Abstract
This review provides an overview of the state and future directions of development and pathology in the craniofacial complex in the context of Cranial Neural Crest Cells (CNCC). CNCC are a multipotent cell population that is largely responsible for forming the vertebrate head. We focus on findings that have increased the knowledge of gene regulatory networks and molecular mechanisms governing CNCC migration and the participation of these cells in tissue formation. Pathology due to aberrant migration or cell death of CNCC, termed neurocristopathies, is discussed in addition to craniosynostoses. Finally, we discuss tissue engineering applications that take advantage of recent advancements in genome editing and the multipotent nature of CNCC. These applications have relevance to treating diseases due directly to the failure of CNCC, and also in restoring tissues lost due to a variety of reasons.
Keywords: craniofacial abnormality, cranial neural crest cell, gene regulatory networks, tissue engineering
What are Neural Crest Cells and in Particular Cranial Neural Crest Cells?
Neural crest cells (NCC) are a transient group of multipotent cells that are specified along the dorsal aspect of the neural tube, delaminate from the neural tube via an epithelial-mesenchymal transition (EMT), migrate in streams along their body segment, and subsequently differentiate under the guide of many signaling pathways throughout their journey. A subset of NCC, termed cranial neural crest cells (CNCC), delaminate from the more anterior portions of the folded neural tube and migrate in a single wave to give rise to many vertebrate head structures, including a majority of the skull and face (Le Douarin and Kalcheim, 1999; Mishina and Snider, 2014). As the CNCC are induced and begin to migrate, they are influenced by their physical contact with one another and also respond to reciprocal signals sent to one another. Local molecular gradients are also known to play a role in their migration (Theveneau and Mayor, 2012).
Initial investigations into the nature of NCC migration took advantage of an avian model consisting of Quail–Chick (QC) chimeras. This model allowed for easy visualization of NCC, based on chromosomal differences visible during interphase between the two species (Le Douarin, 1973). QC chimera experiments have formed an experimental basis to test which genes and gene products effect CNCC behavior and ability to form skeletogenic components of the craniofacial complex. For example, Fgf8 is indispensable for the survival of cells that make up the first branchial arch, as shown by the fact that knock down of this gene in the ectoderm and neuroepithelium results in a lack of craniofacial structures due to a failure of CNCC migration. This phenotype can be rescued by exogenous FGF8 application which promotes CNCC proliferation (Creuzet et al., 2004). Today, a variety of model systems, including turtle and lamprey, but mainly mice and avian models, are used to study CNCC (Santagati and Rijli, 2003; Green and Bronner, 2014; Nagashima et al., 2014; Young et al., 2014).
A number of molecular events must take place to orchestrate this induction, migration, and eventual fate determination of these multipotent cells (Trainor, 2013). Alterations in any of these processes can have a devastating array of effects on the developing embryo. In particular, the cranium and face are involved in a disproportionate number of birth defects, nearly a third, likely due to the intricacies involved in the genesis of a diverse collection of tissues present in a relatively small volume. Extensive collections of genes have been identified or are theorized to take part during the normal development of the craniofacial complex. A recent study reported collecting chick embryos electroporated with the Sox10e2:eGFP to mark migrating CNCC, and then GFP1CNCC were sorted via FACS. Subsequent extensive bioinformatic analysis of the transcritptome of migrating CNCC revealed SOX9 and ETS1 gene expression may be critically important to kick off a cascade of events that helps initiate and then guide CNCC along their migration (Simoes-Costa et al., 2014).
How do NCC Begin Their Migration, and What Structures do They Make?
Several signaling pathways have been shown to be involved in the physiologic and pathologic behavior of NCC, such as the FGF, WNT, TGFβ, and BMP pathways (Mishina and Snider, 2014). Many animal models have been generated to investigate each of these pathways that have been shown to be relevant to human disease. Here, we focus on the BMP and TGFβ signaling pathways and relate advances in knowledge of those pathways in CNCC to increased understanding of craniofacial development.
CNCC are initially specified at the neural plate border and then migrate from the dorsal portion of the neural tube to populate developing cranial structures. An EMT is necessary prior to CNCC commencing their migration. LSox5, p53, and ETS1 are expressed by CNCC (Perez-Alcala et al., 2004; Theveneau et al., 2007; Rinon et al., 2011). It is interesting, and also important for potential therapeutic measures, that apoptosis mediated by p53 is required for normal outcomes in CNCC migration (Rinon et al., 2011). BMP, bone morphogenetic protein, signaling is important to help regulate the cell cycle, which is important for induction and EMT to occur properly (Burstyn-Cohen et al., 2004). MSX2, a downstream target of the BMP signaling pathway, plays a role in the development of the craniofacial complex and also in the apoptosis of CNCC. It has been demonstrated that BMP4 is necessary to induce apoptosis in CNCC (Graham et al., 1994; Winograd et al., 1997; Takahashi et al., 2001).
The first evidence that the TGFβ signaling pathway is quite important during craniofacial development, in particular in palatogenesis, came with the knowledge that global knock out of the TGF-β3 gene results in cleft palate in mice (Kaartinen et al., 1995). Tissue specific interactions resulting from TGFβ signaling were further elucidated with the use of epithelium specific Keratin14-Cre and NCC specific Wnt1-Cre. Tissue specific deletions using mice with the type 1 TGFβ receptor flanked by loxP sites, Alk5fl/fl, revealed that both Wnt1-Cre/Alk5fl/fl mice as well as K14-Cre/Alk5fl/fl displayed cleft palate; however, the deletion of Alk5 in NCC also revealed pathology of the nasal cavity as well as the skull vault (Dudas et al., 2006). Although the underlying mechanism of cleft palate formation between the K14-Cre/Alk5fl/fl and Wnt1-Cre/Alk5fl/fl may be different, as suggested by increased proliferation and cell death in the mesenchyme of the Wnt1-Cre/Alk5fl/fl mice, the indispensible nature of TGFβ signaling within both ectodermal and ectomesenchymal cell types during normal craniofacial development was strongly suggested.
Using Wnt1-Cre to delete a type two TGFβ receptor Tgfbr2, another component in the TGFβ signaling pathway, results in craniofacial development with a phenotype partially overlapping that of the Wnt1-Cre/Alk5fl/fl mice. When mice carrying the Wnt1-Cre transgene are bred with Tgfbr2fl/fl mice, the resulting pups have a cleft palate due to decreased proliferation of CNCC derived palatal mesenchyme, as well as severe skull malformation (Ito et al., 2003). The similarity of the skull vault malformation between Wnt1-Cre/Tgfbr2fl/fl and Wnt1-Cre/Alk5 reveals potential redundancies in TGFβ receptor function in CNCC during skull vault formation.
As Osterix-Cre (Osx-Cre) is expressed by osteoblasts as well as odontoblasts, Tgfbr2fl/fl mice have also been bred with mice carrying the Osx-Cre transgene to investigate the function of Tgfbr2 in cells critical to mineralized tissue formation. Osteoblasts are critical to cranial bone formation and odontoblasts form a majority of the molar tooth root, so using Osx-Cre deletes Tgfbr2 in the skeletogenic progeny of CNCC, thus testing the effects of TGFβ signaling as CNCC become more committed. The palate of Osx-Cre/Tgfbr2fl/fl mice develops normally, whereas the molar root formation and dental mesenchyme are adversely affected (Wang et al., 2013). The absence of a drastic skull phenotype found in Osx-Cre/Tgfbr2fl/fl, compared to those described in Wnt1-Cre/Tgfbr2fl/fl, and Wnt1-Cre/Alk5, indicates that TGFβ signaling in CNCC is required during early skull formation, but becomes dispensable in this process as CNCC continue to differentiate into more committed cells.
Together, these studies underscore the effect of spatiotemporal expression of TGFβ signaling components in CNCC and their derivatives during craniofacial development. These findings also reinforce the notion that the promoter used to drive Cre expression can result in information critical to dissecting out which combinations of genes expressed by unique cell populations are indispensible for a given tissues formation. These TGFβ pathway studies in mice have been shown to be a good model for human disease. The finding of 10 families presenting with craniofacial anomalies and concomitant mutations in either TGFβR1 or TGFβR2 provided strong support that TGFβ signaling is critical for craniofacial development in humans as well (Loeys et al., 2005).
Which Birth Defects in Particular are Rooted in CNCC Defects?
Many birth defects often result in growth and developmental delays at sites in the craniofacial complex as well as involving other anatomical locations. The broad phenotypic display presented in these syndromes obviates the difficulty in ascertaining which findings are primary and which arise secondarily to the etiopathology. For example, aberrant mandibular growth may persist for years in several syndromes which have a broad phenotypic display present from birth (Boutros et al., 2007). Although it is desirable to move away from discussing syndromes based on the names of the discoverer, and instead refer to them by a more informative name indicative of either the etiology or phenotype, eponyms are referred to here to be complete. Improper migration or reduced survival of CNCC is implicated in several syndromes and congenital conditions termed neurocristopathies. Craniofacial anomalies are seen in neurocristopathies, such as DiGeorge syndrome, Waardenburg syndrome (WS), CHARGE syndrome, Treacher Collins syndrome (TCS), and craniofacial microsomia (CFM). (Table 1 provides a summary of neurocristopathies with additional information.)
TABLE 1.
Neurocristopathies with Potential Causes and Clinical Characteristics
| Condition | Causes | Characteristics |
|---|---|---|
| Velocardiofacial syndrome | 22q11.2 locus deletion | Prevalence: 1: 4,000, spontaneous mutation in 90% |
| COMT, TBX1 | Etiopathophysiology: Deletion disrupts neural crest cells during organogenesis | |
| Other names: DiGeorge syndrome, Shprintzen syndrome, CATCH22 | ||
| Key Characteristics: pharyngeal dysfunction, cardiac anomalies (most common is ventriculoseptal defect), dysmorphic facies | ||
| Affects thymus, parathyroid, arteries to face | ||
| Hypocalcemia and subsequent epileptic events | ||
| Low set ears, micrognathia, CP (usually soft palate or submucous), velopharyngeal insufficiency induced feeding difficulties, otitis media, immunodeficiency, vertical maxillary excess | ||
| Cognitive/learning problems | ||
| Psychiatric illness in 10% (bipolar, schizophrenia) | ||
| Diagnosis verified by symptoms & and FISH test (genetics test) | ||
| VPI repair occurs at 3–4 years of age | ||
| Waardenburg syndrome | SOX10 PAX3 | Prevalence: 1:40,000 |
| Hypopigmentation in the eyes and skin | ||
| White tuft of hair present on the anterior scalp | ||
| CHARGE syndrome | CHD7 | Prevalence: 1:8,500–1:10,000 |
| Complex diagnosis based on major and minor characteristics | ||
| Facial anomalies: potential for vision problems due to coloboma of the eye and microphthalmia, choanal atresia, increased likelihood of cranial nerve abnormalities | ||
| Variable presentation between patients | ||
| Treacher collins syndrome | TCOF1 (90%) | Prevalence: 1:50,000 60% spontaneous mutation |
| POLR1C, POLR1D | Autosomal Dominant inheritance | |
| Other names: Mandibulofacial Dysostosis | ||
| TCOF1 (treacle protein); key role in neural crest cell proliferation | ||
| Airway compromise, hearing loss, sleep apnea (25%), delayed motor and & speech development, CP (35%), velopharyngeal insufficiency (35%) | ||
| Facial Abnormalities: downward slanting eyes, colobomas, mandibular retrognathia, midface hypoplasia, malformed, or absent ears | ||
| Dental Anomalies: tooth agenesis, enamel defects, anterior open bite, mouth breathing, ectopic eruption | ||
| Feeding problems, language problems | ||
| Can be detected on ultrasound | ||
| Craniofacial microsomia | Genetics | Prevalence: 1:5000, M > F |
| Teratogens that cause hematoma of arteries of 1st & and 2nd branchial arches leading to vascular problems in utero. | Other names: Hemifacial microsomia, oral-mandibular-auricular syndrome, 1st and 2nd branchial arch syndrome | |
| Key Characteristic: Absence or underdevelopment of structures that arise from 1st & and 2nd pharyngeal arches | ||
| Mandible, maxilla, ear, facial soft tissue and & mm, CN VII | ||
| Disruption during first 6 weeks gestation | ||
| Vascular problem in utero affecting clotting and poor reduced facial blood supply | ||
| 2nd most common congenital facial anomaly (to CLP) | ||
| Facial Abnormalities: CLP (7–22%), malar hypoplasia, facial asymmetry, mandibular mirognathia, absence or malformed TMJ, facial clefts, facial palsy | ||
| Dental Abnormalities: delayed development, occlusal cant, impacted or missing teeth, velopharyngeal insufficiency, | ||
| Ear abnormalities: microtia, accessory auricles, abnormal ossicles | ||
| 55% also have extracranial anomalies –vertebral fusion, trismus, kidney dysfunction, cardiac abnormalities | ||
| OMENS system used to categorize disease presentation (ocular/orbital, mandibular, ear, nerve, soft tissue) | ||
| Goldenhar syndrome | Severe form of CFM | Other names: Oculoauriculovertebral syndrome (OVA) |
| Form of CFM with increased incidence of ear malformation. | ||
| OMENS system used to categorize disease presentation |
DiGeorge (velocardiofacial) syndrome is a classical example of a disease due to changes in CNCC migration due to genetic mutation. The 22q11.2 deletion syndrome (22q11DS) occurs in approximately 1 in 4000 births and typically includes a wide range of defects involving the initial formation and subsequent development of the craniofacial complex. In particular, cardiac defects, cognitive-behavioral problems, speech-language disorders, velopharyngeal insufficiency (VPI), and dysmorphic facial appearance have been well documented. 22q11DS is recognized as the most frequently occurring syndrome associated with VPI and anomalies affecting the palate (Shprintzen et al., 1978; Bassett et al., 2011). Hemizygosity at the TBX1 gene locus is believed to underlie the pathology, although other genes are deleted when this portion of chromosome 22 is deleted (Papangeli and Scambler, 2013).
WS is a neurocristopathy with heterogeneous presentation; four forms have been identified. The most common findings include hypopigmentation of the eyes, isolated patches of white hair on the anterior scalp, and sensori-neural hearing loss, ranging from total deafness to a progressive loss of hearing. Mutations in SOX10 have been implicated to be a potential cause of the hypopigmentation of the skin, hair, and eyes commonly found in patients with WS (Hou and Pavan, 2008). In support of this notion, mice haploinsufficient for Sox10 show similar symptoms, including isolated patches of white fur reminiscent of the hypopigmentation found in WS patients (Southard-Smith et al., 1999). A labor-intensive mutagenesis screening of mice haploinsufficient for Sox10 has also been used to identify genomic loci that exacerbate the hypopigmentation phenotype. Three new loci were identified as potential modifiers of Sox10. A compound heterozygote formed between Sox10 and one of the modifiers led to a decrease in NCC ability to form melanoblasts (Matera et al., 2008). Several other genes have also been identified that lead to specific forms of WS (Pingault et al., 2010).
CHARGE syndrome is diagnosed by the appearance of several distinct features. All of the features may not be present, but CHARGE usually presents with the following findings: coloboma of the eye, choanal atresia, heart defects, retardation of growth, genital malformations, and abnormal ears. CHD7 mutations have been identified as causes of the syndrome (Lalani et al., 2006)
TCS results from a failure of migration of CNCC. Underdeveloped facial bones resulting from autosomal dominant mutations in the TCOF1 gene phenotypically characterize TCS. A mouse model haploinsufficient for Tcof1 and recapitulating the characteristics of TCS has been generated. This allowed the demonstration that lack of formation and proliferation of CNCC were responsible for the craniofacial phenotype (Dixon et al., 2006). Although there is much work yet to be done, showing that inhibition of p53, either with a chemical inhibitor or through genetic deletion of a p53 allele, rescues the TCS phenotype in mice was a huge step toward being able to treat this disease clinically (Jones et al., 2008). Interestingly, the TCS phenotype was corrected without resolving the lack of ribosome biogenesis observed, which suggests that it makes sense to target downstream events in the treatment of neurocristopathies resulting from increased cell death of CNCC.
CFM is a congenital disorder of the face with an estimated prevalence of 1:3500 to 1:5500. CFM is characterized by asymmetric underdevelopment of structures originating from the first and second branchial arches, including the orbit, mandible, nerve, soft tissue, and muscles of mastication. Goldenhar syndrome is one of the forms of CFM and was first described in the 1950s. Goldenhar syndrome presents with the same constellation of symptoms in addition to auricular malformation in the form of partial duplication being noted (Ashokan et al., 2014). In mouse embryos, Hoxa2 expression in the neural crest is restricted to only the second branchial arch which contributes to pinna development, and ectopic expression of Hoxa2 in the first branchial arch neural crest results in the recapitulation of partial duplication of the pinna, a characteristic finding in Goldenhar Syndrome (Minoux et al., 2013).
CNCC and Craniosynostosis
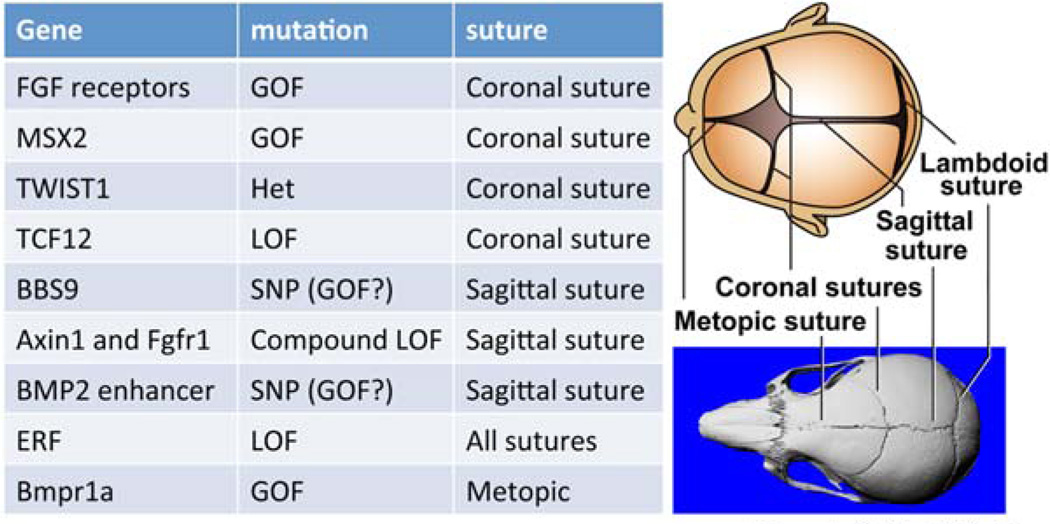
Normal skull growth occurs as the neural tissues expand and appositional bone growth occurs at the border of the bony plates of the skull (Fig. 1). Sutures separate the bony plates, and normal murine suture formation requires epithelial-mesenchymal interactions between CNCC derived mesenchyme, as well as mesoderm-derived mesenchyme. Generally, the more anterior portions of the murine skull are derived from the CNCC, with the exception of the interparietal bone (Noden and Trainor, 2005; Mishina and Snider, 2014). As neural tissue growth slows the pliable infant skull matures, and the sutures begin to fuse, leading to the formation of the more protective adult skull. In the event these sutures fuse prematurely, a condition known as craniosynostosis occurs, resulting in abnormal skull growth resulting in increased intracranial pressure (Fig. 1). The dura mater envelops the brain and is continuous with the calvarial periosteum at the suture. The underlying dura mater is known to interact molecularly with the developing suture and influence patency in rabbit models of craniosynostosis (Cooper et al., 2012; Mishina and Snider, 2014). Dura mater formation is compromised in Wnt1-Cre/Tgfbr2fl/fl mice, leading to skull vault anomalies (Ito et al., 2003). These observations imply that the signaling events from the dura mater are critically important to normal growth and development of the calvaria as neural tissues expand.
FIGURE 1.
Illustration of human and murine skull sutures and a chart of gene mutations resulting in suture-specific craniosynostosis. This figure outlines the diverse collection of genes that can result in craniosynostosis.
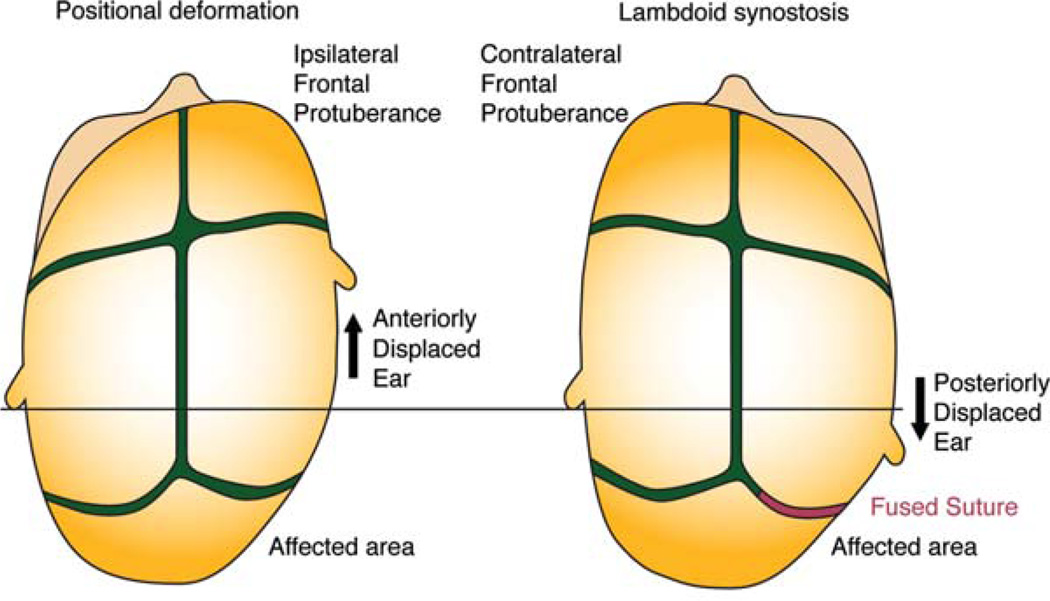
The FGF receptor family as well as the BMP signaling pathway is known to be involved in syndromic forms of craniosynostosis (Table 2). Several forms of craniosynostosis may occur, affecting a single suture or as part of a phenotypically diverse syndrome. There are several molecules that are known to be involved in a suture-specific manner (Roscioli et al., 2013; Fig. 1). Craniosynostosis is well documented to occur in isolation but is also known to present within a constellation of symptoms in syndromes with diverse clinical presentation. For example, gain of function mutations of receptors in the FGF pathway can result in Crouzon syndrome, as well as the closely related Apert syndrome (Jabs et al., 1994; Reardon et al., 1994). Both syndromes include craniosynostosis with Apert syndrome also presenting with syndactyly. Pfeiffer syndrome also presents with craniosynostosis and mutations in FGF receptors (Ibrahimi et al., 2004). Any individual suture as well as combinations of sutures may be affected each with a characteristic defect in cranial morphology (Fig. 1). For example, sagittal synostosis inhibits the lateral growth of the skull and results in an unusually narrow skull with increased anterior–posterior dimension. Figure 2 illustrates the differences between the cranial defect produced by either positional head deformation or craniosynostosis affecting the right, lambdoid suture. The key to differentiating these two ostensibly similar phenotypes is by noting the finding of an ipsilateral frontal protuberance and anteriorly displaced ear in positional head deformation.
TABLE 2.
Summary of Craniosynostoses with Causes and Characteristics
| Condition | Causes | Characteristics |
|---|---|---|
| Positional head deformation | Extrinsic factors | Head usually flat on one side |
| Ear & and forehead on ipsilateral side are rotated forward | ||
| Frontal bossing on ipsilateral side | ||
| Flatness of forehead on opposite side | ||
| 1:2,000 births | ||
| Isolated craniosynostosis | Unknown | Premature fusion of one or more sutures |
| Skull cannot expand perpendicularly to the fused suture so excessive growth occurs in a direction parallel to the fused suture | ||
| Crouzon syndrome | FGFR2 | Autosomal dominant; 1:25,000–60,000 |
| Craniosynostosis, midface hypoplasia, hypertelorism (wideset eyes), proptosis, beak nose | ||
| Apert syndrome | FGFR2 | Autosomal dominant; 1:160,000 |
| Craniosynostosis, midface hypoplasia, hypertelorism, symmetric syndactyly, mental handicap (50%), CP (30%) | ||
| Pfeiffer syndrome | FGFR1 & and 2 | Autosomal dominant; 1:100,000 |
| Craniosynostosis, broad features, cloverleaf skull, syndactyly, elbow ankylosis | ||
| Saethre-Chotzen syndrome | TWIST | Autosomal dominant |
| Craniosynostosis, broad features, syndactyly, beak nose | ||
| Carpenter syndrome | RAB23, MEGF8 | Autosomal Recessive |
| Craniosynostosis, midface hypoplasia, mental handicap, syndactyly |
FIGURE 2.
Diagram of positional head deformation versus lambdoid synostosis. The ability to differentiate between similar phenotypic presentations is key. Noting ear and frontal protuberance position is critical to differentiate these diagnoses. Red indicates a fused suture whereas green indicates patency.
Premature fusion of any single suture or combinations of sutures results in an increase of intracranial pressure, necessitating a series of surgery to offset the pressure increase and allow for normal growth; however, these surgeries are rarely fully corrective. This state of the art allows surgeons to remove the affected suture endoscopically in some instances, thus encouraging normal growth to resume (Sanger et al., 2014). The premature fusion of sutures has wide reaching consequences with regards to the other components of the craniofacial complex. Craniosynostosis affecting the metopic suture results in trigonocephaply with subsequent orbital dysmorphology. The effect on the orbit is directly proportional to the extent of the premature fusion (Ezaldein et al., 2014). The options available to correct the tissue deficits remaining after surgical intervention are currently limited.
Tissue Engineering with CNCC
A complete molecular catalog of the events shaping the craniofacial complex will yield the potential for tissue engineering approaches to regenerate tissues lost to a variety of pathologies. The ability to easily collect and manipulate less differentiated cells offers major advantages for patients in terms of restoring lost tissues. In fact, these areas are currently being investigated with a number of possible sources found in the oral cavity (Achilleos and Trainor, 2012). Numerous sources of purported “stem” cell populations have been identified in the craniofacial region: dental pulp stem cells, stem cells from human exfoliated deciduous teeth, stem cells from apical papilla, periodontal ligament stem cells, as well as progenitor sources that have been reported from the gingiva (Miura et al., 2003; Huang et al., 2009; Xu et al., 2013). A connexin-43 enriched human cell population isolated from periodontal ligament cells from extracted third molars has been shown to be multipotent in both in vitro culture and in vivo teratoma formation assays (Pelaez et al., 2013). This is particularly exciting, due to the fact that third molar extraction is routinely done and would provide a readily accessible source of postnatal stem cells that could be used to regenerate craniofacial tissues lost for a variety of reasons.
It is rapidly becoming obvious that the CRISPR-Cas9 system allows multiple genes to be edited rather quickly and their combinatorial effects assessed in a variety of settings (Hsu et al., 2014). A small guide RNA molecule with base pair complementarity to a particular locus in the genome allows an enzyme that cleaves DNA, Cas9, to locate and remove certain sites in the genome, or the CRISPR-Cas9 system can be used to insert new genetic elements. It will be informative to the field to simultaneously edit combinations of genes in CNCC from a variety of model organisms. The continued characterization of the stem cell nature of CNCC obtained from early mouse embryos (Ishii et al., 2012) and relative ease in obtaining anatomically unique CNCC populations after birth make it attractive to investigate them as a relevant tool for tissue engineering. Moreover, bioinformatic approaches that identify genes and gene networks coupled with the ability to test these networks using CRISPR-Cas9 to simultaneously edit the proposed genes in CNCC culture is a promising strategy to reveal the complex molecular interplay in CNCC.
Once the gene networks are identified, many potential avenues of treatment can be investigated. As ectomesenchymal tissues in the craniofacial complex are generated by CNCC, the knowledge of which gene networks result in lineage specification to either osteoblasts or chondrocytes provides an avenue to pursue regenerating these tissues. This may improve treatment options by allowing for therapies where patient derived cells are collected and used to regenerate lost bone or cartilage. In particular, one could isolate CNCC progenitors from a patient and use the identified gene network to promote bone formation in culture. This cultured bone could then be used to treat bone loss due to periodontal disease. Many hurdles remain in making this a reality, but progress is definitely being made.
Discussion
A complete understanding of CNCC behavior in terms of the molecular events involved initially in their migration and those that end their journey will allow for targeted therapies involving these pathways. The wide ranging nature of the phenotypes displayed by birth defects affecting the head and neck have made ascertaining the root cause of these defects extremely difficult. Increased ability to manipulate the genomes of many different model organisms will allow researchers to identify responsible gene regulatory networks and eventually to intervene to either prevent the phenotype altogether or halt its progression.
The wide range of potential research uses makes studying CNCC particularly appealing. For instance, in addition to the many advantages outlined in this review, consider the following: given the deleterious migration of metastasizing cancer cells which also often undergo EMT, knowing exactly how this extensive migration of CNCC is orchestrated could possibly be extrapolated to halt unwanted migration of oncogenic cell populations (Rogers et al., 2013).
In light of recent reduction in the expense of bioinformatics techniques coupled with their increasing power, it is becoming feasible to check many tissues and gather a snapshot of the differential gene expression patterns at play in them. This will illuminate therapeutic targets that can be tested in model systems to translate genomic readouts into information to treat patients. The knowledge gleaned from these studies will not only be applicable to patients suffering from diseases directly caused by failure of CNCC but also to those patients requiring regeneration of tissue lost due to trauma or other disease processes.
Although CRISPR-Cas9 is a new technology with many potential obstacles yet to be encountered, one can see how clinically useful such a technique could be. It is not out of reach to imagine the following scenario: a craniosynostic patient undergoes endoscopic surgery to resect their prematurely fused suture, stem cells are isolated from the resected tissue, CRISPR-Cas9 is used to excise the defective sequence, and the edited cells are reintroduced into the patient to correct the defect.
Acknowledgments
We thank Drs. Satoru Hayano and Honghao Zhang for critical reading.
Supported by a grant from the National Institutes of Health (R01DE020843) Supported by a grant from the Department of Defense (W81XWH-11-2-0073; to Y.M.).
Supported by a grant from the Tissue Engineering and Regeneration Grant (T32) at the University of Michigan (to T.N.S.).
References
- Achilleos A, Trainor PA. Neural crest stem cells: discovery, properties and potential for therapy. Cell Res. 2012;22:288–304. doi: 10.1038/cr.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashokan CS, Sreenivasan A, Saraswathy GK. Goldenhar syndrome–review with case series. J Clin Diagn Res. 2014;8:ZD17–ZD19. doi: 10.7860/JCDR/2014/7926.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, McDonald-McGinn DM, Devriendt K, et al. Practical guidelines for managing patients with 22q11.2 deletion syndrome. J Pediatr. 2011;159:332–339. doi: 10.1016/j.jpeds.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros S, Shetye PR, Ghali S, et al. Morphology and growth of the mandible in Crouzon, Apert, and Pfeiffer syndromes. J Craniofac Surg. 2007;18:146–150. doi: 10.1097/01.scs.0000248655.53405.a7. [DOI] [PubMed] [Google Scholar]
- Burstyn-Cohen T, Stanleigh J, Sela-Donenfeld D, Kalcheim C. Canonical Wnt activity regulates trunk neural crest delamination linking BMP/noggin signaling with G1/S transition. Development. 2004;131:5327–5339. doi: 10.1242/dev.01424. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Durham EL, Cray JJ, Jr, et al. Tissue interactions between craniosynostotic dura mater and bone. J Craniofac Surg. 2012;23:919–924. doi: 10.1097/SCS.0b013e31824e645f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creuzet S, Schuler B, Couly G, Le Douarin NM. Reciprocal relationships between Fgf8 and neural crest cells in facial and forebrain development. Proc Natl Acad Sci USA. 2004;101:4843–4847. doi: 10.1073/pnas.0400869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci USA. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas M, Kim J, Li WY, et al. Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Dev Biol. 2006;296:298–314. doi: 10.1016/j.ydbio.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaldein HH, Metzler P, Persing JA, Steinbacher DM. Three-dimensional orbital dysmorphology in metopic synostosis. J Plast Reconstr Aesthet Surg. 2014;67:900–905. doi: 10.1016/j.bjps.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Graham A, Francis-West P, Brickell P, Lumsden A. The signalling molecule BMP4 mediates apoptosis in the rhombencephalic neural crest. Nature. 1994;372:684–686. doi: 10.1038/372684a0. [DOI] [PubMed] [Google Scholar]
- Green SA, Bronner ME. The lamprey: a jawless vertebrate model system for examining origin of the neural crest and other vertebrate traits. Differentiation. 2014;87:44–51. doi: 10.1016/j.diff.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Pavan WJ. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res. 2008;18:1163–1176. doi: 10.1038/cr.2008.303. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Pelaez D, Dominguez-Bendala J, et al. Plasticity of stem cells derived from adult periodontal ligament. Regen Med. 2009;4:809–821. doi: 10.2217/rme.09.55. [DOI] [PubMed] [Google Scholar]
- Ibrahimi OA, Zhang F, Eliseenkova AV, et al. Proline to arginine mutations in FGF receptors 1 and 3 result in Pfeiffer and Muenke craniosynostosis syndromes through enhancement of FGF binding affinity. Hum Mol Genet. 2004;13:69–78. doi: 10.1093/hmg/ddh011. [DOI] [PubMed] [Google Scholar]
- Ishii M, Arias AC, Liu L, et al. A stable cranial neural crest cell line from mouse. Stem Cells Dev. 2012;21:3069–3080. doi: 10.1089/scd.2012.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yeo JY, Chytil A, et al. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- Jabs EW, Li X, Scott AF, et al. Jackson-Weiss and Crouzon syndromes are allelic with mutations in fibroblast growth factor receptor 2. Nat Genet. 1994;8:275–279. doi: 10.1038/ng1194-275. [DOI] [PubMed] [Google Scholar]
- Jones NC, Lynn ML, Gaudenz K, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartinen V, Voncken JW, Shuler C, et al. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- Lalani SR, Safiullah AM, Fernbach SD, et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am J Hum Genet. 2006;78:303–314. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N. A biological cell labeling technique and its use in expermental embryology. Dev Biol. 1973;30:217–222. doi: 10.1016/0012-1606(73)90061-4. [DOI] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The neural crest. Cambridge, UK: Cambridge University Press; 1999. [Google Scholar]
- Loeys BL, Chen J, Neptune ER, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 2005;37:275–281. doi: 10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- Matera I, Watkins-Chow DE, Loftus SK, et al. A sensitized mutagenesis screen identifies Gli3 as a modifier of Sox10 neurocristopathy. Hum Mol Genet. 2008;17:2118–2131. doi: 10.1093/hmg/ddn110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M, Kratochwil CF, Ducret S, et al. Mouse Hoxa2 mutations provide a model for microtia and auricle duplication. Development. 2013;140:4386–4397. doi: 10.1242/dev.098046. [DOI] [PubMed] [Google Scholar]
- Mishina Y, Snider TN. Neural crest cell signaling pathways critical to cranial bone development and pathology. Exp Cell Res. 2014;325:138–147. doi: 10.1016/j.yexcr.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura M, Gronthos S, Zhao M, et al. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003;100:5807–5812. doi: 10.1073/pnas.0937635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima H, Shibata M, Taniguchi M, et al. Comparative study of the shell development of hard- and soft-shelled turtles. J Anat. 2014;225:60–70. doi: 10.1111/joa.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papangeli I, Scambler P. The 22q11 deletion: DiGeorge and velocardiofacial syndromes and the role of TBX1. Wiley Interdiscip Rev Dev Biol. 2013;2:393–403. doi: 10.1002/wdev.75. [DOI] [PubMed] [Google Scholar]
- Pelaez D, Huang CY, Cheung HS. Isolation of pluripotent neural crest-derived stem cells from adult human tissues by connexin-43 enrichment. Stem Cells Dev. 2013;22:2906–2914. doi: 10.1089/scd.2013.0090. [DOI] [PubMed] [Google Scholar]
- Perez-Alcala S, Nieto MA, Barbas JA. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–4465. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- Pingault V, Ente D, Dastot-Le Moal F, et al. Review and update of mutations causing Waardenburg syndrome. Hum Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- Reardon W, Winter RM, Rutland P, et al. Mutations in the fibroblast growth factor receptor 2 gene cause Crouzon syndrome. Nat Genet. 1994;8:98–103. doi: 10.1038/ng0994-98. [DOI] [PubMed] [Google Scholar]
- Rinon A, Molchadsky A, Nathan E, et al. p53 coordinates cranial neural crest cell growth and epithelial-mesenchymal transition/delamination processes. Development. 2011;138:1827–1838. doi: 10.1242/dev.053645. [DOI] [PubMed] [Google Scholar]
- Rogers CD, Saxena A, Bronner ME. Sip1 mediates an E-cadherin-to-N-cadherin switch during cranial neural crest EMT. J Cell Biol. 2013;203:835–847. doi: 10.1083/jcb.201305050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscioli T, Elakis G, Cox TC, et al. Genotype and clinical care correlations in craniosynostosis: findings from a cohort of 630 Australian and New Zealand patients. Am J Med Genet C Semin Med Genet. 2013;163C:259–270. doi: 10.1002/ajmg.c.31378. [DOI] [PubMed] [Google Scholar]
- Sanger C, David L, Argenta L. Latest trends in minimally invasive synostosis surgery: a review. Curr Opin Otolaryngol Head Neck Surg. 2014;22:316–321. doi: 10.1097/MOO.0000000000000069. [DOI] [PubMed] [Google Scholar]
- Santagati F, Rijli FM. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci. 2003;4:806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg RB, Lewin ML, et al. A new syndrome involving cleft palate, cardiac anomalies, typical facies, and learning disabilities: velo-cardio-facial syndrome. Cleft Palate J. 1978;15:56–62. [PubMed] [Google Scholar]
- Simoes-Costa M, Tan-Cabugao J, Antoshechkin I, et al. Transcriptome analysis reveals novel players in the cranial neural crest gene regulatory network. Genome Res. 2014;24:281–290. doi: 10.1101/gr.161182.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southard-Smith EM, Angrist M, Ellison JS, et al. The Sox10(Dom) mouse: modeling the genetic variation of Waardenburg-Shah (WS4) syndrome. Genome Res. 1999;9:215–225. [PubMed] [Google Scholar]
- Takahashi K, Nuckolls GH, Takahashi I, et al. Msx2 is a repressor of chondrogenic differentiation in migratory cranial neural crest cells. Dev Dyn. 2001;222:252–262. doi: 10.1002/dvdy.1185. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Neural crest migration: interplay between chemorepellents, chemoattractants, contact inhibition, epithelial-mesenchymal transition, and collective cell migration. Wiley Interdiscip Rev Dev Biol. 2012;1:435–445. doi: 10.1002/wdev.28. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Duband JL, Altabef M. Ets-1 confers cranial features on neural crest delamination. PLoS One. 2007;2:e1142. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor P. In: Neural crest cells: evolution, development and disease. Trainor P, editor. London, UK: Academic Press; 2013. p. 488. [Google Scholar]
- Wang Y, Cox MK, Coricor G, et al. Inactivation of Tgfbr2 in Osterix-Cre expressing dental mesenchyme disrupts molar root formation. Dev Biol. 2013;382:27–37. doi: 10.1016/j.ydbio.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winograd J, Reilly MP, Roe R, et al. Perinatal lethality and multiple craniofacial malformations in MSX2 transgenic mice. Hum Mol Genet. 1997;6:369–379. doi: 10.1093/hmg/6.3.369. [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Akiyama K, et al. Gingivae contain neural-crest- and mesoderm-derived mesenchymal stem cells. J Dent Res. 2013;92:825–832. doi: 10.1177/0022034513497961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Hu D, Lainoff AJ, et al. Embryonic bauplans and the developmental origins of facial diversity and constraint. Development. 2014;141:1059–1063. doi: 10.1242/dev.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]