Abstract
CPAF is a conserved and secreted protease from obligate intracellular bacteria of the order Chlamydiae. Recently, it was demonstrated that most of its host targets are an artifact of inaccurate methods. This review aims to summarize key features of CPAF and propose new approaches for evaluating its role in chlamydial pathogenesis.
Keywords: CPAF, Substrates, Chlamydial virulence factors and pathogenesis
CPAF discovery and biochemical properties
CPAF was first identified as a factor responsible for a chlamydial protease/proteasome-like activity (CPA) in the cytosol of Chlamydia-infected cells [1]. The CPA degraded RFX5 in a cell-free assay and was only inhibited by the irreversible proteasome inhibitor lactacystin. After screening thousands of fractions derived from Chlamydia-infected cell cytosol for a RFX5 degradation activity, two protein bands resolved in a SDS-polyacrylamide gel were correlated with the RFX5 degradation activity. Trypsin digestion and tandem mass spectrometry analyses revealed that the two protein bands represented a single protein with one representing the N- and the other representing the C-terminal half of the protein that is encoded by a hypothetical open reading frame CT858.
Further structural and biochemical characterization of CPAF has laid the foundation for understanding the biological significance of CPAF. Following the observation that native CPAF was detected in C- and N-terminal fragments, the cleavage site was mapped and shown to be necessary and sufficient for CPAF proteolytic activity [2]. Furthermore, it was determined that intramolecular dimerization of these two fragments was necessary for CPAF to cleave its substrates [3]. Both CPAF activity and activation were found to be highly conserved among chlamydial species [4]. The most unique feature of CPAF was realized when CPAF was found to autoprocess and self-activate [5], effectively classifying it as a zymogen. Introducing the missense mutation E558A disrupted one of three proposed water-based catalytic triad residues (H105, S499, and E558) and destroyed its ability to participate in self-activation [6]. The crystal structure analyses and careful biochemical assays of CPAF revealed that autoprocessing was a sequential process occurring at three specific sites between the C- and N-terminal fragments [7]. The cleavage and removal of an internal inhibitory peptide occupying the substrate binding groove were necessary for CPAF to gain proteolytic activity. The crystal structural analyses further suggest that proximity-dependent homodimerization between two independent CPAF proteins allows them to trigger the autoprocessing by carrying out the first trans-cleavage at one of the 3 cleavage sites. The remaining 2 cleavages may take place via a cis-cleavage mechanism. Had it not been for the discovery of the internal inhibitory peptide, the CPAF inhibitory peptide, which was engineered from this internal peptide, would not be used today for specifically inhibiting CPAF during infection. The crystal structural study has also revealed the structural basis for CPAF to possess broad substrate specificity since CPAF cleavage of the inhibitory peptide tolerated multiple substitution mutations.
The chlamydial organisms that produce CPAF
The order Chlamydiae is a unique group of gram-negative obligate intracellular bacteria. Members of this order range from endosymbionts of single-celled eukaryotes to very successful human pathogens, which speaks to their ability to adapt to and parasitize their hosts. Chlamydia trachomatis, the most clinically-relevant of all chlamydial organisms, is divided into biovars that are characterized by the disease they cause, with further subdivision into serovars by the distinct humoral response they elicit [8]. The ocular strains of trachoma biovar, comprised of serovars A through C, cause preventable blindness while the genital strains, consisting of serovars D to K, cause sexually transmitted infections (STIs). Of similar concern are serovars L1 through L3 of the lymphogranuloma venereum (LGV) biovar, which are responsible for a systemic STI in men who have sex with men in Europe [9]. Aside from C. trachomatis, Chlamydia pneumoniae mainly infect the human respiratory tract, causing community-acquired pneumonia [10]. Although Chlamydia psittaci is an animal pathogen, it can cause life-threatening pneumonia in humans when inhaling aerosolized feces from infected birds [11].
Despite the wide variety of diseases they cause, all chlamydial organisms follow the same biphasic lifecycle whereby small (~0.2 μm) metabolically inactive elementary bodies (EBs) enter the cell and transform into large (~1 μm) metabolically active and dividing reticulate bodies (RBs). Intracellular growth is supported and protected by a parasitophorous vacuole, termed the inclusion, in which RBs begin constructing shortly after cell entry. Following around 48 hours of replication and inclusion growth, RBs convert back into EBs and the tightly packed inclusion bursts, releasing EBs into the extracellular environment to infect new host cells. Another common feature of chlamydial organisms is their genomes, which are strikingly similar to one another. How the almost undistinguishable lifestyles and genomes of Chlamydia can contribute to pathogenesis in diverse tissues is of great interest to the Chlamydia field.
Of special importance is the pathogenesis of the serovars with a tropism for urogenital epithelial tissues. While sexually-transmitted infection can be asymptomatic, however if not treated in time, ascension of the chlamydial organisms can lead to complications such as pelvic inflammatory disease, ectopic pregnancy and tubal factor infertility [12]. Clearly, the pathogenicity of C. trachomatis depends on both how successfully chlamydial organisms reproduce inside cells and how efficiently they spread from cell to cell and ascend to the upper genital tract. Chlamydia must have evolved strategies for evading both intra- and extracellular host defense mechanisms so that chlamydial organisms can safely infect the next target cells and start new rounds of intracellular propagation. Identifying chlamydial virulence factors that contribute to the above processes will not only advance our knowledge on the chlamydial pathogenic mechanisms but also provide essential information for developing new intervention and prevention approaches for controlling chlamydial infection and diseases.
CPAF as a chlamydial virulence factor
Behind every successful pathogen is a set of successful virulence factors, and the same is becoming true of C. trachomatis. Thus far, few have been discovered in Chlamydia, most likely due to the fact that a system for targeted genetic manipulation of the chromosome does not exist at the time of writing. However, removal of the chlamydial cryptic plasmid of C. muridarum, the murine equivalent and model of the C. trachomatis urogenital serovars, resulted in reduced upper genital tract pathologies [13]. Mice that spontaneously cleared the plasmid-deficient C. muridarum infection displayed reduced bacterial load and greatly diminished pathologies upon challenge with the wild type organism [13], suggesting that plasmid-free Chlamydia can act as a live attenuated vaccine. Similarly, a plasmid-free C. trachomatis ocular serovar protected half of non-human primates from severe ocular infection when challenged with the wild type agent [14]. It is still unknown how the plasmid contributes to pathogenesis, but likely factors include: Pgp3, an immunodominant antigen [15, 16], secreted protein [17], and component of the chlamydial outer membrane complex, and Pgp4, a master transcriptional regulator of plasmid and chromosomal genes [18, 19]. In addition to these two plasmid genes, the hypothetical protein CT135 has been implicated in pathogenesis due to the fact that the intact gene allows for prolonged urogenital tract infection in mice, but a nonsense mutation resulting in a truncated protein product shortens time to clearance in vivo without affecting in vitro growth dynamics [20].
Multiple approaches have been used for searching for chlamydial virulence factors, including bioinformatics analyses [21], surrogate secretion systems [22, 23] and microscopic localization [24]. These approaches have led to the discovery of many putative effectors. However, chlamydial proteins that have been visualized in the cytosol of Chlamydia-infected cells are CPAF [1], cHtrA [25], CT621 [26], CT622 [27], CT311 [28, 29], CT795 [30], GlgA [31], the C-terminus of OmcB [32] & Pgp3 [17]. Nevertheless, despite the definitive evidence for their localization in the host cell cytosol, the precise roles of these factors in chlamydial pathogenesis remain largely unknown.
Over the course of a decade, the secreted protease CPAF was shown to cleave or degrade a wide range of host proteins [33], including those involved in Golgi apparatus restructuring (golgin-84; ref: [34, 35], apoptosis (Puma, Bik, and Bim; ref: [36]), immune functions (RFX5, USF-1, NFκB p65, HMBG1, USF-1, and CD1d), cell cycle (cyclin B1), cell structure (keratin-8, keratin-18, and vimentin; ref: [37]), cellular adhesion (nectin-1), hypoxia signaling (HIF-1), and DNA repair (PARP). RIPA buffer, which stands for radioimmunoprecipitation assay buffer, was the primary cell lysis solution used to collect infected cell samples to then probe for the disruption of host proteins. However, RIPA buffer is specifically designed to keep proteins intact while releasing them from within cells and organelles. Additionally, CPAF is an atypical serine protease that is not susceptible to inhibition by protease inhibitors [1] used in RIPA buffer. Together with the fact that purified recombinant CPAF has a broad and strong activity against the listed proteins in vitro, these ingredients helped produce the artifacts.
Upon reevaluation of CPAF substrates using new techniques to prevent its activity in vitro, it was discovered that all of those tested were not noticeably cleaved or degraded during infection [38]. Interestingly, the specific proteasome inhibitor lactacystin has the unique ability to inhibit CPAF proteolytic activity[1]. In hindsight, it is not surprising that CPAF displays such broad activity given that an inhibitor of the proteasome, the most notoriously broad class of protease known, is the only known protease inhibitor capable of preventing it from cleaving nearly any proteins. After the procedure was modified to include treatment of infected cells with 150 μm of lactacystin for one hour prior to harvesting, followed by lysis in 8 M urea, there was no change between protein levels or banding patterns in Western Blots for many of the host substrates. In a similar fashion, sodium dodecyl sulfate (SDS) can be used to disrupt all protein activity during cell harvesting without the requirement for specific inhibition of CPAF one hour beforehand.
Despite negative findings, CPAF does have two chlamydial substrates: itself [5, 7] and the 60kDa cysteine-rich outer membrane protein B (OmcB; ref: [39]). Only the cleaved forms of these proteins can be detected in cells lysed in 8 M urea, suggesting that observations of cleavage by CPAF in cell-free assays are indeed true. For OmcB, it has been known that its C-terminus (OmcBc) serves as an immunodominant CD8+ T cell antigen, but not until recently was it shown that this portion of the protein is released into the host cell cytosol [32]. The advent of another CPAF inhibitor, a peptide that mimics an inhibitory domain of the native protein and gains access to the host cell through a polyarginine cell-penetrating peptide tag, has allowed for the further characterization of CPAF-specific cleavage events. When recombinant OmcB is incubated with C. trachomatis-infected HeLa cell lysate, the lysate maintains the ability to completely process the full-length protein into the OmcBc fragment. However, addition of the CPAF inhibitory peptide completely blocks such activity. Given that the inhibitory peptide is derived from CPAF and likely highly specific, these observations have demonstrated that CPAF does cleave OmcB into OmcBc.
The ability of the CPAF-specific inhibitor to inhibit chlamydial growth makes it difficult to determine whether a reduction in OmcBc protein level is due to specific inhibition of CPAF proteolytic activity or lack of chlamydial growth. On the other hand, this phenomenon is promising in that it shows that CPAF is necessary for a normal infection phenotype, as normality is lost upon its inhibition. It is also necessary to determine whether these activities are due to CPAF alone and if other contributing proteases can also be inhibited by lactacystin and the inhibitory peptide. The annotated chlamydial genome contains hundreds of hypothetical proteins and uncharacterized proteases that have developed alongside CPAF, any number of which could serve redundant roles in CPAF activation, OmcB processing, and may also be repressed by CPAF inhibitors. The ability of CPAF to cleave or degrade other chlamydial proteins, such as IncC, IncD, IncE, TARP, CT005, CT288, CT694, and CT813 [37], remains to be validated. Time and effort will eventually answer these inquiries, but more pressing questions on the minds of Chlamydia researchers are: what are the true substrates of CPAF and does it contribute to pathogenesis?
CPAF as an immunogen
Regardless of what substrates CPAF targets, the fact that CPAF is highly immunogenic during chlamydial urogenital and ocular infections in humans [16, 40–43]. Since CPAF-specific human antibodies neutralized CPAF enzymatic activity [44], CPAF was tested as a vaccine candidate antigen in a mouse model and a significant protection against chlamydial challenge infection was induced by CPAF immunization [45]. It turned out that the protection induced by CPAF in mice was dependent on CD4+ T cells [46] and IFNg [47] and the role of anti-CPAF antibodies in protection against chlamydial infection was limited [48]. Although the CPAF-induced murine protection mechanism [49] is consistent with the knowledge on immunity of chlamydial infection in mice [50], it remains unknown whether CPAF-specific immune responses play any roles in chlamydial infection and pathogenesis in humans and what immune mechanisms are involved. Thus, it has now become necessary to evaluate whether humans produce CD4+ T cell responses to CPAF during chlamydial infection and whether the CPAF-specific cellular responses are protective. At the same time, it is equally important to evaluate whether the in vitro neutralization of CPAF enzymatic activity by human antibodies can translate into protection against chlamydial infection and pathology during chlamydial infection in humans. Since CPAF is not associated with the infectious particle EB, anti-CPAF antibodies may not directly affect chlamydial infectivity during chlamydial spreading. If the robust and sustained anti-CPAF antibody response indeed does not play any protective roles, what can it do to human susceptibility to chlamydial infection and pathogenicity? One can reasonably hypothesize that the robust antibody response to CPAF during chlamydial infection in humans may steer the human immune responses away from mounting strong responses to the EB-surface-exposed protective antigens such as the major outer membrane protein (MOMP). Test of this hypothesis may allow us to dig up additional mysteries hidden in the CPAF treasure box.
Biological characteristics of CPAF
Aside from the biochemical and immunological characteristics of CPAF, there are many biologic principles governing it that will also be important for future studies. Most notably, it is a large protein (1827bp, or 609aa in serovar D) for such a small genome (~1Mb). Real estate in this genome is tightly packed and controlled; thus, any protein contained within it is likely to have a very necessary function. Not only does it cost Chlamydia resources in the form of nucleotides and amino acids, but CPAF also requires metabolic energy to maintain its repair, replication, transcription, and translation. Similarly, CPAF is secreted from Chlamydia via a Sec-dependent manner [51], the biological machinery of which also requires cellular resources and a significant amount of energy to construct and maintain. Secretion of any factor by an intracellular pathogen may suggest its role in pathogenesis.
The adaptation of chlamydial organisms and their CPAF over the course of a billion years to host environments from single-celled eukaryotes to the complex human body systems is truly amazing. Although the entire CPAF protein is not well conserved, from environmental Protochlamydia that infect amoebas all the way to the human pathogen C. trachomatis, residues in and surrounding the catalytic domain are [52]. Because CPAF and its secretion has been maintained within diverse organisms, it likely has functions that are as basic as the links between their two vastly different eukaryotic hosts. During the evolution of CPAF, the genome of ancient Chlamydiae experienced over a two-fold reduction in genome size to arrive at the modern C. trachomatis genome, but CPAF has not been lost or greatly changed along the way. These characteristics suggest an essential role for CPAF in chlamydial biology.
Although CPAF is delivered to the host cell cytoplasm, the precise location of CPAF in the cytoplasm remains unknown. Immunofluorescence labeling of CPAF reveals that it is distributed throughout the entire cytoplasm. However, these results do not agree with the lack of valid cytoplasmic host substrates. If CPAF were to be located within the cytoplasm itself, it should gain access to a wide array of host proteins. However, given that it has such broad activity, releasing CPAF directly into the cytoplasm could be so damaging to the host cell as to make it uninhabitable for Chlamydia. Instead, some immunofluorescence images suggest that CPAF may be stored within vesicles in the host cell cytosol [24]. This strategy would not only keep CPAF from detrimentally cleaving many host proteins, but also keep it from being cleaved by the host proteasome and detected by the antigen presentation machinery. Although the secretion of CPAF into the host cell cytosol requires a sec-dependent pathway [51], the sec-secretion pathway alone may only send CPAF into the periplasmic region but may not be sufficient for transporting CPAF all the way into the host cell cytosol. Interestingly, some bacterial exotoxins are known to exit the bacterial periplasmic region via a mechanism of outer membrane vesicle budding (OMVs; ref: [53]. In fact, The OMV-trapped toxins can travel far to reach their targets [54]. It is not known how CPAF exits the chlamydial periplasmic region. One can imagine that the periplasmic CPAF may also use an outer membrane budding mechanism to exit the chlamydial organisms. In this way, CPAF molecules are sequestered within the budding vesicles when the OMVs are in the lumen of the chlamydial inclusions or further pass through the inclusion membrane to reach the host cell cytosol. This hypothesis is supported by the observation that the chlamydial RB outer membrane can undergo extensive vesiculation under certain culture conditions [55], chlamydial organism-free vesicles are detected inside the lumen of chlamydial inclusions [56], and the release of vesicles from chlamydial inclusions can be induced [57]. Thus, it is possible that Chlamydia may use the OMV mechanism to store CPAF in the host cell cytosol for neutralizing host extracellular defense mechanisms during chlamydial cell-to-cell spreading. This hypothesis is consistent with the fact that CPAF is expressed late in the Chlamydia developmental cycle. Therefore, CPAF may be important during the extracellular phase of infection when EBs are exposed to the harsh immunological environment of the human mucosal surface.
New strategies for CPAF research
A number of emerging techniques and methods that are becoming available to the Chlamydia research community could also be used for uncovering the role of CPAF in infection. Foremost, it is critical to continue to carefully examine the way in which CPAF substrates are characterized. The protocol for examining CPAF substrates should include specifically the addition of 150 μM lactacystin to cell culture one hour prior to cell harvesting and lysis in 8 M urea or SDS for host proteins, and continual CPAF inhibition via either lactacystin or the CPAF inhibitory peptide. If these methods and similar strategies are not adhered to, it will be much more difficult to accept their validity given the controversy surrounding the subject.
Recent advancements in genetic manipulation of the chlamydial plasmid [18, 58, 59] and genome [60, 61] have provided essential tools for experimentally defining the roles of putative virulence factors in chlamydial pathogenesis that were previously identified via biochemical, microscopic or computational approaches. Though hindered by its own limitations, broad genomic mutagenesis using the guanine to cytosine chemical conversion agent, ethyl methanesulfonate (EMS), has the ability to introduce spontaneous missense and nonsense mutations in the CPAF gene and other genes necessary for its maintenance and secretion. Efforts are currently underway to develop these mutants. In parallel, the chlamydial plasmid-based shuttle vector transformation system may allow the introduction of dominant negative CPAF or CPAF-specific inhibitory peptide into chlamydial organisms to suppress CPAF function. Though such a system has not been engineered for Chlamydia in the past, it may also be possible to introduce CPAF antisense RNA-expressing constructs that block the translation of CPAF and other important virulence factors. Of course, there remain challenges with these new genetic techniques. For example, EMS often simultaneously generates multiple mutations in multiple genes, which makes it difficult to define the genetic basis of phenotypes detected. Obviously, obtaining CPAF-deficient mutants with little collateral damage to other genes in the process would allow for direct in vivo analysis and answer the most important question of all: what is the contribution and role of CPAF in pathogenesis? Alternatively, the combination of the EMS approach for generating CPAF null mutants with plasmid expression of CPAF for complementing the loss of CPAF may accelerate the progress in revealing the role of CPAF in chlamydial pathogenesis.
Conclusion
Despite the difficulties in searching for CPAF intracellular substrates [38], hypotheses centered on the possible role of CPAF in the extracellular environment are proposed (see Fig. 1). Since CPAF is secreted into and stored in the host cell cytosol during chlamydial intracellular growth, CPAF can be rapidly released into extracellular environment during inclusion bursting. Given its strong proteolytic activity and wide spectrum of substrate specificities, the extracellular CPAF may have the ability to cleave multiple mucosal immunity effectors such as mucin, which is present as a physical barrier to reinfection. Additionally, other more overt immunological effectors, such as dimeric IgA or cationic antimicrobial peptides (AMPs), which have the ability to neutralize or kill chlamydial EBs, may also be targeted by CPAF. Once again, the fact that CPAF is expressed late in the chlamydial developmental cycle supports these hypotheses, as the extracellular phase of the lifecycle is the point at which Chlamydia is most susceptible to the powerful host mechanisms. One should also not discount the possibility that CPAF could function to “prime” the next host cell for infection or aid EBs in the active process of adhering to and entering these cells. Nonetheless, the broad activity of CPAF might allow it to contribute to any combination of these proposed functions. It is worth reminding that although these hypotheses seem to be well rationalized, they are just hypotheses in the absence of reproducible data support. In the past decade, we watched the CPAF stories evolved. The rapidly discovered substrates should have raised the awareness about the authenticity of the substrates. Instead, the warning sign was not only ignored but also triggered the imagination of connecting correlative/indirect data and eagerness to tell “mechanism-driven stories” in high profile journals. Thanks to the careful and brave work by Chen et al [38], a brake was finally landed to stop the CPAF research from going the wrong direction. This lesson should make the chlamydiologists wiser when approaching the next chlamydial problems.
Fig. 1. A hypothetical model for CPAF function during urogenital tract infection with C. trachomatis.
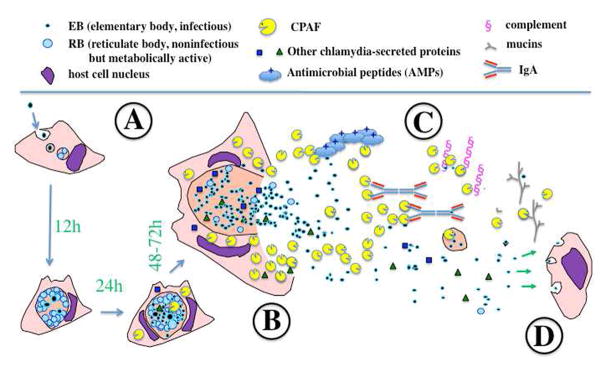
After chlamydial organisms complete their intracellular replication following an initial infection (A), CPAF is accumulated in the cytosol of the infected cell that is ready for releasing the intra-vacuolar chlamydial organisms. When the infected cell bursts open (B), CPAF and other chlamydial components along with the infectious chlamydial organisms (EBs) are released into the extracellular environments where there are numerous host defense molecules with the abilities to kill or inactivate EBs (C). CPAF may target these host defense molecules for degradation or inactivation so that EBs can safely infect the next host cell (D). Such a successful cell-to-cell spreading may promote chlamydial ascending infection and pathogenicity in the upper genital tract.
Though most known intracellular host targets of CPAF have proven to be false positives, there is no reason to believe that none exist. In fact, such an attitude could be detrimental to discovering the genuine substrates. It has yet to be determined how CPAF exits the inclusion and where its ultimate destination may be. Instead, it is wise to begin with the only known substrate: OmcB. Specific inhibition of CPAF using its own inhibitory peptide resulted in greatly reduced OmcB processing and infectivity, suggesting an essential role of CPAF in maintaining chlamydial infectivity. Although OmcB and other intracellular chlamydial proteins could serve as evolutionarily conserved CPAF targets, other functions may have arisen along the way. For example, the C-terminal OmcB fragment cleaved by CPAF is highly immunogenic for cellular and humoral responses, but it could also be a decoy to distract the immune system from more relevant chlamydial antigens. Because it is expected to cleave any number of substrates, identifying the precise host cellular compartment where CPAF is delivered may hold the key to uncovering how CPAF manipulates the host cell or host tissue. Though the endeavor to discover the true substrates of CPAF has historically been subject to Murphy’s Law, the combined knowledge, hypotheses, and novel methods presented herein are the ingredients for a new recipe of success in the field of chlamydial pathogenesis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhong G, Fan P, Ji H, Dong F, Huang Y. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J Exp Med. 2001;193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong F, Pirbhai M, Zhong Y, Zhong G. Cleavage-dependent activation of a chlamydia-secreted protease. Mol Microbiol. 2004;52:1487–1494. doi: 10.1111/j.1365-2958.2004.04072.x. [DOI] [PubMed] [Google Scholar]
- 3.Dong F, Sharma J, Xiao Y, Zhong Y, Zhong G. Intramolecular dimerization is required for the chlamydia-secreted protease CPAF to degrade host transcriptional factors. Infect Immun. 2004;72:3869–3875. doi: 10.1128/IAI.72.7.3869-3875.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong F, Zhong Y, Arulanandam B, Zhong G. Production of a proteolytically active protein, chlamydial protease/proteasome-like activity factor, by five different Chlamydia species. Infect Immun. 2005;73:1868–1872. doi: 10.1128/IAI.73.3.1868-1872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D, Lei L, Flores R, Huang Z, Wu Z, Chai J, Zhong G. Autoprocessing and self-activation of the secreted protease CPAF in Chlamydia-infected cells. Microb Pathog. 2010;49:164–173. doi: 10.1016/j.micpath.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D, Chai J, Hart PJ, Zhong G. Identifying catalytic residues in CPAF, a Chlamydia-secreted protease. Arch Biochem Biophys. 2009;485:16–23. doi: 10.1016/j.abb.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Z, Feng Y, Chen D, Wu X, Huang S, Wang X, Xiao X, Li W, Huang N, Gu L, Zhong G, Chai J. Structural basis for activation and inhibition of the secreted chlamydia protease CPAF. Cell Host Microbe. 2008;4:529–542. doi: 10.1016/j.chom.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Wang SP, Grayston JT. Serotyping of Chlamydia trachomatis by indirect fluorescent-antibody staining of inclusions in cell culture with monoclonal antibodies. J Clin Microbiol. 1991;29:1295–1298. doi: 10.1128/jcm.29.7.1295-1298.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stary G, Stary A. Lymphogranuloma venereum outbreak in Europe. J Dtsch Dermatol Ges. 2008;6:935–940. doi: 10.1111/j.1610-0387.2008.06742.x. [DOI] [PubMed] [Google Scholar]
- 10.Kuo CC, Jackson LA, Campbell LA, Grayston JT. Chlamydia pneumoniae (TWAR) Clin Microbiol Rev. 1995;8:451–461. doi: 10.1128/cmr.8.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewardson AJ, Grayson ML. Psittacosis. Infect Dis Clin North Am. 2010;24:7–25. doi: 10.1016/j.idc.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Sherman KJ, Daling JR, Stergachis A, Weiss NS, Foy HM, Wang SP, Grayston JT. Sexually transmitted diseases and tubal pregnancy. Sex Transm Dis. 1990;17:115–121. doi: 10.1097/00007435-199007000-00001. [DOI] [PubMed] [Google Scholar]
- 13.O’Connell CM, Ingalls RR, Andrews CW, Jr, Scurlock AM, Darville T. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol. 2007;179:4027–4034. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 14.Kari L, Whitmire WM, Olivares-Zavaleta N, Goheen MM, Taylor LD, Carlson JH, Sturdevant GL, Lu C, Bakios LE, Randall LB, Parnell MJ, Zhong G, Caldwell HD. A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J Exp Med. 2011;208:2217–2223. doi: 10.1084/jem.20111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Zhong Y, Lei L, Wu Y, Wang S, Zhong G. Antibodies from women urogenitally infected with C. trachomatis predominantly recognized the plasmid protein pgp3 in a conformation-dependent manner. BMC Microbiol. 2008;8:90. doi: 10.1186/1471-2180-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Zhang Y, Lu C, Lei L, Yu P, Zhong G. A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J Immunol. 2010;185:1670–1680. doi: 10.4049/jimmunol.1001240. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Chen D, Zhong Y, Wang S, Zhong G. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect Immun. 2008;76:3415–3428. doi: 10.1128/IAI.01377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun. 2013;81:636–644. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlson JH, Whitmire WM, Crane DD, Wicke L, Virtaneva K, Sturdevant DE, Kupko JJ, 3rd, Porcella SF, Martinez-Orengo N, Heinzen RA, Kari L, Caldwell HD. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun. 2008;76:2273–2283. doi: 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sturdevant GL, Kari L, Gardner DJ, Olivares-Zavaleta N, Randall LB, Whitmire WM, Carlson JH, Goheen MM, Selleck EM, Martens C, Caldwell HD. Frameshift mutations in a single novel virulence factor alter the in vivo pathogenicity of Chlamydia trachomatis for the female murine genital tract. Infect Immun. 2010;78:3660–3668. doi: 10.1128/IAI.00386-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samudrala R, Heffron F, McDermott JE. Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 2009;5:e1000375. doi: 10.1371/journal.ppat.1000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fields KA, Hackstadt T. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol Microbiol. 2000;38:1048–1060. doi: 10.1046/j.1365-2958.2000.02212.x. [DOI] [PubMed] [Google Scholar]
- 23.Bao X, Nickels BE, Fan H. Chlamydia trachomatis protein GrgA activates transcription by contacting the nonconserved region of sigma66. Proc Natl Acad Sci U S A. 2012;109:16870–16875. doi: 10.1073/pnas.1207300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong G. Chlamydia trachomatis secretion of proteases for manipulating host signaling pathways. Front Microbiol. 2011;2:14. doi: 10.3389/fmicb.2011.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X, Lei L, Gong S, Chen D, Flores R, Zhong G. The chlamydial periplasmic stress response serine protease cHtrA is secreted into host cell cytosol. BMC Microbiol. 2011;11:87. doi: 10.1186/1471-2180-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobolt-Pedersen AS, Christiansen G, Timmerman E, Gevaert K, Birkelund S. Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol Med Microbiol. 2009;57:46–58. doi: 10.1111/j.1574-695X.2009.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong S, Lei L, Chang X, Belland R, Zhong G. Chlamydia trachomatis secretion of hypothetical protein CT622 into host cell cytoplasm via a secretion pathway that can be inhibited by the type III secretion system inhibitor compound 1. Microbiology. 2011;157:1134–1144. doi: 10.1099/mic.0.047746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei L, Dong X, Li Z, Zhong G. Identification of a Novel Nuclear Localization Signal Sequence in Chlamydia trachomatis-Secreted Hypothetical Protein CT311. PLoS One. 2013;8:e64529. doi: 10.1371/journal.pone.0064529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei L, Qi M, Budrys N, Schenken R, Zhong G. Localization of Chlamydia trachomatis hypothetical protein CT311 in host cell cytoplasm. Microb Pathog. 2011;51:101–109. doi: 10.1016/j.micpath.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi M, Lei L, Gong S, Liu Q, DeLisa MP, Zhong G. Chlamydia trachomatis secretion of an immunodominant hypothetical protein (CT795) into host cell cytoplasm. J Bacteriol. 2011;193:2498–2509. doi: 10.1128/JB.01301-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu C, Lei L, Peng B, Tang L, Ding H, Gong S, Li Z, Wu Y, Zhong G. Chlamydia trachomatis GlgA is secreted into host cell cytoplasm. Plos One. 2013:xx. doi: 10.1371/journal.pone.0068764. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi M, Gong S, Lei L, Liu Q, Zhong G. A Chlamydia trachomatis OmcB C-terminal fragment is released into the host cell cytoplasm and is immunogenic in humans. Infect Immun. 2011;79:2193–2203. doi: 10.1128/IAI.00003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong G. Killing me softly: chlamydial use of proteolysis for evading host defenses. Trends Microbiol. 2009;17:467–474. doi: 10.1016/j.tim.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christian JG, Heymann J, Paschen SA, Vier J, Schauenburg L, Rupp J, Meyer TF, Hacker G, Heuer D. Targeting of a chlamydial protease impedes intracellular bacterial growth. PLoS Pathog. 2011;7:e1002283. doi: 10.1371/journal.ppat.1002283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heuer D, Rejman Lipinski A, Machuy N, Karlas A, Wehrens A, Siedler F, Brinkmann V, Meyer TF. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- 36.Fischer SF, Vier J, Kirschnek S, Klos A, Hess S, Ying S, Hacker G. Chlamydia inhibit host cell apoptosis by degradation of proapoptotic BH3-only proteins. J Exp Med. 2004;200:905–916. doi: 10.1084/jem.20040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorgensen I, Bednar MM, Amin V, Davis BK, Ting JP, McCafferty DG, Valdivia RH. The Chlamydia protease CPAF regulates host and bacterial proteins to maintain pathogen vacuole integrity and promote virulence. Cell Host Microbe. 2011;10:21–32. doi: 10.1016/j.chom.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen AL, Johnson KA, Lee JK, Sutterlin C, Tan M. CPAF: a Chlamydial protease in search of an authentic substrate. PLoS Pathog. 2012;8:e1002842. doi: 10.1371/journal.ppat.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou S, Lei L, Yang Z, Qi M, Liu Q, Zhong G. Chlamydia trachomatis outer membrane complex protein B (OmcB) is processed by the protease CPAF. J Bacteriol. 2013;195:951–957. doi: 10.1128/JB.02087-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma J, Bosnic AM, Piper JM, Zhong G. Human antibody responses to a Chlamydia-secreted protease factor. Infect Immun. 2004;72:7164–7171. doi: 10.1128/IAI.72.12.7164-7171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma J, Zhong Y, Dong F, Piper JM, Wang G, Zhong G. Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins. Infect Immun. 2006;74:1490–1499. doi: 10.1128/IAI.74.3.1490-1499.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu C, Holland MJ, Gong S, Peng B, Bailey RL, Mabey DW, Wu Y, Zhong G. Genome-wide identification of Chlamydia trachomatis antigens associated with trachomatous trichiasis. Invest Ophthalmol Vis Sci. 2012;53:2551–2559. doi: 10.1167/iovs.11-9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skwor T, Kandel RP, Basravi S, Khan A, Sharma B, Dean D. Characterization of humoral immune responses to chlamydial HSP60, CPAF, and CT795 in inflammatory and severe trachoma. Invest Ophthalmol Vis Sci. 2010;51:5128–5136. doi: 10.1167/iovs.09-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma J, Dong F, Pirbhai M, Zhong G. Inhibition of proteolytic activity of a chlamydial proteasome/protease-like activity factor by antibodies from humans infected with Chlamydia trachomatis. Infect Immun. 2005;73:4414–4419. doi: 10.1128/IAI.73.7.4414-4419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murthy AK, Cong Y, Murphey C, Guentzel MN, Forsthuber TG, Zhong G, Arulanandam BP. Chlamydial protease-like activity factor induces protective immunity against genital chlamydial infection in transgenic mice that express the human HLA-DR4 allele. Infect Immun. 2006;74:6722–6729. doi: 10.1128/IAI.01119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphey C, Murthy AK, Meier PA, Neal Guentzel M, Zhong G, Arulanandam BP. The protective efficacy of chlamydial protease-like activity factor vaccination is dependent upon CD4+ T cells. Cell Immunol. 2006;242:110–117. doi: 10.1016/j.cellimm.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murthy AK, Chambers JP, Meier PA, Zhong G, Arulanandam BP. Intranasal vaccination with a secreted chlamydial protein enhances resolution of genital Chlamydia muridarum infection, protects against oviduct pathology, and is highly dependent upon endogenous gamma interferon production. Infect Immun. 2007;75:666–676. doi: 10.1128/IAI.01280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murthy AK, Chaganty BK, Li W, Guentzel MN, Chambers JP, Seshu J, Zhong G, Arulanandam BP. A limited role for antibody in protective immunity induced by rCPAF and CpG vaccination against primary genital Chlamydia muridarum challenge. FEMS Immunol Med Microbiol. 2009;55:271–279. doi: 10.1111/j.1574-695X.2008.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murthy AK, Guentzel MN, Zhong G, Arulanandam BP. Chlamydial protease-like activity factor--insights into immunity and vaccine development. J Reprod Immunol. 2009;83:179–184. doi: 10.1016/j.jri.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morrison RP, Caldwell HD. Immunity to murine chlamydial genital infection. Infect Immun. 2002;70:2741–2751. doi: 10.1128/IAI.70.6.2741-2751.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen D, Lei L, Lu C, Flores R, DeLisa MP, Roberts TC, Romesberg FE, Zhong G. Secretion of the chlamydial virulence factor CPAF requires the Sec-dependent pathway. Microbiology. 2010;156:3031–3040. doi: 10.1099/mic.0.040527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Griffiths E, Ventresca MS, Gupta RS. BLAST screening of chlamydial genomes to identify signature proteins that are unique for the Chlamydiales, Chlamydiaceae, Chlamydophila and Chlamydia groups of species. BMC Genomics. 2006;7:14. doi: 10.1186/1471-2164-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mashburn-Warren LM, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol. 2006;61:839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- 54.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, Stanton BA. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsumoto A, Manire GP. Electron Microscopic Observations on the Fine Structure of Cell Walls of Chlamydia psittaci. J Bacteriol. 1970;104:1332–1337. doi: 10.1128/jb.104.3.1332-1337.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen I, Valdivia RH. Pmp-like proteins Pls1 and Pls2 are secreted into the lumen of the Chlamydia trachomatis inclusion. Infect Immun. 2008;76:3940–3950. doi: 10.1128/IAI.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giles DK, Whittimore JD, LaRue RW, Raulston JE, Wyrick PB. Ultrastructural analysis of chlamydial antigen-containing vesicles everting from the Chlamydia trachomatis inclusion. Microbes Infect. 2006;8:1579–1591. doi: 10.1016/j.micinf.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 2011;7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong S, Yang Z, Lei L, Shen L, Zhong G. Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J Bacteriol. 2013:xx. doi: 10.1128/JB.00511-13. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, Nelson DE, Caldwell HD. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci U S A. 2011;108:7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen BD, Valdivia RH. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci U S A. 2012;109:1263–1268. doi: 10.1073/pnas.1117884109. [DOI] [PMC free article] [PubMed] [Google Scholar]


