SUMMARY
Cell penetrating peptides (CPPs) like nona-arginine (9R) poorly translocate siRNA into cells. Our studies demonstrate that attaching 9R to ligands that bind cell-surface receptors quantitatively increases siRNA uptake and importantly, allows functional delivery of complexed siRNA. The mechanism involved accumulation of ligand-9R:siRNA microparticles on the cell membrane, which induced transient membrane inversion at the site of ligand-9R binding and rapid siRNA translocation into the cytoplasm. siRNA release also occurred late after endocytosis when the ligand was attached to the L isoform of 9R, but not the protease-resistant 9DR, prolonging mRNA knockdown. This critically depended on endosomal proteolytic activity implying partial CPP degradation is required for endosome to cytosol translocation. The data demonstrate that ligand attachment renders simple polycationic CPPs effective for siRNA delivery by restoring their intrinsic property of translocation.
INTRODUCTION
Cell penetrating peptides (CPPs) can translocate into cells in an autonomous and receptor-independent manner with low cytotoxicity and immunogenicity [reviewed in (Fonseca, et al., 2009; Gump and Dowdy, 2007)]. Many primary cell membranes that constitute an impermeable barrier to reagents that easily transfect cell lines are permeable to CPPs. CPPs are thus promising delivery vehicles for a vast range of biologically-functional cargo (Du, et al., 2011; Eguchi and Dowdy, 2009; El-Sayed, et al., 2009; Fawell, et al., 1994; Jones, et al., 2005; Looi, et al., 2011; Schwarze, et al., 1999). How CPPs enter cells and access the cytoplasm are areas of active investigation (Erazo-Oliveras, et al., 2012; Schwarze and Dowdy, 2000). Depending on CPP composition, concentration, cell type, and the experimental conditions used, energy-independent as well as endocytic pathways are involved in CPP uptake and translocation (Duchardt, et al., 2007; Fischer, et al., 2004; Fretz, et al., 2007; Futaki, et al., 2007; Hirose, et al., 2012; Madani, et al., 2011; Payne, et al., 2007; Rothbard, et al., 2005; Verdurmen, et al., 2011; Wadia, et al., 2004). Cationic arginine-rich CPPs (R-CPPs) can non-covalently complex with small interfering RNA (siRNA). Despite their typical ability to effectively translocate biological macromolecules, R-CPPs are poor vehicles for cytoplasmic delivery of siRNA. For measurable mRNA knockdown, a huge excess of CPP molecules and high siRNA concentrations (above the therapeutic range) and/or association with reagents that disrupt endosomes is generally necessary (Akita, et al., 2010; Cantini, et al., 2013; El-Sayed, et al., 2009; Endoh and Ohtsuki, 2009; Erazo-Oliveras, et al., 2012; Lee, et al., 2008; Margus, et al., 2012; van Asbeck, et al., 2013; Zhang, et al., 2014). Imaging studies reveal the vast majority of CPP-siRNA complexes trapped for extended time periods in intracellular vesicles with little or no cytoplasmic localization (Al-Taei, et al., 2006; El-Sayed, et al., 2009; Erazo-Oliveras, et al., 2012; Fretz, et al., 2007; Fuchs and Raines, 2004; Maiolo, et al., 2005; Verdurmen, et al., 2011). R-CPPs have thus been utilized more as co-formulants for enhancing transfection efficiencies, rather than as primary components of synthetic siRNA delivery systems (Beloor, et al., 2012; Cheng and Saltzman, 2011; Kim, et al., 2010; Margus, et al., 2012; Nakamura, et al., 2007).
We previously used the homopolymeric R-CPP nona-D-arginine (9DR) for delivering electrostatically-complexed siRNA by covalently coupling 9DR to peptide/protein ligands that bind cell-surface receptors (Kumar, et al., 2008; Kumar, et al., 2007; Subramanya, et al., 2010). This ‘ligand-9R’ approach not only targeted siRNA specifically to cells bearing a receptor for the ligand, but also induced potent gene silencing (Kumar, et al., 2008; Kumar, et al., 2007). This raised the question of how ligand-attachment to 9R elicited functional siRNA delivery, which has been the formidable challenge to realizing the potential of siRNA therapeutics.
By correlating live-cell microscopy observations of siRNA localization with measurements of siRNA bioactivity, we determined that while native 9R peptides (D and L isoforms) lost their intrinsic ability to translocate upon siRNA complexation, ligand-9R enabled cytoplasmic siRNA delivery - (i) at the cell surface by tethering microparticles on the plasma membrane in a receptor-dependent manner, which led to membrane inversion at the site of binding and rapid siRNA translocation (ii) from late endosomes utilizing mechanisms that required endosomal protease activity. The latter occurred only when the L isoform of 9R was used (ligand-9LR) and prolonged the dynamics of gene silencing. Our results demonstrate that attachment to ligands restores the fundamental property of CPP translocation eliciting effective delivery of siRNA.
RESULTS
9R peptides effectively translocate covalently-attached molecules but not siRNA
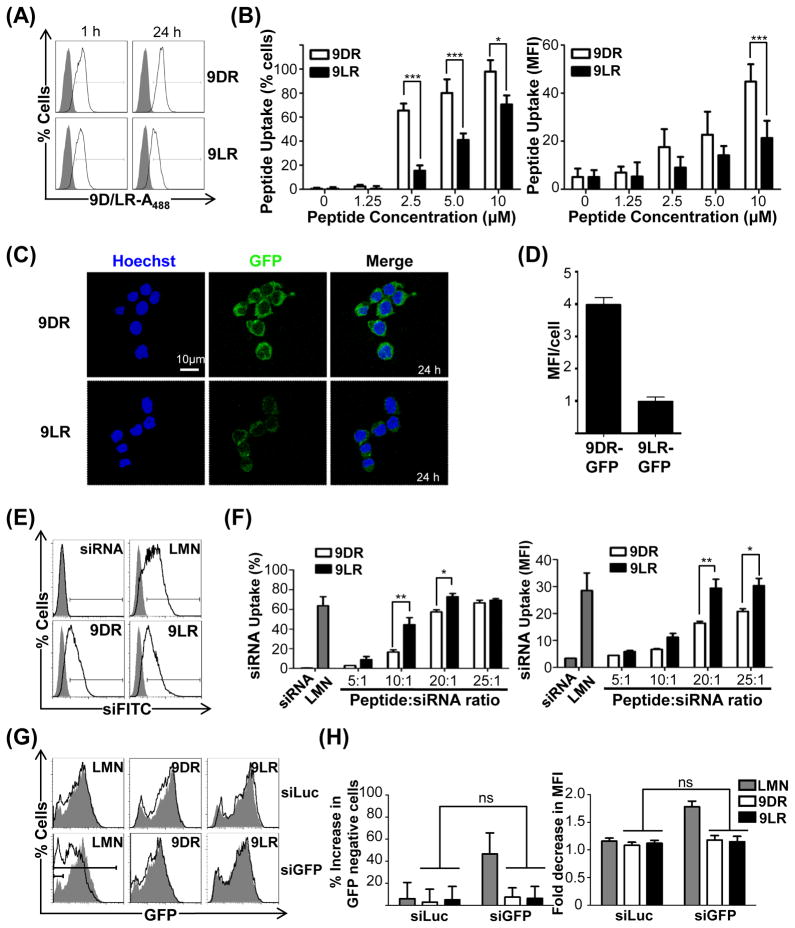
In preliminary analyses of 9R translocation, the murine neuroblastoma cell line, Neuro2a, was exposed to Alexa488 (~700 Da) labeled 9DR or 9LR. Both peptides became cell-associated within 1h and a significantly higher uptake occurred with 9DR by 24h (Figure 1A) in terms of cell numbers and levels per cell (Figure 1B, left and right panels respectively). 9D/LR peptides also translocated covalently-conjugated recombinant GFP protein (Figure 1C) and 9DR-GFP again accumulated to higher levels at 24h (Figure 1D). Thus covalent attachment of low molecular weight or macromolecular cargo to 9R did not hamper cellular uptake and 9DR displayed better translocation properties, keeping with previous reports documenting superior cell penetrating activity of protease-resistant D-oligomers of arginine (Kamei, et al., 2008; Tünnemann, et al., 2008; Verdurmen, et al., 2011; Wender, et al., 2000).
Figure 1. 9DR and 9LR ineffectively translocate siRNA.
(A/B) Flow cytometric analysis of Neuro2a cells after exposure to 10μM 9D/LR-A488. Representative histograms are shown in (A), and cumulative data in (B) depicting transfection efficiencies as percent cells (left panel) and mean fluorescence intensities (MFI, right panel). Filled histograms in (A) represent untreated cells. In (B) cells were scored as positive for uptake using the marker gate (black line) depicted in (A). (C) Live confocal microscopy of Neuro2a cells at 24h after incubation with 9D/LR-GFP conjugates. The nucleus is stained with Hoechst 33342 (blue). (D) Quantification of GFP fluorescence intensities in 9D/LR-GFP treated Neuro2A cells using the Volocity software (sample size −~50 cells). (E/F) Flow cytometric analysis of Neuro2a cells 24h after exposure to 9D/LR complexed with FITC-labeled siRNA (siFITC). Representative histograms for a peptide:siRNA molar ratio of 25:1 are shown in (E), and cumulative data depicting siRNA transfection efficiencies as percent cells (left panel) and average MFI (right panel) in (F). The filled histograms in (E) correspond to cells transfected with siFITC alone. In (F), cells were scored as positive for uptake using the marker gate (black line) depicted in (E). (G/H) Flow cytometric analysis of Neuro2a cells expressing GFP 72h after exposure to 9D/LR complexed with siGFP (100pmol siRNA, 10:1 peptide:siRNA). Representative histograms are shown in (G). The filled histograms correspond to cells transfected with siGFP alone. Cumulative data in (H) depict GFP silencing as percent increase in GFP-negative cells compared to cells treated with siRNA alone scored using the solid marker gate in (E) (left panel) and fold decrease in GFP MFI scored using the dashed marker gate in (E) (right panel).
LMN- Lipofectamine 2000, siLuc and siGFP- siRNAs targeting firefly luciferase and GFP mRNA. In all cases, error bars indicate SEM, *P < 0.05, **P < 0.01, ***P < 0.001 and ns- non-significant. See also Figure S1.
We next evaluated siRNA delivery by the cationic 9R peptide. siRNA was effectively complexed by 9D/LR peptides forming nanoparticles at peptide molar excesses of 10 and above (Figures S1A and S1B). However, transfection efficiencies in Neuro2A cells were relatively poor and comparable with the commercial transfection reagent Lipofectamine 2000 only at peptide:siRNA molar ratios ≥20:1 (Figures 1E and 1F). GFP siRNA complexed to 9DR or 9LR did not silence GFP expression in Neuro2a cells stably expressing the protein (Figures 1G and 1H). Therefore, 9R peptides poorly translocated siRNA, unless used at a high molar excess [>50, (Cantini, et al., 2013; Wang, et al., 2007; Zhang, et al., 2014)].
9R peptides effectively translocate siRNA when attached to a cell-binding ligand
RVG, a 32 amino acid peptide derived from the Rabies virus glycoprotein, binds nicotinic acetylcholine receptor (nAchR) subunits and directs tropism towards receptor-bearing cells (Kumar, et al., 2008; Kumar, et al., 2007; Subramanya, et al., 2010). When chimeric with RVG, both 9R peptides bound siRNA and formed nanoparticles (Figures S1C and S1D). Fusing 9R to another targeting ligand, a single chain antibody, also did not significantly change siRNA binding (data discussed below, Figures S1E and S1F).
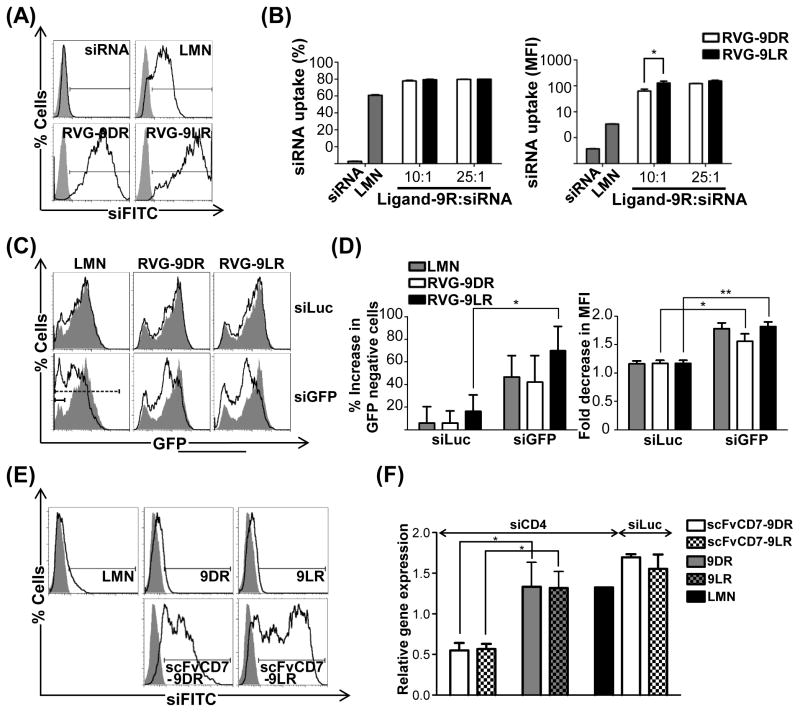
Attachment of 9R to RVG dramatically enhanced uptake of complexed siRNA in Neuro2a cells that express nAchR. Mean fluorescence intensities (MFIs) were at least 2 logs higher 24h following treatment [627±124 and 1527±121 for RVG-9DR and RVG-9LR respectively (Figures 2A and 2B) compared to 6.7±0.2 and 11.2±1.4 for 9D/LR peptides at a 10:1 peptide:siRNA ratio and 81±0.66 with Lipofectamine (Figures 1E and 1F)]. Importantly, RVG-9R:siRNA complexes effectively silenced gene expression (Figures 2C, 2D and S2) and here, siRNA complexed to RVG-9LR silenced GFP better than RVG-9DR and Lipofectamine (GFP-negative cells increased by 69.8 ± 2.7%, 42.1 ± 23.4% and 46.6 ± 19% respectively, Figure 2D). Thus, attachment of 9R to RVG likely enabled delivery of siRNA to cytoplasm, which is key for RNA interference [RNAi, (Wei, et al., 2011)].
Figure 2. 9D/LR effectively translocate siRNA when attached to a cell-binding ligand.
(A/B) Flow cytometric analysis of Neuro2a cells 24h after exposure to RVG-9D/LR:siFITC. Representative histograms at a peptide:siRNA ratio of 10:1 are shown in (A), and cumulative data for siRNA transfection efficiencies depicted as percent cells (left panel) and MFI (right panel) in (B). In (B), cells were scored as positive for uptake using the marker gate (black line) depicted in (A). (C/D) Flow cytometric analysis of Neuro2a cells expressing GFP 72h after exposure to siGFP complexed with RVG-9D/LR (100pmol siRNA, 10:1 peptide:siRNA). Representative histograms are shown in (C). Cumulative data for GFP silencing in (D) depict percent increase in GFP-negative cells (left panel) and fold decrease in GFP MFI (right panel) compared to cells treated with siRNA alone scored respectively using the solid and dashed marker gates in (C). (E) Flow cytometric analysis of Jurkat cells 24h after exposure to siFITC complexed to the indicated reagents. (F) QPCR analysis of CD4 mRNA levels in Jurkat cells 4h after treatment with scFvCD7-9D/LR:siCD4 (200pmol siRNA, 10:1 protein:siRNA).
In (A), (C) and (E), the filled histograms correspond to cells transfected with siRNA alone. LMN- Lipofectamine 2000, siLuc, siGFP and siCD4- siRNAs targeting firefly luciferase, GFP and human CD4 mRNAs respectively. In all cases, error bars indicate SEM, *P < 0.05, **P < 0.01. See also Figures S1 and S2.
Coupling 9D/LR to another targeting ligand - a murine single chain antibody scFvCD7 (Figures S1E and S1F) that binds the CD7 molecule expressed on human T cells (Kumar, et al., 2008), similarly enhanced siRNA uptake into the human T cell line Jurkat (MFIs of 70.83±26.05 and 104.6±12.27 with scFvCD7-9D/LR versus 18.60±0.82 and 20.97±0.99 for 9D/LR and 3.81±0.17 for Lipofectamine, Figure 2E) and appreciably silenced target CD4 mRNA levels as early as 4h post treatment (64.5±5.9% and 65.6±3.6% with siCD4 in comparison to siLuc complexed with scFvCD7-9D/LR, Figure 2F). Native 9D/LR peptides as well as Lipofectamine failed to elicit target mRNA knockdown, in keeping with the highly resistant nature of Jurkat cells to transfection. Thus, the observations were not unique to RVG-9R but a feature of the ligand-9R delivery platform. Attachment to a cell-binding ligand therefore transforms 9R, a poor siRNA delivery agent, into a highly effective siRNA carrier for gene silencing, even in cells resistant to conventional transfection methods.
RVG-9R-mediates cytoplasmic translocation of siRNA
The effective siRNA uptake and gene knockdown attained with RVG-9R unlike with 9R peptides suggests a trafficking pathway that ultimately resulted in cytoplasmic siRNA delivery. Two mechanisms are implicated in the cellular entry of CPPs and their attached cargo: energy-independent direct translocation at CPP concentrations of ≥ 10μM (Nakamura, et al., 2007; Rothbard, et al., 2005; Rydstrom, et al., 2011) and endocytosis mechanisms such as macropinocytosis, at ≤ 5μM (Duchardt, et al., 2007; Fischer, et al., 2004; Madani, et al., 2011; Payne, et al., 2007; Verdurmen, et al., 2011; Wadia, et al., 2004). The final concentration of the RVG-9R peptides in siRNA translocation experiments ranged between 2 and 5 μM, which is at or below the threshold value that elicits direct translocation into the cytosol. The requirement of a ligand for cellular entry was indicative of receptor binding and therefore receptor-mediated endocytosis. Incubation at 4°C reduced, but did not abolish, cellular uptake of RVG-9R:siFITC complexes indicating that temperature-independent mechanisms also play a role in ligand:9R uptake (Figure S3A).
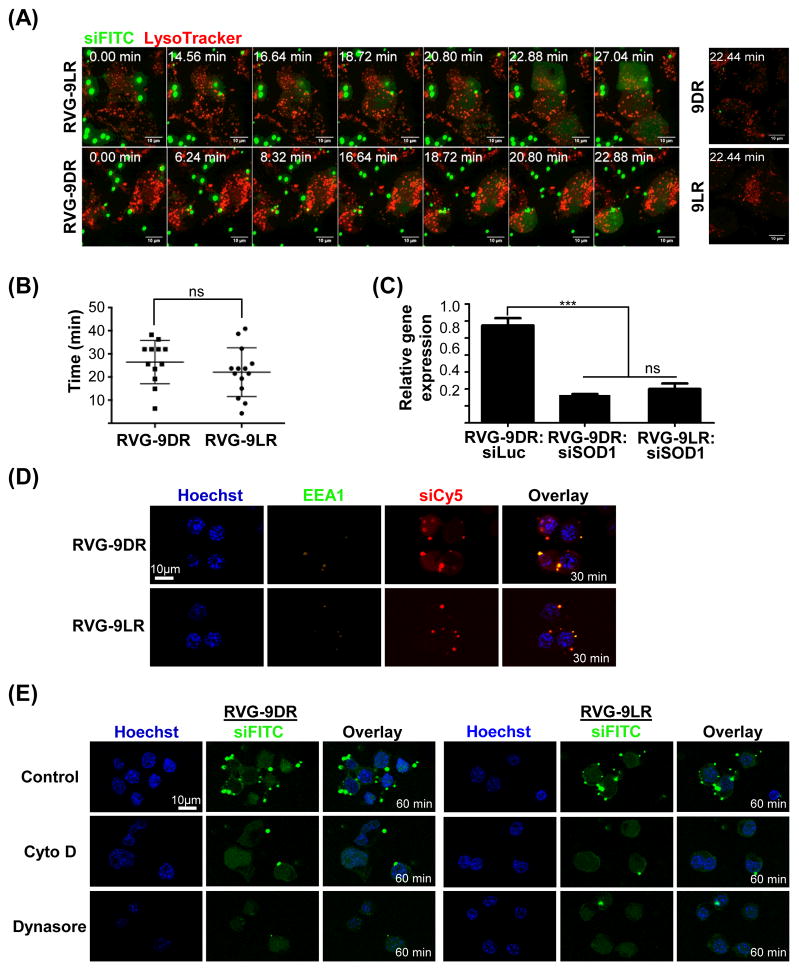
Time-lapse imaging of Neuro2A cells exposed to RVG-9R:siFITC complexes revealed multiple small particle-like structures ~1–3 μm in diameter accumulating and tethering on the plasma membrane (Figure 3A, see also Videos S1 and S2). Strong fluorescence (green) spread throughout the cytoplasm after influx of these particles, typically by about 20 minutes after treatment (26.4 ± 2.7 min and 22.1 ± 2.8 min with RVG-9DR/LR respectively, Figures 3A and 3B). This translated to significant target mRNA knockdown within an hour after transfection confirming early delivery of functional siRNA (Figure 3C). Some siRNA (red) colocalized with early EEA1+ endosomes (green, Figure 3D), however, late endosomes (labeled with the red LysoTracker dye) were completely devoid of siRNA (green, Figure 3A) at these early times. Pre-treatment with the actin depolymerizing agent cytochalasin D or the dynamin inhibitor dynasore did not significantly affect cytoplasmic siRNA fluorescence at 1h post-exposure to peptide-siRNA complexes (green, Figure 3E) although it reduced intracellular punctate fluorescence in cells exposed to transferrin (red, Figure S3B). These data indicate that RVG-9R:siRNA delivery to the cytoplasm occurs rapidly in an actin/dynamin independent manner across the plasma membrane or from very early endosomes.
Figure 3. Early cytoplasmic entry of RVG-9R:siRNA is actin/dynamin independent.
(A) Time-lapse images of Neuro 2A cells pre-stained with LysoTracker (red) acquired within 1h of exposure to peptide:siFITC complexes. Fluorescent and differential interference contrast (DIC) images in the same fields were merged. Time 0: images captured immediately after the addition of siRNA complexes. (B) Quantification of time (minutes) to appearance of cytoplasmic fluorescence in Neuro2a cells treated with RVG-9D/LR:siFITC. Each dot represents a cell. Mean and SD are depicted. (B) QPCR analysis of SOD-1 mRNA levels in Neuro2a cells 1h after treatment with RVG-9D/LR:siRNA. Error bars indicate SEM. (D) Confocal microscopy of Neuro2a cells 30 min after incubation with RVG-9D/LR:siCy5 (red) and stained for the early endosome marker EEA1 (green). Nuclei were stained with Hoechst 33342 (blue). (E) Live confocal microscopy images of Neuro2a cells pre-treated with Cytochalasin D (Cyto D) or Dynasore 1h after incubation with RVG-9D/LR:siFITC complexes (green). Nuclei were stained with Hoechst 33342 (blue). 100pmol siRNA and 10:1 peptide:siRNA was used in all cases. See also Figure S3 and Videos S1–S4.
RVG-9R:siRNA complexes induce topical membrane inversion
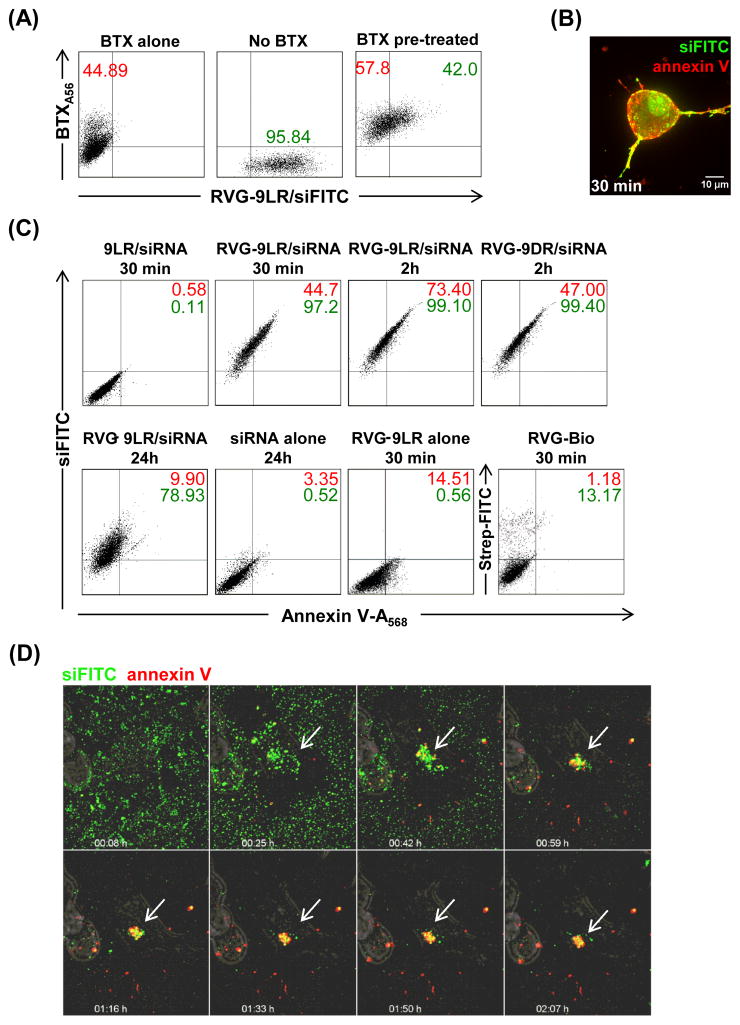
As 9D/LR:siRNA complexes did not associate with the plasma membrane for longer times (Figure 3A- extreme right, no microparticles visualized by 22 minutes) or lead to vesicular or cytoplasmic fluorescence (See Videos S3 and S4), ligand-receptor binding appeared critical for initiating cellular entry of RVG-9R:siRNA complexes. Indeed, pretreatment of Neuro2A with α-bungarotoxin, a competitive inhibitor of RVG (Kumar, et al., 2007) significantly reduced uptake of RVG-9LR/siFITC (Figure 4A). We investigated if plasma membrane inversion, a mechanism identified in the energy-independent entry of 9R into cells (Hirose, et al., 2012), occurred at the site of ligand-9R/siRNA-receptor binding. Exposure of cells treated with RVG-9LR:siRNA complexes resulted in strong co-staining with Alexa568-labeled annexin V within 30 minutes indicating membrane inversion (Figures 4B and 4C). Strong annexin V positivity persisted for upto and beyond 2h in cells treated with RVG9D/LR:siRNA (Figure 4C); surprisingly however, this did not result in a loss of membrane integrity or cell cytotoxicity (measured by a lack of lactate dehydrogenase in culture medium and the Alamar Blue cytotoxicity assay respectively, data not shown). Accordingly, most cells were no longer annexin V positive at 24h, but continued to remain positive for siFITC signifying membrane inversion was temporary (Figure 4C). Annexin V signals were not detected on cells treated with RVG peptide lacking 9R or 9LR:siRNA complexes indicating that ligand attachment as well as the presence of the CPP was essential for membrane inversion in the context of complexed siRNA. Cells exposed to RVG-9LR alone were also positive for annexin V, albeit to a lesser extent, indicating that polyplex formation in the presence of siRNA increased exposure of the inner membrane.
Figure 4. Ligand-9R induces localized membrane inversion.
(A) Flow cytometric analysis of Neuro2a cells pre-treated with A568-labeled α-bungarotoxin (BTX) 30 min after exposure to RVG-9LR:siFITC (100pmol siRNA, 10:1 peptide:siRNA). Numbers in red and green represent cell percentages positive for BTX and siRNA respectively. (B) Live confocal microscopy of Neuro2a cells 30 minutes after exposure to RVG9D/LR:siFITC (green) and Alexa568-labeled annexin V (red). Fluorescent images in the same fields were merged. (C) Flow cytometric analysis of Neuro2a cells treated with A568-labeled annexin V after exposure to the indicated reagents for the indicated times. Numbers in red and green represent cell percentages positive for annexin V and siFITC respectively. (D) Time-lapse images of Neuro2A cells pre-incubated with A568-labeled annexin V and siFITC at the indicated times after exposure to RVG-9LR. Fluorescent and DIC images in the same fields were merged. Arrows indicate sites of peptide:siRNA binding. See also Video S5.
We next resorted to generating siRNA complexes in situ by adding RVG-9R to Neuro2A cells in medium containing siRNA rather than using pre-formed siRNA complexes, as the kinetics of membrane inversion with the latter were too rapid to be captured in movie format. This strategy enabled clear visualization of peptide/siRNA complexes assembling at a reduced rate (~1–2 h) but also significantly reduced subsequent cytoplasmic fluorescence. For these experiments, we restricted our investigations to RVG-9LR as the two peptides appeared to display comparable dynamics of cellular entry and similarly induced annexin, at least at early time points. Annexin V staining (red) was highly localized to the site of RVG-9LR/siFITC binding (green), and appeared to occur even from vesicle-like structures within the cell (Figure 4D - arrows, see also Video S5). No annexin V-staining was detected in the presence of α-bungarotoxin, indicating that ligand-receptor binding and not direct CPP interactions with the cell membrane initiates the mechanism even with siRNA complexes formed in situ (data not shown). These observations provide evidence that ligand-9R induces transient membrane inversion, at the regions corresponding to the ligand-9R binding sites and suggests that receptor binding by ligand-9R increases CPP concentration in a localized manner to above the threshold required for eliciting membrane flipping.
The L-isoform of 9R enables cytoplasmic siRNA delivery by RVG-9R from endosomes
Energy-requiring mechanisms such as receptor-mediated endocytosis also possibly contribute to cellular uptake (Figure S3A). We investigated if ligand-binding induced receptor aggregation. It is not known which of the nAchR subunits the RVG peptide binds. We therefore expressed the α6-subunit fused with mCherry, which in recombinant form is known to localize, traffic and associate with α4 and β2 subunits of the nAchR expressed in Neuro2A cells (Drenan, et al., 2008). Clearly, membrane assembly of ligand-9R:siRNA complexes, generated by adding RVG-9LR into medium containing siFITC, aggregated membrane receptors evidenced by labeled nAchR receptor subunits (red) coalescing with RVG-9LR:siRNA complexes (green) (Figure S4A, see also Video S6).
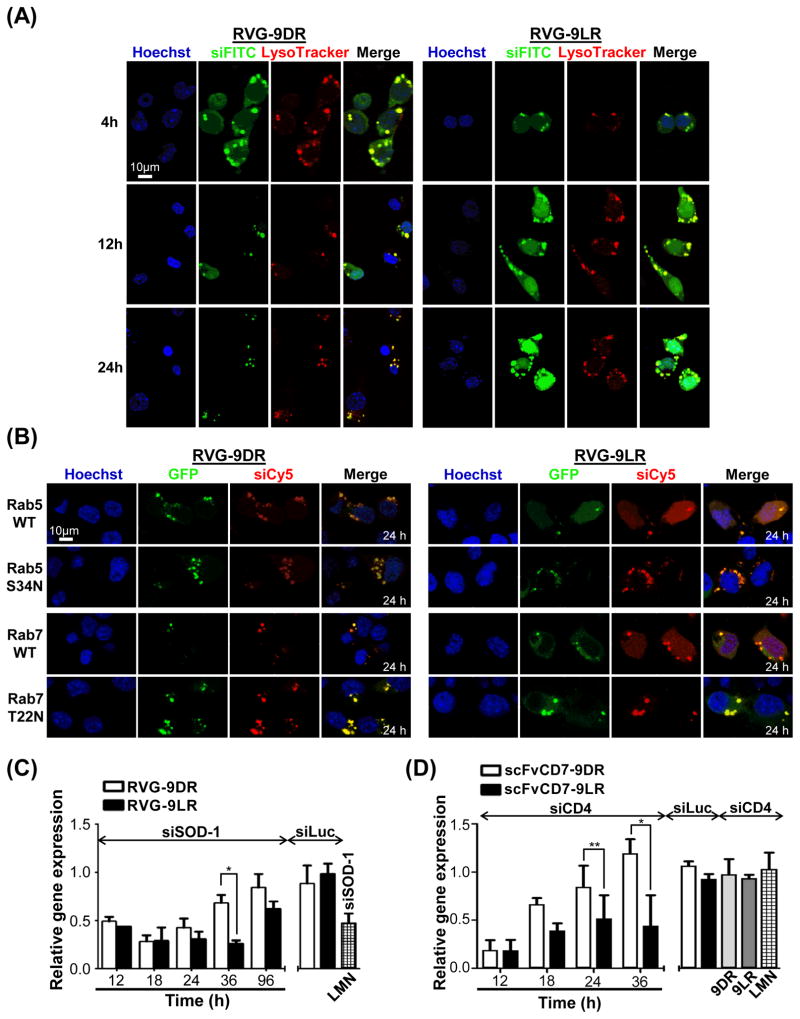
Live cell microscopy at later time points after transfection revealed siRNA (green) in the cytoplasm as well as in LysoTracker-positive (red) endocytic vesicles indicating that RVG-9R-siRNA complexes also get endocytosed and traffic within vesicles that mature into late endosomes (Figure 5A). Even nuclear fluorescence was evident in keeping with previous reports of cytoplasm to nuclear transport of 9R (Zaro, et al., 2009). 9D/LR:siRNA complexes were also endocytosed but completely resident within vesicles at these times (Figure S4B) accounting for the uptake but insignificant target mRNA knockdown observed in Figure 1. At 24h, RVG-9DR:siRNA treated cells had a markedly punctate fluorescence distribution with increased ‘arrest’ of green siRNA within late endosomal compartments with little/no cytoplasmic staining (Figure 5A). On the other hand, prominent cytoplasmic fluorescence was still evident in RVG-9LR:siRNA transfected cells. Image analysis revealed cytoplasmic siRNA in about 40% of RVG-9LR transfected cells in contrast to <10% of RVG-9DR treated cells at 24h (Figure S4C).
Figure 5. Ligand-9LR enables siRNA release from endosomes prolonging mRNA knockdown.
(A) Live confocal microscopy of Neuro2a cells pre-stained with LysoTracker (red) at 4, 12 and 24h after exposure to RVG9D/LR:siFITC (green). (B) Neuro2a cells over-expressing wild type (WT) or dominant negative (S34N, T22N) Rab5- or Rab7-GFP (green) were analyzed 24h after exposure to RVG-9D/LR:siCy5 (red). Nuclei are stained with Hoechst 33342 (blue). (C) QPCR analysis of SOD-1 mRNA levels in Neuro2a cells treated with RVG-9D/LR:siSOD1 at the indicated times post transfection. (D) QPCR analysis of CD4 mRNA levels in Jurkat cells treated with scFvCD7-9D/LR:siCD4 at the indicated times post transfection In all cases 100 pmol siRNA, 10:1 peptide:siRNA was used except in (D) where siRNA amounts equaled 200 pmol. LMN- Lipofectamine 2000, siLuc- siRNA targeting firefly luciferase. Error bars indicate SEM, *P < 0.05, **P < 0.01. See also Figure S4 and Video S6.
These data suggested that siRNA release continues to occur after endosome maturation from intracellular vesicles with RVG-9LR:siRNA. To explore this, we expressed GFP-reporter tagged wild-type and dominant mutant Rab GTPases, Rab5 S34N the Rab7 T22N that block early and late endosome maturation (Barbieri, et al., 2000; Feng, et al., 1995). Expression of the mutant GFP-Rab proteins did not alter the dominantly endosomal localization of siRNA (red) at 24h in Neuro2a cells treated with RVG-9DR:siCy5 (Figures 5B and S4D). On the other hand, in RVG-9LR:siCy5 exposed cells, inhibition of endosomal maturation resulted in an almost complete accumulation of red fluorescence within vesicles and a marked reduction in cytoplasmic fluorescence when compared with cells expressing wild-type Rab5 or Rab7. Thus, while cytoplasmic siRNA delivery by RVG-9DR is restricted to a limited time early after transfection by the membrane-flipping mechanisms, RVG-9D/LR:siRNA extends siRNA release from endosomes and vesicle maturation actively contributes to this.
We carried out a temporal analysis of gene knockdown in Neuro2a cells to determine if the clear-cut differences in cytoplasmic siRNA fluorescence with the two RVG-9R peptides at later time points after transfection was reflected in siRNA bioactivity. RVG-9R:siSOD1 effectively reduced target mRNA levels (SOD-1) for up to 24h, and a marginally higher knockdown was elicited with RVG-9LR:siRNA (Figure 5C). While cells treated with RVG-9DR:siSOD1 began to re-accumulate SOD-1 mRNA at 36h, significant knockdown persisted in RVG-9LR:siSOD1 treated cells in accordance with the persisting cytoplasmic siRNA fluorescence at 24 h (Figure 5B). siRNA effects ceased by about 96 h given the rapid doubling time of Neuro2A cells (~0.8 day), which governs intracellular siRNA concentration and thus duration of gene silencing (Bartlett and Davis, 2006). A similarly prolonged silencing activity to beyond 36 h was also noted with scFvCD7-9LR delivered siCD4 in Jurkat cells while mRNA knockdown with scFvCD7-9DR:siCD4 was short-lived (Figure 5D). Thus, although early siRNA translocation is dependent on both ligand and CPP functions, the CPP, through its stereochemistry, critically influences siRNA release from late endosomes.
siRNA release from endosomes is dependent on proteolytic activity
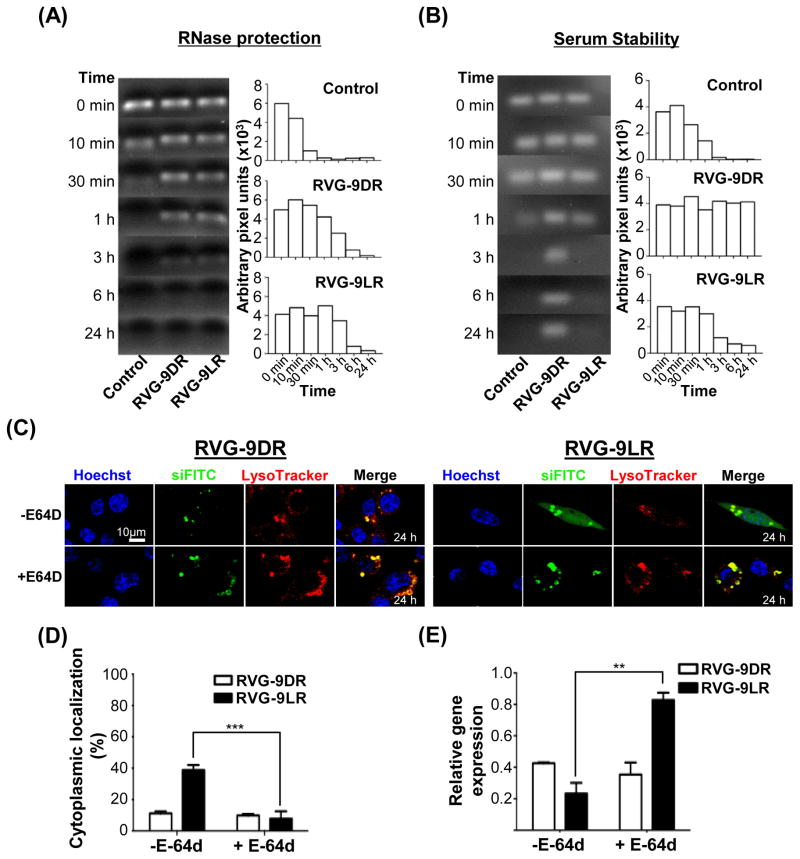
The above observations imply that sensitivity of 9LR, but not 9DR, to late endosomal proteases may contribute to siRNA release into the cytoplasm. siRNA complexed with the two RVG-9R peptides was stable at 37°C for up to 3h of exposure to RNase A in serum-free medium while naked siRNA was degraded rapidly within 30 minutes demonstrating shielding of complexed siRNA from nucleases (Figure 6A). However, recovery of intact siRNA from RVG-9LR:siRNA complexes incubated in human serum containing medium was significantly reduced by 3h while siRNA associated with RVG-9DR was intact for as long as 24h (Figure 6B). This suggested that serum protease-mediated degradation of 9LR in RVG-9LR may have exposed siRNA to serum nucleases while the protease resistant 9DR isoform continued to protect complexed siRNA. We explored whether a similar scenario existed in the late endosome. We treated Neuro2a cells with the membrane permeable inhibitor E-64d that blocks endosomal cysteine peptidase activity. This dramatically accumulated fluorescence in vesicles and reduced cytoplasmic siRNA localization in RVG-9LR/siRNA treated cells in comparison to untreated cells at 24h post transfection (Figures 6C and 6D). Further, E-64d treatment also selectively and significantly affected mRNA knockdown by RVG-9LR/siRNA indicating that some level of degradation of the 9R carrier in the late endosome may be a dominant factor contributing to the overall efficiencies of RNAi achieved by ligand-9LR delivery systems (Figure 6E).
Figure 6. Endosomal proteolysis is required for siRNA release from late endosomes.
(A and B) Stability of RVG-9D/LR-siRNA complexes after exposure to RNase A (A) and 50% human AB serum (B) for the indicated periods of time. The bar graphs represent corresponding band intensities estimated as arbitrary pixel units using the Image J software. Control refers to siRNA treated identically in the absence of ligand-9R. (C and D) Fluorescence distribution in E-64d-treated Neuro2a cells 24h after exposure to RVG-9D/LR:siFITC. Nuclei were stained with Hoechst 33342 (blue) and endosomal vesicles with LysoTracker (red). Data in (D) were calculated using the Volocity software and a sample size of 50 cells. (E) Analysis of murine SOD-1 mRNA levels in Neuro2a cells 36h after exposure to RVG9D/LR-siSOD1 (100pmol siRNA, 10:1 peptide:siRNA). Error bars indicate SEM. **P<0.01, ***P<0.001.
Collectively, these data demonstrate that cell-binding ligands can transform 9R peptides into effective siRNA delivery agents by eliciting trafficking mechanisms that result in cytoplasmic delivery of siRNA (Figure 7).
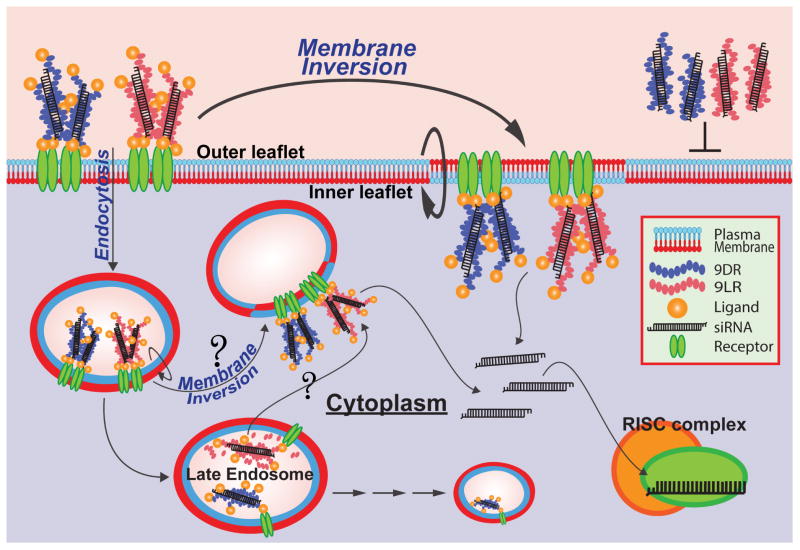
Figure 7. Scheme illustrating the mechanism of ligand-9R transloaction.
The path taken by ligand-9R from the cell exterior to the cytosol begins with binding of the ligand to its cell-surface receptor. Cellular entry then proceeds by localized membrane inversion at the site of ligand-9R microparticle binding and accumulation on the plasma membrane and/or within early endosomes. siRNA then quickly translocates across the membrane and diffuses in the cytoplasm. This pathway is the major route of cytosolic delivery for both ligand-9D/LR:siRNA complexes. siRNA translocation from late endosomes after endocytosis is a second, more slow and controlled route that results only when siRNA is complexed to ligand-9LR. Blocking vesicle maturation or inhibiting endosomal proteases interferes with the process indicating that some level of proteolytic degradation of the ligand-9LR carrier is required. Late endosome to cytosol translocation could occur by ‘membrane-flipping’ or other such mechanisms. 9D/LR complexes are not capable of effectively engaging either route for cellular entry unless used at very high concentrations.
DISCUSSION
Cationic CPPs like 9R form nano-sized particles with negatively charged siRNA. However, when used in this manner for ‘actively’ transporting siRNA into cells, gene knockdown is not very effective as siRNA is poorly transferred to the cytoplasm (Endoh and Ohtsuki, 2009; Margus, et al., 2012). We investigated mechanisms whereby attachment to cell-binding ligands restores CPP function transforming 9R peptides into potent agents for siRNA delivery.
Ligand attachment to 9R increased siRNA uptake by more than two orders of magnitude. Ligand-receptor binding was vital for uptake, however mechanistic translocation of siRNA critically depended on 9R. Live confocal microscopy revealed fluorescent ligand-9R:siRNA complexes tethering as microparticles on the plasma membrane whose influx resulted in rapid cytoplasmic distribution of siRNA fluorescence and mRNA knockdown within 1h. The process had little dependence on actin/dynamin function. Annexin V staining revealed topical inversion of cell membrane focused at the site of ligand-9R:siRNA binding. These observations are strikingly consistent with a study that found dodeca-arginine (R12) induced microparticle formation on cell membranes and subsequent inversion of the lipid bilayer at these spots influxed R12 directly into the cell cytoplasm (Hirose, et al., 2012). The process was energy/temperature-independent and not affected by cytochalasin D or dynasore. The requirement of the ligand for 9R-induced membrane inversion when 9R is associated with siRNA suggests that tethering of the CPP in close proximity to the cell surface through receptor binding, which in turn increases CPP concentration locally, is required for eliciting membrane inversion. Alternatively, conjugation of hydrophobic moieties to R12 increased affinity for the plasma membrane favoring the formation of multimeric particles at the cell surface (Hirose, et al., 2012). Thus, ligand attachment, in addition to mediating cell-binding, could be promoting amphiphilicity of 9R to induce translocation through similar mechanisms.
Ligand-9R:siRNA complexes also trafficked through Rab5+ and Rab7+ endosomes. Octa-arginine (R8), at <5μM concentrations, is known to enter cells by macropinocytosis (Appelbaum, et al., 2012) after binding to membrane proteoglycans and phospholipids (Erazo-Oliveras, et al., 2012; Verdurmen, et al., 2011). However, the evidence of ligand-induced receptor-aggregation suggests that the endocytic trafficking pathway elicited is a consequence of receptor-mediated endocytosis. The nuclear siRNA fluorescence is also in agreement with observations that strongly endocytosed CPPs, which can result from attachment to endocytosed ligands, result in nuclear transport (Zaro, et al., 2009). The endocytic route also contributed to cytosolic delivery of siRNA at later times, but only when ligand-9LR was used for delivery. It is possible that fluorophore released by nuclease-degraded siRNA that has dissociated from ligand-9LR due to protease action could have contributed to cytosolic fluorescence. However, prolonged mRNA knockdown indicated functional siRNA activity in the cytosol and this was not the case with ligand-9DR:siRNA despite its accumulation within LysoTracker-positive late endosomes. Therefore, endosomal sequestration, the major limiting factor for CPP-mediated siRNA delivery was appreciably overcome using ligand-9LR.
Our investigations into siRNA binding by RVG-9R peptides reveal strong association and tight complex formation between the two at low pH (Figure S4E). In addition, the ligand-CPP-siRNA complex is no longer tethered to the membrane and dissociated from the receptor at low pH within late endosomal compartents. This likely hinders membrane flipping or other mechanisms like ‘pore formation’, which require multidentate hydrogen bonds between the guanidinium side group in 9R and the negatively charged phospholipids enriched in the intraluminal lipid bilayers of late endosomes. (Kobayashi, et al., 2001; Matsuo, et al., 2004; Mishra, et al., 2011; Yang, et al., 2010; Ziegler, 2008). Disruption/destabilization of ligand-9LR:siRNA by late endosomal proteases could be requisite to promote CPP-membrane interactions for continued release of siRNA. In keeping with our data, substitution of R-CPPs and oligo-L-arginines with D-amino acids, at the terminal positions, obstructed degradation by endosomal proteases reducing cytoplasmic localization of CPP, bioactivity of attached cargo, and transfection efficiencies (Abes, et al., 2008; Fischer, et al., 2010; Mason, et al., 2007; Verdurmen, et al., 2011; Wu, et al., 2007). The association of RISC machinery components with intraluminal vesicles and membranes of the late endosome (Lee, et al., 2009; Saleh, et al., 2006; Siomi and Siomi, 2009) also raises the possibility that ligand-9LR instability, in combination with ionic competition for nucleic acid binding at lower pH, may displace siRNA for direct loading onto the endosomal RISC enhancing the dynamics of RNA silencing in the cytoplasm.
Hydrophobic components like stearyl and cholesteryl groups, lipid membranes and other agents that promote endosomal escape have conventionally been used to improve cytosolic delivery of R-CPPs and their cargo (El-Sayed, et al., 2009; Endoh and Ohtsuki, 2009; Hayashi, et al., 2011; Kim, et al., 2010; Kim, et al., 2006; Nakamura, et al., 2007; Nakase, et al., 2004; Tonges, et al., 2006). Our data demonstrates that attachment of 9R and possibly other CPPs to cell-binding ligands can overcome the key rate-limiting step of delivery to the cytosol as depicted schematically in Figure 7. We hypothesize that ligand-receptor binding induces localized accumulation of microparticle-like structures on the plasma membrane, concentrating CPPs to a threshold level that induces siRNA translocation into the cytosol across the membrane from the exterior and/or from endosomes. The mechanism involved is CPP-mediated membrane flipping, which is actin/dynamin independent when it occurs at the plasma membrane. This constitutes the major route of cytosolic delivery. Late endosome to cytosol is a second, more controlled route, that extends the kinetics of gene silencing through proteolytic disruption of ligand-9LR:siRNA complexes promoting membrane-CPP interactions. 9D/LR:siRNA complexes are not capable of effectively engaging either route for cellular entry unless used at very high concentrations.
As the ligand-9R strategy relies on surface receptor expression, it can be effectively employed for primary and suspension cells independent of confluence under conditions identical to those established in cell lines. Through the use of a ligand that confers cell-specificity, the approach has the benefit of concentrating siRNA activity within select cells enabling reduced dosing, key for in vivo treatment potency. Alternatively, ligands that bind receptors on multiple cell types, e.g., epidermal growth factor transferrin and those of the integrin family, can expand applicability to whole cell populations (Ming, 2011). This flexibility makes the ligand-9R a compelling approach for harnessing the therapeutic potential of siRNA.
SIGNIFICANCE
Cationic cell penetrating peptides (CPPs) like nona-arginine (9R) that translocate covalently-attached biomolecules into the cellular cytoplasm poorly deliver electrostatically-complexed siRNA. Our study highlights the proficiency of 9R in functional siRNA delivery when chimeric with peptide and protein ligands (ligand-9R) that bind cellular receptors. By correlating siRNA bioactivity with live-cell confocal and time-lapse microscopy observations, we conclude that ligand-9R primarily enables rapid siRNA translocation into the cytosol through microparticles that accumulate on cell membranes upon ligand attachment to the receptor inducing localized topical membrane inversion. siRNA is also released into the cytosol from late endosomes after receptor-mediated endocytosis of ligand-9R complexes, but only with the L isoform of 9R (ligand-9LR), and is critically dependent on endosomal protease activity. siRNA complexed to ligand-9LR efficiently prolongs target gene mRNA knockdown even in cells resilient to transfection like human T cells. siRNAs are an indispensable analytical platform for biological target validation in vitro. siRNAs have however been under-exploited as a therapeutic platform as effective siRNA delivery systems are lacking. Our studies highlight that 9R and possibly other CPPs can be harnessed for effective siRNA-transport by routing them to receptors using cell-binding ligands as this initiates a pathway that synergizes cellular uptake with mechanisms that lead to cytosolic release- a critical requirement for the functional outcome of siRNA.
EXPERIMENTAL PROCEDURES
siRNA transfection experiments
9D/LR or RVG-9D/LR was incubated with siRNA (100 pmol) in serum-free DMEM at molar ratios ranging between 5 and 25 for 20min at room temperature and added to Neuro2a cells cultured in DMEM-10 at final concentrations between 1 and 5μM. Experiments with scFvCD7-9D/LR used 5μM protein on human Jurkat T cells. Carrier to siRNA ratio was maintained at 10:1 unless otherwise mentioned. Lipofectamine 2000 (Invitrogen) was used according to manufacturer’s instructions. For inhibiting actin/dynamin activity, Neuro2a cells were pre-treated for 1h with 5μM Cytochalasin D (Sigma) or 80μM Dynasore (Sigma). For inhibiting endosomal proteases, Neuro2a cells were pre-treated for 1h with 40μM E-64d (Sigma) and chased in medium containing E-64d after addition of peptide:siRNA complexes. For blocking nAchR, Neuro2a cells were pre-treated for 30 min on ice with 10−5M A568- labeled α-bungarotoxin prior to addition of peptide:siRNA complexes. Mammalian expression plasmids encoding (i) wild-type Rab5, Rab7, Rab5(S34N) and Rab7(T22N) fused in frame to eGFP (ii) α4, β2 and α6 (the latter fused to mCherry) subunits of the mammalian nAchR were transfected into Neuro2a cells using Lipofectamine 2000 48h prior to treatment with RVG-9R:siRNA complexes.
Gene silencing was analyzed by flow cytometry for reporter GFP expression or qPCR for murine SOD1 and human CD4 mRNA levels (Kumar, et al., 2008; Kumar, et al., 2007) normalizing to murine GAPDH and human beta-actin respectively.
Confocal Microscopy
Neuro2a cells seeded and cultured in complete medium on poly-L-lysine (Sigma-Aldrich) treated coverslips were incubated with 9D/LR-GFP (1μM) or peptide:siRNA complexes (300pmol siRNA, final concentration- 3μM, 10:1 molar ratio). Confocal microscopy was performed in most cases, without washing or fixing, using Volocity spinning disc Nikon TE2000 confocal microscope equipped with an environmental chamber (LIVE CELL; Pathology Devices) and a Nikon Perfect Focus with a 60X Nikon objective (ND 1.4) or a Leica TCS SP5 Spectral Confocal Microscope at 63X magnification. Hoechst 33342 was used to distinguish nuclei, LysoTracker Red to stain late endosomal vesicles. For experiments visualizing membrane-interactions of ligand-9R:siRNA complexes, siFITC (300 pmol, final concentration- 3μM) and/or Alexa568-labeled annexin V was first added to Neuro2A cells followed by addition of RVG-9LR (10:1 peptide:siRNA ratio). Early endosomes were detected in cells fixed with 4% paraformaldehyde by staining with rabbit anti-EEA1 and mouse anti-rabbit FITC (green) antibodies. All images were processed using Volocity 6.2.1 software (Improvision/PerkinElmer). Quantification of image intensities was performed with Image J 1.49b software.
Ribonuclease A and serum protection assays
Peptide:siRNA complexes (100 pmol siRNA, 10:1 ratio) were incubated at 37°C in 100ng RNase A or 50% human AB serum (not heat inactivated) contained in DMEM. Timed aliquots were treated with proteinase K (50μg/ml, 10min at 25°C) and RNA extracted with phenol chloroform-isoamyl alcohol (25:24:1 v/v). All samples were subjected to electrophoresis on a 2% agarose gel with ethidium bromide. Band intensities estimated using the Image J software.
Statistical Analysis
Results are presented as mean ± SD or mean ± SEM as indicated. Statistical significance was calculated using data from three independent experiments using one-way ANOVA with Bonferroni’s multiple-comparison test for pairwise comparisons within groups. Otherwise, comparisons were made using a two-tailed t test.
Supplementary Material
Figure S1, related to Figures 1 and 2. 9R and RVG-9R bind siRNA to form nanoparticles. Electrophoretic gel mobility shift assay of 100pmol siRNA complexed to 9D/LR (A), RVG-9D/LR (C) and scFvCD7-9D/LR (F) at the indicated molar excess of peptide/antibody. Note: the RVG peptide by itself does not bind siRNA in (C). (B/D) Average particle diametric size distribution in nanometer (ZAve) and surface charge distribution (ζ) of peptide-siRNA complexes (100 pmol siRNA, 10:1 peptide:siRNA) measured by dynamic light scattering. (E) MALDI-TOF MS spectrum of scFvCD7-9D/LR conjugates.
Figure S2, related to Figure 2. Functional siRNA delivery by RVG9R. QPCR analysis of SOD-1 mRNA levels in Neuro2a cells 4h after exposure to siRNA complexes (100 pmol siRNA, 10:1 peptide:siRNA).
Figure S3, related to Figure 3. Analysis of cellular uptake mechanisms of RVG-9R:siRNA. (A) Flow cytometric analysis of Neuro2a cells exposed to RVG9D/LR:siFITC (10:1 peptide:siRNA molar ratio) after incubation at the indicated temperatures. The filled histograms correspond to cells transfected with siFITC alone and the values represent MFIs of the open histograms. The graph on the right is a quantitation of reduction in siRNA uptake measured as percent fold reduction in MFIs at 4°C compared to that at 37°C. (B) Confocal images of Neuro2a cells demonstrating pretreatment with inhibitors cytochalasin-D or dynasore inhibits internalization of A568-labeled transferrin.
Figure S4, related to Figure 5 and Video S6. (A) Time-lapse images of Neuro2A cells co-expressing α6-mCherry, α4 and β2 subunits of nAchR pre-incubated in medium containing siFITC at the indicated times after exposure to RVG-9LR. Fluorescent and DIC images in the same fields were merged. Arrows indicate site of peptide:siRNA binding and receptor aggregation. (B) Confocal microscopy of Neuro2a cells 24h after incubation with 9D/LR:siFITC complexes. Nuclei are stained with Hoechst 33342 (blue). (C and D) Analysis of intracellular fluor distribution in Neuro2A cells in Figures 5A and 5B at 24h. Data shown is cumulative of 3 independent experiments and represents cell numbers as percentages calculated with a sample size of ~50 cells. Error bars depict SEM. ***P<0.001. (E) Ethidium bromide (EtBr) exclusion assay conducted with peptide:siRNA complexes (400 pmol siRNA, 2.5:1 peptide:siRNA ratio) in phosphate buffers at the indicated pH values. The plot shows the amount of free siRNA (not complexed with peptides) detected in solution using EtBr. The fluorescence value for free siRNA without peptide at each pH value tested was set to 100%. Note: The 9LR peptide associates more strongly with siRNA than RVG-9LR suggesting poor availability to interact with cellular membranes. This was also reflected in the observation that ligand attachment lowered the average zeta potential of siRNA complexes suggesting a weaker association of siRNA in the ligand-9R format (Figure S1). Reducing the pH to 5.0 increased association of RVG-9LR with siRNA making it comparable to 9LR at pH 6.5 in this assay. Thus, under low pH conditions in late endosomes, RVG-9R-siRNA complex may need complex destabilization by action of endosomal proteases to promote membrane interaction and translocation of siRNA into the cytosol.
Supplementary Videos S1-S4, related to Figure 3. RVG-9R induces rapid cytosolic translocation of siRNA. Representative time-lapse videos of Neuro2a cells incubated with RVG9DR:siFITC (Movie S1), RVG9LR:siFITC (Movie S2), 9DR:siFITC (Movie S3) and 9LR:siFITC (Movie S4) complexes. LysoTracker (red) was used to label late endosomes and a 10:1 peptide:siRNA ratio was used.
Representative time-lapse videos of Neuro2A cells pre-incubated with A568-labeled annexin V and siFITC followed by exposure to RVG-9LR. Upper left and right panels show fluorescent images of RVG-9LR:siFITC and A568-labeled annexin V in the same fields respectively. The lower left and right panels are merged images of fluorescence and DIC images in the same fields.
Representative time-lapse videos of Neuro2A cells co-expressing α6-mCherry, α4 and β2 subunits of nAchR pre-incubated in medium containing siFITC and then exposed to RVG-9LR. Upper left and right panels show fluorescent images of RVG-9LR:siFITC and mCherry-fused nAchR in the same fields respectively. The lower left and right panels are merged images of fluorescence and DIC images in the same fields.
Acknowledgments
We thank Andrew Jackson for assisting with purification of recombinant scFvCD7. We thank Dr. Craig Roy (Yale University) for the kind gift of the wild type and mutant Rab5- and Rab7-GFP fusion constructs. This work was supported by National Institutes of Health [R21AI088484 to P.K.] and the Korea National Research Foundation [2009-0079989 and 2009-0081880 to S.K.L.]. C.C. was supported by NRF-2012R1A6A1029029.
Footnotes
AUTHOR CONTRIBUTION
Skye Zeller and Changseon Choi are co-first authors. S.Z. and C.C. designed and performed the main experiments. P.D.U, H.B., and A.S. performed and/or contributed additional experiments. T.M.F and W.M. contributed essential analytic tools. P.D.U. edited the manuscript. P.K. and S.K.L conceived and supervised the study, interpreted the results and wrote the paper. All authors discussed and interpreted the results.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abes R, Moulton HM, Clair P, Yang ST, Abes S, Melikov K, Prevot P, Youngblood DS, Iversen PL, Chernomordik LV, et al. Delivery of steric block morpholino oligomers by (R-X-R)4 peptides: structure–activity studies. Nucleic Acids Res. 2008;36:6343–6354. doi: 10.1093/nar/gkn541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akita H, Kogure K, Moriguchi R, Nakamura Y, Higashi T, Nakamura T, Serada S, Fujimoto M, Naka T, Futaki S, et al. Nanoparticles for ex vivo siRNA delivery to dendritic cells for cancer vaccines: programmed endosomal escape and dissociation. J Control Release. 2010;143:311–317. doi: 10.1016/j.jconrel.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Al-Taei S, Penning NA, Simpson JC, Futaki S, Takeuchi T, Nakase I, Jones AT. Intracellular traffic and fate of protein transduction domains HIV-1 TAT peptide and octaarginine. Implications for their utilization as drug delivery vectors. Bioconjug Chem. 2006;17:90–100. doi: 10.1021/bc050274h. [DOI] [PubMed] [Google Scholar]
- Appelbaum JS, LaRochelle JR, Smith BA, Balkin DM, Holub JM, Schepartz A. Arginine topology controls escape of minimally cationic proteins from early endosomes to the cytoplasm. Chemistry & biology. 2012;19:819–830. doi: 10.1016/j.chembiol.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri MA, Roberts RL, Gumusboga A, Highfield H, Alvarez-Dominguez C, Wells A, Stahl PD. Epidermal growth factor and membrane trafficking. EGF receptor activation of endocytosis requires Rab5a. The Journal of cell biology. 2000;151:539–550. doi: 10.1083/jcb.151.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloor J, Choi CS, Nam HY, Park M, Kim SH, Jackson A, Lee KY, Kim SW, Kumar P, Lee SK. Arginine-engrafted biodegradable polymer for the systemic delivery of therapeutic siRNA. Biomaterials. 2012;33:1640–1650. doi: 10.1016/j.biomaterials.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Cantini L, Attaway CC, Butler B, Andino LM, Sokolosky ML, Jakymiw A. Fusogenic-oligoarginine peptide-mediated delivery of siRNAs targeting the CIP2A oncogene into oral cancer cells. PloS one. 2013;8:e73348. doi: 10.1371/journal.pone.0073348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CJ, Saltzman WM. Enhanced siRNA delivery into cells by exploiting the synergy between targeting ligands and cell-penetrating peptides. Biomaterials. 2011;32:6194–6203. doi: 10.1016/j.biomaterials.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenan RM, Nashmi R, Imoukhuede P, Just H, McKinney S, Lester HA. Subcellular trafficking, pentameric assembly, and subunit stoichiometry of neuronal nicotinic acetylcholine receptors containing fluorescently labeled alpha6 and beta3 subunits. Molecular pharmacology. 2008;73:27–41. doi: 10.1124/mol.107.039180. [DOI] [PubMed] [Google Scholar]
- Du L, Kayali R, Bertoni C, Fike F, Hu H, Iversen PL, Gatti RA. Arginine-rich cell-penetrating peptide dramatically enhances AMO-mediated ATM aberrant splicing correction and enables delivery to brain and cerebellum. Human molecular genetics. 2011;20:3151–3160. doi: 10.1093/hmg/ddr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic (Copenhagen, Denmark) 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- Eguchi A, Dowdy SF. siRNA delivery using peptide transduction domains. Trends in pharmacological sciences. 2009;30:341–345. doi: 10.1016/j.tips.2009.04.009. [DOI] [PubMed] [Google Scholar]
- El-Sayed A, Futaki S, Harashima H. Delivery of macromolecules using arginine-rich cell-penetrating peptides: ways to overcome endosomal entrapment. The AAPS journal. 2009;11:13–22. doi: 10.1208/s12248-008-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh T, Ohtsuki T. Cellular siRNA delivery using cell-penetrating peptides modified for endosomal escape. Advanced Drug Delivery Reviews. 2009;61:704–709. doi: 10.1016/j.addr.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Erazo-Oliveras A, Muthukrishnan N, Baker R, Wang TY, Pellois JP. Improving the Endosomal Escape of Cell-Penetrating Peptides and Their Cargos: Strategies and Challenges. Pharmaceuticals (Basel) 2012;5:1177–1209. doi: 10.3390/ph5111177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Press B, Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. The Journal of cell biology. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R, Hufnagel H, Brock R. A Doubly Labeled Penetratin Analogue as a Ratiometric Sensor for Intracellular Proteolytic Stability. Bioconjugate Chemistry. 2010;21:64–73. doi: 10.1021/bc900301k. [DOI] [PubMed] [Google Scholar]
- Fischer R, Kohler K, Fotin-Mleczek M, Brock R. A stepwise dissection of the intracellular fate of cationic cell-penetrating peptides. The Journal of biological chemistry. 2004;279:12625–12635. doi: 10.1074/jbc.M311461200. [DOI] [PubMed] [Google Scholar]
- Fonseca SB, Pereira MP, Kelley SO. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Advanced Drug Delivery Reviews. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Fretz MM, Penning NA, Al-Taei S, Futaki S, Takeuchi T, Nakase I, Storm G, Jones AT. Temperature-, concentration- and cholesterol-dependent translocation of L-and D-octa-arginine across the plasma and nuclear membrane of CD34+ leukaemia cells. Biochem J. 2007;403:335–342. doi: 10.1042/BJ20061808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Raines RT. Pathway for polyarginine entry into mammalian cells. Biochemistry. 2004;43:2438–2444. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futaki S, Nakase I, Tadokoro A, Takeuchi T, Jones AT. Arginine-rich peptides and their internalization mechanisms. Biochem Soc Trans. 2007;35:784–787. doi: 10.1042/BST0350784. [DOI] [PubMed] [Google Scholar]
- Gump JM, Dowdy SF. TAT transduction: the molecular mechanism and therapeutic prospects. Trends Mol Med. 2007;13:443–448. doi: 10.1016/j.molmed.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Yamauchi J, Khalil IA, Kajimoto K, Akita H, Harashima H. Cell penetrating peptide-mediated systemic siRNA delivery to the liver. Int J Pharm. 2011;419:308–313. doi: 10.1016/j.ijpharm.2011.07.038. [DOI] [PubMed] [Google Scholar]
- Hirose H, Takeuchi T, Osakada H, Pujals S, Katayama S, Nakase I, Kobayashi S, Haraguchi T, Futaki S. Transient focal membrane deformation induced by arginine-rich peptides leads to their direct penetration into cells. Mol Ther. 2012;20:984–993. doi: 10.1038/mt.2011.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SW, Christison R, Bundell K, Voyce CJ, Brockbank SM, Newham P, Lindsay MA. Characterisation of cell-penetrating peptide-mediated peptide delivery. British journal of pharmacology. 2005;145:1093–1102. doi: 10.1038/sj.bjp.0706279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei N, Morishita M, Eda Y, Ida N, Nishio R, Takayama K. Usefulness of cell-penetrating peptides to improve intestinal insulin absorption. J Control Release. 2008;132:21–25. doi: 10.1016/j.jconrel.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Kim HK, Davaa E, Myung CS, Park JS. Enhanced siRNA delivery using cationic liposomes with new polyarginine-conjugated PEG-lipid. International Journal of Pharmaceutics. 2010;392:141–147. doi: 10.1016/j.ijpharm.2010.03.047. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Christensen LV, Jo S, Yockman JW, Jeong JH, Kim YH, Kim SW. Cholesteryl Oligoarginine Delivering Vascular Endothelial Growth Factor siRNA Effectively Inhibits Tumor Growth in Colon Adenocarcinoma. Mol Ther. 2006;14:343–350. doi: 10.1016/j.ymthe.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Startchev K, Whitney AJ, Gruenber J. Localization of lysobisphosphatidic acid-rich membrane domains in late endosomes. Biol Chem. 2001;382:483–485. doi: 10.1515/BC.2001.059. [DOI] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- Lee SH, Choi SH, Kim SH, Park TG. Thermally sensitive cationic polymer nanocapsules for specific cytosolic delivery and efficient gene silencing of siRNA: swelling induced physical disruption of endosome by cold shock. J Control Release. 2008;125:25–32. doi: 10.1016/j.jconrel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim DH, Cho IS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looi CY, Imanishi M, Takaki S, Sato M, Chiba N, Sasahara Y, Futaki S, Tsuchiya S, Kumaki S. Octa-arginine mediated delivery of wild-type Lnk protein inhibits TPO- induced M-MOK megakaryoblastic leukemic cell growth by promoting apoptosis. PloS one. 2011;6:e23640. doi: 10.1371/journal.pone.0023640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani F, Lindberg S, Langel U, Futaki S, Graslund A. Mechanisms of cellular uptake of cell-penetrating peptides. J Biophys. 2011;2011:414729. doi: 10.1155/2011/414729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiolo JR, Ferrer M, Ottinger EA. Effects of cargo molecules on the cellular uptake of arginine-rich cell-penetrating peptides. Biochimica et biophysica acta. 2005;1712:161–172. doi: 10.1016/j.bbamem.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Margus H, Padari K, Pooga M. Cell-penetrating Peptides as Versatile Vehicles for Oligonucleotide Delivery. Mol Ther. 2012;20:525–533. doi: 10.1038/mt.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason AJ, Leborgne C, Moulay G, Martinez Al, Danos O, Bechinger B, Kichler A. Optimising histidine rich peptides for efficient DNA delivery in the presence of serum. Journal of Controlled Release. 2007;118:95–104. doi: 10.1016/j.jconrel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- Ming X. Cellular delivery of siRNA and antisense oligonucleotides via receptor-mediated endocytosis. Expert Opinion on Drug Delivery. 2011;8:435–449. doi: 10.1517/17425247.2011.561313. [DOI] [PubMed] [Google Scholar]
- Mishra A, Lai GH, Schmidt NW, Sun VZ, Rodriguez AR, Tong R, Tang L, Cheng J, Deming TJ, Kamei DT, et al. Translocation of HIV TAT peptide and analogues induced by multiplexed membrane and cytoskeletal interactions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16883–16888. doi: 10.1073/pnas.1108795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Kogure K, Futaki S, Harashima H. Octaarginine-modified multifunctional envelope-type nano device for siRNA. Journal of Controlled Release. 2007;119:360–367. doi: 10.1016/j.jconrel.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Nakase I, Niwa M, Takeuchi T, Sonomura K, Kawabata N, Koike Y, Takehashi M, Tanaka S, Ueda K, Simpson JC, et al. Cellular uptake of arginine-rich peptides: roles for macropinocytosis and actin rearrangement. Mol Ther. 2004;10:1011–1022. doi: 10.1016/j.ymthe.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Payne CK, Jones SA, Chen C, Zhuang X. Internalization and trafficking of cell surface proteoglycans and proteoglycan-binding ligands. Traffic (Copenhagen, Denmark) 2007;8:389–401. doi: 10.1111/j.1600-0854.2007.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbard JB, Jessop TC, Wender PA. Adaptive translocation: the role of hydrogen bonding and membrane potential in the uptake of guanidinium-rich transporters into cells. Adv Drug Deliv Rev. 2005;57:495–504. doi: 10.1016/j.addr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Rydstrom A, Deshayes S, Konate K, Crombez L, Padari K, Boukhaddaoui H, Aldrian G, Pooga M, Divita G. Direct translocation as major cellular uptake for CADY self-assembling peptide-based nanoparticles. PloS one. 2011;6:e25924. doi: 10.1371/journal.pone.0025924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze SR, Dowdy SF. In vivo protein transduction: intracellular delivery of biologically active proteins, compounds and DNA. Trends in pharmacological sciences. 2000;21:45–48. doi: 10.1016/s0165-6147(99)01429-7. [DOI] [PubMed] [Google Scholar]
- Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. RISC hitches onto endosome trafficking. Nat Cell Biol. 2009;11:1049–1051. doi: 10.1038/ncb0909-1049. [DOI] [PubMed] [Google Scholar]
- Subramanya S, Kim SS, Abraham S, Yao J, Kumar M, Kumar P, Haridas V, Lee SK, Shultz LD, Greiner D, et al. Targeted delivery of small interfering RNA to human dendritic cells to suppress dengue virus infection and associated proinflammatory cytokine production. J Virol. 2010;84:2490–2501. doi: 10.1128/JVI.02105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonges L, Lingor P, Egle R, Dietz GPH, Fahr A, Bahr M. Stearylated octaarginine and artificial virus-like particles for transfection of siRNA into primary rat neurons. RNA. 2006;12:1431–1438. doi: 10.1261/rna.2252206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tünnemann G, Ter-Avetisyan G, Martin RM, Stöckl M, Herrmann A, Cardoso MC. Live-cell analysis of cell penetration ability and toxicity of oligo-arginines. Journal of Peptide Science. 2008;14:469–476. doi: 10.1002/psc.968. [DOI] [PubMed] [Google Scholar]
- van Asbeck AH, Beyerle A, McNeill H, Bovee-Geurts PH, Lindberg S, Verdurmen WP, Hallbrink M, Langel U, Heidenreich O, Brock R. Molecular parameters of siRNA--cell penetrating peptide nanocomplexes for efficient cellular delivery. ACS nano. 2013;7:3797–3807. doi: 10.1021/nn305754c. [DOI] [PubMed] [Google Scholar]
- Verdurmen WP, Bovee-Geurts PH, Wadhwani P, Ulrich AS, Hallbrink M, van Kuppevelt TH, Brock R. Preferential uptake of L- versus D-amino acid cell-penetrating peptides in a cell type-dependent manner. Chemistry & biology. 2011;18:1000–1010. doi: 10.1016/j.chembiol.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nature medicine. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- Wang YH, Hou YW, Lee HJ. An intracellular delivery method for siRNA by an arginine-rich peptide. Journal of biochemical and biophysical methods. 2007;70:579–586. doi: 10.1016/j.jbbm.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Wei J, Jones J, Kang J, Card A, Krimm M, Hancock P, Pei Y, Ason B, Payson E, Dubinina N, et al. RNA-induced silencing complex-bound small interfering RNA is a determinant of RNA interference-mediated gene silencing in mice. Molecular pharmacology. 2011;79:953–963. doi: 10.1124/mol.110.070409. [DOI] [PubMed] [Google Scholar]
- Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: Peptoid molecular transporters. Proceedings of the National Academy of Sciences. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RP, Youngblood DS, Hassinger JN, Lovejoy CE, Nelson MH, Iversen PL, Moulton HM. Cell-penetrating peptides as transporters for morpholino oligomers: effects of amino acid composition on intracellular delivery and cytotoxicity. Nucleic Acids Res. 2007;35:5182–5191. doi: 10.1093/nar/gkm478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ST, Zaitseva E, Chernomordik LV, Melikov K. Cell-penetrating peptide induces leaky fusion of liposomes containing late endosome-specific anionic lipid. Biophysical journal. 2010;99:2525–2533. doi: 10.1016/j.bpj.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaro JL, Vekich JE, Tran T, Shen WC. Nuclear Localization of Cell-Penetrating Peptides Is Dependent on Endocytosis Rather Than Cytosolic Delivery in CHO Cells. Molecular Pharmaceutics. 2009;6:337–344. doi: 10.1021/mp800239p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kollmer M, Buhrman JS, Tang MY, Gemeinhart RA. Arginine-rich, cell penetrating peptide-anti-microRNA complexes decrease glioblastoma migration potential. Peptides. 2014;58:83–90. doi: 10.1016/j.peptides.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A. Thermodynamic studies and binding mechanisms of cell-penetrating peptides with lipids and glycosaminoglycans. Adv Drug Deliv Rev. 2008;60:580–597. doi: 10.1016/j.addr.2007.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figures 1 and 2. 9R and RVG-9R bind siRNA to form nanoparticles. Electrophoretic gel mobility shift assay of 100pmol siRNA complexed to 9D/LR (A), RVG-9D/LR (C) and scFvCD7-9D/LR (F) at the indicated molar excess of peptide/antibody. Note: the RVG peptide by itself does not bind siRNA in (C). (B/D) Average particle diametric size distribution in nanometer (ZAve) and surface charge distribution (ζ) of peptide-siRNA complexes (100 pmol siRNA, 10:1 peptide:siRNA) measured by dynamic light scattering. (E) MALDI-TOF MS spectrum of scFvCD7-9D/LR conjugates.
Figure S2, related to Figure 2. Functional siRNA delivery by RVG9R. QPCR analysis of SOD-1 mRNA levels in Neuro2a cells 4h after exposure to siRNA complexes (100 pmol siRNA, 10:1 peptide:siRNA).
Figure S3, related to Figure 3. Analysis of cellular uptake mechanisms of RVG-9R:siRNA. (A) Flow cytometric analysis of Neuro2a cells exposed to RVG9D/LR:siFITC (10:1 peptide:siRNA molar ratio) after incubation at the indicated temperatures. The filled histograms correspond to cells transfected with siFITC alone and the values represent MFIs of the open histograms. The graph on the right is a quantitation of reduction in siRNA uptake measured as percent fold reduction in MFIs at 4°C compared to that at 37°C. (B) Confocal images of Neuro2a cells demonstrating pretreatment with inhibitors cytochalasin-D or dynasore inhibits internalization of A568-labeled transferrin.
Figure S4, related to Figure 5 and Video S6. (A) Time-lapse images of Neuro2A cells co-expressing α6-mCherry, α4 and β2 subunits of nAchR pre-incubated in medium containing siFITC at the indicated times after exposure to RVG-9LR. Fluorescent and DIC images in the same fields were merged. Arrows indicate site of peptide:siRNA binding and receptor aggregation. (B) Confocal microscopy of Neuro2a cells 24h after incubation with 9D/LR:siFITC complexes. Nuclei are stained with Hoechst 33342 (blue). (C and D) Analysis of intracellular fluor distribution in Neuro2A cells in Figures 5A and 5B at 24h. Data shown is cumulative of 3 independent experiments and represents cell numbers as percentages calculated with a sample size of ~50 cells. Error bars depict SEM. ***P<0.001. (E) Ethidium bromide (EtBr) exclusion assay conducted with peptide:siRNA complexes (400 pmol siRNA, 2.5:1 peptide:siRNA ratio) in phosphate buffers at the indicated pH values. The plot shows the amount of free siRNA (not complexed with peptides) detected in solution using EtBr. The fluorescence value for free siRNA without peptide at each pH value tested was set to 100%. Note: The 9LR peptide associates more strongly with siRNA than RVG-9LR suggesting poor availability to interact with cellular membranes. This was also reflected in the observation that ligand attachment lowered the average zeta potential of siRNA complexes suggesting a weaker association of siRNA in the ligand-9R format (Figure S1). Reducing the pH to 5.0 increased association of RVG-9LR with siRNA making it comparable to 9LR at pH 6.5 in this assay. Thus, under low pH conditions in late endosomes, RVG-9R-siRNA complex may need complex destabilization by action of endosomal proteases to promote membrane interaction and translocation of siRNA into the cytosol.
Supplementary Videos S1-S4, related to Figure 3. RVG-9R induces rapid cytosolic translocation of siRNA. Representative time-lapse videos of Neuro2a cells incubated with RVG9DR:siFITC (Movie S1), RVG9LR:siFITC (Movie S2), 9DR:siFITC (Movie S3) and 9LR:siFITC (Movie S4) complexes. LysoTracker (red) was used to label late endosomes and a 10:1 peptide:siRNA ratio was used.
Representative time-lapse videos of Neuro2A cells pre-incubated with A568-labeled annexin V and siFITC followed by exposure to RVG-9LR. Upper left and right panels show fluorescent images of RVG-9LR:siFITC and A568-labeled annexin V in the same fields respectively. The lower left and right panels are merged images of fluorescence and DIC images in the same fields.
Representative time-lapse videos of Neuro2A cells co-expressing α6-mCherry, α4 and β2 subunits of nAchR pre-incubated in medium containing siFITC and then exposed to RVG-9LR. Upper left and right panels show fluorescent images of RVG-9LR:siFITC and mCherry-fused nAchR in the same fields respectively. The lower left and right panels are merged images of fluorescence and DIC images in the same fields.