea31. GUIDE-Seq might also be adapted to assess the genome-wide specificities of nucleases such as dimeric ZFNs, TALENs, and CRISPR RNA-guided FokI nucleases (RFNs) 32, 33 that generate 5′ overhangs or paired Cas9 nickases34, 35 that generate 5′ or 3′ overhangs. In preliminary experiments, we have already shown that blunt dsODN can be captured into ZFN-, TALEN- and RFN-induced breaks (data not shown); however, extending GUIDE-Seq to detect these other types of DSBs will undoubtedly require additional modification of the dsODN to optimize its efficient capture into such breaks.
We expect that our overall approach using GUIDE-Seq and AMP-based sequencing will prove to be very useful for the evaluation of off-target mutations and genomic rearrangements induced by RGNs. GUIDE-Seq can most likely be extended for use in any cell in which NHEJ is active and into which the required components can be efficiently introduced; for example, we have already achieved efficient dsODN integration in human K562 and mouse embryonic stem cells (data not shown) and it will be of great interest in future experiments to perform the method in non-transformed primary cells. The strategies outlined here can be used as part of a rigorous pre-clinical pathway for objectively assessing the potential off-target effects of any RGNs proposed for therapeutic use, thereby substantially improving the prospects for eventual translation of these reagents to the clinic.
ONLINE METHODS
Human cell culture and transfection
U2OS and HEK293 cells were cultured in Advanced DMEM (Life Technologies) supplemented with 10% FBS, 2 mM GlutaMax (Life Technologies), and penicillin/streptomycin at 37 °C with 5% CO2. U2OS cells (program DN-100) and HEK293 cells (program CM-137) were transfected in 20 μl Solution SE on a Lonza Nucleofector 4-D according to the manufacturer’s instructions. dsODN integration rates were assessed by restriction fragment length polymorphism (RFLP) assay using NdeI. Cleavage products were run and quantified by a Qiaxcel capillary electrophoresis instrument (Qiagen) as previously described32.
dsODN for GUIDE-Seq
The blunt-ended dsODN used in our GUIDE-Seq experiments was prepared by annealing two modified oligonucleotides of the following compositions: 5′-P-G*T*TTAATTGAGTTGTCATATGTTAATAACGGT*A*T-3′ and 5′-P-A*T*ACCGTTATTAACATATGACAACTCAATTAA*A*C-3′ where P represents a 5′ phosphorylation and * indicates a phosphorothioate linkage.
Isolation and preparation of genomic DNA for GUIDE-Seq
Genomic DNA was isolated using solid-phase reversible immobilization magnetic beads (Agencourt DNAdvance), sheared with a Covaris S200 instrument to an average length of 500 bp, end-repaired, A-tailed, and ligated to half-functional adapters, incorporating a 8-nt random molecular index. Two rounds of nested anchored PCR, with primers complementary to the oligo tag, were used for target enrichment. Full details of the GUIDE-Seq protocol can be found in Supplementary Methods.
Processing and consolidation of sequencing reads
Reads that share the same six first bases of sequence as well as identical 8-nt molecular indexes were binned together because they are assumed to originate from the same original pre-PCR template fragment. These reads were consolidated into a single consensus read by selecting the majority base at each position. A no-call (N) base was assigned in situations with greater than 10% discordant reads. The base quality score was taken to be the highest among the pre-consolidation reads. Consolidated reads were mapped to human genome reference (GrCh37) using BWA-MEM36.
Identification of off-target cleavage sites
Start mapping positions for reads with mapping quality ≥50 were tabulated, and regions with nearby start mapping positions were grouped using a 10 bp sliding window. Genomic windows harboring integrated dsODNs were identified by one of the following criteria: 1) two or more unique molecular-indexed reads mapping to opposite strands in the reference sequence or 2) two or more unique molecular-indexed reads amplified by forward and reverse primers. 25 bp of reference sequence flanking both sides of the inferred breakpoints were aligned to the intended target site and RGN off-target sites with eight or fewer mismatches from the intended target sequence were retained. SNPs and indels were called in these positions by a custom bin-consensus variant-calling algorithm based on molecular index and SAMtools, and off-target sequences that differed from the reference sequence were replaced with the corresponding cell-specific sequence.
AMP-based sequencing
For AMP validation of GUIDE-Seq detected DSBs, primers were designed to regions flanking inferred double-stranded breakpoints as described previously37, with the addition of an 8-nt molecular molecular index. Where possible, we designed two primers to flank each DSB.
Analysis of AMP validation data
Reads with average quality scores greater than 30 were analyzed for insertions, deletions, and integrations that overlapped with the GUIDE-Seq inferred DSB positions using Python. 1 bp indels were included only if they were within 1 bp of the predicted DSB site to minimize the introduction of noise from PCR or sequencing error. Integration and indel frequencies were calculated on the basis of consolidated molecular indexed reads. Sites with background indel frequencies >1% were excluded from the analysis.
Structural variation
Translocations, large deletions, and inversions were identified using a custom algorithm based on split BWA-MEM alignments. Candidate fusion breakpoints within 50 bases on the same chromosome were grouped to accommodate potential resection around the Cas9 cleavage site. A fusion event was called with at least 3 uniquely mapped split reads, a parameter also used by the segemehl tool to minimize false positives38. Information on the strand to which reads mapped (plus or minus) was maintained in order to identify reciprocal fusions between different ends of the same DSBs, and for determining deletion or inversion. Deletions of less than 1 kb in size were excluded from this analysis as they may arise from a single DSB, end-resection, and canonical NHEJ. The remaining DSBs involved in fusions were classified into four categories: ‘on-target’, ‘off-target’, ‘hotspot’ or ‘other’.
Comparison of GUIDE-Seq with computational prediction methods
We used the MIT CRISPR Design Tool to identify potential off-target sites for all ten RGNs. This tool assigns each potential off-target site a corresponding percentile. We then grouped these percentiles into quintiles for visualization purposes. The E-CRISP tool does not rank off-target sites and so we simply used the program to identify these sites for each RGN.
Analysis mismatches, DNA accessibility and local PAM density on off-target cleavage rate
We assessed the impact of mismatch position, mismatch type and DNA accessibility on specificity using linear regression models fit to estimated cleavage rates at potential off-target sites with four or fewer mismatches. Mismatch position covariates were defined as the number of mismatched bases within each of five non-overlapping 4 bp windows upstream of the PAM. Mismatch type covariates were defined as i) the number mismatches resulting in wobble pairing (target T replaced by C, target G replaced by A), ii) the number of mismatches resulting in a non-wobble purine-pyrimidine base-pairing (target C replaced by T, target A replaced by G), and iii) the number as mismatches resulting in purine-purine or pyrimidine-pyrimidine pairings.
Each of the three factors was used in a separate model as a predictor of relative cleavage rates, estimated by log2(1 + GUIDE-Seq read count). The effect size estimates were adjusted for inter-target site variability. The proportion of intra-site cleavage rate variability explained by each factor was assessed by the partial eta-squared statistic based on the regression sums of squares (SS): 2p = SSfactor/(SSfactor + SSerror). In addition to the single-factor models, we also fit a combined linear regression model including all three factors, expression level, and PAM density in a 1 kb window to assess their independent contribution to off-target cleavage probability.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Angstman, B. Kleinstiver, Y. Fu, J. Gehrke, and R. Cottman for helpful comments on the manuscript and M. Maeder and J. Foden for technical assistance. This work was funded by a National Institutes of Health (NIH) Director’s Pioneer Award (DP1 GM105378), NIH R01 GM088040, NIH R01 AR063070, and the Jim and Ann Orr Massachusetts General Hospital (MGH) Research Scholar Award. S.Q.T. was supported by NIH F32 GM105189. This material is based upon work supported by, or in part by, the US Army Research Laboratory and the US Army Research Office under grant number W911NF-11-2-0056. Plasmids described in this work will be deposited with and made available through the nonprofit plasmid distribution service Addgene (http://www.addgene.org/crispr-cas).
Footnotes
AUTHOR CONTRIBUTIONS
S.Q.T. and J.K.J. conceived of the GUIDE-Seq method. S.Q.T., Z.Z., A.J.I., L.P.L, and J.K.J. planned experiments. S.Q.T., Z.Z., N.T.N., M.L., N.W., and C.K. performed experiments. S.Q.T., Z.Z., V.V.T., V.T., and M.J.A. performed bioinformatics and computational analysis of the data. S.Q.T. and J.K.J. wrote the paper.
COMPETING FINANCIAL INTERESTS
J.K.J. is a consultant for Horizon Discovery. J.K.J. has financial interests in Editas Medicine and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
REFERENCES
- 1.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghezraoui H, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gostissa M, et al. IgH class switching exploits a general property of two DNA breaks to be joined in cis over long chromosomal distances. Proc Natl Acad Sci U S A. 2014;111:2644–2649. doi: 10.1073/pnas.1324176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai SQ, Joung JK. What’s changed with genome editing? Cell Stem Cell. 2014;15:3–4. doi: 10.1016/j.stem.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Marx V. Gene editing: how to stay on-target with CRISPR. Nat Methods. 2014;11:1021–1026. doi: 10.1038/nmeth.3108. [DOI] [PubMed] [Google Scholar]
- 14.Veres A, et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith C, et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan J, et al. Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res. 2014;24:1009–1012. doi: 10.1038/cr.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 19.Cencic R, et al. Protospacer Adjacent Motif (PAM)-Distal Sequences Engage CRISPR Cas9 DNA Target Cleavage. PLoS One. 2014;9:e109213. doi: 10.1371/journal.pone.0109213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlando SJ, et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt M, et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 22.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 23.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 27.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crosetto N, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10:361–365. doi: 10.1038/nmeth.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osborn MJ, et al. TALEN-based gene correction for epidermolysis bullosa. Mol Ther. 2013;21:1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sander JD, et al. In silico abstraction of zinc finger nuclease cleavage profiles reveals an expanded landscape of off-target sites. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fonfara I, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Z, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014 doi: 10.1038/nm.3729. advance online publication. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann S, et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15:R34. doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.