Abstract
Background
Directly Acting Antivirals (DAAs) are predicted to transform hepatitis C (HCV) therapy, yet little is known about the prevalence of naturally occurring resistance mutations in recently acquired HCV. This study aimed to determine the prevalence and frequency of drug resistance mutations in the viral quasispecies among HIV positive and negative individuals with recent HCV.
Methods
The NS3 protease, NS5A and NS5B polymerase genes were amplified from fifty genotype 1a participants of the Australian Trial in Acute Hepatitis C. Amino acid variations at sites known to be associated with possible drug resistance were analysed by ultra-deep pyrosequencing.
Results
Twelve percent of individuals harboured dominant resistance mutations, while 36% demonstrated non dominant resistant variants below that detectable by bulk sequencing (ie < 20%) but above a threshold of 1%. Resistance variants (< 1%) were observed at most sites associated with DAA resistance from all classes, with the exception of sofosbuvir.
Conclusions
Dominant resistant mutations were uncommonly observed in the setting of recent HCV. However, low level mutations to all DAA classes were observed by deep sequencing at the majority of sites, and in most individuals. The significance of these variants and impact on future treatment options remains to be determined.
Keywords: recent hepatitis C, directly acting antivirals, resistance mutations, ultra-deep pyrosequencing, viral quasispecies
Introduction
The rapid development of directly acting antiviral drugs (DAAs) for the treatment of hepatitis C virus (HCV) should revolutionise therapeutic options. However, despite the significantly greater potency offered with these agents, antiviral resistance may remain a potential challenge to their use.
The development of resistance to DAAs was demonstrated in early clinical trials involving prolonged monotherapy (1, 2). Using both phenotypic and genotypic methodology, several key signature mutations for both protease and polymerase inhibitors were described, many of which indicate cross-class resistance (3). In the context of triple interferon-based therapy (one DAA plus interferon and ribavirin), resistance mutations often emerge during the first few weeks of therapy as the viral quasispecies changes, but are subsequently brought under control (2). A proportion of these patients, however, will go on to experience viral breakthrough and subsequent treatment failure (4). Whether the presence, and frequency, of naturally occurring mutations in the viral quasispecies prior to the commencement of treatment impacts on subsequent treatment response is unclear.
The naturally occurring prevalence of resistance mutations in untreated patients with chronic HCV has been examined in a number of studies (5–8). Standard bulk (Sanger) sequencing techniques estimate the prevalence of variations within a population to be low at 0.2–2.2%, although one study demonstrated up to 21.5% of individuals infected with subtype 1a exhibit genetic variation to at least one known drug resistance site (9). However, by more sensitive techniques such as ultra-deep pyrosequencing (UDPS) variation at sites associated with resistance can be shown to pre-exist in most individuals ranging from 0.01 – 99% of the total viral population (10–17). The frequency of resistance mutations in recently acquired HCV is not understood. In this paper, we aimed to characterise the prevalence and frequency of resistance mutations in a unique cohort of HIV positive and HIV negative individuals with recently acquired untreated HCV infection using UDPS methodology.
Methods
Study design and participants
All participants were enrolled within the Australian Trial in Acute Hepatitis C (ATAHC). ATAHC was a multicenter, prospective cohort study of the natural history and treatment of recent HCV, as described elsewhere (18). Recent infection with either acute or early chronic HCV infection was diagnosed with the following eligibility criteria: First positive anti-HCV antibody within 6 months of enrolment; and either
Acute clinical hepatitis C infection, defined as symptomatic seroconversion illness or alanine aminotransferase (ALT) level greater than 10 times the upper limit of normal (>400 IU/L) with exclusion of other causes of acute hepatitis, at most 12 months before the initial positive anti-HCV antibody; or
Asymptomatic hepatitis C infection with seroconversion, defined by a negative anti-HCV antibody in the two years prior to the initial positive anti-HCV antibody.
All study participants provided written informed consent. The study protocol was approved by St Vincent’s Hospital Human Research Ethics Committee and local ethics committees at sites. The study was registered with clinicaltrials.gov registry (NCT00192569). Both HIV positive and negative participants were enrolled. Due to the relationship between HCV genotype and protease inhibitor-based resistance, only genotype 1a (GT1a) participants with a stored screening or pre-treatment sample available, with an HCV RNA> 1000 IU/ml were included.
Detection and quantification of HCV RNA
Qualitative and quantitative HCV RNA testing was performed using the Versant TMA assay [Bayer, Australia; <10 IU/ml] and the Versant HCV RNA 3.0 (Bayer, Australia; <615 IU/ml), respectively.
Sequencing methodology
Viral RNA extraction
Viral RNA was extracted from plasma samples using the COBAS AMPLICOR HCV Specimen Preparation Kit v2.0 (Roche Applied Science) according to manufacturers’ instructions.
Generation of NS3 protease, NS5A and NS5B polymerase amplicons
Two initial RT-PCRs were performed to cover the non-structural regions of HCV (19). The first-round products were then used as templates in nested second round PCRs using generic or genotype specific primers as previously described (19). Alternative generic and genotype/subtype specific primers targeting the NS3 protease, NS5A and NS5B regions were also used. The primer sequences (5′ to 3′) were as follows: for NS3 protease 3337F GGCYTGCCYGTCTCYGC and 4035R GTGCTCTTRCCGCTRCCNGT (modified from (7)); for NS5A/B 6158F ATGCWGCTGCCCGCGTCAC and 6940R AGCTGGCTGGCCGAAGAGCT, 6821F TCCCTTGCGAGCCCGAACCGGA and 7522R TGAGATCCGGGTCCCCAGGCTC, 7370F CCTTGGCCGAGCTTGCCAC and 8080R CCCCCAGGTCAGGGTACACAAT, 8011F ACCATCATGGCGAAGAAYGA and 8683R GAGGAGCAAGATGTTATGAGCTC. Thermocycling conditions were: one cycle for 94°C for 2 minutes followed by 20 cycles at 94°C for 15 seconds then annealing temperature (Tm) 1 for 30 seconds then 72°C for 1 minute. This was followed by another 20 cycles at 94°C for 15 seconds then Tm2 for 30 seconds then 72°C for 1 minute followed by a single cycle at 72°C for 1 minute. For the primer pair 3337F/4035R Tm1 was 56°C and Tm2 was 54°C. For the primer pair 8011F/8683R Tm1 was 53°C and Tm2 was 51°C. For the primer pairs 6158F/6940R, 6821F/7522R and 7370F/8080R Tm1 was 61°C and Tm2 was 59°C.
FLX 454 UDPS generation and analysis
UDPS was carried-out using the 454 Life Sciences platform (GS-FLX, Roche Applied Science) for all participants. PCR products (as described above) were quantified and then pooled for each individual. These products were ligated to adaptors and clonally amplified on capture beads in water-in-oil emulsion micro-reactors. The resultant libraries were sequenced using a PicoTiterPlate and nucleotide data collected and quality filtered using the Roche 454 software (default settings). Following the removal of primer sequences (using an in-house program), sequence reads were aligned using the Softgenetics NextGENe software to the H77 genotype 1a sequence (GenBank accession number: NC004102). To ensure quality of sequence reads, an 80% nucleotide identity threshold against the reference sequence was used for inclusion of a read for analysis. Sequences were then imported as fasta files into the program BioEdit. Likely homopolymer errors relative to the reference sequence and sequence errors near read ends were removed (20). Nucleotide alignment was then translated to appropriate amino acid sequence. Sites associated with DAA resistance were identified and sorted based on amino acid composition. The number of reads for each amino acid at each site associated with drug resistance was tabulated. A variation was included if it was present on at least three sequence reads.
Results
Participants
Fifty GT1a ATAHC participants had samples available for testing. Twenty six (52%) participants were HIV positive. Median HCV RNA was 5.3 logs (IU/ml) and the median estimated duration of HCV infection at time of analysis was 29 weeks. Further participant characteristics at baseline, stratified by HIV status, infection can be found in Table 1.
Table 1.
Participant characteristics stratified by HIV infection.
| Overall HCV GT1a (n=50) | HIV uninfected (n=26) | HIV infected (n=24) | |
|---|---|---|---|
| Sex, n (%) | |||
| Female | 11 (22%) | 11 (42%) | 0 (0%) |
| Male | 39 (78%) | 15 (58%) | 24 (100%) |
| Age (yrs), mean/SD | 34 (9) | 31 (10) | 38 (7) |
| Mode of HCV acquisition, n (%) | |||
| IDU | 34 (68%) | 20 (77%) | 14 (58%) |
| Sexual transmission | 11 (22%) | 2 (8%) | 9 (38%) |
| Other | 5 (10%) | 4 (15%) | 1 (4%) |
| IL28B genotype (rs12979860) | |||
| TT | 1 (2%) | 1 (4%) | 0 (0%) |
| CT | 23 (46%) | 13 (50%) | 10 (42%) |
| CC | 25 (50%) | 12 (46%) | 13 (54%) |
| Estimated duration of infection | |||
| Weeks, mean/SD | 29 (19) | 35 (19) | 23 (18) |
| <26weeks | 29 (58%) | 13 (50%) | 16 (67%) |
| ≥26 weeks | 21 (42%) | 13 (50%) | 8 (33%) |
| Median HCV RNA (log IU/mL)*, 25%, 75% | 5.3 (3.9, 6.1) | 5.3 (4.0, 5.9) | 5.3 (3.9, 6.4) |
NS3/4 protease sequencing
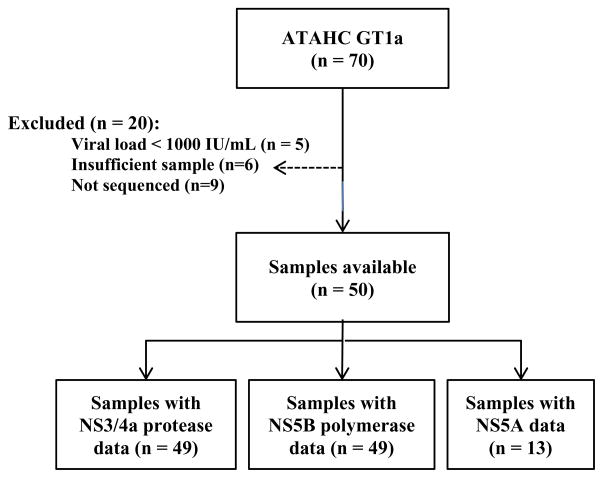
Among those with available samples (n=50), sequencing of the NS3/4 protease gene was successfully performed in 49 particpants with GT1a (Figure 1). Eleven sites known to be associated with possible drug resistance were examined for evidence of amino acid variation including the following site changes: V36A/M, Q41R, F43C/S/L, T54A/S, V55A, Q80K/R, S138T, R155K/Q/T, A156S/T/V, D168A/T/V and V170A/T (Table 2). The mean coverage at each site varied from 3918–5239 reads. Only four (8%) participants had evidence of a dominant resistant variant, one with a V36M variation present at 99.4% and a Q80K at 99.2%, one with a sole Q80K present at 98.5%, and two with a V55A present at 89.6% and 99.9% of total sequences, respectively. Seven additional participants (14%) had evidence of a resistance variant present in sequences at a level above 1%, but in all cases the prevalence of the variant was very low and never exceeded 2%. Two of these participants had more than one resistant variant present, however, the presence of more than one variant within a single 500bp viral sequence (present at low frequency) was not observed.
Figure 1.
Flow chart describing the selection of ATAHC participants for inclusion in this analysis.
Table 2.
Participants with resistance mutation present at > 1% of total quasispecies population
| Amino acid mutation | Proportion of mutation present, % | Subject number | HIV positive | Estimated duration of infection, weeks |
|---|---|---|---|---|
| Protease (n = 49) | ||||
| Mutation at >50 % | ||||
| V36M | 99.4 | 127 | Y | 25 |
| V55A | 89.6 | 609 | N | 58 |
| V55A | 99.9 | 664 | N | 9 |
| Q80K | 98.5 | 804 | Y | 58 |
| Q80K | 99.2 | 127 | Y | 25 |
| Mutation >1–50 % | ||||
| Q41R | 1.02 | 2403 | Y | 16 |
| V55A | 1.03 | 634 | N | 31 |
| V55A | 1.10 | 628 | Y | 12 |
| Q80R | 1.27 | 2016 | N | 20 |
| A156S | 1.99 | 501 | N | 44 |
| I170T | 1.10 | 2605 | N | 60 |
| I170T | 1.54 | 501 | N | 44 |
| V170A | 1.08 | 120 | Y | 9 |
|
| ||||
| Polymerase (n = 49) | ||||
| Mutation at >50 % | ||||
| M423I | 98 | 803 | N | 57 |
| Mutation >1–50 % | ||||
| S365T | 1.46 | 653 | N | 20 |
| S365T | 2.62 | 620 | Y | 64 |
| S365T | 1.24 | 2003 | N | 29 |
| S365T | 1.38 | 120 | Y | 9 |
| L419M | 1.81 | 1402 | N | 21 |
| L419M | 1.19 | 1001 | N | 74 |
| L419V | 1.47 | 302 | N | 21 |
| M423T | 1.56 | 125 | N | 23 |
| M423T | 1.78 | 609 | N | 58 |
| V494A | 1.10 | 501 | N | 44 |
| V494A | 1.03 | 2402 | Y | 15 |
| P496S | 2.60 | 630 | Y | 30 |
| V499A | 18.8 | 120 | Y | 9 |
|
| ||||
| NS5A (n = 13) | ||||
| Mutation >50 % | ||||
| M28V | 99.5 | 302 | N | 21 |
| Mutation >1–50 % | ||||
| Q30R | 2.29 | 120 | Y | 9 |
| Q30R | 1.33 | 627 | ||
| H58P | 1.10 | 120 | Y | 9 |
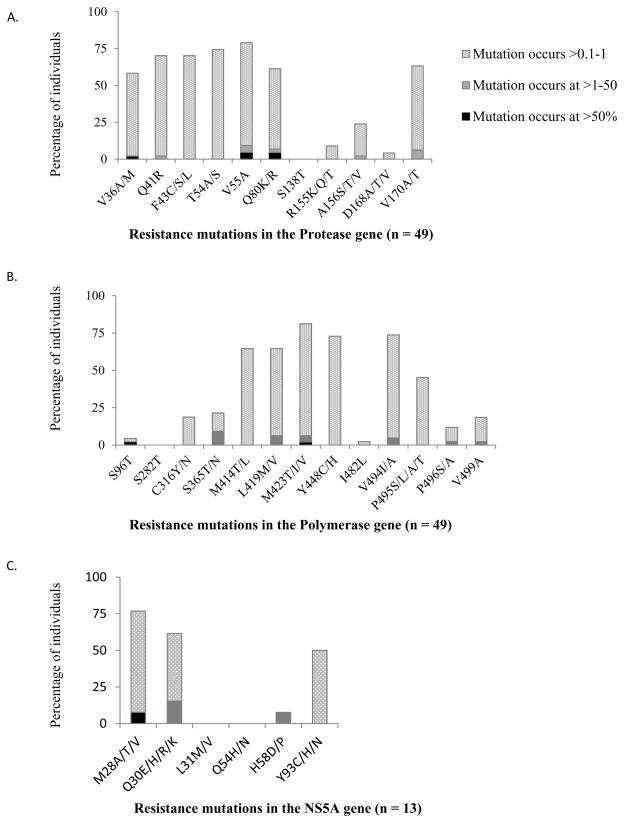
Variation above 0.1% and below 1% was seen at a much greater frequency (Figure 2a). Evidence of resistant variants occurring at this frequency was observed at all sites except the S138T position (< 0.1% in all participants). Some sites however were relatively conserved with low proportions of patients with variance at this level, notably D168A/T/V (4%), and R155K/Q/T (9%), whereas other sites such as T54A/S, V55A and Q41R demonstrated far greater variance with 74%, 70% and 68% of participants respectively demonstrating low level resistance variants (0.1–1%) at these sites.
Figure 2. Percentage of individuals with resistance mutations present in the (A) NS3 protease gene (n=49), NS5B polymerase gene (n=49) and (C) NS5A gene (n=13).
Resistance mutations are represented as occuring at >50% (“Dominant”, solid bars), >1–50% (grey hatched bars), >0.1–1% (light grey hatched bars). All mutations known to confer resistance at each position were catergorised together, irrespective of the amino acid change. For particpants who had evidence of multiple mutations at one site (eg. Both M423T andM423V), the mutation seen at higher % was reported. No mutations were found to occur in the range between 3 and 88%. Dominant resistant mutations generally occurred at > 98%.
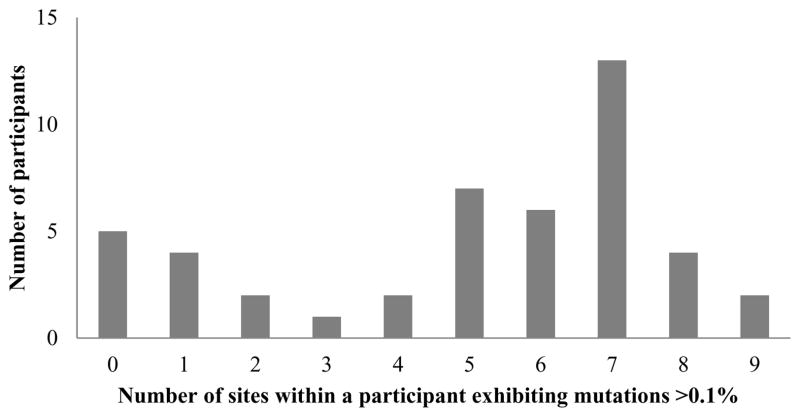
The spread of resistance variation within individual participants did not follow any particular pattern. Around one quarter of participants (n=11) demonstrated either no or very minimal variation (two or fewer mutations) whilst others had variation at most sites (Figure 3), however, there were no significant differences observed in the amount of variation by either HIV status or duration of infection. The median number of sites per subject with evidence of RAVS > 0.1% was 6. In those with 6 or more sites affected the proportion of HIV positivity was similar to those with less than 6 sites affected (42% vs 52%, p=0.490), and although the duration of infection was greater at 24.5 weeks versus 16 weeks, this was non-significant, p=0.11.
Figure 3. Number of resistance mutations in the NS3 protease gene, within each participant, exhibited at > 0.1% frequency.
The occurrence of at least one acid change known to confer resistance was included as varation at that site. Individuals demonstrated a range of the number of resistance mutations found >0.1% (0 – 9 sites).
NS5B polymerase sequencing
Among those with available samples (n=50), sequencing of the NS5B polymerase gene was possible in 49 participants (Figure 1). Variation in amino acid composition was studied at 12 sites known to be associated with resistance to polymerase inhibitors. These included the positions S282T and S96T, P496A/S, P495S/L/A/T C316Y/N, S365 T/AL419M/V, M243 T/I/V and Y448C/H. Mean coverage at sites ranged from 2102–6722 reads. Only one (2%) participant had evidence of a potential dominant resistant variant, a M423I change in 98% of the viral population. A further 12 participants had evidence of a potential resistant variant present as a minority variant at a level above 1% (Table 2). These changes predominately involved low level variation between 1–3% at the following sites S365T (n=4), L419M/V (n=3), M423T (n=2), V494A (n=2) and P496S (n=1). Only one participant had a non-dominant variant at > 3% prevalence – this participant had a V499A change present at 18.8% of viral quasispecies. The same participant also had an S365T resistant variant present at 1.38%. The presence of multiple resistant mutations on a single viral strain was not observed (based on approximately 500bp sequence reads).
As with the protease gene, variation at a level below 1% but above 0.1% was observed far more frequently (Figure 2b). The amount of variation differed depending on the site. For example no participant demonstrated any resistance variant at the S282T site and only 2% of participants’ demonstrated resistance variant at the S96T site at a level above 0.1%. At other sites variation was much more frequent. Over two thirds of participants exhibited low level resistance variants at a prevalence of between 0.1% and 1% at M423T, Y448C/H and Y494I/A sites (Figure 2b).
NS5A sequencing
A subgroup of thirteen genotype 1a participants also underwent sequencing of the NS5A region in which mutations conferring resistance to the first generation NS5A inhibitors including daclatasvir and ledipasvir are found. Six sites of interest were examined for mutational change including M28 A/T/V, Q30 E/H/R/K, L31 M/V, Q54 H/N, H58 D/P and Y93 C/H/N (Figure 2c). Mean coverage at sites ranged from 2583–2984 reads. Only one participant had a dominant resistance variant with a M28V change in 99.52% of quasispecies. Two participants had variants present at between 1 and 3% of viral quasispecies; one with a Q30R at 1.33% and an H58P change at 1.10%, while the second had only a Q30R mutation at 2.29% (Table 2). As with protease and polymerase regions however resistance variants were commonly present at between 0.1 and 1% of the total virus population, observed at the following frequencies at sites: M28V/A/T (64%), Q30R/E/R/K (46%), and Y93 C/H/N (50%). Only the L31M/V site was highly conserved with no evidence of resistance variants at a level above 0.01% found in any participants.
Effect of immune pressure on frequency of resistance mutations
Overlap between host T-cell immune pressure, as directed by the Human Leucocyte Antigen (HLA) repertoire of the host, and drug pressure at specific sites in the viral genome can influence an individual’s risk to develop drug resistance (8, 21, 22). We investigated whether subjects with specific HLA alleles, corresponding to overlapping HLA-restricted epitopes with resistance sites, correlated with detectable circulating mutations at >1% across the three proteins of interest.
In the NS3 protease region, the HLA-A2-restricted epitope from 1073–1081 (CINGVCWTV) in the polyprotein includes position 54 and 55 in the protease protein. Of those subjects with HLA typing available (50% of cohort), only one had a resistant variant at 1% at position 55 and that subject carried HLA-A2 (1/8 with HLA-A2). The HLA-A24-restricted epitope from position 1100–1108 (MYTNVDQDL) includes position 80. Of the four subjects with HLA-A24, one had lysine (K) as the dominant amino acid whereas two out of 13 subjects without HLA-A24 had a different dominant amino acid to glutamine (Q). In the NS5A region, only four out of 14 individuals had HLA typing. For the HLA-A3-restricted epitope from 2017–2026 (GVWRGDGIMH) including position 54 in the NS5A protein, only one subject had a variant at 1.6% and carried HLA-A3 (1/2 with HLA-A2). For the NS5B region, there were five HLA-restricted epitopes that also spanned a RAV: HLA-A3-restricted epitope 2510–2518 (SLTPPHSAK) including position 96; HLA-B27-restricted epitope from 2841–2849 (ARMILMTHF) including position 423; HLA-A1-restricted epitope from 2858–2868 (QLEQALDCEIY) including position 448; HLA-B55-restricted epitope from 2898–2907 (SPGEINRVAA) including position 482; and HLA-B57-restricted epitope from 2912–2921 (LGVPPLRAWR) including positions 494, 495, 496 and 499. For the HLA-A3-restricted epitope, four of 18 subjects carried HLA-A3 but no subject with HLA typing had a variant at >1%. For the HLA-B27-restricted epitope, only two subjects carried HLA-B27 and neither showed variation at position 423 at >1% or any of the other subjects. For the HLA-A1-restricted epitope, of the nine subjects that carried HLA-A1 none had a variant at >1% or any other of the subjects. For the HLA-B55-restricted epitope, no subjects had a variant at greater than 1% and only one subject carried HLA-B55. For the HLA-B57-restricted epitope, only one subject carried HLA-B57 and did not have a variant at >1% for positions any variant sites. One subject had a variant at 2.6% at position 496 but did not carry HLA-B57 and one subject had a variant at 18.8% at 499 but did not carry HLA-B57.
Discussion
This study provides the largest and most comprehensive analysis of the prevalence of naturally occurring DAA resistance in subjects with recent HCV. Dominant resistant variants (at levels of 50–100%) were observed, albeit infrequently, in the setting of relatively short duration of infection. Overall, six of 50 (12%) individuals demonstrated evidence of dominant resistance variants conferring reduced sensitivity to one DAA treatment class. Ten percent of individuals had a dominant variant conferring protease inhibitor resistance, one individual had a dominant variant conferring polymerase inhibitor resistance and another had a variant conferring resistance to the NS5A inhibitor class. An additional 18 (36% of those sequenced) demonstrated evidence of resistant variants at a level below that detectable by bulk sequencing (ie < 20%) but above a threshold of 1%. At a lower threshold level of between 0.1 and 1%, resistant variants were almost universal, although notably absent at a few sites including S282T/S96T (sofosbuvir) and the L31M (daclatasavir).
Both the prevalence and significance of naturally occurring drug resistance mutations in chronic HCV have been debated. A number of studies have examined the presence of pre-treatment drug resistance, generally to the protease inhibitor class, with varying results (5, 8, 23–25). A large cohort of 507 individuals from US, Germany and Switzerland examined using bulk (population) based sequencing identified either protease or polymerase dominant mutants present in 8.6% of subjects (5). A further large study composed of 405 individuals from the UK, Australia and Switzerland suggested that up to 21.5% of genotype 1a subjects may have drug resistance mutations on bulk sequencing (8). Pre-existing mutations have been shown to influence treatment outcome as in the PROVE1/2 studies, where all of the four participants with the R155K mutation present as the dominant strain at baseline failed to achieve SVR (23). Similarly, population sequencing of participants in the Boceprevir registration studies demonstrated that the SVR rate in participants with both baseline mutations and a <1 log decline during four weeks lead-in was 0% (compared to 28–38% in those with wild type) (26, 27). Recently, the presence of Q80K prior to treatment with the second generation protease inhibitor Simeprevir has been shown to negatively impact on treatment outcome, bringing SVR rates down to 58%, similar to that of PEG/RBV alone (28). Q80K has been demonstrated to occur in up to 47% of untreated GT1a participants in North America (28, 29), but geographical variability is observed and the prevalence in the chronic Australian population is unknown. In our study of recent infection Q80K was infrequent, observed in just two (4%) of untreated participants. To date, there is insufficient data to determine if the prevalence of the Q80K in recently acquired HCV occurs less frequently than in chronic HCV.
Given that a variant can only be detected by population sequencing when it comprises > 20% of the viral quasispecies, there obviously exists the potential for minor drug resistant variants to be present at levels not detected by this method. Under drug pressure these minor variants may have the capacity to be selected due to their survival advantage in the presence of drug and fill the replication space to become the dominant species. This phenomenon has been extensively described using clonal analysis in longitudinal studies of antiviral monotherapy and/or treatment failures (1, 2, 30–33).
The spectrum and prevalence of pre-existing variants present using sensitive clonal or deep sequencing methodology in treatment naïve individuals with chronic HCV has been explored in a number of small studies (10–17). In a study of 18 participants from the PROVE2 study who underwent deep sequencing at baseline almost every participant had evidence of the R155K/T/Q (levels between 0.1%-7.8%), A156 S/T/V (0.l2% to 3.2%) and I170 A/T (0.1–0.5%) although evidence of other mutations (V36 A/M, T54 A/S, V55A, Q80 R/K) were less common (10). Other studies have also demonstrated a low prevalence of variations (<1%) in treatment-naïve subjects (14, 34). Bartolini et al (2013) explored the prevalence of resistance mutations in the NS3 protease in both HCV monoinfected (n = 10) and HIV coinfected subjects (n=18), and identified pre-existing resistance mutations to occur at a higher frequency in some individuals; V36A (26%), Q41H (45%, 19%), Q80L (40%) (15), although this did not differ between the two groups.
The period after newly acquired HCV infection is unique in several ways. During this time spontaneous clearance of HCV is possible, changes in the viral quasispecies occur as immune pressure drives viral adaption, and treatment is likely to be successful (20, 35–39). Very few studies have examined the composition of the viral quasispecies at this time, particularly with relevance to the sites associated with drug resistance. One recent small study in 38 HIV infected individuals with acute HCV examined regions in the protease gene by both population ultra-deep sequencing (40). In this study16 % of individuals had a ‘dominant’ resistance variant, but deep sequencing down to a level of <1% demonstrated every site in all samples, with the exception of T54M/L and R155Q. The authors postulated that their higher level of resistance detection compared to monoinfected studies may be due to a higher replication rate in HIV positive individuals, a higher mutation rate in recent HCV or a founder effect. Our study, performed on a larger number of individuals with recently acquired HCV infection, and including both HIV positive and negative participants, does not support any of these hypotheses. No association was observed with either HIV status or length of infection, either when considering the presence of dominant mutations alone or the presence of lower frequency mutations.
The data also suggest that some individuals may be more prone to variation. This could in part be explained by the hosts own immune environment such as the influence of Human Leucocyte Antigen (HLA) type. However, although this study was limited by the number of subjects with HLA typing available (only 50% of cohort) and the number of subjects with each HLA type, there was no suggestion that an individual’s HLA type either enriched, or influenced the prevalence for overlapping DAA resistance mutation sites in these subjects.
The infrequency of mutations present at a level of >20% in our study undoubtedly reflects the reduced fitness of most of these variants and suggests that these sites require either compensatory mutations or a strong selective pressure (drug) to emerge. This is supported by the fact that very few participants had more than one dominant resistance mutation and there was little evidence of multiple mutations on single strains occurring at low frequency. However the low prevalence of resistance mutations in this study reduced the power to detect the presence of compensatory and false negative results could not be excluded.
The relevance of viral strains present at between 1–20% (close to limit of Sanger or bulk sequencing detection) and their impact on therapeutic outcome is unknown. In the setting of HIV infection viral strains present at as low as 1% of the viral quasispecies can impact on drug efficacy. In a study of the CCR5 antagonist vicriviroc, drug resistant strains emerged from levels of < 1% to become the dominant virus within just two weeks, indicating that even the presence of very low frequency variants may have significant implications (41). At a level of between 0.1 and 1% the clinical significance of the presence of viral variants is even less well established. As HCV replication is both rapid and highly error prone it can be calculated that all single and double mutants are generated multiple times a day, many of which will not survive due to inherent weakness or lack of fitness (42). It is interesting in our study to note that although at many sites resistance mutant variants are observed at very low levels in the majority of individuals, in a few sites no evidence of resistance variants in any participants were observed, even down to a threshold of 0.1%. In particular, this phenomenon was observed at site 282 in the polymerase gene, known to be associated in vitro with resistance to the nucleotide inhibitors including sofosbuvir, and at site 31 in the NS5A region associated with daclatasvir resistance. This data supports the likelihood that combination regimens with these classes of drugs are likely to be highly effective in the setting of recently acquired HCV, without the issues of resistance which have troubled the HIV treatment field. Given that repeated courses of short duration treatments may be required for those at ongoing risk of HCV reinfections, these data are reassuring.
Our data has several limitations. Despite the fact that it is the largest study of HCV resistance performed in the setting of recently acquired HCV to date, the numbers of participants included are smaller than those studied in chronic HCV using bulk sequencing, thus the confidence with which the prevalence of dominant variants can be predicted is reduced. Secondly, the certainty around the presence of resistance variants between 0.1 and 1% is less definite due to potential sequencing errors creating background noise. However, the pattern of variants observed at this level, for example from 70% for some of the more common protease inhibitor mutations to 0% for the S282T variant, argues for a true reflection of the viral quasispecies rather than technique errors simply overestimating diversity. In addition, although the methodology utilised here did not incorporate primer ID type technology (43) low frequency variants observed in a smaller subsets of participants was similar to data from a second aliquot from the same time-point and overlapped with data from a second time-point tested, approximately after 12 weeks of the initial time-point.
In summary, this dataset provides the largest and most comprehensive analysis of the pattern of DAA resistant variants present in the viral quasispecies at the time of recently acquired HCV from both HIV positive and negative individuals. Our study finds a very low occurrence of dominant resistance mutations although a high prevalence of resistant variants present at low levels of < 1%, the significance of which are unknown. Resistance variants were observed at most of the known sites associated with resistance to DAAs from all classes with the exception of the nucleotide agent sofosbuvir for which no evidence of resistant variants was observed.
Acknowledgments
We gratefully acknowledge the participants, nurses, trial coordinators and investigators of the ATAHC study for their ongoing support. We also greatly appreciate the technical help and laboratory support provided by Brendan Jacka, Francois Lamoury, Sofia Bartlett, Austin Butcher from the Viral Hepatitis and Clinical Research Program and staff at the Institute for Immunology and Infectious Diseases, Murdoch University. We would also like to acknowledge support from Pip Marks throughout the study.
Funding
This work was supported by the National Health and Medical Research Council (NHMRC) Project Grant [grant number 1006331]. The Kirby Institute for Infection and Immunity in Society is funded by the Australian Government Department of Health and Ageing and is affiliated with the Faculty of Medicine, UNSW Australia. The views expressed in this publication do not necessarily represent the position of the Australian Government. Roche Pharmaceuticals supplied financial support for pegylated IFN-alfa-2a/ribavirin for the ATAHC from which participant samples were used in this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
- The International Liver Congress 46th annual meeting of the European Association for the Study of the Liver (April 2012), Berlin, Germany.
- 20th International HIV and Hepatitis Virus Drug Resistance Workshop (June 2011), Los Cabos, Mexico.
- 62nd Annual Meeting of the American Association for the Study of Liver Diseases (Nov 2011), San Francisco, CA.
- 18th International Symposium on Hepatitis C and Related Viruses (Sept 2011), Seattle, WA.
- Australian Centre for HIV & Hepatitis Virology Research, 7th annual workshop (June 2011), Sunshine Coast, Queensland, Australia.
Conflict of interest statement
Dr. Applegate reports grants from National Health and Medical Research Council and from National Health and Medical Research Council during the conduct of the study; Dr. Gaudieri reports grants from National Health and Medical Research Council of Australia during the conduct of the study; Ms. Plauzolles has nothing to disclose; Dr. Chopra has nothing to disclose. Dr. Grebely reports grants and personal fees from Merck, grants from Gilead, grants from Janssen, grants from Abbvie outside the submitted work; Associate Professor Lucas has nothing to disclose; Dr. Hellard reports grants from null outside the submitted work; Dr. Luciani has nothing to disclose; Professor Dore reports grants, personal fees and non-financial support from Roche, grants, personal fees and non-financial support from Merck, grants and personal fees from Janssen, grants, personal fees and non-financial support from Gilead, grants, personal fees and non-financial support from Bristol-Myers Squibb, grants and personal fees from Abbvie, grants from Vertex, grants from Boeringher Ingelheim outside the submitted work. Dr. Matthews reports grants from NHMRC during the conduct of the study; grants and other from Gilead Inc, grants and other from Janssen, other from Roche, other from BMS, grants and other from MSD outside the submitted work.
References
- 1.Kieffer TL, Sarrazin C, Miller JS, Welker MW, Forestier N, Reesink HW, et al. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology. 2007 Sep;46(3):631–9. doi: 10.1002/hep.21781. [DOI] [PubMed] [Google Scholar]
- 2.Sarrazin C, Kieffer TL, Bartels D, Hanzelka B, Muh U, Welker M, et al. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology. 2007 May;132(5):1767–77. doi: 10.1053/j.gastro.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 3.Thompson AJ, McHutchison JG. Antiviral resistance and specifically targeted therapy for HCV (STAT-C) Journal of viral hepatitis. 2009 Jun;16(6):377–87. doi: 10.1111/j.1365-2893.2009.01124.x. [DOI] [PubMed] [Google Scholar]
- 4.Rong L, Ribeiro RM, Perelson AS. Modeling quasispecies and drug resistance in hepatitis C patients treated with a protease inhibitor. Bulletin of mathematical biology. 2012 Aug;74(8):1789–817. doi: 10.1007/s11538-012-9736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuntzen T, Timm J, Berical A, Lennon N, Berlin AM, Young SK, et al. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology. 2008 Dec;48(6):1769–78. doi: 10.1002/hep.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim AY, Timm J, Nolan BE, Reyor LL, Kane K, Berical AC, et al. Temporal dynamics of a predominant protease inhibitor-resistance mutation in a treatment-naive, hepatitis C virus-infected individual. The Journal of infectious diseases. 2009 Mar 1;199(5):737–41. doi: 10.1086/596657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colson P, Brouk N, Lembo F, Castellani P, Tamalet C, Gerolami R. Natural presence of substitution R155K within hepatitis C virus NS3 protease from a treatment-naive chronically infected patient. Hepatology. 2008 Feb;47(2):766–7. doi: 10.1002/hep.22122. [DOI] [PubMed] [Google Scholar]
- 8.Gaudieri S, Rauch A, Pfafferott K, Barnes E, Cheng W, McCaughan G, et al. Hepatitis C virus drug resistance and immune-driven adaptations: relevance to new antiviral therapy. Hepatology. 2009 Apr;49(4):1069–82. doi: 10.1002/hep.22773. [DOI] [PubMed] [Google Scholar]
- 9.Alves R, Queiroz AT, Pessoa MG, da Silva EF, Mazo DF, Carrilho FJ, et al. The presence of resistance mutations to protease and polymerase inhibitors in Hepatitis C virus sequences from the Los Alamos databank. Journal of viral hepatitis. 2013 Jun;20(6):414–21. doi: 10.1111/jvh.12051. [DOI] [PubMed] [Google Scholar]
- 10.Chevaliez S, Rodriguez C, Soulier A, Ahmed-Belkacem A. Molecular characterization of HCV resistance to telaprevir by means of ultra-deep pyrosequencing: preexisting resistant variants and dynamics of resistant populations. Journal of Hepatology. 2011;54(Supplement 1):S30. [Google Scholar]
- 11.Nasu A, Marusawa H, Ueda Y, Nishijima N, Takahashi K, Osaki Y, et al. Genetic heterogeneity of hepatitis C virus in association with antiviral therapy determined by ultra-deep sequencing. PloS one. 2011;6(9):e24907. doi: 10.1371/journal.pone.0024907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauck M, Alvarado-Mora MV, Becker EA, Bhattacharya D, Striker R, Hughes AL, et al. Analysis of hepatitis C virus intrahost diversity across the coding region by ultradeep pyrosequencing. Journal of virology. 2012 Apr;86(7):3952–60. doi: 10.1128/JVI.06627-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirst ME, Li EC, Wang CX, Dong HJ, Liu C, Fried MW, et al. Deep sequencing analysis of HCV NS3 resistance-associated variants and mutation linkage in liver transplant recipients. PloS one. 2013;8(7):e69698. doi: 10.1371/journal.pone.0069698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svarovskaia ES, Martin R, McHutchison JG, Miller MD, Mo H. Abundant drug-resistant NS3 mutants detected by deep sequencing in hepatitis C virus-infected patients undergoing NS3 protease inhibitor monotherapy. Journal of clinical microbiology. 2012 Oct;50(10):3267–74. doi: 10.1128/JCM.00838-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartolini B, Giombini E, Zaccaro P, Selleri M, Rozera G, Abbate I, et al. Extent of HCV NS3 protease variability and resistance-associated mutations assessed by next generation sequencing in HCV monoinfected and HIV/HCV coinfected patients. Virus research. 2013 Nov 6;177(2):205–8. doi: 10.1016/j.virusres.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Trimoulet P, Pinson P, Papuchon J, Foucher J, Vergniol J, Chermak F, et al. Dynamic and rapid changes in viral quasispecies by UDPS in chronic hepatitis C patients receiving telaprevir-based therapy. Antiviral therapy. 2013 May 23; doi: 10.3851/IMP2632. [DOI] [PubMed] [Google Scholar]
- 17.Franco S, Casadella M, Noguera-Julian M, Clotet B, Tural C, Paredes R, et al. No detection of the NS5B S282T mutation in treatment-naive genotype 1 HCV/HIV-1 coinfected patients using deep sequencing. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2013 Oct 3; doi: 10.1016/j.jcv.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 18.Dore GJ, Hellard M, Matthews GV, Grebely J, Haber PS, Petoumenos K, et al. Effective treatment of injecting drug users with recently acquired hepatitis C virus infection. Gastroenterology. 2010 Jan;138(1):123–35. e1–2. doi: 10.1053/j.gastro.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rauch A, James I, Pfafferott K, Nolan D, Klenerman P, Cheng W, et al. Divergent adaptation of hepatitis C virus genotypes 1 and 3 to human leukocyte antigen-restricted immune pressure. Hepatology. 2009 Oct;50(4):1017–29. doi: 10.1002/hep.23101. [DOI] [PubMed] [Google Scholar]
- 20.Bull RA, Luciani F, McElroy K, Gaudieri S, Pham ST, Chopra A, et al. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS pathogens. 2011 Sep;7(9):e1002243. doi: 10.1371/journal.ppat.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.John M, Moore CB, James IR, Mallal SA. Interactive selective pressures of HLA-restricted immune responses and antiretroviral drugs on HIV-1. Antiviral therapy. 2005;10(4):551–5. [PubMed] [Google Scholar]
- 22.Tschochner M, Chopra A, Maiden TM, Ahmad IF, James I, Furrer H, et al. Effects of HIV type-1 immune selection on susceptability to integrase inhibitor resistance. Antiviral therapy. 2009;14(7):953–64. doi: 10.3851/IMP1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartels DJ, Zhou Y, Zhang EZ, Marcial M, Byrn RA, Pfeiffer T, et al. Natural prevalence of hepatitis C virus variants with decreased sensitivity to NS3.4A protease inhibitors in treatment-naive subjects. The Journal of infectious diseases. 2008 Sep 15;198(6):800–7. doi: 10.1086/591141. [DOI] [PubMed] [Google Scholar]
- 24.Paolucci S, Fiorina L, Piralla A, Gulminetti R, Novati S, Barbarini G, et al. Naturally occurring mutations to HCV protease inhibitors in treatment-naive patients. Virology journal. 2012;9:245. doi: 10.1186/1743-422X-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vicenti I, Rosi A, Saladini F, Meini G, Pippi F, Rossetti B, et al. Naturally occurring hepatitis C virus (HCV) NS3/4A protease inhibitor resistance-related mutations in HCV genotype 1-infected subjects in Italy. The Journal of antimicrobial chemotherapy. 2012 Apr;67(4):984–7. doi: 10.1093/jac/dkr581. [DOI] [PubMed] [Google Scholar]
- 26.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, et al. Boceprevir for untreated chronic HCV genotype 1 infection. The New England journal of medicine. 2011 Mar 31;364(13):1195–206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merck and Co. I. FDA Antiviral Drugs Advisory Committee Meeting Boceprevir Capsules (NDA 202–258) Briefing Document. 2011. [Google Scholar]
- 28.Jacobson I, Dore GJ, Foster GR, Fried MW, MPM, Marcellin P, et al. Simeprevir (TMC435) with peginterferon/ribavirin for treatment of chronic HCV genotype 1 infection in treatment-naïve patients: efficacy in difficult-to-treat patient sub-populations in the QUEST 1 and 2 phase III trial. 64th Annual Meeting of the American Association for the Study of Liver diseases,; Washington, USA. 2013 November 1–5; 2013. p. Abstract 1122. [Google Scholar]
- 29.Berger KL, Triki I, Cartier M, Marquis M, Massariol MJ, Bocher WO, et al. Baseline HCV NS3 Polymorphisms and their Impact on Treatment Response in Clinical Studies of the HCV NS3 Protease Inhibitor Faldaprevir. Antimicrobial agents and chemotherapy. 2013 Nov 11; doi: 10.1128/AAC.01976-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Susser S, Vermehren J, Forestier N, Welker MW, Grigorian N, Fuller C, et al. Analysis of long-term persistence of resistance mutations within the hepatitis C virus NS3 protease after treatment with telaprevir or boceprevir. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2011 Dec;52(4):321–7. doi: 10.1016/j.jcv.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 31.Barnard RJ, McHale CM, Newhard W, Cheney CA, Graham DJ, Himmelberger AL, et al. Emergence of resistance-associated variants after failed triple therapy with vaniprevir in treatment-experienced non-cirrhotic patients with hepatitis C-genotype 1 infection: a population and clonal analysis. Virology. 2013 Sep 1;443(2):278–84. doi: 10.1016/j.virol.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Le Pogam S, Yan JM, Chhabra M, Ilnicka M, Kang H, Kosaka A, et al. Characterization of hepatitis C virus (HCV) quasispecies dynamics upon short-term dual therapy with the HCV NS5B nucleoside polymerase inhibitor mericitabine and the NS3/4 protease inhibitor danoprevir. Antimicrobial agents and chemotherapy. 2012 Nov;56(11):5494–502. doi: 10.1128/AAC.01035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Sun JH, O’Boyle DR, 2nd, Nower P, Valera L, Roberts S, et al. Persistence of resistant variants in hepatitis C virus-infected patients treated with the NS5A replication complex inhibitor daclatasvir. Antimicrobial agents and chemotherapy. 2013 May;57(5):2054–65. doi: 10.1128/AAC.02494-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fonseca-Coronado S, Escobar-Gutierrez A, Ruiz-Tovar K, Cruz-Rivera MY, Rivera-Osorio P, Vazquez-Pichardo M, et al. Specific detection of naturally occurring hepatitis C virus mutants with resistance to telaprevir and boceprevir (protease inhibitors) among treatment-naive infected individuals. Journal of clinical microbiology. 2012 Feb;50(2):281–7. doi: 10.1128/JCM.05842-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nature reviews Gastroenterology & hepatology. 2013 Sep;10(9):553–62. doi: 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- 36.Grebely J, Matthews GV, Dore GJ. Treatment of acute HCV infection. Nature reviews Gastroenterology & hepatology. 2011 May;8(5):265–74. doi: 10.1038/nrgastro.2011.32. [DOI] [PubMed] [Google Scholar]
- 37.Feld JJ. Treatment indication and response to standard of care with peginterferon and ribavirin in acute and chronic HCV infection. Best practice & research Clinical gastroenterology. 2012 Aug;26(4):429–44. doi: 10.1016/j.bpg.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Plauzolles A, Lucas M, Gaudieri S. Hepatitis C virus adaptation to T-cell immune pressure. The Scientific World Journa. 2013:673240. doi: 10.1155/2013/673240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Stoddard MB, Wang S, Blair LM, Giorgi EE, Parrish EH, et al. Elucidation of hepatitis C virus transmission and early diversification by single genome sequencing. PLoS pathogens. 2012;8(8):e1002880. doi: 10.1371/journal.ppat.1002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leggewie M, Sreenu VB, Abdelrahman T, ECML, Wilkie GS, Klymenko T, et al. Natural NS3 resistance polymorphisms occur frequently prior to treatment in HIV-positive patients with acute hepatitis C. Aids. 2013 Sep 24;27(15):2485–8. doi: 10.1097/QAD.0b013e328363b1f9. [DOI] [PubMed] [Google Scholar]
- 41.Tsibris AM, Korber B, Arnaout R, Russ C, Lo CC, Leitner T, et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PloS one. 2009;4(5):e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rong L. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Science Translational Medicine. 2010;(30):30ra2. doi: 10.1126/scitranslmed.3000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proceedings of the National Academy of Sciences of the United States of America. 2011 Dec 13;108(50):20166–71. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]