Abstract
Background
The kinetochore is a multiprotein machine that couples chromosome movement to microtubule (MT) polymerization and depolymerization. It uses numerous copies of at least three MT-binding proteins to generate bidirectional movement. The nanoscale organization of these proteins within the kinetochore plays an important role in shaping the mechanisms that drive persistent, bidirectional movement of the kinetochore.
Results
We used Förster Resonance Energy Transfer (FRET) between genetically encoded fluorescent proteins fused to kinetochore subunits to reconstruct the nanoscale organization of the budding yeast kinetochore. We performed > 60 FRET and high resolution colocalization measurements involving the essential MT-binding kinetochore components: Ndc80, Dam1, Spc105, and Stu2. These measurements reveal that neighboring Ndc80 complexes within the kinetochore are narrowly distributed along the length of the MT. Dam1 complex molecules are concentrated near the MT-binding domains of Ndc80. Stu2 localizes in high abundance within a narrowly defined territory within the kinetochore centered ~ 20 nm on the centromeric side of the Dam1 complex.
Conclusions
Our data show that the microtubule attachment site of the budding yeast kinetochore is well-organized. Ndc80, Dam1 and Stu2 are all narrowly distributed about their average positions along the kinetochore-MT axis. The relative organization of these components, their narrow distributions, and their known MT-binding properties together elucidate how their combined actions generate persistent, bidirectional kinetochore movement coupled to MT polymerization and depolymerization.
Introduction
Understanding the molecular mechanism of bidirectional chromosome movement during cell division is an enduring challenge in cell biology. Chromosome movement is driven by the kinetochore, a multiprotein, microtubule-based force coupler [1]. The properties of individual MT-binding proteins in the kinetochore are known. However, a cohesive understanding of how these proteins cooperate to achieve bidirectional chromosome movement is lacking.
A distinguishing feature of the kinetochore is its ability to generate chromosome movement in the absence of external energy sources by generating a motile force from the mechanochemical changes accompanying MT polymerization and depolymerization [2]. The mechanisms underlying such force generation depend on both, the MT-binding properties of kinetochore proteins and their nanoscale organization. For example, force coupling properties of the MT-binding Dam1 complex and the biophysical mechanism of Dam1-coupled motility depend on its oligomerization state [3, 4]. Similarly, the persistence of motility mediated by the kinetochore complex Ndc80 depends on its copy number and distribution relative to the MT tip [5, 6]. Thus, the nanoscale organization of MT-binding proteins within the kinetochore must be known in order to define the mechanisms underlying kinetochore motility.
The budding yeast kinetochore is an ideal model for studying how the organization of MT-binding proteins shapes the mechanisms of kinetochore movement. Key data regarding the MT-binding machinery of the yeast kinetochore are known. This machinery consists of three components: Ndc80 complex, Dam1 complex, and the protein Spc105. In addition, the MT-associated protein Stu2 is a functional component that is necessary for chromosome dynamics [7, 8]. The structures of these proteins are known [9–11]. It is also known that the yeast kinetochore incorporates an invariant copy number of each protein: at least 16–20 molecules of Dam1, and 5–8 copies of Spc105 and Ndc80, positioned at well-defined average locations along the kinetochore-MT attachment (Figure 1A top, [12–16]). Two critical facets of kinetochore organization remain unknown: (1) the distribution of multiple copies of each protein about its average position, and (2) their distribution around the MT circumference (Figure 1A). These data are necessary to define the organization of the yeast kinetochore, and to understand how this organization generates movement.
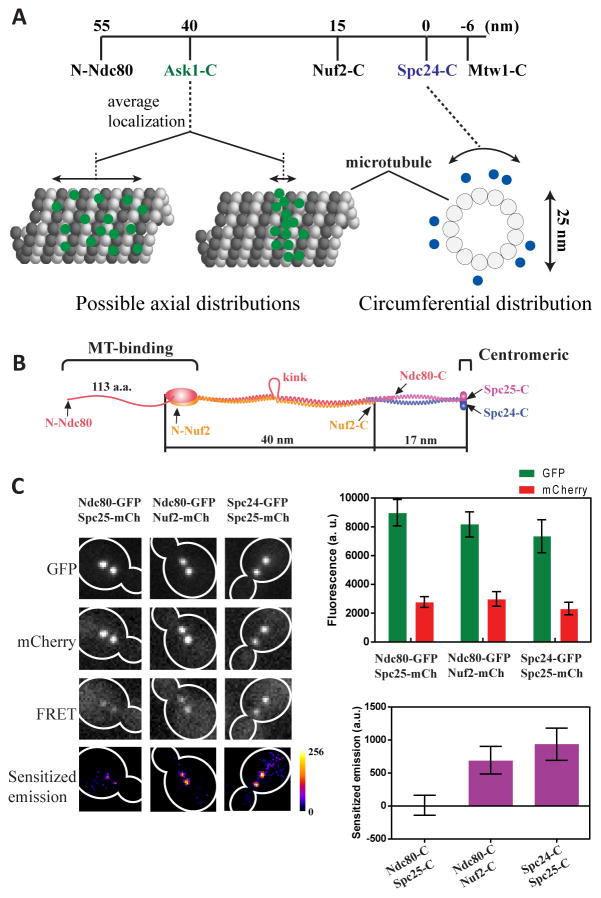
Figure 1. Quantification of FRET from metaphase kinetochore clusters.
(A) Top: The known average positions of kinetochore proteins along the kinetochore-MT attachment [14]. Nanoscale protein distributions along the MT axis (possibilities indicated in the cartoon) or around the circumference of the MT are unknown. (B) Physical dimensions and subunit organization of the Ndc80 complex. (C) Metaphase cells expressing two labeled Ndc80 subunits (indicated the top) as observed in the GFP, mCherry, and FRET channel. Heat maps of sensitized emission intensity were calculated by subtracting contributions of GFP bleed-through and mCherry cross-excitation (estimated using the GFP and mCherry signals measured in the respective images), and cellular auto-fluorescence from the FRET image. Quantification of the GFP, mCherry and sensitized emission intensity per kinetochore cluster (mean ± s. d.) shown on the right. Reduction in the GFP signal is due to FRET [17].
We previously developed a FRET-based technique to reconstruct nanoscale distributions of kinetochore proteins [17, 18]. To measure FRET, selected proteins, e.g. subunits of the Ndc80 complex (Figure 1B), are fused to either GFP(S65T) (the donor) or mCherry (the acceptor). FRET is quantified as the sensitized emission intensity, which is the acceptor fluorescence due to FRET, emanating from the two kinetochore clusters, each containing 16 kinetochores, seen in haploid cells in metaphase (Fig. 1C). This intensity is calculated by subtracting the GFP bleed-though and mCherry cross-excitation intensity from the fluorescence measured in the FRET channel [17]. We ensure that the sensitized emission intensity for each kinetochore cluster depends only on: (1) average FRET efficiency for all FRET pairs and (2) total number of FRET pairs within the cluster [17]. Furthermore, we construct strains wherein the labeled kinetochore subunits generate the same number of FRET pairs per kinetochore cluster. In such strains, the measured sensitized emission intensity depends only on the FRET efficiency, and hence the average donor-acceptor separation. Therefore, comparison of sensitized emission intensities can reveal relative proximities between labeled proteins.
Here we report the organization of the MT-binding machinery in the budding yeast kinetochore. For each protein, we obtained two types of FRET measurements. To elucidate axial protein distribution (i.e. along the length of the kinetochore-MT attachment), we measured FRET between the labeled protein and suitable reference points in the kinetochore. To obtain circumferential protein distribution, we measured FRET between neighboring copies of the same protein in heterozygous diploid strains. Importantly, each budding yeast kinetochore binds to exactly one MT tip. Therefore, we could combine these FRET data with known kinetochore protein structures, copy numbers, and localizations to reconstruct the in vivo architecture of the metaphase kinetochore-MT attachment.
Results
Subunit organization of the Ndc80 complex
First, we used the known structure of the Ndc80 complex to validate our methodology (Figure 1B, ref. [9, 19, 20]). Since FRET quantitation is obtained from fluorescence intensity, it is affected by cell-to-cell variation in GFP and mCherry maturation, and the distance of the kinetochore cluster from the coverslip [12] [17]. To minimize the effects of this experimental variation, we normalized the sensitized emission intensity for each cluster by dividing it with the sum of GFP bleed-through fluorescence and mCherry fluorescence due to cross-excitation (see Supplemental Experimental Procedures, also [17]). This normalized sensitized emission, termed as ‘Proximity Ratio’, is 0 when FRET efficiency is negligible, and directly proportional to non-zero FRET efficiency values [17].
As predicted by the known organization of the Ndc80 complex, we recorded the highest FRET between the carboxyl termini of Spc24 and Spc25, abbreviated as Spc24-C and Spc25-C respectively (Figure 2A, ref. [9, 20]). Ndc80-C and Nuf2-C produced a lower proximity ratio. For Ndc80-C and Spc25-C, which are ~ 10–15 nm apart, the proximity ratio was 0. We also detected FRET between the amine terminus of Ndc80 (N-Ndc80) and N-Nuf2, which are separated by a 113 amino acid long unstructured tail of Ndc80 (Figure 2A). This observation reveals that at least some of the tails are located within 10 nm of N-Nuf2 either belonging to the same Ndc80 complex or an adjacent complex. FRET increased when the tail was deleted (Δ113-Ndc80, Figure 2A), suggesting that some N-Ndc80 termini extend away from N-Nuf2. Thus, the in vivo FRET data confirm the known Ndc80 subunit organization.
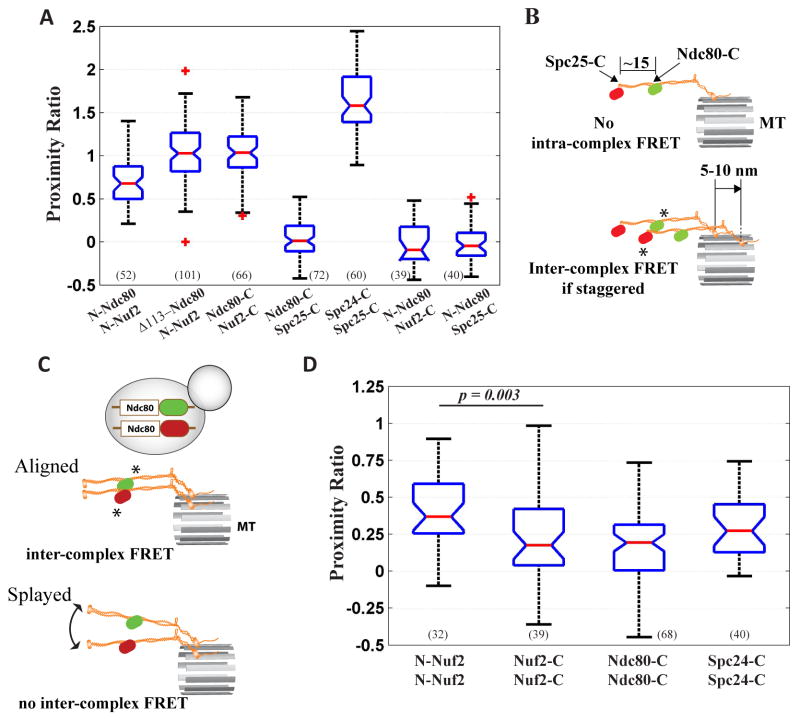
Figure 2. Distribution of Ndc80 complex molecules within the kinetochore.
(A) Box and whisker plot for proximity ratios quantifying FRET between Ndc80 complex subunits. The horizontal blue lines of each box represent the 25th and 75th percentile values, and the whiskers display the extreme values. Red line in each box indicates the median; red crosses display outliers. Non-overlapping notches on the box plots signify statistically significant differences in mean values (p < 0.05 using the Wilcoxon rank-sum test). The number of measurements for each dataset is indicated at the bottom. (B) If two Ndc80 complexes are staggered by distances > 10 nm along the length of the MT, then inter-complex FRET can occur between a GFP and mCherry on adjacent complexes (marked with asterisks). (C) Inter-complex FRET can be measured in heterozygous strains expressing one copy of the gene of interest labeled with GFP and the other with mCherry only if adjacent Ndc80 complexes are aligned with each other (FRET pair denoted by asterisks) and if they do not splay away from one another. (D) Proximity ratio quantification for inter-complex FRET between the labeled domains of neighboring Ndc80 complexes.
Neighboring Ndc80 complexes are aligned along their entire length
We next assessed the distribution of adjacent Ndc80 complexes within the kinetochore. The rigid structure and subunit organization of the Ndc80 complex precludes FRET between subunits belonging to the same Ndc80 complex, i.e. intra-complex FRET. However, inter-complex FRET can occur if neighboring Ndc80 complexes are staggered along the length of the MT (Figure 2B). Therefore, we specifically tested whether domains of the Ndc80 complex that are separated by > 10 nm within one complex, e.g. Ndc80-C and Spc25-C, nonetheless exhibit FRET. FRET between Ndc80-C and Spc25-C was negligible implying that neighboring Ndc80 complexes are rarely staggered by 5–10 nm. FRET was also absent between N-Ndc80 and either Nuf2-C or Spc25-C, which are separated from N-Ndc80 by 37 and > 57 nm respectively, confirming that the staggering does not exceed the 10 nm detection limit for FRET. Thus, neighboring Ndc80 complexes are rarely staggered along the length of the kinetochore.
The lack of FRET between Ndc80-C and Spc25-C could also indicate that the average spacing between neighboring Ndc80 complexes around the MT circumference is > 10 nm. To test this, we quantified inter-complex FRET in heterozygous diploid strains that express two versions of a selected subunit: one labeled with GFP and the other with mCherry (Figure 2C). The kinetochores in such strains incorporate GFP and mCherry labeled molecules randomly. For accurate comparison of proximity ratios, it is essential that the average number of GFP and mCherry labeled molecules per kinetochore cluster in each strain is equal [17]. We verified this by comparing the average GFP and mCherry fluorescence per kinetochore cluster in diploid strains with the kinetochore cluster fluorescence in haploid strains that express only GFP or only mCherry labeled subunits (Figure S1).
In diploid strains, FRET can occur only if adjacent complexes labeled with GFP and mCherry are located within 10 nm. Furthermore, if the neighboring complexes are parallel to and aligned with each other, then FRET will be similar along their entire length (Fig. 2C). Accordingly, modest inter-molecular FRET could be detected at three points positioned along the length of the Ndc80 complex (Figure 2D), indicating that at least a subset of neighboring Ndc80 molecules are within 10 nm. FRET between adjacent N-Nuf2 was slightly higher than the FRET between Nuf2-C, Ndc80-C or Spc24-C. Since N-Nuf2 is proximal to the MT-binding Calponin Homology (CH) domain of Ndc80, higher FRET indicates that the MT-binding domains of adjacent Ndc80 molecules are closer to one another than the centromere-binding domains. FRET between adjacent Nuf2-C, Ndc80-C, or Spc24C domains was statistically indistinguishable revealing that adjacent complexes run parallel to one another.
The Ndc80 complex bends in metaphase
The Ndc80 complex bends freely through 90° at the flexible kink in the Nuf2/Ndc80 dimer in vitro [21]. Therefore, we tested whether the Ndc80 complex also bends in vivo by measuring its length projected along the spindle axis using high resolution colocalization, and then comparing the measured length with its contour length [14]. Any reduction in the projected length reveals bending of the Ndc80 complex perpendicular to the axis of the kinetochore-MT attachment. This inference assumes that all Ndc80 complexes are aligned along the microtubule, which is supported by MT-binding and unbinding rates measured for single human Ndc80 molecules [22].
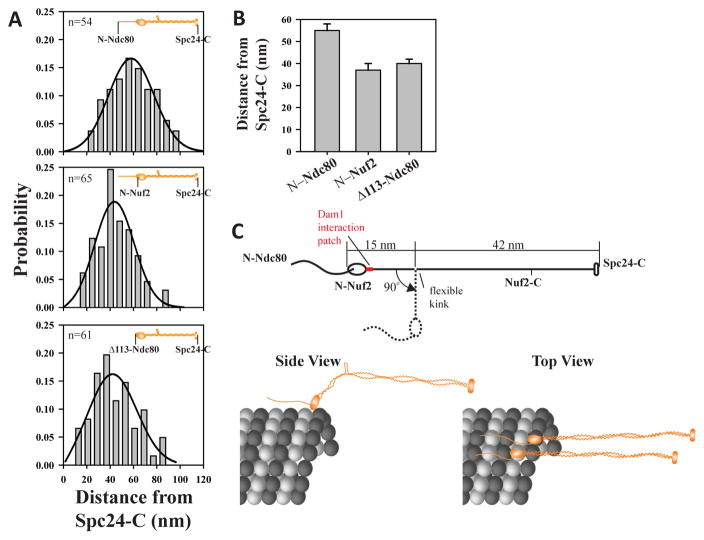
Using high-resolution colocalization of N-Nuf2 and Spc24-C, we found that these termini are separated by 37 ± 2 nm along the length of the kinetochore-MT attachment (Figure 3A–B, mean ± 95% confidence intervals predicted by maximum likelihood estimation, ref. [23]). This distance is significantly smaller than the extended contour length of 57 nm for Ndc80 [9]. Previous measurements also show that the projected length of the Spc24/Spc25 dimer in vivo equals its contour length [14]. Therefore, the observed reduction in the projected length is due to the bending of the Ndc80/Nuf2 dimer at the kink. The measured separation of 40 ± 3 nm between Δ113-Ndc80 and Spc24-C confirmed that the tail extends by ~ 15 nm away from the CH-domains adding this distance to the measured separation between N-Ndc80 and Spc24-C (also see Figure S2).
Figure 3. The Ndc80 complex bends in metaphase.
(A) Frequency distribution of the separation between the centroids of Spc24-mCherry and GFP-Ndc80, GFP-Nuf2, and GFP-Δ113-Ndc80 (indicated in the insets). Black curve is the non-Gaussian maximum likelihood fit [23]. (B) Average separation predicted by a non-Gaussian maximum likelihood fit (mean ± s. e. m.). (C) Metaphase architecture of the Ndc80 complex deduced from FRET and colocalization data.
The FRET and high-resolution colocalization measurements together define the distribution and conformation respectively of the Ndc80 complex in the metaphase kinetochore (Figure 3C). This organization establishes a spatial frame of reference that is useful for elucidating the distributions of other kinetochore components.
Dam1 molecules are concentrated near Ndc80/Nuf2 CH-domains
Two facets of Dam1 organization within the kinetochore are necessary for defining its function. First, the orientation of the 10-subunit Dam1 complex monomer within the kinetochore is needed to understand the contributions of individual Dam1 subunits to Dam1 complex function and regulation. Second, the oligomerization state of Dam1 molecules must be known to determine the properties and mechanism of Dam1-mediated force coupling [3, 4].
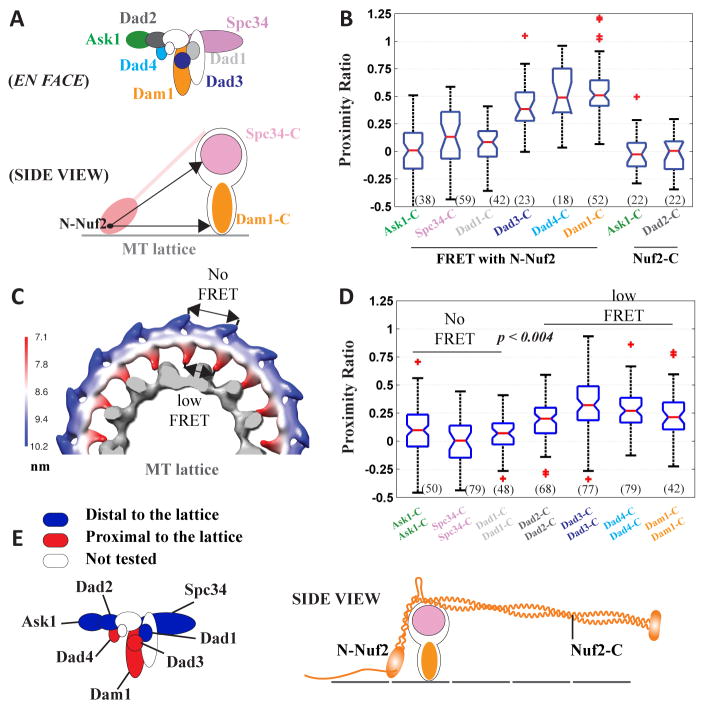
To understand the orientation of the Dam1 complex, we measured the proximity of Dam1 subunits relative to N-Nuf2, which is proximal to the MT lattice (Figure 4A). FRET between Ask1-C, Spc34-C, or Dad1-C with N-Nuf2 was negligible, suggesting that these C-termini are distal from N-Nuf2 (category 1). In contrast, significantly higher FRET detected for Dad3-C, Dad4-C, and Dam1-C suggests a closer proximity of these subunits to N-Nuf2 (category 2, Figure 4B). The same classification emerged from FRET measured between Dam1 subunits and Δ113-Ndc80 (Figure S2).
Figure 4. Distribution of Dam1 molecules in the metaphase kinetochore.
(A) The subunit organization of the Dam1 complex [10], and the schematic of our FRET-based approach to position each subunit relative to N-Nuf2, which is proximal to the MT. (B) Proximity ratio quantification for FRET between Dam1 subunits and either N-Nuf2 or Nuf2-C. (C) Pseudo-colored Dam1 ring EM density map (based on EMDB 5254, [24]) displays the separation between equivalent points on adjacent monomers of the ring. The MT lattice is displayed in gray. (D) Proximity ratio quantification for neighboring molecules of Dam1 subunits. (E) The proximity of the tested subunits to the MT lattice and the metaphase architecture of the Dam1 complex relative to Ndc80.
The categorization of Dam1 subunits above may reflect subunit placement either along the MT or radially away from it. To distinguish between these possibilities, we calculated the distance between copies of a Dam1 subunit in adjacent complexes as a function of its radial position (Figure 4C displays a pseudo-colored density map based on ref. [24]). According to this calculation, neighboring copies of a MT-proximal Dam1 subunit should allow FRET, while the copies of a subunit distal to the MT should not (indicated in Figure 4C). To measure this dependence in vivo, we assessed the separation between adjacent copies of the Dam1 subunits. FRET between adjacent C-termini of subunits in category 1 was minimal, which indicates a spacing ≥ 10 nm. In contrast, C-termini of subunits in category 2 generated modest FRET revealing that their spacing is < 10 nm (Figure 4D). These measurements suggest that Dam1 subunits in category 1 extend away from the MT lattice and from N-Nuf2. Thus, the Dam1 complex is positioned in the kinetochore with shoulders containing Dad1-C, Spc34-C, and Ask1-C distal to the MT, and Dam1-C, Dad3-C, and Dad4-C proximal to the MT lattice (Figure 4E).
The analysis above does not reveal whether Dam1 molecules oligomerize, because Dam1 rings and dimers predict the same subunit proximity (if the dimer fits into the ring without any conformational change). Lack of Dam1 oligomerization should correspond to a wide distribution of at least 8–10 Dam1 dimers, each of ~ 15 nm width, ~ 10 nm height, and ~ 5 nm thickness [10]. In this case, at least some of the Dam1 dimers should be proximal to Nuf2-C situated ~ 20 nm away from the CH-domains (Fig. 3C). However, we did not detect any FRET between Ask1-C or Dad2-C, and Nuf2-C. Since the average position of all the Dam1 molecules is also in close vicinity of the CH-domains [14], the absence of FRET suggests that most of the Dam1 molecules are concentrated here (Figure 4E). As discussed in the next section, localization of Dam1 molecules near N-Nuf2 is confirmed by the lack of FRET between Dam1 subunits and Stu2, which is concentrated at Nuf2-C.
An experimental issue with the Dam1 complex data in heterozygous diploid strains was that the apparent number of Dam1 molecules was unexpectedly low for all of the subunits tested (Figure S1). Quantitative Western blots revealed that the lower kinetochore recruitment was not due to lower protein expression (Figure S1). The lower fluorescence may be due to hypomorphic chimeras. Importantly, the lower abundance was observed for all the Dam1 subunits. Therefore, our conclusions regarding the proximity between adjacent copies of a Dam1 subunit based on comparative FRET analysis should not be affected.
Stu2 is narrowly distributed in the interior of the kinetochore
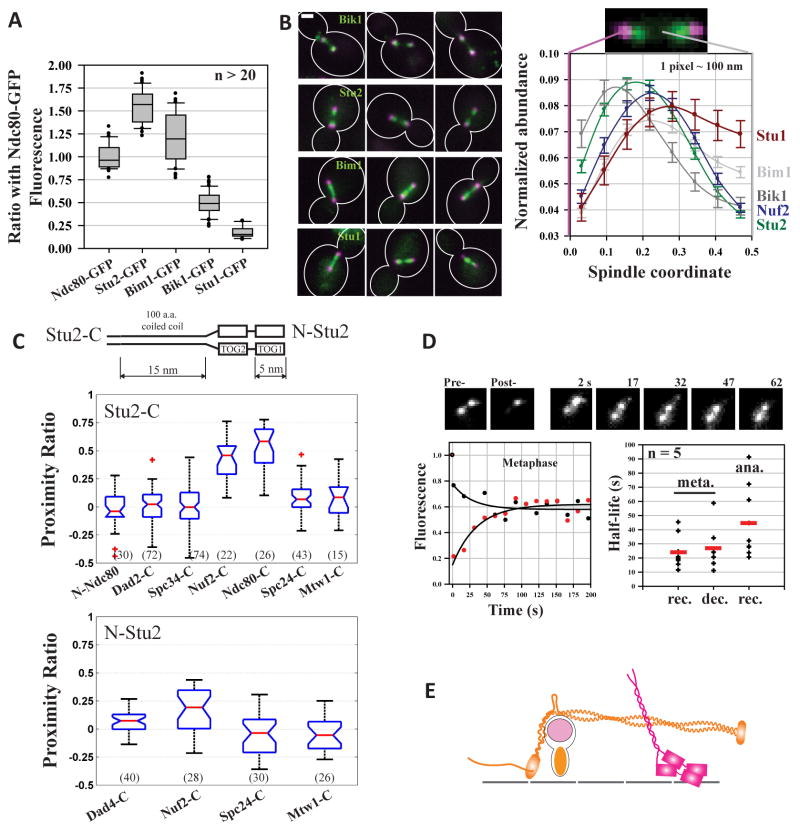
Microtubule Associated Proteins (MAPs) also contribute to kinetochore motility, either through a kinetochore-specific function or by controlling MT dynamics and spindle stability. Therefore, we examined the spindle abundance and distribution of four MAPs that bind the MT tip: Stu1 (CLASP), Bik1 (CLIP-170), Bim1 (EB1) and Stu2 (XMAP215/chTOG1), to assess whether they perform kinetochore-specific functions.
We first quantified the abundance of each MAP using quantitative fluorescence microscopy. Stu2 was the most abundant, followed by Bim1 (Figure 5A, [25]). The measured Stu2 abundance translates into an average of ~ 6–7 Stu2 dimers per kinetochore (assuming 8 Ndc80 molecules per kinetochore [13]). Bik1 abundance was significantly lower. Stu1, which functions mainly at unattached kinetochores [26], was the least abundant MAP. We next quantified the distribution of each MAP along the spindle using a previously established method [27, 28]. Stu1 distribution was skewed towards the spindle mid-zone, whereas Bik1 distribution was skewed towards the spindle pole (Figure 5B). Bim1 distribution was more or less uniform along the length of the spindle. Strikingly, Stu2 formed two distinct clusters that colocalized with Nuf2, a Ndc80 complex subunit. The large number of Stu2 molecules concentrating within the kinetochore suggests a kinetochore-specific function for Stu2.
Figure 5. Abundance and spindle distribution of MAPs.
(A) Relative abundance of Stu2, Bik1, Bim1, and Stu1. In comparison with Stu2, other MAPs have a significantly lower abundance (p-value < 1e-9 from a two-sided Student’s t-test) and a significantly larger cell-to-cell variation in abundance (p-value < 0.02 from a two-sided F-test). (B) Representative micrographs of MAP distribution (spindle extremities marked by Spc97-mCherry, a spindle pole body protein; scale bar ~ 1 μm). Normalized distributions of Bik1, Bim1, Stu1, Stu2, and Nuf2 in metaphase arrested cells (mean +\− s.e.m.). (C) Top: schematic of the Stu2 dimer. Stu2 length was estimated by adding 5 nm length of its two TOG domains [42], and the estimated 15 nm contour length of its 100 amino acid long α-helical coiled coil domain (3.6 residues per turn and a pitch of 0.54 nm). FRET quantification for Stu2-C (upper graph) or N-Stu2 (lower graph) and kinetochore subunits. (D) Fluorescence Recovery after Photobleaching (FRAP) of Stu2-GFP. Red circles display the fluorescence recovery, and black circles display the concurrent fluorescence decay of the unbleached cluster in the same cell. The initial, steep decrease in the intensity of the unbleached cluster is due to inadvertent photobleaching. Black lines display single exponential fit of the data. The scatter plot displays the half-life for fluorescence recovery (rec.) and decay (dec.) in metaphase and only recovery in anaphase. Red line indicates the mean value. (E) Stu2 localization in the metaphase kinetochore.
Stu2 is necessary for chromosome dynamics and for keeping spindle MTs dynamic [7, 29]. Therefore, we characterized its distribution within the kinetochore using FRET. We found that Stu2-C and N-Stu2 both localize close to Nuf2-C and Ndc80-C, as evidenced by high FRET (Figure 5C). Stu2 was also narrowly distributed, because Stu2-C or N-Stu2 were not proximal to any other kinetochore subunits that are centromere proximal (Mtw1-C) or located in the outer kinetochore (N-Ndc80, Dad2-C or Dad4-C). Thus, Stu2 molecules occupy a distinct territory within the kinetochore. High-resolution colocalization also placed the centroid of Stu2-C fluorescence in close vicinity of Nuf2-C, confirming that the majority of Stu2 molecules are concentrated near Nuf2-C or Ndc80-C (Figure S4). FRET between Ndc80 subunits and either Bim1 or Bik1 was negligible, consistent with their skewed spindle distributions (Figure S4).
Unlike other kinetochore proteins, Stu2 localization in the kinetochore was highly dynamic [12]. Stu2-GFP fluorescence recovered completely within a minute after photobleaching (Figure 5D). The turn-over rate is > 2-fold faster than tubulin turn-over (t1/2 = 24 ± 5 s in metaphase and 45 ± 5 s in anaphase, mean ± s.e.m., n > 5 for both, [30]). Thus, Stu2 emerges as the only tip-interacting protein that occupies a well-defined position within the yeast kinetochore (Figure 5E).
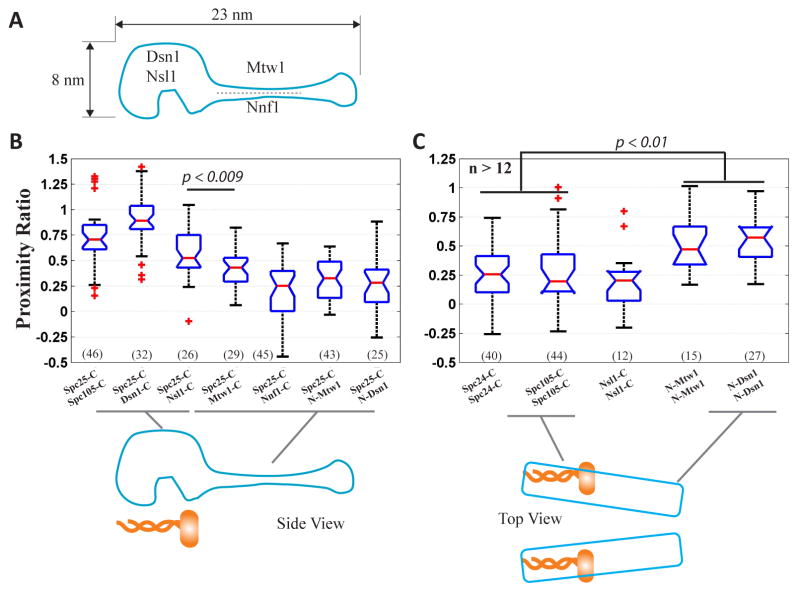
Organization of the Mtw1 complex relative to the Ndc80 complex
Finally, we investigated the organization of the Mtw1 complex, and the kinetochore protein Spc105. Mtw1 complex connects Ndc80 to the centromere via physical interaction between the Mtw1 subunit and the centromeric protein CENP-C [31–33]. High resolution colocalization data place the C-termini of Mtw1 complex subunits within ~ 10 nm of Spc25-C [14]. Therefore, we selected Spc25-C as the reference point to measure the distribution of Mtw1 subunits. Nsl1-C and Dsn1-C as well as Spc105-C are proximal to Spc25-C, as evidenced by the high FRET between these termini and Spc25-C (Figure 6B). FRET between Spc25-C and Nnf1-C, Mtw1-C, N-Dsn1 or N-Mtw1 was lower, indicating a larger separation. These data are largely consistent with the subunit organization of the Mtw1 complex predicted by structural studies [34, 35]. Direct binding between Mtw1 and Ndc80 complex predicts that the circumferential distribution of Mtw1 subunits proximal to Ndc80 should be the same as Ndc80 complexes. This was the case for Nsl1-C and Spc105-C (Figure 6C). Surprisingly, inter-complex FRET was significantly higher for both N-Mtw1 and N-Dsn1 indicating a narrower spacing of these two termini (schematic, Fig. 6C).
Figure 6. Metaphase architecture of the Mtw1 complex.
(A) Architecture of the Mtw1 complex based on ref. [34, 35]. (B) FRET between Spc105-C or Mtw1 subunits, and Spc25-C. (C) FRET between neighboring Mtw1 complexes.
Kinetochore subunit organization is maintained in late anaphase\telophase
In late anaphase, the kinetochore architecture is expected to change due to partial dissociation of the Dam1 complex and also the centromere-bound CBF3 complex [12, 36]. Despite these changes in kinetochore composition and behavior, the organization of kinetochore proteins relative to one another did not change in late anaphase/telophase. However, the kinetochore was more compact as evidenced by a systematic increase in FRET (Figure S5).
Discussion
The FRET and high-resolution colocalization data provide novel insights into the physiological organization of the kinetochore-MT attachment. We combined these insights with structural data and previously reported protein counts to reconstruct a 3-D visualization of the metaphase kinetochore-MT attachment (Figure 7A). Although the large size of fluorescent proteins can be problematic when using FRET for deducing protein architecture, our experimental design ensures that it has minimal effect on the main conclusions (Supplementary Note 1). We used FRET only to compare: (a) proximity between different subunits of the same heteromeric complex and one reference point, or (b) proximity between adjacent copies of subunits of the same heteromeric complex. This design ensures that each experiment involves similar numbers of FRET pairs and differences in proximity ratios result from differences in FRET efficiencies. We also confirmed each key conclusion with multiple measurements using similarly situated kinetochore subunits (by virtue of their known average position or because they belong to the same complex). Therefore, the organization of MT-binding proteins revealed by the FRET data can be used to elucidate protein functions.
Figure 7. Visualization of the budding yeast kinetochore-MT attachment.
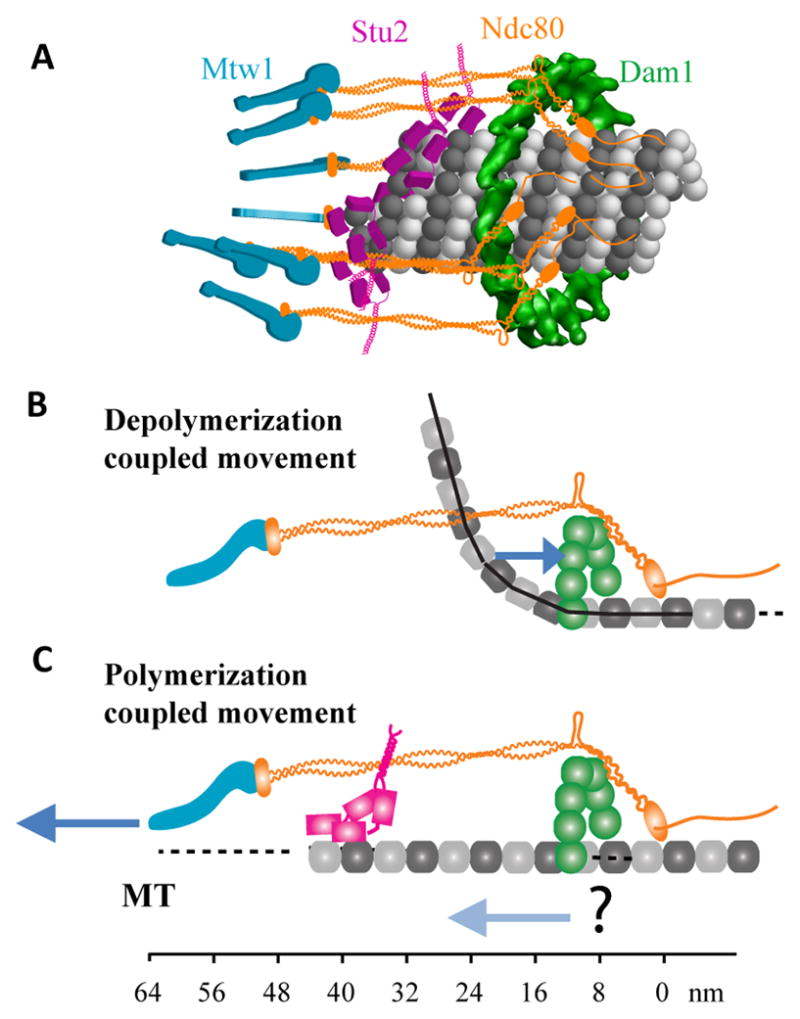
(A) Visualization of the kinetochore-MT attachment in metaphase. Spc105 is not shown. (B) Proposed mechanism of MT depolymerization-coupled motility (only one protofilament of the MT is shown). Black line displays the protofilament profile for a kinetochore MT from Fig. 4b in [49]. Blue arrow indicates force coupling by the Dam1 complex. (C) Proposed mechanisms for MT polymerization-coupled motility. The dark blue arrow represents centromeric tension, while the light blue arrow represents higher Dam1 affinity for GTP-tubulin [50].
We find that the Ndc80 complex bends even in metaphase, likely at the kink within the Ndc80/Nuf2 dimer. A previous study depicted the Ndc80 molecule running parallel to the MT along its entire length [14]. However, this study assumed that the unstructured tail at the N-terminus of Ndc80 does not contribute to the in vivo, projected length of the Ndc80 complex. We find that the projected length measured from N-Nuf2 to Spc24-C is ~ 40 nm, and that the unstructured tail adds 15 nm to this length. This Ndc80 conformation is also consistent with the average position of Dam1 molecules ~ 15 nm on the centromeric side of N-Ndc80. This position implies that Dam1 molecules should be proximal to N-Nuf2. This is confirmed by detected FRET between Dam1 subunits and N-Nuf2. The bent Ndc80 conformation will facilitate optimal binding between the CH-domains and the MT [37]. Based on the location of the Dam1-interacting residues proximal to the CH-domain of Ndc80, it may also promote binding of Ndc80 to Dam1 [38].
We also find that at least a fraction of Ndc80 complexes have a neighbor within 10 nm. Together with the lack of staggering along the length of the kinetochore, this observation suggests that the Ndc80 complexes are narrowly distributed along the length of the MT. The narrow Ndc80 complex distribution along the length of the MT is not well-suited for persistent attachment with a depolymerizing MT tip, especially under opposing forces [6]. Therefore, the Ndc80 complex is unlikely to be the major force coupler during MT depolymerization-driven motility. The circumferential distribution of Ndc80 complexes cannot be determined directly. However, detectable inter-complex FRET suggests that it cannot be symmetric, because symmetric placement of 8 Ndc80 complexes over a circle > 25 nm in diameter translates into an inter-complex spacing > 10 nm. As the simplest case, we depict Ndc80 complexes as randomly distributed around the MT circumference (Fig. 7A).
The Dam1 complex acts as a processivity factor for the Ndc80 complex in vitro [39, 40]. Our data show that Dam1 molecules are concentrated in close vicinity of the CH-domains of the Ndc80 complex. This narrow Dam1 distribution and its strong tendency to oligomerize when bound to the MT suggest that the metaphase kinetochore may contain a Dam1 ring or ring-like oligomer. Such an oligomeric Dam1 assembly can efficiently generate force coupled to MT depolymerization and drive kinetochore motility [3, 4]. The organization of Dam1 subunits relative to the MT lattice is generally consistent with the subunit organization suggested by structural studies [10]. However, one notable difference is the opposite orientations of Dam1-C suggested by the two studies. FRET data suggest that Dam1-C is oriented towards the MT lattice (consistent with the localization of the GFP-density observed in Dam1 rings incorporating Dam1-GFP [41]), whereas a recent study suggests that Dam1-C points away from the MT lattice [10]. Further work is necessary to resolve this discrepancy.
The narrow distribution of a large number of Stu2 molecules within the kinetochore is surprising, because Stu2 is capable of binding to MTs anywhere along the spindle. We hypothesize that Stu2 localizes within the kinetochore by autonomously binding at the tip of the MT. The TOG domains of Stu2 bind specifically to tubulin dimer conformations present only the MT tip [42–44]. This tip-specific binding has been observed for Stu2 homologs in vivo [45] and for Stu2 in vitro [43, 44]. Furthermore, MT tips, when stabilized with taxol, extend up to Spc24-C in vertebrate kinetochores [46]. Stu2 is narrowly distributed in the same region in the yeast kinetochore. It is unlikely that the Ndc80 complex, which is proximal to Stu2, recruits it, because physical interaction between the two is undetectable [8]. Together, these observations suggest that the dynamically localizing Stu2 molecules likely reveal the position of growing MT tips in the yeast kinetochore.
When considered together, the distributions of MT-binding proteins described here suggest an elegant, integrative mechanism that corrals the dynamic MT tip within the kinetochore. In this mechanism, the Ndc80-Dam1 assembly couples kinetochore movement to MT depolymerization, and ensures persistent attachment by implementing tension-dependent rescue [47]. MT destabilizing activity of Stu2 located in the interior of the kinetochore ensures that the growing MT tip remains within the kinetochore. Although the MT-binding machinery in yeast, with the exception of the Ndc80 complex, is not conserved in higher eukaryotes, the overall organization of MT-binding proteins is maintained [14, 46, 48]. Therefore, the mechanism of corralling the dynamic MT tip may be conserved by the organization of MT-binding proteins in the eukaryotic kinetochore.
Experimental Procedures
A complete description of the imaging and image analysis methodology is provided in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Ndc80 complexes are narrowly distributed within the budding yeast kinetochore
Dam1 complex molecules concentrate near microtubule-binding domains of Ndc80
Stu2 molecules localize dynamically proximal to the centromeric end of the Ndc80
This organization suggests a cohesive model of bidirectional kinetochore movement
Acknowledgments
We would like to dedicate this study to the memory of the late Prof. Alan Hunt. We would like to thank Arshad Desai, David Drubin, and Sue Biggins for sharing reagents, and Jennifer Deluca and Mara Duncan for critical comments on the manuscript. APJ is supported by the Career Award at the Scientific Interface from the Burroughs-Wellcome Fund. This work was supported by R01-GM-105948.
Footnotes
Supplemental information includes five figures, one supplementary note, a table of strains used in the study, and supplemental experimental procedures.
The authors have no financial interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Santaguida S, Musacchio A. The life and miracles of kinetochores. Embo J. 2009 doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, Barnes G, Ataullakhanov FI, McIntosh JR. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Efremov A, Grishchuk EL, McIntosh JR, Ataullakhanov FI. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19017–19022. doi: 10.1073/pnas.0709524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joglekar AP, Bloom KS, Salmon ED. Mechanisms of force generation by end-on kinetochore-microtubule attachments. Curr Opin Cell Biol. 2010;22:57–67. doi: 10.1016/j.ceb.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K. Yeast Kinetochores Do Not Stabilize Stu2p-dependent Spindle Microtubule Dynamics. Molecular Biology of the Cell. 2003;14:4181–4195. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maure JF, Komoto S, Oku Y, Mino A, Pasqualato S, Natsume K, Clayton L, Musacchio A, Tanaka TU. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol. 2011;21:207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramey VH, Wong A, Fang J, Howes S, Barnes G, Nogales E. Subunit organization in the Dam1 kinetochore complex and its ring around microtubules. Mol Biol Cell. 2011;22:4335–4342. doi: 10.1091/mbc.E11-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Bassam J, van Breugel M, Harrison SC, Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. The Journal of cell biology. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravamudhan P, Felzer-Kim I, Joglekar AP. The budding yeast point centromere associates with two Cse4 molecules during mitosis. Curr Biol. 2013;23:770–774. doi: 10.1016/j.cub.2013.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. The Journal of cell biology. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coffman VC, Wu P, Parthun MR, Wu JQ. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. The Journal of cell biology. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joglekar AP, Chen R, Lawrimore JG. A sensitized emission based calibration of FRET efficiency for probing the architecture of macromolecular machines. Cell and Molecular Bioengineering. 2013;6:369–382. doi: 10.1007/s12195-013-0290-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller EG, Snydsman BE, Novik I, Hailey DW, Gestaut DR, Niemann CA, O’Toole ET, Giddings TH, Jr, Sundin BA, Davis TN. The organization of the core proteins of the yeast spindle pole body. Mol Biol Cell. 2005;16:3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaytsev AV, Ataullakhanov FI, Grishchuk EL. Highly Transient Molecular Interactions Underlie the Stability of Kinetochore-Microtubule Attachment During Cell Division. Cell Mol Bioeng. 2013;6 doi: 10.1007/s12195-013-0309-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Churchman LS, Flyvbjerg H, Spudich JA. A non-Gaussian distribution quantifies distances measured with fluorescence localization techniques. Biophys J. 2006;90:668–671. doi: 10.1529/biophysj.105.065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramey VH, Wang HW, Nakajima Y, Wong A, Liu J, Drubin D, Barnes G, Nogales E. The Dam1 ring binds to the E-hook of tubulin and diffuses along the microtubule. Mol Biol Cell. 2011;22:457–466. doi: 10.1091/mbc.E10-10-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang PJ, Huffaker TC. Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body. The Journal of cell biology. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ortiz J, Funk C, Schäfer A, Lechner J. Stu1 inversely regulates kinetochore capture and spindle stability. Genes & Development. 2009;23:2778–2791. doi: 10.1101/gad.541309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sprague BL, Pearson CG, Maddox PS, Bloom KS, Salmon ED, Odde DJ. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys J. 2003;84:3529–3546. doi: 10.1016/S0006-3495(03)75087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimogawa MM, Graczyk B, Gardner MK, Francis SE, White EA, Ess M, Molk JN, Ruse C, Niessen S, Yates JR, III, et al. Mps1 Phosphorylation of Dam1 Couples Kinetochores to Microtubule Plus Ends at Metaphase. Current Biology. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolyniak MJ, Blake-Hodek K, Kosco K, Hwang E, You L, Huffaker TC. The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol Biol Cell. 2006;17:2789–2798. doi: 10.1091/mbc.E05-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westermann S, Cheeseman IM, Anderson S, Yates JR, 3rd, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. The Journal of cell biology. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrovic A, Mosalaganti S, Keller J, Mattiuzzo M, Overlack K, Krenn V, De Antoni A, Wohlgemuth S, Cecatiello V, Pasqualato S, et al. Modular Assembly of RWD Domains on the Mis12 Complex Underlies Outer Kinetochore Organization. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 33.Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornung P, Maier M, Alushin GM, Lander GC, Nogales E, Westermann S. Molecular Architecture and Connectivity of the Budding Yeast Mtw1 Kinetochore Complex. Journal of Molecular Biology. 2010;405:548–559. doi: 10.1016/j.jmb.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maskell DP, Hu XW, Singleton MR. Molecular architecture and assembly of the yeast kinetochore MIND complex. The Journal of cell biology. 2010;190:823–834. doi: 10.1083/jcb.201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouck DC, Bloom KS. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5408–5413. doi: 10.1073/pnas.0405925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. The Journal of cell biology. 2013;200:21–30. doi: 10.1083/jcb.201210091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. The Journal of cell biology. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. The Journal of cell biology. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang HW, Ramey VH, Westermann S, Leschziner AE, Welburn JP, Nakajima Y, Drubin DG, Barnes G, Nogales E. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007;14:721–726. doi: 10.1038/nsmb1274. [DOI] [PubMed] [Google Scholar]
- 42.Ayaz P, Ye X, Huddleston P, Brautigam CA, Rice LM. A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–860. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maurer SP, Cade NI, Bohner G, Gustafsson N, Boutant E, Surrey T. EB1 Accelerates Two Conformational Transitions Important for Microtubule Maturation and Dynamics. Curr Biol. 2014 doi: 10.1016/j.cub.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Breugel M, Drechsel D, Hyman A. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. The Journal of cell biology. 2003;161:359–369. doi: 10.1083/jcb.200211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura S, Grigoriev I, Nogi T, Hamaji T, Cassimeris L, Mimori-Kiyosue Y. Dissecting the nanoscale distributions and functions of microtubule-end-binding proteins EB1 and ch-TOG in interphase HeLa cells. PLoS One. 2012;7:e51442. doi: 10.1371/journal.pone.0051442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. The Journal of cell biology. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McIntosh JR, O’Toole E, Zhudenkov K, Morphew M, Schwartz C, Ataullakhanov FI, Grishchuk EL. Conserved and divergent features of kinetochores and spindle microtubule ends from five species. The Journal of cell biology. 2013;200:459–474. doi: 10.1083/jcb.201209154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.