Abstract
Diet composition may affect energy metabolism in a tissue-specific manner. Using C57Bl/6J mice, we tested the effect of ketosis-inducing and non-inducing high fat diets on genes relevant to brain bioenergetic infrastructures, and on proteins that constitute and regulate that infrastructure. At the end of a one-month study period the two high fat diets appeared to differentially affect peripheral insulin signaling, but brain insulin signaling was not obviously altered. Some bioenergetic infrastructure parameters were similarly impacted by both high fat diets, while other parameters were only impacted by the ketogenic diet. For both diets, mRNA levels for CREB, PGC1α, and NRF2 increased while NRF1, TFAM, and COX4I1 mRNA levels decreased. PGC1β mRNA increased and TNFα mRNA decreased only with the ketogenic diet. Brain mtDNA levels fell in both the ketogenic and non-ketogenic high fat diet groups, although TOMM20 and COX4I1 protein levels were maintained, and mRNA and protein levels of the mtDNA-encoded COX2 subunit were also preserved. Overall, the pattern of changes observed in mice fed ketogenic and non-ketogenic high fat diets over a one month time period suggests these interventions enhance some aspects of the brain’s aerobic infrastructure, and may enhance mtDNA transcription efficiency. Further studies to determine which diet effects are due to changes in brain ketone body levels, fatty acid levels, glucose levels, altered brain insulin signaling, or other factors such as adipose tissue-associated hormones are indicated.
Keywords: bioenergetics, brain, high fat diet, ketogenic diet, mitochondria
Introduction
Brain bioenergetic dysfunction is observed in several neurodegenerative diseases, and may contribute to their onset or progression (Swerdlow 2009, Swerdlow 2012b, Swerdlow 2012a). Because diet impacts bioenergetics, the ability of diet interventions to exacerbate or mitigate certain neurodegenerative disease states, at either a disease modifying or symptom level, is increasingly being considered (Swerdlow 2011, Swerdlow 2014).
Diet manipulations may have tissue or organ-specific bioenergetic effects (McKhann et al. 2011). Because of our interest in brain bioenergetics, in this study we used C57Bl/6J mice to test the impact of two high-fat diet protocols on selected brain bioenergetics-relevant parameters. In one diet we increased fat content while maintaining a high carbohydrate percentage (Western diet). For the other, we increased fat content while reducing carbohydrate and protein (ketogenic diet). We found these diet protocols affected brain bioenergetic infrastructures in both similar and unique ways.
Materials and Methods
Animal procedures and diets
All animal protocols and procedures conformed to the NIH guidelines for the care and use of laboratory animals, and were approved by the University of Kansas Medical Center Animal Care and Use Committee. Five-month old male C57Bl/6J mice (Jackson Laboratory, #000664) were randomly assigned to be fed ad libitum one of three diets. For mice on the standard diet (n=11), caloric content (3.0 kcal/g) derived from chow containing 24.3% protein, 4.7% fat (soybean oil), and 40.2% carbohydrate by weight (Teklad Rodent diet #8604). For mice on the high-fat “Western” diet (n=15) caloric content (5.51 kcal/g) derived from chow containing 20.5% protein, 36% fat (lard), and 36% carbohydrate by weight (BioServ #F1850). For mice on the high-fat ketogenic diet, caloric content (7.2 kcal/g) derived from chow containing 8.6% protein, 75.1% fat (lard), and 3.2% carbohydrate by weight (BioServ #F3666). After 4 weeks on these respective diets, mice were fasted for 4–6 hours and ~50 μL of blood was collected via tail snip for blood glucose readings (AccuCheck) and serum insulin assay (ALPCO #80-INSMSU-E01). Homeostatic model assessment of insulin resistance (HOMA-IR) values were calculated as previously described (Matthews et al. 1985). On day 30 mice were decapitated and tissues rapidly harvested and snap-frozen in liquid nitrogen. During euthanasia, additional blood was collected and used to measure serum β-hydroxybutyrate content (β-Hydroxybutyrate Assay Kit, Pointe Scientific #H7587).
RNA Isolation and Gene Expression/qPCR
RNA was isolated from ~20 mg of cerebral hemisphere-derived brain tissue using a standard protocol. Briefly, cell extracts were collected using TRI reagent (Sigma-Aldrich #T9424). RNA was separated by chloroform, precipitated with isopropanol, and washed with 75% ethanol. cDNA was generated using an RT-PCR cDNA kit (Applied Biosystems #4368814). qPCR reactions were performed on an ABI Prism 384-well Real Time PCR System using Bio-Rad iTaq Master Mix (Bio-Rad #172–5131) and Applied Biosystems TaqMan gene expression primers. Primer sets assessed expression of cyclic AMP response element binding protein 1 (CREB) (Mm00501607_m1), nuclear respiratory factor 1 (NRF1) (Mm01135606_m1), nuclear respiratory factor 2 (NRF2) (Mm00484598_m1), transcription factor A of the mitochondria (TFAM) (Mm00447485_m1), tumor necrosis factors α (TNFα) (Mm00443260_g1), cytochrome oxidase subunit 4 isoform 1 (COX4I1) (Mm01250094_m1), cytochrome oxidase subunit 2 (COX2) (Mm03294838_m1), peroxisome proliferator activated receptor gamma coactivator 1α (PGC1α) (Mm01208835_m1), and peroxisome proliferator activated receptor gamma coactivator 1β (PGC1β) (Mm00504720_m1). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Mm99999915_g1) was used as an internal reference.
mtDNA content
Total tissue DNA was isolated using phenol-chloroform-isoamyl alcohol as previously described (Guo et al. 2009). 2 ng of total DNA was loaded into each well of a 384-well plate with Applied Biosystems TaqMan primers for 18S nuclear (Mm03928990_g1) or 16S mitochondrial (Mm04260181_s1) DNA. Reactions were performed on an ABI Prism 384-well Real Time PCR System.
Immunochemistry
Approximately 50 mg of frozen brain tissue was gently homogenized using a Teflon-glass homogenizer in NE-PER kit cytoplasmic extraction buffer (Pierce #78835) containing protease and phosphatase inhibitor cocktail (Pierce #78444) and EDTA. NE-PER kit protocol instructions were followed for cytoplasmic protein extractions. 20 μg of total protein were loaded into Criterion TGX SDS-PAGE gels (Bio-Rad #567–1085, #567–1084), subjected to electrophoresis, and transferred to nitrocellulose membranes (Whatman Protran #10401196). Blots were blocked in 5% BSA (for antibodies to phosphorylated proteins) or 5% milk in phosphate buffered saline with Tween 20 (PBST) for one hour, incubated in primary antibody overnight, washed three times with PBST, incubated for one hour in secondary antibody (1:2000, Cell Signaling #7074), and then washed three times in PBST. The primary antibodies used were to COX4I1 (1:1000, Cell Signaling #4850), COX2 (1:1000, Invitrogen #A-6404), mammalian target of rapamycin (mTOR) (1:1000, Cell Signaling #2983), phospho-mTOR Ser2448 (1:1000, Cell Signaling #5536), Akt (1:1000, Cell Signaling #5373), phospho-Akt Ser473 (1:1000, Cell Signaling #4060), glycogen synthase kinase 3 beta (GSK3β) (1:1000, Cell Signaling #9315), phospho-GSK3β Ser9 (1:1000, Cell Signaling #9322), and translocase of the outer mitochondrial membrane 20 (TOMM20) (1:500, Santa Cruz Biotechnology #sc-136211). To assess loading of these cytosolic proteins, antibodies to GAPDH (1:2000, Cell Signaling #2118) were used. Bands were visualized using SuperSignal West Femto Chemiluminescent Substrate (Thermo Pierce #34096) on a ChemiDoc XRS system (Bio-Rad).
Statistical analysis
One-way ANOVA with Least Significant Difference (LSD) post-hoc analysis was performed to determine significance between the data means of the three groups using IBM SPSS Statistics software. Statistical significance was considered for p < 0.05.
Results
Weight, beta-hydroxybutyrate, glucose, and insulin resistance
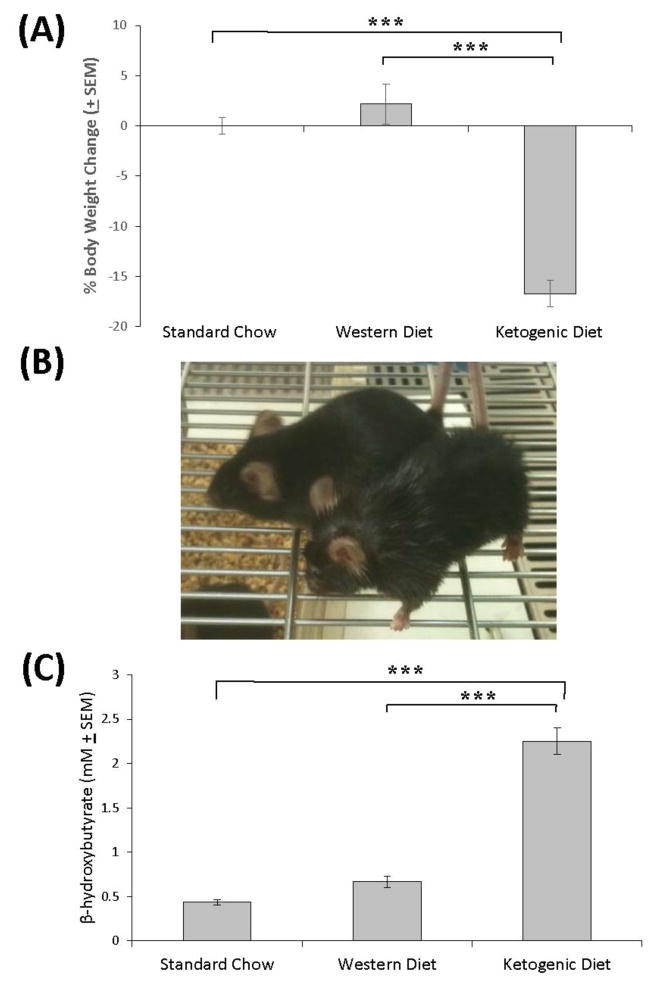
Immediately prior to initiation of the ketogenic and Western diets, during which time all mice were on standard chow, the mean weight was 31.4 ± 1.9 g. After four weeks weights did not appreciably change in mice on the standard and Western diet chows, while mice on the ketogenic diet lost approximately 17% of their body weight (reduced by 5.7 g, to 25.6 ± 2.3 g) (Figure 1A–B). When we measured serum β-hydroxybutyrate at the time of euthanasia, levels were significantly elevated (approximately five-fold) in mice on the ketogenic diet but not in mice on the Western diet (Figure 1C).
Figure 1. Effect of the diets on body weight and β-hydroxybutyrate.
(A) Compared to mice on the standard chow and Western high fat diets, mice on the ketogenic diet lost weight. (B) The mouse in the front of the picture was on the ketogenic diet, and is shown next to a mouse that was on the standard chow diet. The mouse on the ketogenic diet is noticeably thinner. (C) The ketogenic but not the Western high fat diet induced a robust increase in the serum β-hydroxybutyrate concentration. Error bars show SEM. * indicates p<0.05; ***indicates p<0.001.
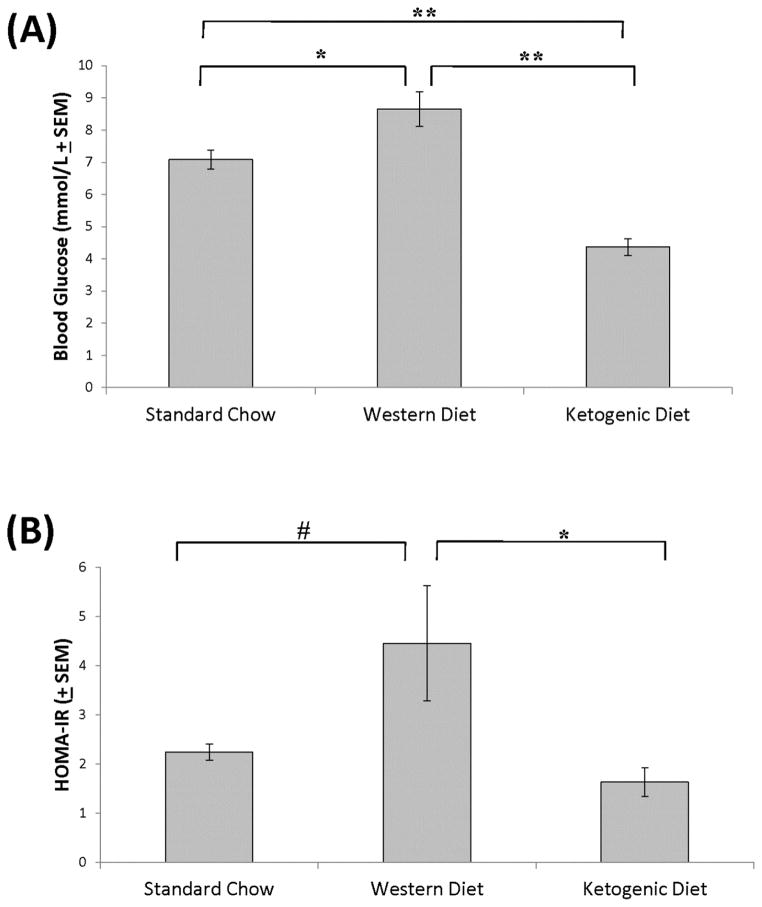
At the end of week 4, compared to mice on the standard chow mice on the ketogenic diet also showed lower fasting blood glucose levels, while mice on the high fat diet showed elevated fasting blood glucose (standard diet=7.09 ± 0.96 mmol/L, Western diet=8.65 ± 2.07mmol/L, ketogenic diet=4.37 ± 0.99 mmol/L) (Figure 2A). While fasting insulin levels were not significantly changed after four weeks for any of the diets (data not shown), mice on the ketogenic diet had significantly less insulin resistance as calculated by the HOMA-IR method than mice on the Western high fat diet (Figure 2B). This was largely due to an upward trend in the Western diet-fed mouse mean HOMA-IR value (p=0.07 when compared to the standard chow-fed mice), in conjunction with a slight concomitant downward trend in the ketogenic diet-fed mice.
Figure 2. Effect of the diets on blood glucose and HOMA-IR.
(A) Compared to mice on the standard chow diet, fasting blood glucose levels were higher in mice on the Western high fat diet and lower in mice on the ketogenic high fat diet. Compared to mice on the Western high fat diet, blood glucose levels were lower in mice on the ketogenic high fat diet. (B) Compared to mice on the standard chow, a trend towards a higher HOMA-IR was observed for mice on the Western diet. The HOMA-IR was lower in mice on the ketogenic diet than it was in mice on the high fat diet. Error bars show SEM. # indicates p=0.07; * indicates p<0.05; **indicates p<0.01.
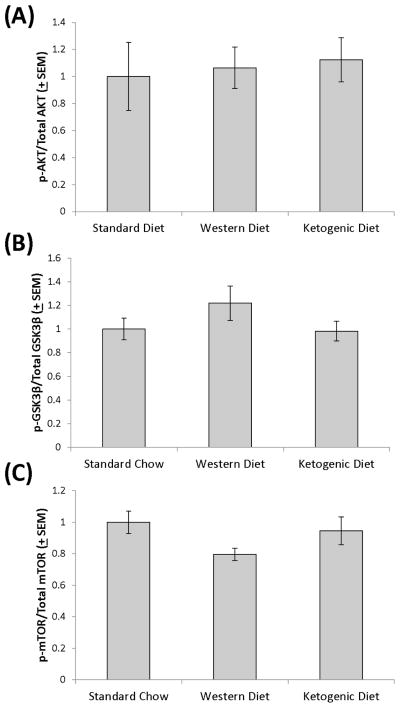
Akt phosphorylation at Ser473 is mediated to a large extent by insulin signaling and Akt, when phosphorylated at Ser473, phosphorylates GSK3β at Ser9 (Manning & Cantley 2007). mTOR Ser2448 phosphorylation levels are also considered to positively correlate with the amount of insulin signaling that is present (Jamart et al. 2012, Reynolds et al. 2002, Chiang & Abraham 2005). Despite the apparent opposite effects on peripheral insulin signaling indicated by the HOMA-IR calculation, brain phospho-Akt/total Akt, phospho-GSK3β/total GSK3β, and phospho-mTOR/total mTOR ratios from mice on the three diets were comparable (Figure 3). Normalizing levels of phosphorylated Akt, GSK3β, or mTOR to GAPDH similarly did not reveal any inter-group differences (data not shown). Total Akt, GSK3β, and mTOR levels were also equivalent across the groups (data not shown).
Figure 3. Brain Akt, GSK3β, and mTOR phosphorylation.
In brain protein lysates, Akt Ser473 (A), GSK3β Ser9 (B), and mTOR Ser2448 phosphorylation (C) levels were comparable across the standard chow, Western high fat, and ketogenic high fat diet-fed groups.
Bioenergetics-related gene expression
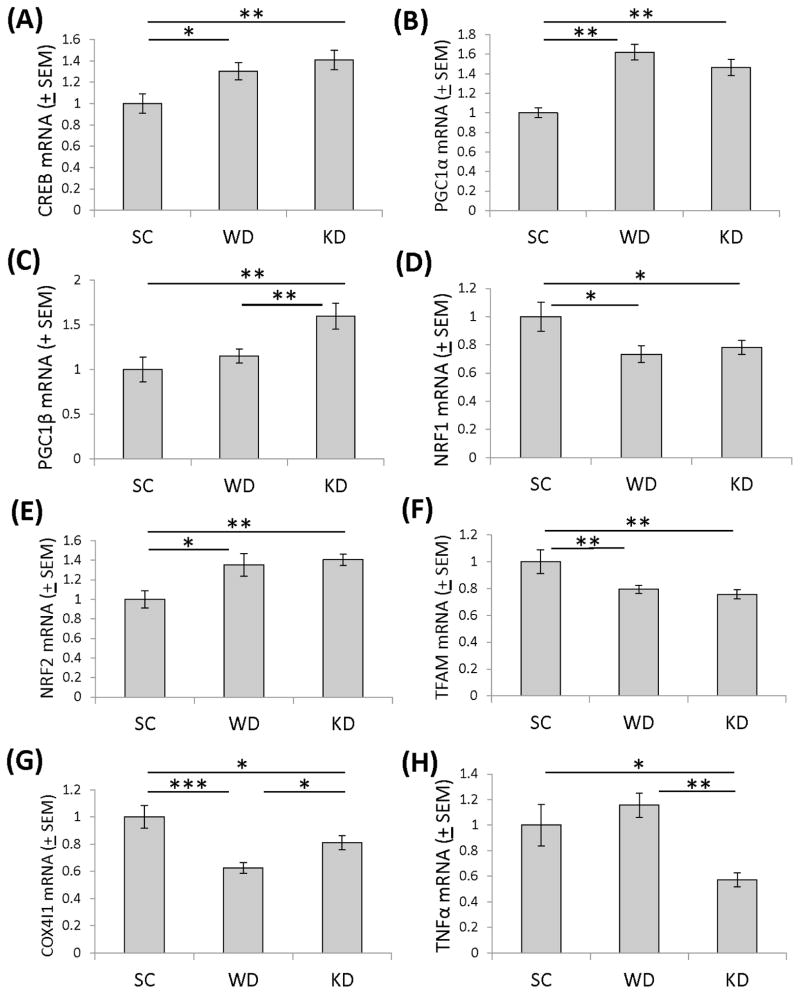
To evaluate the impact of the different diets on brain bioenergetics, we measured the expression levels of genes that monitor, modify, and contribute to cell energy supplies. Brains from mice on Western and ketogenic high fat diets showed a significant increase in CREB gene expression (30% and 41% increases, respectively) (Figure 4A). CREB is responsive to cell energy states (Altarejos & Montminy 2011), and can induce expression of PGC1α, a transcriptional coactivator that facilitates the expression of genes whose products localize to the mitochondria (Herzig et al. 2001, Puigserver et al. 1999). PGC1α gene expression was significantly elevated in brains from mice on Western and ketogenic high fat diets (62% and 46% increases, respectively) (Figure 4B). mRNA levels of another PGC1 family member, PGC1β, were elevated (by 60%) only in mice on the ketogenic diet (Figure 4C). NRF1 mRNA was significantly reduced in brains from mice on both a Western high fat diet (26.7% decrease) and a ketogenic diet (21.8% decrease) (Figure 4D), while NRF2 gene expression increased significantly in the brains of mice on a Western high fat diet (35.1% increase) and a ketogenic diet (41.0% increase) (Figure 4E). TFAM expression was significantly reduced in brains from mice on a Western diet (21.0% reduction) and a ketogenic diet (24.3% reduction) (Figure 4F). Compared to mice on the standard diet COX4I1 mRNA levels were 37% lower in mice on the Western diet and 18% lower in mice on the ketogenic diet (Figure 4G).
Figure 4. Expression levels of bioenergetics-relevant genes in the brains of mice on the standard chow, Western, and ketogenic diets.
(A) Compared to mice on the standard chow diet, CREB mRNA levels were higher in mice on Western and ketogenic diets. (B) Compared to mice on the standard chow diet, PGC1α mRNA levels were higher in mice on Western and ketogenic diets. (C) PGC1β mRNA levels were higher in the brains of mice on the ketogenic diet than they were in mice on the standard chow and high fat diets. (D) Compared to mice on the standard chow diet, NRF1 mRNA levels were lower in mice on Western and ketogenic diets. (E) Compared to mice on the standard chow diet, NRF2 mRNA levels were higher in mice on Western and ketogenic diets. (F) Compared to mice on the standard chow diet, TFAM mRNA levels were lower in mice on Western and ketogenic diets. (G) Compared to mice on the standard chow diet, COX4I1 mRNA levels were lower in mice on Western and ketogenic diets. (H) TNFα mRNA levels were lower in the brains of mice on the ketogenic diet than they were in mice on the standard chow and high fat diets. Error bars show SEM. *indicates p<0.05; **indicates p<0.01; ***indicates p<0.001. SC=standard chow, WD=Western high fat diet, KD=ketogenic high fat diet.
Because bioenergetic phenomena are known to influence inflammation pathways and Western diets have been linked to pro-inflammatory effects (Lopez-Armada et al. 2013, Barbaresko et al. 2013), we also measured TNFα mRNA levels. Mice on the ketogenic diet, but not on the Western diet, showed a significant (42.6%) reduction in brain TNFα expression (Figure 4H).
mtDNA and mitochondrial protein levels
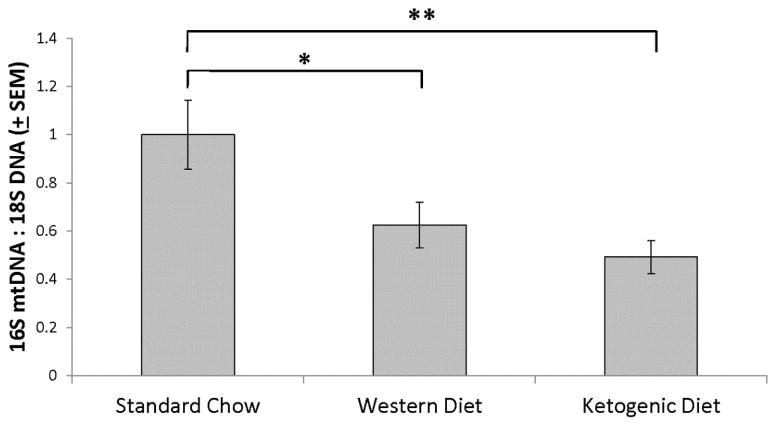
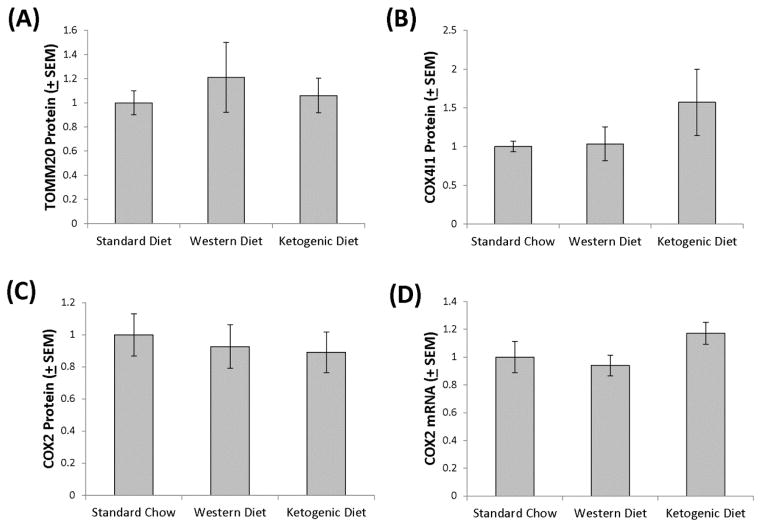
Using a qPCR-based approach to measure mtDNA levels, we found that brains of mice on ketogenic and Western diets contained less PCR-amplifiable mtDNA than the brains of mice on the standard diet (Figure 5). Relative to the standard chow-fed mice, in the brains of the Western high fat diet-fed mice the mtDNA level was reduced by 37.5% and in the brains of the ketogenic diet-fed mice the mtDNA level was reduced by 50.8%. Since reduced mtDNA could indicate less mitochondrial mass, and ketogenic diets reportedly increase hippocampal, skeletal muscle, and liver mitochondrial mass (Eagles & Chapman 2007, Bough et al. 2006, Ahola-Erkkila et al. 2010, Noh et al. 2004) we further assessed brain mitochondrial mass by measuring TOMM20, COX4I1, and COX2 protein levels. TOMM20, COX4I1, and COX2 protein levels were statistically comparable across the groups (Figure 6). Indeed, the ketogenic diet-fed group exhibited a possible trend towards increased COX4I1 protein.
Figure 5. Brain mtDNA levels.
qPCR yielded less mtDNA amplification when genomic DNA was prepared from brain homogenates of mice maintained on the Western and ketogenic diets than it did when genomic DNA was prepared from brain homogenates of mice maintained on the standard chow diet. Error bars show SEM. *indicates p<0.05; **indicates p<0.01.
Figure 6. Protein mitochondrial mass markers and COX2 mRNA in brains of mice on the Western and ketogenic high fat diets.
In brain protein lysates, TOMM20 (A), COX4I1 (B), and COX2 (C) levels were comparable across the standard chow, Western high fat, and ketogenic high fat diet-fed groups. COX2 mRNA levels were also comparable across groups (D).
COX2 is encoded by an mtDNA gene. Since COX2 protein levels were comparable across groups but mtDNA copy number was lower in the Western and ketogenic high fat diet groups, we measured COX2 mRNA. COX2 mRNA levels were comparable across the groups (Figure 6). This suggests transcription of this mtDNA gene, when normalized to the amount of mtDNA present, was enhanced in the Western and ketogenic high fat diet groups.
Discussion
In mice, ketogenic and non-ketogenic high fat diets affect brain mtDNA levels and the expression of genes that respond to or regulate bioenergetic infrastructures. Some changes, at least at face value, occur independently of the presence or absence of ketosis, although the effects of ketogenic and non-ketogenic high fat diets were not equivalent on all the parameters we tested.
Our intent was to maintain diet interventions long enough to induce stable brain bioenergetic infrastructure changes. The life-cycle of a rodent brain mitochondrion is estimated at 2–4 weeks (Menzies & Gold 1971, Beattie et al. 1967, Khan & Wilson 1965, Gross et al. 1969, Rajwade et al. 1975), so we assumed that a one-month diet would suffice. Consistent with this strategy, a prior ketogenic diet study performed using rats showed that after three weeks, hippocampal oxidative stress-related parameters were altered and nuclear factor-like 2 (Nrf2; not to be confused with NRF2) protein levels were elevated (Milder et al. 2010).
During this time, however, diet-related physiologic changes also occurred, especially for mice on the ketogenic diet. As expected (Kennedy et al. 2007), mice on that diet lost considerable weight. Also, after a 4–6 hour fast blood glucose levels fell in the ketogenic diet mice, which potentially could reflect reduced liver glycogen reserve (Conlee et al. 1990, Colle & Ulstrom 1964). In any case, at the end of the fast glucose levels were lower in the ketogenic diet-treated mice at a time when insulin levels were comparable between groups. The HOMA-IR calculation, therefore, suggests the ketogenic diet enhanced peripheral insulin sensitivity, at least when compared to the Western high fat diet, which over an adequate duration would itself predictably induce peripheral insulin resistance in C57Bl/6J mice (Surwit et al. 1988, Sandu et al. 2005, Guilford et al. 2011).
Despite an apparent difference in peripheral insulin sensitivity between mice on the ketogenic high fat and Western high fat diets, differences in brain insulin signaling were not observed. Brain Akt Ser473 phosphorylation, a post-translational modification driven by insulin binding to its surface receptor (Manning & Cantley 2007), was comparable between the diets. GSK3β Ser9 phosphorylation, an Akt-mediated post-translational modification (Grimes & Jope 2001), was also comparable. mTOR Ser2448 phosphorylation levels were also similar across the groups, which suggests that the mTOR activation state did not change (Jamart et al. 2012, Reynolds et al. 2002, Chiang & Abraham 2005). Taken together, our Akt, GSK3β, and mTOR data argue against the presence of diet-induced differences in brain insulin sensitivity, at least during the one month duration of the diet interventions.
The inability of one-month duration Western or ketogenic high fat diets to pervasively alter the brain insulin signaling pathway in young C57Bl/6 mice could simply reflect a lack of an effect, or at least a lack of uncompensated effects. The possibility that changes to the insulin signaling pathway were induced, but at one month were compensated for, is worth considering since the phosphorylation status of proteins like Akt are additionally influenced by other pathways (Hresko & Mueckler 2005, Sarbassov et al. 2005). Compensations due to changes in these other pathways could potentially mitigate insulin-induced changes to Akt and, secondarily, GSK3β or mTOR phosphorylation. This negative finding could also imply a poor correlation exists between peripheral and central insulin resistance, which certainly could be the case if, as reported, insulin transport across the blood-brain barrier is affected by peripheral insulin resistance (Heni et al. 2013). Whether or not longer diet durations will ultimately alter brain insulin signaling remains an important question.
The ketogenic high fat diet, but not the Western high fat diet, induced robust β-hydroxybutyrate production. β-hydroxybutyrate itself, therefore, may not uniquely account for molecular changes that occurred with both the ketogenic and Western high fat diets (increased CREB, PGC1α, and NRF2 mRNA; decreased NRF1 mRNA, TFAM mRNA, COX4I1 mRNA, and mtDNA). The simplest explanation for these concordant changes would be the increased presence of dietary fat. It is generally reported, however, that at least some fatty acids do not freely penetrate the blood-brain barrier (Edmond 2001, Pardridge & Mietus 1980), which would suggest plasma fatty acid levels alone should minimally affect brain metabolism. On the other hand, diffusion of some fatty acids across the blood-brain barrier does occur and blood-brain barrier fatty acid transporters do exist (Mitchell et al. 2011, Mitchell & Hatch 2011), fasting increases cerebrospinal fluid fatty acid levels (Davis et al. 2008), and omega-3 fatty acid deficiency exacerbates mitochondrial dysfunction in a rat traumatic brain injury model (Agrawal et al. 2013). These findings alternatively argue dietary fat could in fact directly impact brain bioenergetics.
Adipose tissue itself has endocrine functions, and produces hormones that enter the brain and affect energy metabolism (Harwood 2012, Gustafson 2010). Ketogenic and Western high fat diets, however, affect adipose tissue in dramatically different ways. Diet-induced shifts in insulin levels, peripheral insulin resistance, and blood glucose were also discordant. Because of this, endocrine changes probably cannot by themselves account for similar ketogenic and Western high-fat diet induced shifts in CREB, PGC1α, NRF1, NRF2, TFAM, and COX4I1 mRNA levels and in mtDNA levels.
It is possible that even for the concordant molecular changes we observed between the ketogenic and non-ketogenic high fat diets, the basis underlying those changes nevertheless differed. In support of this, we did find molecular-level differences between ketogenic and Western high fat diets. Specifically, brain PGC1β gene expression rose and TNFα gene expression fell only with the ketogenic diet. Which of these changes were due to shifts in ketone body levels, brain glucose levels, brain insulin levels, brain insulin resistance differences we failed to detect, or other endocrine factors is unclear.
Western diets reportedly reduce mitochondrial mass in some tissues, while ketogenic diets reportedly enhance it (Bough et al. 2006, Sparks et al. 2005). We were, therefore, surprised to see mouse brain mtDNA levels decline after one month on a ketogenic diet. This decrease was observed despite the fact that expression of the genes that encode CREB and PGC1α increased in ketogenic diet-fed mice. Protein markers of mitochondrial mass including TOMM20, COX4I1, and COX2 protein levels, however, did not fall. PGC1β, which may specifically favor aerobic energy production more than PGC1α does (Lin et al. 2003, St-Pierre et al. 2003) also showed increased gene expression in the ketogenic diet-fed mice. For these reasons, we believe both diets, and in particular the ketogenic diet, are more likely to have increased the efficiency of brain respiration than they are to have actually reduced brain respiration.
Consistent with this view, despite the fact that both high fat diets decreased brain mtDNA levels, COX2 mRNA levels did not decrease. This suggests the high fat diets increased mtDNA transcription efficiency. It is possible, therefore, that mtDNA levels declined in mice on the high fat diets because the high fat diets ultimately enhanced mtDNA transcription.
One potential mechanistic or possibly alternative explanation to our finding that brain mtDNA levels, but not mitochondrial protein levels, fell in mice on the high fat diets is that these diets could have increased mitochondrial autophagy (mitophagy) rates. There certainly is precedence for this possibility, as a previous study of brains from human subjects with Alzheimer’s disease (AD) reported the presence of less PCR-amplifiable mtDNA but also increased cytochrome oxidase subunit 1 (COX1) protein (Hirai et al. 2001). In these same AD brains, increased amounts of mtDNA were observed within autophagosomes. This suggests mtDNA within degraded mitochondria may be rendered less available to PCR, while the ability to detect mitochondrial proteins within degraded mitochondria via immunohistochemistry remains relatively preserved. Regardless, pruning a cell’s less effective mtDNA molecules through mitophagy could potentially enhance mtDNA transcription efficiency. As we did not directly assess the effects of our diet interventions on brain autophagy we cannot confirm or refute this possibility, although it is important to note changes in autophagy rates might be expected to associate with changes in mTOR levels or phosphorylation, neither of which were observed.
A goal of this mouse diet study was to generate brain bioenergetics-relevant data that can be extrapolated to humans. Whether or not our findings would uniformly extend to humans subjected to short-term Western or ketogenic high fat diets, however, is unclear. Also, to simplify our study and reduce the number of mice utilized we evaluated only male mice. It is clear, though, that due to neuroendocrine and perhaps other factors distinct brain energy metabolism-related functions can differ between male and female mice (Fuente-Martin et al. 2013, Brinton 2008). Similar sex differences may also extend to humans. It would be interesting (and useful) to see if repeating our study in female mice yielded equivalent results. Similarly, it would be interesting to see if repeating our study in aged mice yielded equivalent results, since mitochondrial function in aged mice would predictably be less robust than it was in the young mice used in our current study (Navarro & Boveris 2007).
An association between Western diets, peripheral insulin resistance, and type II diabetes is recognized. Insulin resistance and type II diabetes, in turn, associate with specific dysfunctions or diseases of the nervous system (Burns et al. 2012). Perhaps for these reasons it is generally assumed Western diets should adversely rather than beneficially affect brain health. On the other hand, some believe ketone bodies and ketogenic diets may positively impact various brain functions, and data exist to support this view (Swerdlow 2014). In further respect to the ketogenic diet, our observations of potentially enhanced respiratory efficiency as well as a reduction in the expression of TNFα, which reflects levels of brain inflammation (McCoy & Tansey 2008), are encouraging. We feel the ability of a ketogenic diet to mitigate a neuroinflammation biomarker particularly justifies testing this diet in neurodegenerative conditions that feature excess neuroinflammation.
A better understanding of how ketogenic and non-ketogenic high fat diets affect brain bioenergetics specifically and brain health in general may, therefore, provide insights into brain aging, neurodegeneration, and therapeutic development. These insights may have translational implications.
Acknowledgments
This project was supported by the University of Kansas Alzheimer’s Disease Center (NIH P30 AG035982), the University of Kansas Physician Scientist Training Program, the Frank and Evangeline Thompson Alzheimer’s Treatment Program fund, the Hugh and Betty Libby Foundation, the Greater Kansas City Automobile Dealers Association, and the Gene and Marge Sweeney Chair.
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Agrawal R, Tyagi E, Vergnes L, Reue K, Gomez-Pinilla F. Coupling energy homeostasis with a mechanism to support plasticity in brain trauma. Biochim Biophys Acta. 2013;1842:535–546. doi: 10.1016/j.bbadis.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Ahola-Erkkila S, Carroll CJ, Peltola-Mjosund K, Tulkki V, Mattila I, Seppanen-Laakso T, Oresic M, Tyynismaa H, Suomalainen A. Ketogenic diet slows down mitochondrial myopathy progression in mice. Hum Mol Genet. 2010;19:1974–1984. doi: 10.1093/hmg/ddq076. [DOI] [PubMed] [Google Scholar]
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaresko J, Koch M, Schulze MB, Nothlings U. Dietary pattern analysis and biomarkers of low-grade inflammation: a systematic literature review. Nutr Rev. 2013;71:511–527. doi: 10.1111/nure.12035. [DOI] [PubMed] [Google Scholar]
- Beattie DS, Basford RE, Koritz SB. The turnover of the protein components of mitochondria from rat liver, kidney, and brain. J Biol Chem. 1967;242:4584–4586. [PubMed] [Google Scholar]
- Bough KJ, Wetherington J, Hassel B, et al. Mitochondrial biogenesis in the anticonvulsant mechanism of the ketogenic diet. Ann Neurol. 2006;60:223–235. doi: 10.1002/ana.20899. [DOI] [PubMed] [Google Scholar]
- Brinton RD. The healthy cell bias of estrogen action: mitochondrial bioenergetics and neurological implications. Trends Neurosci. 2008;31:529–537. doi: 10.1016/j.tins.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JM, Honea RA, Vidoni ED, Hutfles LJ, Brooks WM, Swerdlow RH. Insulin is differentially related to cognitive decline and atrophy in Alzheimer’s disease and aging. Biochim Biophys Acta. 2012;1822:333–339. doi: 10.1016/j.bbadis.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280:25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- Colle E, Ulstrom RA. KETOTIC HYPOGLYCEMIA. The Journal of pediatrics. 1964;64:632–651. doi: 10.1016/s0022-3476(64)80611-9. [DOI] [PubMed] [Google Scholar]
- Conlee RK, Hammer RL, Winder WW, Bracken ML, Nelson AG, Barnett DW. Glycogen repletion and exercise endurance in rats adapted to a high fat diet. Metabolism: clinical and experimental. 1990;39:289–294. doi: 10.1016/0026-0495(90)90049-i. [DOI] [PubMed] [Google Scholar]
- Davis LM, Rho JM, Sullivan PG. UCP-mediated free fatty acid uncoupling of isolated cortical mitochondria from fasted animals: correlations to dietary modulations. Epilepsia. 2008;49(Suppl 8):117–119. doi: 10.1111/j.1528-1167.2008.01854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagles DA, Chapman GB. A light- and electron-microscope study of hepatocytes of rats fed different diets. Comptes rendus biologies. 2007;330:62–70. doi: 10.1016/j.crvi.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Edmond J. Essential polyunsaturated fatty acids and the barrier to the brain: the components of a model for transport. J Mol Neurosci. 2001;16:181–193. doi: 10.1385/JMN:16:2-3:181. discussion 215–121. [DOI] [PubMed] [Google Scholar]
- Fuente-Martin E, Garcia-Caceres C, Morselli E, Clegg DJ, Chowen JA, Finan B, Brinton RD, Tschop MH. Estrogen, astrocytes and the neuroendocrine control of metabolism. Reviews in endocrine & metabolic disorders. 2013;14:331–338. doi: 10.1007/s11154-013-9263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes CA, Jope RS. The multifaceted roles of glycogen synthase kinase 3beta in cellular signaling. Prog Neurobiol. 2001;65:391–426. doi: 10.1016/s0301-0082(01)00011-9. [DOI] [PubMed] [Google Scholar]
- Gross NJ, Getz GS, Rabinowitz M. Apparent turnover of mitochondrial deoxyribonucleic acid and mitochondrial phospholipids in the tissues of the rat. J Biol Chem. 1969;244:1552–1562. [PubMed] [Google Scholar]
- Guilford BL, Ryals JM, Wright DE. Phenotypic changes in diabetic neuropathy induced by a high-fat diet in diabetic C57BL/6 mice. Experimental diabetes research. 2011;2011:848307. doi: 10.1155/2011/848307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Jiang L, Bhasin S, Khan SM, Swerdlow RH. DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion. 2009;9:261–265. doi: 10.1016/j.mito.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson DR. Adiposity hormones and dementia. J Neurol Sci. 2010;299:30–34. doi: 10.1016/j.jns.2010.08.036. [DOI] [PubMed] [Google Scholar]
- Harwood HJ., Jr The adipocyte as an endocrine organ in the regulation of metabolic homeostasis. Neuropharmacology. 2012;63:57–75. doi: 10.1016/j.neuropharm.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Heni M, Schopfer P, Peter A, Sartorius T, Fritsche A, Synofzik M, Haring HU, Maetzler W, Hennige AM. Evidence for altered transport of insulin across the blood-brain barrier in insulin-resistant humans. Acta diabetologica. 2013 doi: 10.1007/s00592-013-0546-y. [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Hirai K, Aliev G, Nunomura A, et al. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko RC, Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- Jamart C, Francaux M, Millet GY, Deldicque L, Frere D, Feasson L. Modulation of autophagy and ubiquitin-proteasome pathways during ultra-endurance running. J Appl Physiol (1985) 2012;112:1529–1537. doi: 10.1152/japplphysiol.00952.2011. [DOI] [PubMed] [Google Scholar]
- Kennedy AR, Pissios P, Otu H, et al. A high-fat, ketogenic diet induces a unique metabolic state in mice. American journal of physiology. Endocrinology and metabolism. 2007;292:E1724–1739. doi: 10.1152/ajpendo.00717.2006. [DOI] [PubMed] [Google Scholar]
- Khan AA, Wilson JE. STUDIES OF TURNOVER IN MAMMALIAN SUBCELLULAR PARTICLES: BRAIN NUCLEI, MITOCHONDRIA AND MICROSOMES. J Neurochem. 1965;12:81–86. doi: 10.1111/j.1471-4159.1965.tb11942.x. [DOI] [PubMed] [Google Scholar]
- Lin J, Tarr PT, Yang R, Rhee J, Puigserver P, Newgard CB, Spiegelman BM. PGC-1beta in the regulation of hepatic glucose and energy metabolism. J Biol Chem. 2003;278:30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- Lopez-Armada MJ, Riveiro-Naveira RR, Vaamonde-Garcia C, Valcarcel-Ares MN. Mitochondrial dysfunction and the inflammatory response. Mitochondrion. 2013;13:106–118. doi: 10.1016/j.mito.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- McCoy MK, Tansey MG. TNF signaling inhibition in the CNS: implications for normal brain function and neurodegenerative disease. J Neuroinflammation. 2008;5:45. doi: 10.1186/1742-2094-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies RA, Gold PH. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J Biol Chem. 1971;246:2425–2429. [PubMed] [Google Scholar]
- Milder JB, Liang LP, Patel M. Acute oxidative stress and systemic Nrf2 activation by the ketogenic diet. Neurobiol Dis. 2010;40:238–244. doi: 10.1016/j.nbd.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RW, Hatch GM. Fatty acid transport into the brain: of fatty acid fables and lipid tails. Prostaglandins, leukotrienes, and essential fatty acids. 2011;85:293–302. doi: 10.1016/j.plefa.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Mitchell RW, On NH, Del Bigio MR, Miller DW, Hatch GM. Fatty acid transport protein expression in human brain and potential role in fatty acid transport across human brain microvessel endothelial cells. J Neurochem. 2011;117:735–746. doi: 10.1111/j.1471-4159.2011.07245.x. [DOI] [PubMed] [Google Scholar]
- Navarro A, Boveris A. The mitochondrial energy transduction system and the aging process. Am J Physiol Cell Physiol. 2007;292:C670–686. doi: 10.1152/ajpcell.00213.2006. [DOI] [PubMed] [Google Scholar]
- Noh HS, Lee HP, Kim DW, Kang SS, Cho GJ, Rho JM, Choi WS. A cDNA microarray analysis of gene expression profiles in rat hippocampus following a ketogenic diet. Brain Res Mol Brain Res. 2004;129:80–87. doi: 10.1016/j.molbrainres.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Pardridge WM, Mietus LJ. Palmitate and cholesterol transport through the blood-brain barrier. J Neurochem. 1980;34:463–466. doi: 10.1111/j.1471-4159.1980.tb06621.x. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Rajwade MS, Katyare SS, Fatterpaker P, Sreenivasan A. Regulation of mitochondrial protein turnover by thyroid hormone(s) Biochem J. 1975;152:379–387. doi: 10.1042/bj1520379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds THt, Bodine SC, Lawrence JC., Jr Control of Ser2448 phosphorylation in the mammalian target of rapamycin by insulin and skeletal muscle load. J Biol Chem. 2002;277:17657–17662. doi: 10.1074/jbc.M201142200. [DOI] [PubMed] [Google Scholar]
- Sandu O, Song K, Cai W, Zheng F, Uribarri J, Vlassara H. Insulin resistance and type 2 diabetes in high-fat-fed mice are linked to high glycotoxin intake. Diabetes. 2005;54:2314–2319. doi: 10.2337/diabetes.54.8.2314. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sparks LM, Xie H, Koza RA, Mynatt R, Hulver MW, Bray GA, Smith SR. A high-fat diet coordinately downregulates genes required for mitochondrial oxidative phosphorylation in skeletal muscle. Diabetes. 2005;54:1926–1933. doi: 10.2337/diabetes.54.7.1926. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- Surwit RS, Kuhn CM, Cochrane C, McCubbin JA, Feinglos MN. Diet-induced type II diabetes in C57BL/6J mice. Diabetes. 1988;37:1163–1167. doi: 10.2337/diab.37.9.1163. [DOI] [PubMed] [Google Scholar]
- Swerdlow RH. The neurodegenerative mitochondriopathies. J Alzheimers Dis. 2009;17:737–751. doi: 10.3233/JAD-2009-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH. Role and treatment of mitochondrial DNA-related mitochondrial dysfunction in sporadic neurodegenerative diseases. Curr Pharm Des. 2011;17:3356–3373. doi: 10.2174/138161211798072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH. Does mitochondrial DNA play a role in Parkinson’s disease? A review of cybrid and other supportive evidence. Antioxid Redox Signal. 2012a;16:950–964. doi: 10.1089/ars.2011.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH. Mitochondria and cell bioenergetics: increasingly recognized components and a possible etiologic cause of Alzheimer’s disease. Antioxid Redox Signal. 2012b;16:1434–1455. doi: 10.1089/ars.2011.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH. Bioenergetic medicine. Br J Pharmacol. 2014;171:1854–1869. doi: 10.1111/bph.12394. [DOI] [PMC free article] [PubMed] [Google Scholar]