Abstract
The yeast pathogen Candida glabrata is the second most frequent cause of Candida infections. However, from the phylogenetic point of view, C. glabrata is much closer to Saccharomyces cerevisiae than to Candida albicans. Apparently, this yeast has relatively recently changed its life style and become a successful opportunistic pathogen. Recently, several C. glabrata sister species, among them clinical and environmental isolates, have had their genomes characterized. Also, hundreds of C. glabrata clinical isolates have been characterized for their genomes. These isolates display enormous genomic plasticity. The number and size of chromosomes vary drastically, as well as intra- and interchromosomal segmental duplications occur frequently. The observed genome alterations could affect phenotypic properties and thus help to adapt to the highly variable and harsh habitats this yeast finds in different human patients and their tissues. Further genome sequencing of pathogenic isolates will provide a valuable tool to understand the mechanisms behind genome dynamics and help to elucidate the genes contributing to the virulence potential.
Keywords: pathogenic yeast, Candida, virulence genes, chromosome polymorphism, genome rearrangements
Introduction
In humans, during the recent years, several factors have increased the risk of fungal infections, like broad spectrum antibiotics, immune-compromised patients, cytotoxic chemotherapy, and transplantations (Malani et al., 2005; Nucci & Marr, 2005). Among all fungal pathogens, Candida species remain the most common cause of invasive fungal infections (Pfaller & Diekema, 2007). Although Candida glabrata has been considered as a human commensal and can be often found in the oral cavity (Fidel et al., 1999), it has recently emerged as the second most prevalent cause of Candida infections, just after Candida albicans (Pfaller & Diekema, 2004). Roughly, C. glabrata is responsible for 15–25% of disseminated candidiasis (Perlroth et al., 2007).
Phylogenetically, C. glabrata is much closer to Saccharomyces cerevisiae than to C. albicans and its sister species (Dujon et al., 2004). Indeed, C. glabrata belongs to post-WGD (whole genome duplication) yeasts (Fig. 1). Candida glabrata with three sister species, Kluyveromyces delphensis, Candida castellii and Kluyveromyces bacillisporus, have been classified to belong to the Nakaseomyces genus (Kurtzman, 2003). Two additional clinical isolates, Candida nivariensis and Candida bracarensis, have later been added to the same genus (Alcoba-Florez et al., 2005; Correia et al., 2006; Sharma et al., 2013). The Nakaseomyces has been divided into two subgroups, the first, ‘glabrata group’ includes the three pathogenic species and N. delphensis, and the second group includes C. castellii and N. bacillisporus.
Fig. 1.
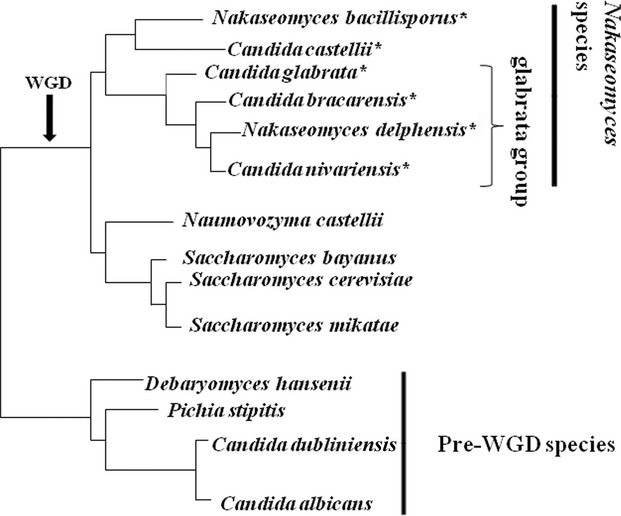
A schematics phylogenetic tree (adopted from Gabaldón et al., 2013) shows the Nakaseomyces species (marked with an asterisk) and some other yeasts. Candida glabrata, C. nivariensis, C. bracarensis are pathogenic fungi within the ‘glabrata group’. The whole-genome duplication event (WGD), which took place app. 100 million years ago, is arrowed.
Unlike S. cerevisiae, which is diploid and sexual, C. glabrata is believed to be obligate haploid and mating has never been observed (Kaur et al., 2005; Muller et al., 2008). Candida albicans can live in two different morphological forms (Sudbery, 2011), but the switching between the yeast form and hyphae has not been observed in C. glabrata (Kaur et al., 2005; Fig. 2). Recently, there has been more focus on C. glabrata genome dynamics and several new sister species genomes have been reported, both providing additional insight into metabolism and virulence potential of the yeast species.
Fig. 2.
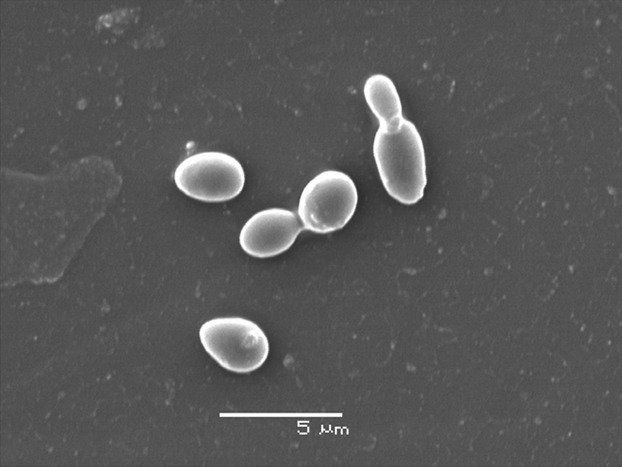
Candida glabrata yeast under electron microscope. The figure shows two single cells and two mother–daughter pairs during the budding process.
Candida glabrata genome potential
Dujon et al. (2004) have reported that C. glabrata CBS 138 has 13 chromosomes with the genome size of 12.3 Mb. In comparison with S. cerevisiae, which has 5807 protein-coding genes (Dujon et al., 2004), C. glabrata has 5283 coding sequences, with an average size of 493 codons. Thus, this yeast lineage has reduced its coding potential upon separation from other post-WGD yeasts. Candida glabrata has two intrachromosomal rDNA repeats loci in subtelomeric regions compared to S. cerevisiae, which has a single intrachromosomal rDNA repeat locus. As S. cerevisiae, C. glabrata has 42 tRNA-encoding genes, which are scattered throughout the genome (Dujon et al., 2004).
In C. albicans, the CUG codon is translated as serine instead of leucine, but this genetic codon alteration is not found in C. glabrata. In general, most genes transcribed by RNA polymerase III and even other noncoding RNA genes do not have well-conserved size among yeasts (Gabaldón et al., 2013). For example, in C. glabrata, RNase P RNA (RPR1) gene is 1149 nucleotides long, whereas in S. cerevisiae it is 568 nucleotides long (Kachouri et al., 2005). This large ncRNA seems to be present in C. glabrata and sister species, and K. delphensis has a 1368 nucleotide long copy (Gabaldón et al., 2013). Candida glabrata and other related species have a lower number of intron containing protein-coding genes than S. cerevisiae (Neuvéglise et al., 2011).
It has been found by Conant & Wolfe (2007) that 551 duplicated gene pairs are conserved between C. glabrata and S. cerevisiae, among them six gene pairs encoding glycolytic enzymes ENO1/ENO2, PYC1/PYC2, GLK1/EMI2, HXK1/HXK2, TDH2/TDH3, and CDC19/PYK2. Therefore, this yeast has a genetic capacity for a high glycolytic flow. Indeed, C. glabrata has recently been reported as an efficient alcohol producing yeast even under aerobic conditions (Hagman et al., 2013). However, it is so far unclear whether this trait plays an important role during yeast pathogenicity. For example, C. albicans and its sister species are Crabtree-negative yeasts and do not produce ethanol under aerobic conditions (Hagman et al., 2013).
Comparative genomics of C. glabrata and phylogenetically related species
Recently, several C. glabrata sister species have been sequenced. All the species in Nakaseomyces have transposon-free genomes that are smaller than S. cerevisiae and contain fewer genes (Gabaldón et al., 2013). Moreover, all species in this group have a haploid genome, the size of roughly 10–12.5 Mb, with the exception of N. bacillisporus which is a diploid. The chromosomal number ranges from 8 in C. castellii to 15 in N. bacillisporus, while the ‘glabrata group’ yeasts have from 10 to 13 chromosomes. Note that chromosome number in C. glabrata isolates varies (Ahmad et al., 2013). The gene-order conservation is very high between N. delphensis, C. bracarensis and C. nivariensis, but in general, it is low compared with S. cerevisiae (Gabaldón et al., 2013). The divergence between orthologous proteins varies from 53% between C. castellii and N. bacillisporus to higher identity level among ‘glabrata group’ species (77–88%).
After the WGD event, many genes have been lost in C. glabrata, for example, galactose metabolism (five genes), phosphate metabolism (four genes), cell defense and virulence (three genes), and nitrogen and sulfur metabolism (three genes; Dujon et al., 2004). Candida glabrata genome has also lost genes involved in de novo biosynthesis of nicotinic acid (BNA), and this has been initially interpreted as a way to adapt to the host environment (Domergue et al., 2005). However, the species among this clade have different habitats, and the loss of BNA has been observed in all of them and likely occurred in the common progenitor when it was still a free-living yeast organism. Another example of gene loss in C. glabrata and ‘glabrata group’ is loss of the six genes cluster involved in allantoin catabolism, DAL, which is conserved in S. cerevisiae (Wong & Wolfe, 2005). On the other hand, the DAL cluster is present in both C. castellii and N. bacillisporus (Gabaldón et al., 2013).
Apparently, similar to C. glabrata, the duplicated genes encoding glycolytic enzymes, for example ENO1/ENO2 and PYC1/PYC2 are conserved in Nakaseomyces (Merico et al., 2007). In contrast to ‘glabrata group’ where the glycolytic enzymes encoding genes are in pairs, pyruvate kinase, enolase, and glyceraldehyde-3-phosphate dehydrogenase encoding genes are present as a single copy in C. castellii and N. bacillisporus. Compared to all Nakaseomyces species that encode four hexokinase (HXK) genes, C. castellii genome encodes two HXK genes. Moreover, all Nakaseomyces genomes encode two copies of ADH genes, which have orthologs in S. cerevisiae, ADH1 and ADH3, and are involved in conversion of acetaldehyde into ethanol. Saccharomyces cerevisiae genome also encodes ADH2, which involves in the conversion of the ethanol into acetaldehyde, and this gene has no ortholog in Nakaseomyces.
The genome of C. glabrata type strain (CBS 138) contains a family of 18 EPA genes (homologues of the FLO genes in S. cerevisiae), coding for glycosylphosphatidylinositol (GPI)-anchored cell-wall proteins (CWP), which are known to be adhered to human epithelium (Cormack et al., 1999; Gabaldón et al., 2013). Similarly, C. bracarensis has 12 members of the EPA family, while C. nivariensis genome harbors nine genes. On the other hand, the nonpathogenic yeasts in Nakaseomyces, C. castellii, has three homologs of the EPA genes, whereas N. bacillisporus has only one homolog, which is distant and clusters together with the PWP (PA-14 containing wall protein adhesion gene) and adhesion-like protein genes in C. glabrata. Although the ‘glabrata group’ has a common ancestor, the genome of N. delphensis has a single copy of the EPA gene (Gabaldón et al., 2013).
Mini and megasatellites
Minisatellites are DNA tandem repeats that show size variation among different yeast species (Richard et al., 2008). In S. cerevisiae, it has been found that several genes which are involved in cell wall formation contain minisatellites with variable sizes. These genes belong to the FLO family of mannoprotein-encoding genes. The mannoprotein-encoding genes show a variable number of repeats among different yeast isolates and the length of the repeat is directly related to flocculation and cell adhesion. For example, strains with FLO1 carrying a longer repeat show better adhesion and increased flocculation (Verstrepen et al., 2005).
The minisatellites within the hemiascomycetous yeasts are not conserved among their host genes. Moreover, the diversity of minisatellite sequences within the orthologous genes may indicate that the acquisition and loss of minisatellites have been fast during evolution (Richard & Dujon, 2006). Despite similar genome size, the number of minisatellites in C. glabrata is significantly higher than in S. cerevisiae. In addition, C. glabrata has two more unusual minisatellites, namely, compound minisatellite, with different alternative repeats and megasatellites, with unusually long motifs (Thierry et al., 2008).
Haber & Louis (1998) suggested that minisatellites originated by strand slippage during replication of two short (5–10 bp) sequences or by unequal crossing over. This replication slippage has been reported in S. cerevisiae (Richard & Dujon, 2006) but not observed in C. glabrata where other mechanisms might be involved (Thierry et al., 2008). Some authors proposed that the megasatellite motifs propagate by motif jumps, which may explain the presence of motifs between nonorthologous gene families (Rolland et al., 2010).
The genome of C. glabrata has 40 megasatellites mapped to 33 genes, which are distributed on 11 chromosomes with strong preference toward the subtelomeric region. The megasatellites have long motifs ranging from 135 up to 417 nucleotides and belong to two large families, ‘SHITT’ and ‘SFFIT’ (the names come from conservation of five amino acid in each motif in the megasatellite; Thierry et al., 2008). In a recent study, which covered 20 completely sequenced fungal genomes, 216 peptide motifs encoded by megasatellites and located in protein-coding genes have been extracted (Tekaia et al., 2013). The detected megasatellites have been characterized according to the motif sequence similarity and their amino acid composition. Based on this analysis, there is a weak match between SHITT and FLO motifs despite their similar size (Tekaia et al., 2013).
The originally sequenced C. glabrata has three EPA genes containing megasatellites (EPA2, EPA11, and EPA13), another three genes (EPA1, EPA3, and EPA15) contain minisatellites, but there are no tandem repeats in EPA6, EPA7, and EPA8 (Thierry et al., 2008). A recent study has shown that the chromosomal translocations that occurred among different strains of C. glabrata are associated with deletions ranging from 130 bp up to 12 kb and two of them were located within megasatellites (Muller et al., 2009). Even translocations and segmental duplications may operate through mini- and megasatellites (Poláková et al., 2009).
Genome rearrangements
Candida glabrata isolates have been reported to show variable karyotypes (Shin et al., 2007). Apparently, the genome of C. glabrata is frequently rearranged and some authors consider these rearrangements as an adaptive mechanism. Muller et al. (2009) have reported that the most frequent changes in karyotype belong to chromosomal length polymorphism (CLP), mainly due to chromosomal translocations and copy number variations within tandem gene repeats. These gene tandem repeats encode putative proteins like yapsins, cell wall aspartyl proteases (Kaur et al., 2007). Muller et al. (2009) have also shown that there is size variation in the subtelomeric EPA genes, which is most likely due to the differences in the number of minisatellites within the genes, as proposed by Thierry et al. (2008). In general, Muller et al. (2009), based on their French strain collection, claim that the C. glabrata genome is comparatively stable. This is in contrast to many other works, which propose a very dynamic nature of the C. glabrata genome.
Poláková et al. (2009) have found that among Danish isolates, chromosomal size variation is associated with either gross intrachromosomal translocations or interchromosomal segmental duplications. Some novel chromosomes have been observed and shown to originate by segmental duplications. These novel chromosomes carry duplicated genes, which are potentially involved in yeast–host interaction, and they include the ABC transporter family genes, which plays a role in multidrug resistance. The formation of new chromosomes has also been reported in a new study analyzing over 200 clinical isolates; Ahmad et al. (2013) have found several extra chromosomes originating through at least two different mechanisms (Fig. 3). Remarkably, even phylogenetically related isolates have shown rearranged chromosomes, implying that C. glabrata frequently remodels its genome. It has been speculated that in this way, the yeast can ‘improve’ its fitness when exposed to new environmental conditions (Ahmad et al., 2013). A striking observation of genome dynamics has been made on recent derivatives from a single laboratory strain. Apparently, among these very closely related strains, different phenotypic groups correlated with specific karyotypic changes (Bader et al., 2012). Thus, genome aberrations and functional adaptations may occur not only during infection, but also under laboratory conditions during a very short time span and without extreme selective pressures.
Fig. 3.
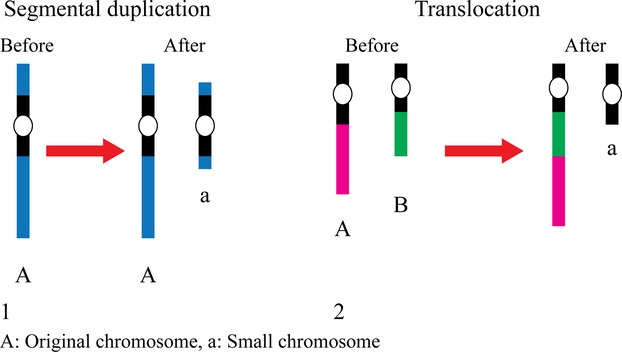
Two possible mechanisms behind the birth of small chromosomes are shown (adapted from Ahmad et al., 2013). (1) The small chromosome (a) originated by segmental duplication within chromosome A. (2) Interchromosomal translocation, where a large segment of chromosome A is translocated to join the chromosome B leaving the rest of chromosome A as a small chromosome.
Three large gene families are associated with the cell wall, the YPS gene cluster, consisting of six genes encoding the yapsins (extracellular GPI-linked aspartyl proteinases), eight alpha-1,3-mannosyltransferase, and five beta-mannosyltransferase gene clusters (Dujon et al., 2004; Dujon, 2010; Jawhara et al., 2012). Apart from the chromosome remodeling, also gene duplications and cluster and gene family formation apparently play an important role in the specialization to specific environmental condition, virulence, and interaction with the host (Butler et al., 2009; Moran et al., 2011).
The genome of C. glabrata encodes Sir complex including Sir2p, Sir3p, and Sir4p. These proteins are responsible for subtelomeric silencing in C. glabrata (Castaño et al., 2005; Maestre-Reyna et al., 2012). Recently, it has been reported that C. glabrata isolates were more adherent to the epithelial cells with overexpression of the EPA1 gene and, moreover, these strains contain several polymorphisms in Sir3p, which might cause reduced silencing (Martinez-Jimenez et al., 2013). On the other hand, it has been shown that the deletion of HST1 (homologue of SIR2) altered the C. glabrata response to stressful conditions with an increase in the fluconazole resistance and decrease in the susceptibility to hydrogen peroxide (Orta-Zavalza et al., 2013).
In short, recent studies on C. glabrata genome polymorphism have pointed out that this property is clearly connected to the virulence potential of this organism. Future genome sequencing of different isolates will likely provide even further insights into the interplay between genome dynamics and virulence.
OMICS aspects
The first genome structure has provided a background to develop different postgenomic tools and approaches. These could now in turn be used to analyze the connection between the genotype variability and different phenotypic traits. For example, the availability of commercial microarrays has provided a powerful tool to study global aspects of gene expression (Fukuda et al., 2013) to identify potential virulence genes, as well as to determine the level of variation in gene expression among different clinical isolates (O.P. Ishchuk, J. Piškur, unpublished data).
Unlike C. albicans, the mechanism of virulence at the molecular level has so far not been well understood in C. glabrata (Silva et al., 2011). The proteomic approach to study protein expression in C. glabrata has confirmed a previous observation that the virulence of C. glabrata increases when Ace2p, transcriptional factor, is inactivated (Stead et al., 2010). In the same study, 32 of 123 proteins were overexpressed in the ace2 mutant (Stead et al., 2010). Moreover, it has been found that the expression of proteins involved in glucose metabolism, the TCA (Krebs) cycle, respiration and protein synthesis is low when C. glabrata grows at pH 7.4 or 8.0, whereas the expression of proteins involved in stress responses and protein catabolism was increased (Schmidt et al., 2008). Proteomic approaches have also been used to study the C. glabrata biofilm. The stress-response protein Trx1p, a component of oxidative stress defenses of C. glabrata, was found to be upregulated (Seneviratne et al., 2008, 2010). Hsp12p is another protein, which has been highly expressed in C. glabrata biofilm. In S. cerevisiae, the expression of this protein is induced by different kinds of stress including heat shock, oxidative stress and osmotic stress (Perez-Torrado et al., 2005), and it is involved in azole antifungal resistance in C. albicans (Karababa et al., 2004). The proteomic studies have also shown that several glycolytic enzymes including Fba1p, Tdh3p, Gpm1p, and Eno1p are downregulated in C. glabrata biofilm (Seneviratne et al., 2010). It would also be interesting to apply the proteome approach on different clinical isolates to see whether the genome variation correlates with the proteome variation.
A new analysis based on high-resolution mass spectrometry has been conducted to identify new peptides, which were previously unknown in C. glabrata (Prasad et al., 2012). By genome search for specific peptides (GSSP) and other comparative genomic strategies, the conservation and absence of protein-coding genes in C. glabrata and other related species has been elucidated. New postgenomic tools might also be helpful for early diagnosis and proper treatment, thereby reducing the mortality rate. To overcome the pathogen misdiagnosis, Perl et al. (2011) have suggested a new sensitive approach to study the volatile organic compounds, multicapillary column (MCC) equipped ion mobility spectrometer (MCC-IMS), to differentiate between different pathogenic fungi and their strains.
Current sequencing projects of tens of different pathogenic isolates, for example at the Cornell and Lund Universities, will provide further background for development of new postgenomic approaches and will help to reveal further genes behind C. glabrata virulence. The sequencing of their genomes will make these strains attractive models for postgenomic tool analysis.
Acknowledgments
We would like to thank the Swedish Research Council (VR-M), the Fysiografen, Lindström and Sörensen Foundations, and NIH (grant NIH-AI085286) for their financial support. We would also like to acknowledge Rita Wallen for providing us with the electron microscope picture (Fig. 2).
References
- Ahmad KM, Ishchuk OP, Hellborg L, Jørgensen G, Skvarc M, Stenderup J, Jørck-Ramberg D, Polakova S, Piškur J. Small chromosomes among Danish Candida glabrata isolates originated through different mechanisms. Antonie Van Leeuwenhoek. 2013;104:111–122. doi: 10.1007/s10482-013-9931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcoba-Florez J, Méndez-Alvarez S, Cano J, Guarro J, Pérez-Roth E, del Pilar Arévalo M. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. Clin Microbiol. 2005;43:4107–4111. doi: 10.1128/JCM.43.8.4107-4111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader O, Schwarz A, Kraneveld EA, et al. Gross karyotypic and phenotypic alterations among different progenies of the Candida glabrata CBS138/ATCC2001 reference strain. PLoS One. 2012;7:e52218. doi: 10.1371/journal.pone.0052218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G, Rasmussen MD, Lin MF, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño I, Pan SJ, Zupancic M, Hennequin C, Dujon B, Cormack BP. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol Microbiol. 2005;55:1246–1258. doi: 10.1111/j.1365-2958.2004.04465.x. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol. 2007;3:129. doi: 10.1038/msb4100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Ghori N, Falkow S. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science. 1999;285:578–582. doi: 10.1126/science.285.5427.578. [DOI] [PubMed] [Google Scholar]
- Correia A, Sampaio P, James S, Pais C. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int J Syst Evol Microbiol. 2006;56:313–317. doi: 10.1099/ijs.0.64076-0. [DOI] [PubMed] [Google Scholar]
- Domergue R, Castaño I, De Las Peñas A, Zupancic M, Lockatell V, Hebel JR, Johnson D, Cormack BP. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science. 2005;308:866–870. doi: 10.1126/science.1108640. [DOI] [PubMed] [Google Scholar]
- Dujon B. Yeast evolutionary genomics. Nat Rev Genet. 2010;11:512–524. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Fidel PL, Jr, Vazquez JA, Sobel JD. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin Microbiol Rev. 1999;12:80–96. doi: 10.1128/cmr.12.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y, Tsai HF, Myers TG, Bennett JE. Transcriptional profiling of Candida glabrata during phagocytosis by neutrophils and in the infected mouse spleen. Infect Immun. 2013;81:1325–1333. doi: 10.1128/IAI.00851-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldón T, Martin T, Marcet-Houben M, et al. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics. 2013;14:623. doi: 10.1186/1471-2164-14-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Louis EJ. Minisatellite origins in yeast and humans. Genomics. 1998;48:132–135. doi: 10.1006/geno.1997.5153. [DOI] [PubMed] [Google Scholar]
- Hagman A, Säll T, Compagno C, Piskur J. Yeast “make-accumulate-consume” life strategy evolved as a multi-step process that predates the whole genome duplication. PLoS One. 2013;8:e68734. doi: 10.1371/journal.pone.0068734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawhara S, Mogensen E, Maggiotto F, Fradin C, Sarazin A, Dubuquoy L, Maes E, Guérardel Y, Janbon G, Poulain D. Murine model of dextran sulfate sodium-induced colitis reveals Candida glabrata virulence and contribution of β-mannosyltransferases. J Biol Chem. 2012;287:11313–11324. doi: 10.1074/jbc.M111.329300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachouri R, Stribinskis V, Zhu Y, Ramos KS, Westhof E, Li Y. A surprisingly large RNase P RNA in Candida glabrata. RNA. 2005;11:1064–1072. doi: 10.1261/rna.2130705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karababa M, Coste AT, Rognon B, Bille J, Sanglard D. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob Agents Chemother. 2004;48:3064–3079. doi: 10.1128/AAC.48.8.3064-3079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Domergue R, Zupancic ML, Cormack BP. A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol. 2005;8:378–384. doi: 10.1016/j.mib.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Kaur R, Ma B, Cormack BP. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. P Natl Acad Sci USA. 2007;104:7628–7633. doi: 10.1073/pnas.0611195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP. Phylogenetic circumscription of Saccharomyces Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 2003;4:233–245. doi: 10.1016/S1567-1356(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Maestre-Reyna M, Diderrich R, Veelders MS, Eulenburg G, Kalugin V, Brückner S, Keller P, Rupp S, Mösch HU, Essen LO. Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. P Natl Acad Sci USA. 2012;109:16864–16869. doi: 10.1073/pnas.1207653109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malani A, Hmoud J, Chiu L, Carver PL, Bielaczyc A, Kauffman CA. Candida glabrata fungemia: experience in a tertiary care center. Clin Infect Dis. 2005;41:975–981. doi: 10.1086/432939. [DOI] [PubMed] [Google Scholar]
- Martinez-Jimenez V, Ramirez-Zavaleta CY, Orta-Zavalza E, de León GD, Gutiérrez-Escobedo G, de Leon AP, Sifuentes-Osornio J, del Valle MB, De Las Penas A, Castaño I. Sir3 Polymorphisms in Candida glabrata clinical isolates. Mycopathologia. 2013;175:207–219. doi: 10.1007/s11046-013-9627-2. [DOI] [PubMed] [Google Scholar]
- Merico A, Sulo P, Piskur J, Compagno C. Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J. 2007;274:976–989. doi: 10.1111/j.1742-4658.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- Moran GP, Coleman DC, Sullivan DJ. Comparative genomics and the evolution of pathogenicity in human pathogenic fungi. Eukaryot Cell. 2011;10:34–42. doi: 10.1128/EC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Hennequin C, Gallaud J, Dujon B, Fairhead C. The asexual yeast Candida glabrata maintains distinct a and alpha haploid mating types. Eukaryot Cell. 2008;7:848–858. doi: 10.1128/EC.00456-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Thierry A, Coppée JY, Gouyette C, Hennequin C, Sismeiro O, Talla E, Dujon B, Fairhead C. Genomic polymorphism in the population of Candida glabrata: gene copy-number variation and chromosomal translocations. Fungal Genet Biol. 2009;46:264–276. doi: 10.1016/j.fgb.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Neuvéglise C, Marck C, Gaillardin C. The intronome of budding yeasts. C R Biol. 2011;334:662–670. doi: 10.1016/j.crvi.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Nucci M, Marr KA. Emerging fungal diseases. Clin Infect Dis. 2005;41:521–526. doi: 10.1086/432060. [DOI] [PubMed] [Google Scholar]
- Orta-Zavalza E, Guerrero-Serrano G, Gutiérrez-Escobedo G, Cañas-Villamar I, Juárez-Cepeda J, Castaño I, De Las Peñas A. Local silencing controls the oxidative stress response and the multidrug resistance in Candida glabrata. Mol Microbiol. 2013;88:1135–1148. doi: 10.1111/mmi.12247. [DOI] [PubMed] [Google Scholar]
- Perez-Torrado R, Bruno-Bárcena JM, Matallana E. Monitoring stress-related genes during the process of biomass propagation of Saccharomyces cerevisiae strains used for wine making. Appl Environ Microbiol. 2005;71:6831–6837. doi: 10.1128/AEM.71.11.6831-6837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl T, Jünger M, Vautz W, Nolte J, Kuhns M, Borg-von Zepelin M, Quintel M. Detection of characteristic metabolites of Aspergillus fumigatus and Candida species using ion mobility spectrometry-metabolic profiling by volatile organic compounds. Mycoses. 2011;54:e828–e837. doi: 10.1111/j.1439-0507.2011.02037.x. [DOI] [PubMed] [Google Scholar]
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45:321–346. doi: 10.1080/13693780701218689. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Twelve years of fluconazole in clinical practice: global trends in species distribution and fluconazole susceptibility of bloodstream isolates of Candida. Clin Microbiol Infect. 2004;1:11–23. doi: 10.1111/j.1470-9465.2004.t01-1-00844.x. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poláková S, Blume C, Zárate JA, Mentel M, Jørck-Ramberg D, Stenderup J, Piskur J. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. P Natl Acad Sci USA. 2009;106:2688–2693. doi: 10.1073/pnas.0809793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TS, Harsha HC, Keerthikumar S, et al. Proteogenomic analysis of Candida glabrata using high resolution mass spectrometry. Proteome Res. 2012;11:247–260. doi: 10.1021/pr200827k. [DOI] [PubMed] [Google Scholar]
- Richard GF, Dujon B. Molecular evolution of minisatellites in hemiascomycetous yeasts. Mol Biol Evol. 2006;23:189–202. doi: 10.1093/molbev/msj022. [DOI] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T, Dujon B, Richard GF. Dynamic evolution of megasatellites in yeasts. Nucleic Acids Res. 2010;38:4731–4739. doi: 10.1093/nar/gkq207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P, Walker J, Selway L, Stead D, Yin Z, Enjalbert B, Weig M, Brown AJ. Proteomic analysis of the pH response in the fungal pathogen Candida glabrata. Proteomics. 2008;8:534–544. doi: 10.1002/pmic.200700845. [DOI] [PubMed] [Google Scholar]
- Seneviratne CJ, Wang Y, Jin L, Abiko Y, Samaranayake LP. Candida albicans biofilm formation is associated with increased anti-oxidative capacities. Proteomics. 2008;8:2936–2947. doi: 10.1002/pmic.200701097. [DOI] [PubMed] [Google Scholar]
- Seneviratne CJ, Wang Y, Jin L, Abiko Y, Samaranayake LP. Proteomics of drug resistance in Candida glabrata biofilms. Proteomics. 2010;10:1444–1454. doi: 10.1002/pmic.200900611. [DOI] [PubMed] [Google Scholar]
- Sharma C, Wankhede S, Muralidhar S, et al. Candida nivariensis as an etiologic agent of vulvovaginal candidiasis in a tertiary care hospital of New Delhi, India. Diagn Microbiol Infect Dis. 2013;76:46–50. doi: 10.1016/j.diagmicrobio.2013.02.023. [DOI] [PubMed] [Google Scholar]
- Shin JH, Chae MJ, Song JW, Jung SI, Cho D, Kee SJ, Kim SH, Shin MG, Suh SP, Ryang DW. Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J Clin Microbiol. 2007;45:2385–2391. doi: 10.1128/JCM.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Adherence and biofilm formation of non-Candida albicans Candida species. Trends Microbiol. 2011;19:241–247. doi: 10.1016/j.tim.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Stead DA, Walker J, Holcombe L, Gibbs SR, Yin Z, Selway L, Butler G, Brown AJ, Haynes K. Impact of the transcriptional regulator, Ace2, on the Candida glabrata secretome. Proteomics. 2010;10:212–223. doi: 10.1002/pmic.200800706. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Tekaia F, Dujon B, Richard GF. Detection and characterization of megasatellites in orthologous and nonorthologous genes of 21 fungal genomes. Eukaryot Cell. 2013;12:794–803. doi: 10.1128/EC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry A, Bouchier C, Dujon B, Richard GF. Megasatellites: a peculiar class of giant minisatellites in genes involved in cell adhesion and pathogenicity in Candida glabrata. Nucleic Acids Res. 2008;36:5970–5982. doi: 10.1093/nar/gkn594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Wolfe KH. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat Genet. 2005;37:777–782. doi: 10.1038/ng1584. [DOI] [PubMed] [Google Scholar]


