Abstract
The Mexican tetra, Astyanax mexicanus, is a unique model system consisting of cave-adapted and surface-dwelling morphotypes that diverged >1 million years (My) ago. This remarkable natural experiment has enabled powerful genetic analyses of cave adaptation. Here, we describe the application of next-generation sequencing technology to the creation of a high-density linkage map. Our map comprises more than 2200 markers populating 25 linkage groups constructed from genotypic data generated from a single genotyping-by-sequencing project. We leveraged emergent genomic and transcriptomic resources to anchor hundreds of anonymous Astyanax markers to the genome of the zebrafish (Danio rerio), the most closely related model organism to our study species. This facilitated the identification of 784 distinct connections between our linkage map and the Danio rerio genome, highlighting several regions of conserved genomic architecture between the two species despite ∼150 My of divergence. Using a Mendelian cave-associated trait as a proof-of-principle, we successfully recovered the genomic position of the albinism locus near the gene Oca2. Further, our map successfully informed the positions of unplaced Astyanax genomic scaffolds within particular linkage groups. This ability to identify the relative location, orientation, and linear order of unaligned genomic scaffolds will facilitate ongoing efforts to improve on the current early draft and assemble future versions of the Astyanax physical genome. Moreover, this improved linkage map will enable higher-resolution genetic analyses and catalyze the discovery of the genetic basis for cave-associated phenotypes.
Keywords: next-generation sequencing, QTL analysis, blind Mexican cave tetra, regressive phenotypic evolution
The blind Mexican cave tetra is a powerful system for understanding the evolutionary mechanisms governing regressive phenotypes. These animals were discovered in 1936 and initially were assigned to a new genus—Anoptichthys (“bony fish without eyes”) (Hubbs and Innes 1936). Breeding studies in the 1940s led to the discovery of viable hybrid offspring resulting from crosses between the (derived) blind cave-dwelling forms and (ancestral) surface-dwelling forms from the same geographical region of northeast Mexico (Breder 1943a, b). Both morphotypes are now regarded as members of the same (or a closely related) species, Astyanax mexicanus. This system has spurred more than half a century of comparative research (Şadoǧlu 1956) focusing on unresolved problems in evolution (Jeffery 2001), development (Pottin et al. 2011), genetics (Schemmel 1980), physiology (Salin et al. 2010), and behavior (Burchards et al. 1985).
Classical and quantitative genetic approaches have provided clear evidence that many troglomorphic (cave-associated) phenotypes evolved through heritable genetic changes. These studies centered on both Mendelian and complex phenotypes, including eye regression (Yamamoto et al. 2004; Protas et al. 2007; Yoshizawa et al. 2012; O’Quin et al. 2013), feeding-related behaviors (Schemmel 1980; Yoshizawa et al. 2012), sleep loss (Duboué et al. 2011), schooling behavior (Kowalko et al. 2013), pigmentation loss (reviewed in Jeffery 2009), and intraspecific aggression (Elipot et al. 2013). QTL studies have identified candidate genes mediating a variety of these traits, such as retinal degeneration (O’Quin et al. 2013), rib number, eye size (Gross et al. 2008), albinism (Oca2) (Protas et al. 2006), and the brown phenotype (Mc1r) (Gross et al. 2009).
Genomic resources for this model system, however, have historically been limited. The first linkage map was calculated based on recombination frequencies of an experimental F1 × Pachón cave backcross pedigree using markers generated from random amplified polymorphic DNA (RAPD) fingerprinting (Borowsky and Wilkens 2002). This map was supplanted by a higher-resolution map using more individuals and markers composed of polymorphic microsatellites identified using CAN dinucleotide repeats (Protas et al. 2006). Using this second-generation linkage map, Protas et al. (2008) discovered a genetic basis for several cave-associated phenotypic changes including pigmentation regression, reduced rib numbers, slower weight loss, and increased chemical sensitivity. Early comparative genomic analyses utilizing this map first demonstrated extensive synteny conserved between Astyanax and Danio rerio, despite ∼150 My of divergence (Gross et al. 2008). The first next-generation sequencing (NGS)-based linkage map using restriction-associated DNA sequencing (RAD-seq) technology was published by O’Quin et al. (2013). This map, comprising 698 markers on 25 linkage groups, strengthened the evidence for vast regions of synteny between the genomes of Astyanax and zebrafish and identified several critical loci associated with retinal degeneration (O’Quin et al. 2013).
Here, we present the most dense, comprehensive linkage map to date using genotyping-by-sequencing (GBS) technology. This technology enables accurate and high-throughput collection of massive amounts of sequence data (Davey et al. 2011), including thousands of single-nucleotide polymorphisms (SNPs) segregating between cave-dwelling and surface-dwelling morphs. GBS utilizes deep Illumina sequencing of restriction enzyme-nicked genomic DNA libraries that are uniquely barcoded for each member of an experimental pedigree. This technique is optimized to avoid inclusion of repetitive portions of the genome and is extremely specific and highly reproducible (Elshire et al. 2011). Fish are well-represented among studies using GBS and other RAD-seq–based methodologies (Rowe et al. 2011). However, a majority of GBS studies in fish have focused on species of commercial (Everett et al. 2012; Houston et al. 2012; Li et al. 2014) or conservational concern (Hecht et al. 2013; Ogden et al. 2013; Hess et al. 2014; Larson et al. 2014). Here, we adapted this technology to construct a high-density linkage map for evolutionary and developmental studies in our emerging model system. The resulting linkage map will enable higher-resolution genomic studies and inform the assignment of chromosomal builds for the ongoing Astyanax genome sequencing project (McGaugh et al. 2014).
Materials and Methods
Pedigree, husbandry, and genomic DNA isolation
Linkage mapping and QTL studies were performed using genotypic and phenotypic data obtained from two separate F2 hybrid mapping populations (n = 129; n = 41) bred from a male surface fish and female cavefish from the Pachón cave. In addition, surface (n = 4), Pachón cave (n = 4), and surface × Pachón F1 hybrid (n = 4) specimens were used to evaluate and code GBS markers for use with JoinMap software (v. 4.1; Kyazma; see below), but were not included in linkage mapping calculations. Parental specimens belonged to laboratory populations originally sourced from the El Abra region of northeastern Mexico and all fish used were generously provided to our laboratory by Dr. Richard Borowsky (New York University). All live fish used in this study were maintained as previously described (see Gross et al. 2013). Every individual from the “Asty66” F2 population (n = 129) was individually reared in a 1-liter tank. All phenotypic data from the “Asty12” F2 population (n = 41) were obtained from paraformaldehyde-preserved specimens.
Genotyping-by-sequencing
Genomic DNA was extracted from caudal tail fin tissue of live surface, cave, and F1 and F2 hybrid Astyanax mexicanus specimens using the DNeasy Blood and Tissue Kit (Qiagen) as previously described (Gross et al. 2013). Twenty genomic samples were digested with EcoRI, subjected to gel electrophoresis and imaged to verify that sample quality, concentration, and restriction fragment size distributions were suitable for use in downstream analyses. DNA samples were then pipetted into individual wells of 96-well plates and diluted to a final volume of 30 µl (100 ng/µl). Samples were processed by the Institute for Genomic Diversity (Cornell University), where genomic libraries were constructed and GBS was performed as described elsewhere (Elshire et al. 2011; Lu et al. 2013).
GBS marker selection
Genotypes for each of 7956 GBS markers (each consisting of a single SNP in a 64-bp-long sequence fragment) were screened in cave and surface (parental) forms to assign the morphotypic origin of each allele. F1 individuals were then evaluated to confirm heterozygosity at each locus. The morphotypic origin of each allele was assigned by consensus—if three or more (out of four) surface or cave individuals had the identical nucleotide at a particular locus, then the genotype was assigned to the consensus parental population. Likewise, a true “hybrid” genotype was assigned if three or more F1 individuals harbored the same heterozygous condition (e.g., M, R, S, W, Y, K SNP code) at a given locus. Those genotypes with an ambiguous morphotypic origin were denoted “NA.”
Markers were then screened for suitability in linkage calculations. Markers were deemed unsuitable and discarded from further analysis if neither parental genotype could be assigned (i.e., both the surface and cave genotypes were scored “NA”) or if the assigned surface and cave genotypes were identical; 6006 genomic markers were deemed suitable and prepared for linkage map calculation using the “cross-pollination” (CP) segregation coding used in JoinMap. At this stage, 107 markers were found to be uninformative (i.e., a single genotype was shared by all F2 individuals) and discarded from further analysis. We screened the remaining set (n = 5899) to identify markers failing to conform to predicted genotypic ratios (e.g., 1:2:1 ratios across the entire pedigree); 2896 markers demonstrated a χ2 value more than 50, implying significant departure from the predicted genotype ratio and were discarded from further analysis. Our final GBS marker set included 3003 markers evaluated in 170 F2 individuals.
Linkage map construction and QTL analysis
Linkage map calculations were performed using JoinMap (v.4.1, Kyazma). Our workflow used program default settings, with the following exceptions: the maximum grouping independence LOD value was set to 50.0; linkage groups were calculated using regression mapping; and linkage mapping was performed using the Kosambi method (Kosambi 1943). Linkage groups were assigned based on independence LOD scores. We increased the maximum grouping independence LOD value to 50.0, because the default value of 10.0 did not allow sufficient subdivision of our data into an appropriate number of groups. Initial groupings identified 29 groups populated with between 10 and 225 markers, with independence LOD scores ranging from 7.0 to 21.0. These groups were then processed for formal mapping calculations.
The first round of mapping produced 28 linkage groups comprising a total map length of 2956 cM. At this stage, one linkage group (comprising 10 markers, independence LOD = 19.0) failed to assemble into a consolidated group and was therefore eliminated from further analysis. The remaining individual linkage groups ranged in length from 27.25 to 187.46 cM, containing between 10 and 225 markers with an average intermarker distance between 0.51 and 6.40 cM. After this initial round of mapping, we further screened existing linkages to target the most optimal 25 groups (Astyanax mexicanus has karyotypic number of 25; Kirby et al. 1977) and reduce the average intermarker distance to a target of ∼1 cM. Accordingly, nine groups (10 ≤ n ≤ 45 markers) were removed because of low marker number and/or unusually high average intermarker distance. The five largest groups (154 ≤ n ≤ 225 markers) were then subdivided at the lowest independence LOD value resulting in two linkage groups comprising 20 or more markers. Throughout mapping, we limited the inflation of the overall map length by eliminating certain markers sparsely populating distal ends of otherwise densely populated linkage groups. This resulted in size reduction of the five longest remaining linkage groups (142.041 ≤ n ≤ 187.458 cM) by splitting them at the lowest independence LOD score at which a group (comprising 10 or more markers) was separated. In these cases, the larger of the two resulting groups was retained. The resulting 25 linkage groups (independence LOD scores 10.0 ≤ n ≤ 24.0) were subjected to additional mapping. Groupings of markers eliminated during this or a subsequent round of mapping were excluded from further analysis.
The second round of mapping produced a 2556.6-cM linkage map composed of 25 linkage groups, each consisting of 25 to 171 markers, ranging in length from 31.18 to 142.78 cM, with mean intermarker distances ranging from 0.47 to 3.66 cM. Using the same criteria described above, an additional group (comprising 25 markers and an average intermarker distance of 3.658) was eliminated. A densely populated group with a high independence LOD (153 markers; 135.73 cM; independence LOD of 24.0) was split and 12 linkage groups (103.982 ≤ n ≤ 142.783 cM) were trimmed.
The result of this third and final round of mapping was then analyzed for genomic synteny shared between Astyanax mexicanus and the zebrafish genome and used to map albinism as a proof of concept. Albinism was scored as a binary phenotype wherein presence of melanin (0) or absence of melanin (1) was assigned to each of the members of our experimental F2 pedigrees. All QTL analyses of albinism were conducted using R/qtl (Broman et al. 2003) run for each of three scan-one mapping methods: marker regression (MR), expect maximum (EM), and Haley-Knott (HK), according to the methodology in Gross et al. (2014).
Assignment of genomic synteny between the Astyanax mexicanus and Danio rerio genomes
At present, physical genome resources for Astyanax mexicanus are in their early draft phases (McGaugh et al. 2014). Therefore, we anchored our GBS-based linkage map to the physical genome of the most closely related fish model system with comprehensive resources, Danio rerio. Astyanax and Danio are members of the superorder Ostariophysii, which diverged ∼150 My ago (Briggs 2005). Despite this distance, significant genome-level synteny remains between these species (Gross et al. 2008; O’Quin et al. 2013). Our GBS marker set was derived from endonuclease restriction site-based libraries and was therefore anonymous. We first identified all GBS markers that could be directly localized to a conserved region in the D. rerio genome. Accordingly, we performed BLAST searches of the 64-bp sequences comprising our marker sequences directly against the Danio genome (downloaded from the Ensembl genome browser; www.ensembl.org).
These and all subsequent searches were performed using a BLASTN script run on the Ohio Supercomputing Cluster (OSC). All quality control defaults, including an expect value (e-value) cutoff of 10, were maintained. The script permitted the return of alignments between a given 64-bp marker sequence and regions of up to three distinct targets (e.g., three different Danio rerio chromosomes). In cases where a single marker sequence aligned multiple times with the same target, raw results were filtered by e-value, retaining the lowest e-value alignment for each marker-target pairing. There are two 64-bp sequences for each GBS marker, differing only in that each contains one of the two alleles for the imbedded SNP. Because both of these sequences were included when BLAST searches using the 64-bp marker sequences were conducted, this filtering step also served to collapse these results into a single set of results, retaining the better of the two alignments for each marker-target pairing.
In some instances, a single queried sequence returned alignments with multiple targets. These instances were resolved by sorting results to determine the “top hit,” which was defined as having the lowest e-value and highest percent identity (in case of an e-value tie) to a particular target sequence. If the target of the top hit (i.e., the alignment with the lowest e-value) for a given marker sequence agreed with the target reported for one or more other markers on the same linkage group that returned only a single robust hit, then the top hit for the marker in question was considered “supported” and retained. If the top hit was not supported in this fashion but a different BLAST result was, then the latter “not top hit, supported” result was retained instead. If none of the results returned for a marker sequence were supported, then the top hit was retained, despite the lack of support. In rare cases, there was no way to resolve which result should be retained. Results for these “unresolved” markers were discarded.
When using BLAST searches to align our 64-bp markers directly to the Danio rerio genome returned relatively few high-quality hits, we developed a strategy whereby we first aligned our GBS marker sequences to the Astyanax mexicanus genome and transcriptome data. This information was then used to identify homologous Danio genomic and transcriptomic sequences. Current genomic resources in Astyanax consist of >10,000 unplaced genomic scaffolds (Bioproject PRJNA89115). The collective sequence data for the Astyanax genome (GenBank Assembly ID GCA_000372685) were downloaded from Ensembl, along with the transcript sequences for 23,042 predicted genes. BLAST searches were used to determine putative locations for the 64-bp sequences of the 2235 GBS markers comprising our final linkage map in the Astyanax genomic and transcriptomic data sets. After initial searches were performed as described, ∼2000-bp stretches of genomic sequence harboring our 64-bp GBS marker sequences were aligned with the Danio genome. Similarly, full sequences for predicted Astyanax transcripts to which our GBS markers aligned were queried against a Danio cDNA database downloaded from Ensembl. Both data sets were then filtered (as described), yielding a single “best” Danio alignment for each informative query. This process enabled us to leverage draft genomic and transcriptomic data to augment the amount of sequence information associated with our 64-bp GBS markers and to identify homologous genomic positions in a well-characterized model system.
After BLAST searches using the direct, genomic, and transcriptomic alignment methods were completed, the filtered results for all three were combined. When multiple methods returned results for the same marker, a single result was chosen and retained using the same filtering process applied to single data sets (above). The Circos program (Krzywinski et al. 2009) was used to visualize comparative genomic positions between our linkage map and the Astyanax and Danio rerio genomes.
Position identification for previously published markers in the Astyanax genome
Previous maps published by Gross et al. (2008) and O’Quin et al. (2013) were used to examine synteny between Astyanax and Danio and to provide a comparison between this study and prior studies. These authors provided predicted Danio positions for the markers used in their analyses, but positions in the draft Astyanax genome were not determined because these studies predated available genomic resources. Our GBS-based map does not share any markers with the two previous maps, so it was necessary to identify positions of previously generated markers in Astyanax to enable comparison between previous mapping efforts and those described here. Accordingly, microsatellite and RAD-seq marker sequences (where available) for each data set were aligned with Astyanax genome scaffolds using the same BLAST and filtering protocols used for our own data (above). Both previous studies included markers located in candidate genes. The locations of Astyanax orthologs of these candidate genes were identified using Ensembl.
GBS marker sequences and genotyping data are available from the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.6s718).
Results and Discussion
A high-density linkage map in Astyanax mexicanus
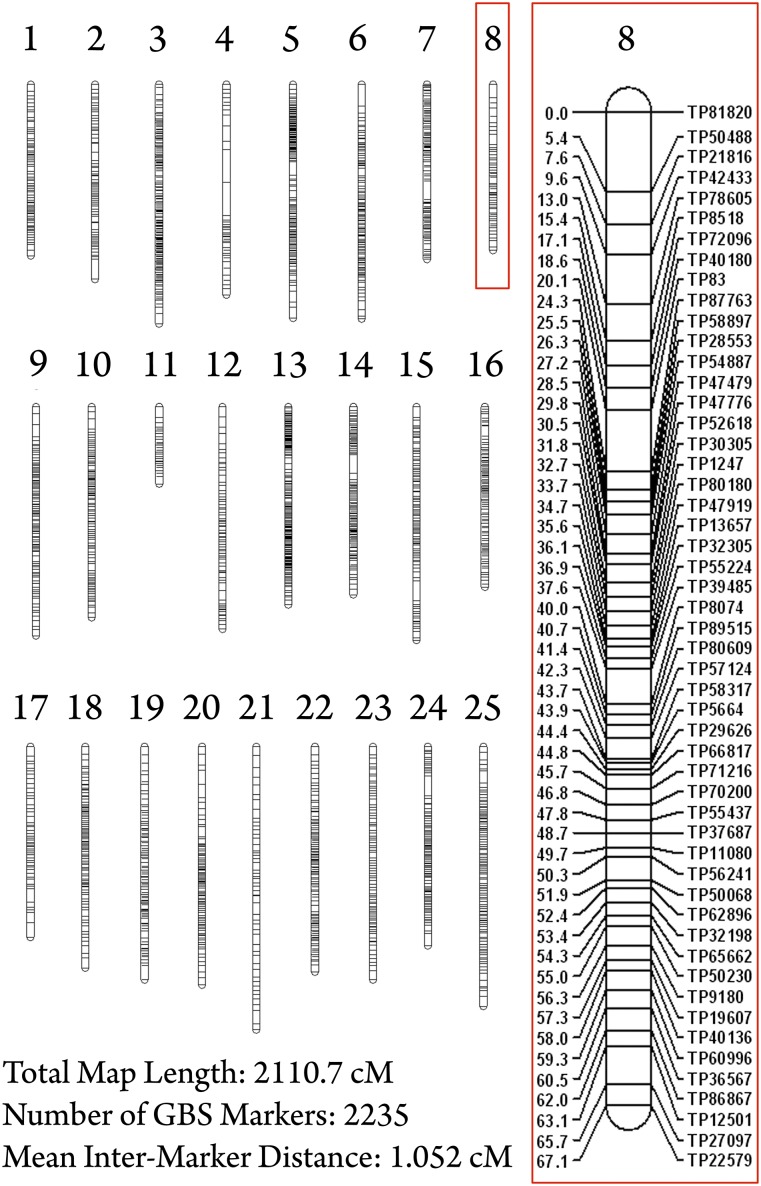
Here, we present a dense linkage map for Astyanax mexicanus generated using genotyping-by-sequencing technology. This map was created using 170 experimental F2 individuals based on genotypic information for 3003 loci. The construction of this map ultimately yielded 25 linkage groups (the karyotypic number for Astyanax) comprising 2235 markers spanning 2110.7 cM, with an average intermarker distance of 1.052 cM (Figure 1, Supporting Information, Table S1). The strategy we used enables application of powerful, cost-effective, next-generation sequencing technology to facilitate genetic studies in emerging or nonmodel systems.
Figure 1.
A GBS-based linkage map in the Mexican cave tetra, Astyanax mexicanus. We analyzed 3003 SNP markers in 170 individuals using genotyping-by-sequencing technology. This linkage map consists of 2235 markers in 25 linkage groups (A. mexicanus karyotype number = 25), spanning a total distance of 2110.7 cM (mean intermarker distance = 1.052 cM). Astyanax linkage group 8 (red box) illustrates typical marker density observed in most groups. This group consists of 52 GBS markers spanning 67.061 cM with a mean intermarker distance of 1.315 cM.
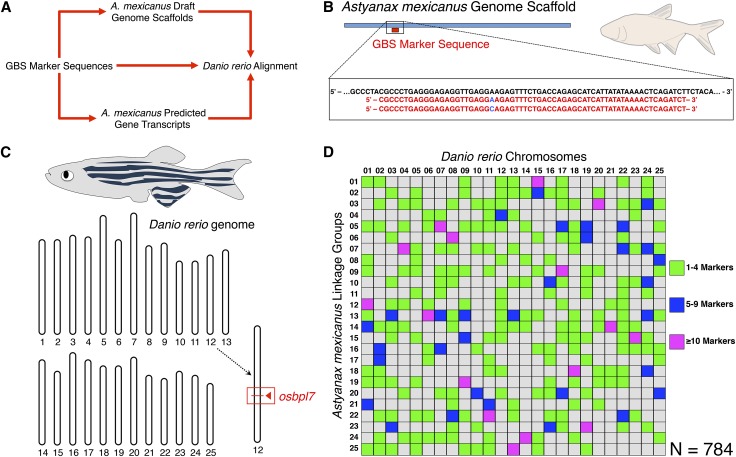
Cross-genera marker identification was greatly facilitated by alignment first to draft Astyanax genomic and transcriptomic resources, followed by searches of the homologous sequences in Danio (Figure 2, A–C). Although direct BLAST searches of our 64-bp GBS marker sequences returned results for few of the markers in our map (1.2%), success rates were much higher when using Astyanax genomic (26.5%) or transcriptomic (13.3%) sequences as an intermediary (Table 1). Each Danio rerio chromosome was represented in our comparative genomic analysis, with Astyanax linkage groups containing 14–52 markers (average = 30.84) comprising ancient syntenic blocks shared with each of 25 zebrafish chromosomes (Figure 2D). Of the 2235 GBS markers that constitute our linkage map, 784 marker sequences (35.1%) were successfully identified in the Danio rerio genome (Figure 3A).
Figure 2.
Short GBS sequences identify syntenic stretches between two Ostariophysian freshwater fish species. To reveal syntenic regions between Astyanax mexicanus and Danio rerio, we first identified stretches of the Danio genome harboring homologous sequences to our anonymous GBS marker sequences (A). Individual 64-bp sequences for the 2235 GBS markers in our linkage map were compared with the Danio genome both directly and by first aligning to larger Astyanax genomic scaffolds and predicted gene transcripts (B), followed by alignment of some or all of the larger sequence to the Danio genome based on BLAST sequence analysis (C). This resulted in identification of homologous sequences for 784 Astyanax GBS markers within the Danio genome. The markers shared between Danio chromosomes and Astyanax linkage groups are represented using an Oxford plot (D).
Table 1. Summary of BLAST results and identification of markers used in Astyanax-to-Danio syntenic analysis.
| GBS Markers to Danio Genomea | GBS Markers to Astyanax Genomeb | Astyanax Genome to Danio Genomec | GBS Markers to Astyanax Transcriptomed | Astyanax Transcriptome to Danio Transcriptomee | |
|---|---|---|---|---|---|
| Total no. of BLAST queries | 2235 | 2235 | 2088 | 2235 | 572 |
| BLAST result categories | |||||
| Single robust hit | 14 (0.6%) | 1838 (82.2%) | 255 (12.2%) | 508 (22.7%) | 110 (19.2%) |
| Top hit, with positional support | 0 (0.0%) | 173 (7.7%) | 92 (4.4%) | 15 (0.7%) | 120 (21.0%) |
| Top hit, without positional support | 10 (0.4%) | 71 (3.2%) | 138 (6.6%) | 60 (2.7%) | 61 (10.7%) |
| Not top hit, with positional support | 2 (<0.1%) | 6 (0.3%) | 108 (5.2%) | 2 (<0.1%) | 7 (1.2%) |
| Unresolved | 4 (0.2%) | 14 (0.6%) | 4 (0.2%) | 12 (0.5%) | 0 (0.0%) |
| No result | 2205 (98.7%) | 133 (6.0%) | 1491 (71.4%) | 1638 (73.3%) | 274 (47.9%) |
| Identified syntenic markers between Astyanax and Danio | 26 | N/A | 593 | N/A | 298 |
Results of 64-bp GBS markers BLASTed directly to the Danio rerio genome.
Results of 64-bp GBS markers BLASTed directly to the Astyanax genome draft assembly.
Results of ∼2-kb genomic intervals harboring 64-bp GBS markers BLASTed to the Danio rerio genome.
Results of 64-bp GBS markers BLASTed directly to the Astyanax predicted transcriptome.
Results of Astyanax transcripts harboring 64-bp GBS markers BLASTed to the Danio rerio transcriptome.
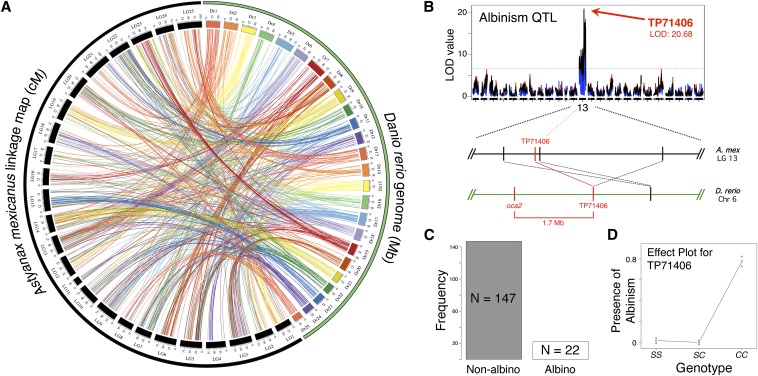
Figure 3.
Whole-genome synteny between Astyanax and Danio and a proof-of-concept analysis of albinism. Syntenic links between our GBS map and the Danio genome were visualized using Circos (A). Each line represents a connection between the position of a particular marker in our linkage map (black; scale in cM) and a homologous sequence in Danio (various colors; scale in Mb). We scored albinism, a Mendelian trait associated with the Oca2 gene in cave-dwelling Astyanax (C), and performed QTL analysis using R/qtl. Each of three mapping methods (MR in red; EM in blue; HK in black) revealed peak LOD scores of ∼20 (LOD at 0.001α threshold = 6.75) at, or adjacent to, GBS marker TP71406 on Astyanax linkage group 13 (B). Homologous sequences to TP71406 and several of its neighbors on Astyanax linkage group 13 are clustered together on Danio chromosome 6 near the Oca2 gene. A phenotypic effect plot for marker TP71406 revealed the predicted association between the homozygous “cave” condition (genotype CC) and albinism in F2 individuals (D).
We performed a proof-of-concept analysis using the albinism phenotype to validate the utility of our GBS-based linkage map (Figure 3, B–D). Accordingly, we mapped the monogenic trait of albinism using the R/qtl package to evaluate phenotypic and genotypic data for the 170 F2 hybrid individuals used to construct our map. We identified a peak LOD score of 20.68 on linkage group 13, associated with marker TP71406. This marker and the surrounding region form a syntenic block within a region of Danio rerio chromosome 6. This genomic interval contains the gene Oca2, previously demonstrated to be the causative locus for albinism in Astyanax cavefish. This supports previous findings of conserved synteny inclusive of significant portions of chromosome 6 in Danio (Gross et al. 2008; O’Quin et al. 2013) and implies our densely populated map will enable future QTL studies of trait evolution in Astyanax.
Conserved genomic architecture between Astyanax and Danio based on GBS markers
Our analysis of synteny between Astyanax and Danio illustrates variable levels of genomic conservation across linkage groups (Figure 2D, Figure 3A). Certain chromosomes, for instance, appear to have changed little since the divergence of these teleost species (e.g., Danio chromosomes 6 and 23 in Astyanax linkage groups 13 and 15, respectively). However, other Danio chromosomes appear scattered across several linkage groups, without a consensus representation for any particular group (e.g., Danio chromosomes 2 and 5).
We believe these findings most likely reflect genomic rearrangements that have occurred since the divergence of these two species. However, this finding could also be attributed to low representation of particular Danio chromosomes within our GBS marker set. We examined this possibility by assessing the number of syntenic links between our GBS-based linkage map and each Danio chromosome. We would anticipate that longer chromosomes would naturally harbor more syntenic links. Values were therefore expressed as a ratio of syntenic links per megabase (mean = 0.59 GBS markers/Mb). Although the mean value for chromosomes that were not strongly represented on any particular linkage group in our map (i.e., had fewer than 10 syntenic links with each linkage group, mean = 0.52 GBS markers/Mb, n = 8) was lower than that for chromosomes demonstrating strong synteny with a particular linkage group (mean = 0.61 GBS markers/Mb, n = 17), there was not a significant difference between the two groups (t23 = 0.5809, P = 0.5670). This leads us to conclude that, although representation of particular chromosomes in our data set may be a contributing factor, it is unlikely that this is the primary cause of the differences in chromosomal representation patterns observed.
Alternatively, BLAST results for Astyanax GBS markers (or the larger Astyanax sequences to which they were aligned) may include paralogous genes or otherwise ambiguous results that could lead to erroneous links between a linkage group and a Danio chromosome. Although we cannot rule out this possibility, we feel our strategy prioritized the “optimal” BLAST result among multiple hits for a single marker leading to alignments that agree with nearby unambiguous results (Table 1). As a result, of the 784 markers in our map for which a putative Danio position was determined, only 15.9% (n = 125) of final calls were unsupported by the results for other markers belonging to the same linkage group (Table S1). Given that chromosomal arrangements have occurred over the ∼150 My since divergence, we feel our systematic approach best identifies paralogous genes and other potential sources of ambiguity.
Erroneous or ambiguous genotyping data may have led to incorrect assignment of “cave” and “surface” alleles for particular markers. These erroneous assignments could have adversely affected downstream efforts, causing markers to be incorrectly placed during the grouping and/or mapping stages of linkage map construction. All efforts were made to ensure allelic identification was accurate using a stringent screening process (see Materials and Methods); however, we relied on a relatively small number of cave, surface, and F1 hybrid individuals (n = 4 each) to identify parental allelic origin. Similarly, the relatively small number of meiotic events represented by the 170 F2 individuals may have resulted in linkage map inaccuracies (Gross et al. 2008). Future comparisons between the map we present here and a finished-grade Astyanax genome will clarify if regions lacking synteny between Astyanax and Danio are attributable to errors in our linkage map or genomic rearrangements that have occurred since the divergence of these taxa.
Unplaced Astyanax genome scaffolds can be anchored to our new linkage map
Positional locations in the current draft of the Astyanax genome were established for 93.6% (n = 2091) of the 2235 GBS markers present in our map. These markers were localized to positions spread across 598 different Astyanax genome scaffolds. Our 25 Astyanax linkage groups contain markers representing between 12 (linkage groups 8 and 22) and 55 (linkage group 3) genome scaffolds each, with a map-wide average of 27.64 scaffolds/linkage group. Individual genome scaffolds contained between 1 and 31 GBS markers appearing in our final map, with an average of 3.50 markers per scaffold. GBS markers located on the same genomic scaffold colocalized to a single linkage group 87.3% of the time. This suggests that our recombination mapping successfully recapitulated the true genomic positions of the markers used to construct our map.
Improved linkage mapping resources in Astyanax
We sought to compare our linkage map with maps previously published by Gross et al. (2008) and O’Quin et al. (2013) that also examined synteny between Astyanax and Danio. Metrics such as the number of linkage groups, total map length, number of markers, and marker density are commonly used to compare linkage maps within species. Both our GBS-based map and the RAD-seq and microsatellite-based map published by O’Quin et al. (2013) consist of 25 linkage groups, matching the Astyanax mexicanus karyotype number of 25. The microsatellite-based map presented by Gross et al. (2008) contains 28 groups (Table 2). Although our map is of comparable length, it represents a dramatic increase in marker number (+559% compared with that of Gross et al. 2008; +320% compared with that of O’Quin et al. 2013) and marker density (+473% compared with that of Gross et al. 2008; +279% compared with that of O’Quin et al. 2013) relative to previously published linkage maps for this system. As a result, we saw a substantial increase in the number of syntenic links between our map and Danio (+506% compared with that of Gross et al. 2008; +453% compared with that of O’Quin et al. 2013) and an increase in the number of unplaced Astyanax scaffolds that can be anchored to our map (+263% compared with that of Gross et al. 2008; +171% compared with that of O’Quin et al. 2013).
Table 2. Comparison of Astyanax linkage maps and syntenic studies with Danio rerio.
| Gross et al. 2008 | O’Quin et al. 2013 | Current Analysis | |
|---|---|---|---|
| Total no. of linkage groups | 28 | 25 | 25 |
| Total no. of genomic markers | 400 | 698 | 2235 |
| Linkage map length | 1783 cM | 1835.5 cM | 2110.7 cM |
| Marker density | 0.224 per cM | 0.380 per cM | 1.06 per cM |
| Marker type | Microsatellite | Microsatellite + RAD-seq | Genotyping-by-sequencing |
| No. of Astyanax genomic scaffolds represented by map | 227 | 350 | 598 |
| No. of syntenic markers identified between Astyanax and Danio | 155 | 173 | 784 |
Our map contains a total of 784 links between our linkage groups and the Danio rerio genome and an average of 30.84 links (minimum = 14, maximum = 52) per Danio rerio chromosome (Table 3). This represents a considerable improvement over the results presented by Gross et al. (2008) (155 total links, average links per Danio chromosome = 6.20, minimum = 0, maximum = 15) and O’Quin et al. (2013) (173 total links, average links per Danio chromosome = 6.92, minimum = 1, maximum = 20). Additionally, although instances of synteny strongly represented in previous maps were also identified in this analysis, our map demonstrated increased representation of certain Danio chromosomes poorly represented in previous maps. For example, Gross et al. (2008) did not identify links between their map and Danio rerio chromosome 11; however, we identified 36 links between our map and chromosome 11. Similarly, Danio chromosomes 17 and 19 are each represented once in the map of O’Quin et al. (2013). We identified substantial links between these chromosomes and our linkage groups 9 (n = 21) and 23 (n = 15), respectively.
Table 3. Comparison of syntenic analyses between Astyanax linkage maps and their association with the Danio rerio genome across multiple studies.
| Gross et al. 2008 | O’Quin et al. 2013 | Current Analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Danio rerio Chromosome | No. of Syntenic Linksa | Represented Linkage Group(s)b | No. of Represented Astyanax Genome Scaffoldsc | No. of Syntenic Links | Represented Linkage Group(s) | No. of Represented Astyanax Genome Scaffolds | No. of Syntenic Links | Represented Linkage Group(s) | No. of Represented Astyanax Genome Scaffolds |
| 1 | 13 | 5, 8, 21 | 7 | 15 | 4, 5, 9, 18, 21, 23 | 11 | 42 | 1, 5, 8, 9, 12, 13, 14, 18, 19, 21, 25 | 26 |
| 2 | 6 | 2, 14, 15, 22 | 2 | 6 | 7, 12, 13, 16, 23 | 5 | 23 | 1, 3, 5, 14, 16, 17, 18, 19, 22, 24, 25 | 20 |
| 3 | 6 | 1, 4, 19 | 6 | 4 | 4, 15, 25 | 3 | 28 | 2, 6, 10, 12, 13, 14, 15, 19, 22, 23, 25 | 23 |
| 4 | 3 | 6, 7 | 2 | 4 | 3 | 4 | 24 | 3, 5, 7, 9, 12, 14, 20, 24 | 17 |
| 5 | 15 | 1, 5, 9, 10, 20 | 9 | 13 | 2, 8, 16, 17, 19 | 11 | 22 | 2, 3, 7, 8, 9, 11, 15, 19, 22, 24, 25 | 17 |
| 6 | 9 | 4, 13 | 4 | 20 | 1, 2, 11, 16, 18 | 16 | 37 | 4, 6, 12, 13, 16, 17, 22, 24 | 23 |
| 7 | 11 | 17, 22, 24, 26 | 10 | 6 | 13, 22, 23, 25 | 6 | 31 | 4, 5, 7, 9, 10, 11, 13, 16, 20, 24 | 25 |
| 8 | 4 | 9, 12 | 4 | 7 | 7, 14, 17 | 6 | 42 | 3, 6, 7, 9, 10, 13, 14, 20, 21, 22, 23, 24 | 29 |
| 9 | 5 | 3, 17 | 4 | 8 | 10, 11 | 7 | 31 | 1, 2, 5, 9, 12, 13, 15, 19, 22, 25 | 23 |
| 10 | 3 | 17, 18 | 3 | 4 | 8, 10, 14 | 4 | 14 | 3, 5, 7, 16, 20, 23, 25 | 12 |
| 11 | 0 | — | 0 | 5 | 14, 17, 22 | 5 | 36 | 3, 4, 5, 7, 9, 15, 17, 18, 21, 22, 24, 25 | 24 |
| 12 | 7 | 10, 16 | 4 | 6 | 24 | 4 | 26 | 1, 2, 3, 4, 5, 8, 11, 12, 13, 14, 15, 18, 20 | 18 |
| 13 | 11 | 1, 5 | 6 | 7 | 4, 12 | 4 | 34 | 1, 2, 4, 7, 10, 13, 14, 21, 22, 25 | 21 |
| 14 | 6 | 6, 7 | 4 | 9 | 3, 6, 15, 19 | 6 | 24 | 2, 5, 8, 10, 13, 14, 24 | 16 |
| 15 | 5 | 2 | 5 | 8 | 1, 7, 12, 14 | 6 | 27 | 1, 2, 6, 9, 13, 16, 20, 24, 25 | 17 |
| 16 | 3 | 13 | 3 | 7 | 8, 19 | 7 | 23 | 2, 8, 10, 11, 16, 17, 19, 20, 22, 23 | 17 |
| 17 | 6 | 3, 23 | 2 | 1 | 20 | 1 | 52 | 1, 2, 3, 5, 6, 9, 10, 12, 14, 15, 20, 22, 23 | 34 |
| 18 | 8 | 11 | 3 | 7 | 5 | 3 | 32 | 3, 5, 6, 7, 13, 14, 15, 16, 18, 19, 21, 24 | 26 |
| 19 | 3 | 19 | 3 | 1 | 25 | 1 | 42 | 5, 6, 9, 10, 11, 15, 17, 20, 23, 25 | 28 |
| 20 | 7 | 1, 2 | 7 | 3 | 1, 2 | 3 | 21 | 2, 3, 9, 13, 14, 18, 19, 21 | 15 |
| 21 | 3 | 15, 17 | 1 | 6 | 2, 7 | 6 | 16 | 12, 14, 18, 19 | 12 |
| 22 | 4 | 12, 20 | 4 | 7 | 14, 18, 22 | 6 | 42 | 1, 3, 5, 6, 7, 9, 10, 11, 12, 13, 14, 15, 16, 18, 19, 22, 23 | 32 |
| 23 | 6 | 26 | 3 | 7 | 14, 16, 18, 25 | 6 | 33 | 3, 4, 7, 10, 14, 15, 16, 22 | 17 |
| 24 | 8 | 1, 13, 15 | 6 | 9 | 2, 3, 8, 11 | 8 | 38 | 1, 2, 3, 7, 10, 11, 13, 15, 16, 18, 23 | 25 |
| 25 | 3 | 6, 7 | 3 | 3 | 3, 6 | 3 | 31 | 3, 5, 7, 8, 9, 13, 16, 17, 20, 24 | 19 |
Bold indicates that a listed linkage group harbors five or more links with a given Danio chromosome.
Indicates the number of syntenic links identified between Astyanax linkage maps and each listed Danio rerio chromosome.
Indicates the identity of Astyanax linkage groups harboring syntenic links with each listed Danio rerio chromosome.
Indicates the number of Astyanax genome scaffolds harboring connections with each listed Danio rerio chromosome.
Our linkage map uses an entirely different marker set than those used in previous maps. Therefore, it was not possible to make direct comparisons with the linkage groups across prior studies. However, we could indirectly compare maps by examining connections between Astyanax genomic scaffolds and each linkage map. We examined the five strongest syntenic links between single linkage groups in our GBS-based map and single Danio chromosomes and then identified analogous connections between those chromosomes and specific linkage groups in the maps presented by Gross et al. (2008) and O’Quin et al. (2013).
Astyanax genomic scaffolds harboring markers associating each linkage group with a particular Danio chromosome were then compared (Table 4). We found that many of the identified Astyanax genomic scaffolds colocalize to putatively analogous linkage groups in both our GBS-based map and those of Gross et al. (2008) and/or O’Quin et al. (2013). However, in every case examined, our linkage groups were inclusive of a much higher number of Astyanax genomic scaffolds compared with prior studies. Thus, while the linkage groups in our map represent genomic intervals similar to those represented in prior maps, our map achieves a higher level of detail and resolution. These results also suggest that future mapping efforts in Astyanax may benefit by combining GBS marker discovery with those markers used by Gross et al. (2008) and O’Quin et al. (2013) to generate the most comprehensive linkage mapping resource.
Table 4. Representative analysis of linkage group equivalence and quality based on highly syntenic chromosomes in Danio rerio and linkage groups in Astyanax mexicanus.
Bold indicates genomic scaffolds containing syntenic markers on the principal represented linkage group in our GBS-based map and one or more previous maps. Italic lettering indicates scaffolds that contain a syntenic marker in the GBS-based map and are associated with the principal linkage group(s) in previous map(s) but do not contain a syntenic marker (and vice versa).
Indicates the most common (i.e., “principal”) linkage group anchoring to the indicated Danio rerio chromosome.
Indicates the number of points of synteny between the principal linkage group from this article and the indicated Danio rerio chromosome.
Lists the identity of Astyanax genomic scaffolds to which each point of synteny identifies.
High-density GBS-based linkage mapping will inform the Astyanax genome sequencing project
Preliminary Astyanax genomic resources enabled us to locate 64-bp, anonymous GBS markers, and to assess the quality and reliability of our Astyanax linkage map. This emerging resource did not allow us to determine how well the 25 Astyanax chromosomes are represented in our map. However, these resources allowed us to determine if markers predicted to occur in the same genome scaffolds also co-occur in our GBS-based linkage map. Overall, we observed a high level of agreement between our linkage groups and one or more unplaced Astyanax genomic scaffolds.
In many cases, markers present on the same scaffold clustered together over a portion of a linkage group with little or no interruption from unplaced markers or markers from other scaffolds (Figure 4). We expect these results will help inform chromosomal positions of scaffolds, given that linkage maps have been successfully used to augment genomic resources in other fish species, including several species of catfish (Liu 2011; Ninwichian et al. 2012), rainbow trout (Palti et al. 2011; Palti et al. 2012), and Atlantic salmon (Lorenz et al. 2010). We believe our high-density GBS-based map resources will both provide a resource for more refined QTL analyses and inform the genomic architecture of the Astyanax genome sequencing project.
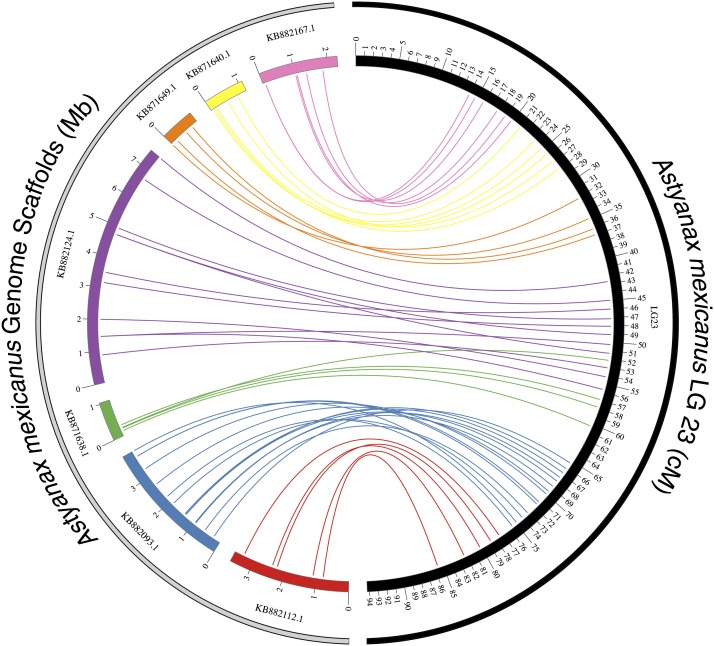
Figure 4.
Colinearity between Astyanax linkage groups and genome scaffolds. We visualized the “anchoring” of seven unplaced Astyanax genome scaffolds (various colors) to linkage group 23 (black) in our Astyanax linkage map. For clarity, only scaffolds harboring four or more GBS markers were included. Scaffolds correspond to discrete, colinear sections of the linkage group with minimal overlap. The linear arrangement of markers is largely preserved between the scaffold and the linkage group. The scale for Astyanax scaffolds is in Mb; the scale for linkage group 23 is shown in cM.
Conclusions
We constructed a high-density linkage map for Astyanax mexicanus based on high-throughput genotyping-by-sequencing data. We leveraged emerging Astyanax genomic and transcriptomic resources and Danio rerio genomic and transcriptomic data to locate syntenic regions shared between our map and the Danio genome. These findings were based on the physical position of homologous (64-bp) GBS marker sequences. As expected, based on the significant divergence between these species, we recovered varying levels of synteny between portions of our Astyanax linkage groups and regions of the Danio genome. As a proof of concept, we successfully mapped a strong QTL associated with albinism and demonstrated significant conserved genomic architecture in the regions surrounding the gene Oca2, between Astyanax and Danio. We successfully anchored emerging Astyanax genomic information to our GBS-based linkage map, identifying the putative location of thousands of anonymous GBS marker sequences within unplaced Astyanax genome scaffolds. This strategy revealed significant colinearity between genomic scaffolds and our linkage map, and it demonstrated the utility of high-density, GBS-based linkage maps to inform and improve nascent genomic resources. Multiple comparisons with previously published maps suggest that our GBS-based map offers a higher level of resolution and a greater number of connections between Astyanax and Danio genomes. We hope that this resource and technology will accelerate the search and identification of genes mediating cave-associated traits in Astyanax, facilitate the genomic assembly for this system, and prove useful to other natural model systems of evolutionary and biomedical relevance.
Supplementary Material
Acknowledgments
The authors thank Amanda Krutzler, Bethany Stahl, and members of the Gross laboratory for valuable effort and input. The authors are also grateful to Wesley Warren, Suzanne McGaugh, and the Genome Institute at Washington University for providing access to the draft genome assembly (Bioproject PRJNA89115 NCBI accession number APWO00000000; supported by NIH grant R24 RR032658-01 to W.W.). Additionally, the authors thank Suzanne McGaugh for providing B.M.C. and other members of the Gross laboratory for instruction in script-based BLAST search methods. This project was supported by National Institutes of Health (National Institute of Dental and Craniofacial Research) grant DE022403 (to J.B.G.) and a Cave Research Foundation Graduate Student Research Grant (to B.M.C.).
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.015438/-/DC1
Communicating editor: A. S. McCallion
Literature Cited
- Borowsky R., Wilkens H., 2002. Mapping a cave fish genome: polygenic systems and regressive evolution. J. Hered. 93: 19–21. [DOI] [PubMed] [Google Scholar]
- Breder C. M., Jr, 1943a Apparent changes in phenotypic ratios of the Characins at the type locality of Anoptichthys jordani Hubbs and Innes. Copeia 1943: 26–30. [Google Scholar]
- Breder C. M., Jr, 1943b Problems in the behavior and evolution of a species of blind cave fish. Trans. N. Y. Acad. Sci. 5: 168–176. [Google Scholar]
- Briggs J., 2005. The biogeography of otophysan fishes (Ostariophysi: Otophysi): A new appraisal. J. Biogeogr. 32: 287–294. [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Burchards H., Dölle A., Parzefall J., 1985. Aggressive behaviour of an epigean population of Astyanax mexicanus (Characidae, Pisces) and some observations of three subterranean populations. Behav. Processes 11: 225–235. [DOI] [PubMed] [Google Scholar]
- Davey J. W., Hohenlohe P. A., Etter P. D., Boone J. Q., Catchen J. M., et al. , 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 12: 499–510. [DOI] [PubMed] [Google Scholar]
- Duboué E. R., Keene A. C., Borowsky R. L., 2011. Evolutionary convergence on sleep loss in cavefish populations. Curr. Biol. 21: 671–676. [DOI] [PubMed] [Google Scholar]
- Elipot Y., Hinaux H., Callebert J., Retaux S., 2013. Evolutionary shift from fighting to foraging in blind cavefish through changes in the serotonin network. Curr. Biol. 23: 1–10. [DOI] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett M. V., Miller M. R., Seeb J. E., 2012. Meiotic maps of sockeye salmon derived from massively parallel DNA sequencing. BMC Genomics 13: 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. B., Protas M., Conrad M., Scheid P. E., Vidal O., et al. , 2008. Synteny and candidate gene prediction using an anchored linkage map of Astyanax mexicanus. Proc. Natl. Acad. Sci. USA 105: 20106–20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. B., Borowsky R., Tabin C. J., 2009. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 5(1): e1000326 DOI: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. B., Furterer A., Carlson B. M., Stahl B. A., 2013. An integrated transcriptome-wide analysis of cave and surface dwelling Astyanax mexicanus. PLoS ONE 8(2): e55659 DOI: 10.1371/journal.pone.0055659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. B., Krutzler A. J., Carlson B. M., 2014. Complex craniofacial changes in blind cave-dwelling fish are mediated by genetically symmetric and asymmetric loci. Genetics 196: 1303–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht B. C., Campbell N. R., Holecek D. E., Narum S. R., 2013. Genome-wide association reveals genetic basis for the propensity to migrate in wild populations of rainbow and steelhead trout. Mol. Ecol. 22: 3061–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, J. E., N. R. Campbell, M. F. Docker, C. Baker, A. Jackson et al., 2014 Use of genotyping-by-sequencing data to develop a high-throughput and multi-functional SNP panel for conservation applications in Pacific lamprey. Mol. Ecol. Resour. Advance online publication. Epub 2014 Jun 5. 10.1111/1755–0998.12283.
- Houston R. D., Davey J. W., Bishop S. C., Lowe N. R., Mota-Velasco J. C., et al. , 2012. Characterisation of QTL-linked and genome-wide restriction site-associated DNA (RAD) markers in farmed Atlantic salmon. BMC Genomics 13: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbs, C. L., and W. T. Innes, 1936 The first known blind fish of the family Characidae: A new genus from Mexico. Occas. Pap. Mus. Zool. Univ. Mich. 342: 1–7. [Google Scholar]
- Jeffery W. R., 2001. Cavefish as a model system in evolutionary developmental biology. Dev. Biol. 231: 1–12. [DOI] [PubMed] [Google Scholar]
- Jeffery W. R., 2009. Regressive evolution in Astyanax cavefish. Annu. Rev. Genet. 43: 25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby R. F., Thompson K. W., Hubbs C., 1977. Karyotypic similarities between the Mexican and blind tetras. Copeia 1977: 578–580. [Google Scholar]
- Kosambi D. D., 1943. The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Kowalko J. E., Rohner N., Rompani S. B., Peterson B. K., Linden T. A., et al. , 2013. Loss of schooling behavior in cavefish through sight-dependent and sight-independent mechanisms. Curr. Biol. 23: 1874–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., et al. , 2009. Circos: An information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson W. A., Seeb L. W., Everett M. V., Waples R. K., Templin W. D., et al. , 2014. Genotyping by sequencing resolves shallow population structure to inform conservation of Chinook salmon (Oncorhynchus tshawytscha). Evol. Appl. 7: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., G. Waldbieser, B. Bosworth, B. H. Beck, W. Thongda et al., 2014 SNP discovery in wild and domesticated populations of blue catfish, Ictalurus furcatus, using genotyping-by-sequencing and subsequent SNP validation. Mol. Ecol. Resour. 14: 1261–1270. [DOI] [PubMed]
- Liu Z., 2011. Development of genomic resources in support of sequencing, assembly, and annotation of the catfish genome. Comp. Biochem. Physiol. D 6: 11–17. [DOI] [PubMed] [Google Scholar]
- Lorenz S., Brenna-Hansen S., Moen T., Roseth A., Davidson W. S., et al. , 2010. BAC-based upgrading and physical integration of a genetic SNP map in Atlantic salmon. Anim. Genet. 41: 48–54. [DOI] [PubMed] [Google Scholar]
- Lu F., Lipka A. E., Glaubitz J., Elshire R., Cherney J. H., et al. , 2013. Switchgrass genomic diversity, ploidy, and evolution: Novel insights from a network-based SNP discovery protocol. PLoS Genet. 9: e1003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh S. E., Gross J. B., Aken B., Blin M., Borowsky R., et al. , 2014. The cavefish genome reveals candidate genes for eye loss. Nat. Commun. 5: 5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninwichian P., Peatman E., Liu H., Kucuktas H., Somridhivej B., et al. , 2012. Second-generation genetic linkage map of catfish and its integration with the BAC-based physical map. G3 (Bethesda) 2: 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Quin K. E., Yoshizawa M., Doshi P., Jeffery W. R., 2013. Quantitative genetic analysis of retinal degeneration in the blind cavefish Astyanax mexicanus. PLoS ONE 8: e57281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden R., Gharbi K., Mugue N., Martinsohn J., Senn H., et al. , 2013. Sturgeon conservation genomics: SNP discovery and validation using RAD sequencing. Mol. Ecol. 22: 3112–3123. [DOI] [PubMed] [Google Scholar]
- Palti Y., Genet C., Luo M. C., Charlet A., Gao G., et al. , 2011. A first generation integrated map of the rainbow trout genome. BMC Genomics 12: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palti Y., Genet C., Gao G., Hu Y., You F. M., et al. , 2012. A second generation integrated map of the rainbow trout (Oncorhynchus mykiss) genome: Analysis of conserved synteny with model fish genomes. Mar. Biotechnol. (NY) 14: 343–357. [DOI] [PubMed] [Google Scholar]
- Pottin K., Hinaux H., Rétaux S., 2011. Restoring eye size in Astyanax mexicanus blind cavefish embryos through modulation of the Shh and Fgf8 forebrain organising centres. Development 138: 2467–2476. [DOI] [PubMed] [Google Scholar]
- Protas M. E., Hersey C., Kochanek D., Zhou Y., Wilkens H., et al. , 2006. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat. Genet. 38: 107–111. [DOI] [PubMed] [Google Scholar]
- Protas M., Conrad M., Gross J. B., Tabin C., Borowsky R., 2007. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr. Biol. 17: 452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas M., Tabansky I., Conrad M., Gross J. B., Vidal O., et al. , 2008. Multi-trait evolution in a cave fish, Astyanax mexicanus. Evol. Dev. 10: 196–209. [DOI] [PubMed] [Google Scholar]
- Rowe H. C., Renaut S., Guggisberg A., 2011. RAD in the realm of next-generation sequencing technologies. Mol. Ecol. 20: 3499–3502. [DOI] [PubMed] [Google Scholar]
- Şadoǧlu P., 1956. A preliminary report on the genetics of the Mexican cave characins. Copeia 1956: 113–114. [Google Scholar]
- Salin K., Voituron Y., Mourin J., Hervant F., 2010. Cave colonization without fasting capacities: An example with the fish Astyanax fasciatus mexicanus. Comp. Biochem. Physiol. A 156: 451–457. [DOI] [PubMed] [Google Scholar]
- Schemmel C., 1980. Studies on the genetics of feeding behaviour in the cave fish Astyanax mexicanus f. anoptichthys. An example of apparent monofactorial inheritance by polygenes. Z. Tierpsychol. 53: 9–22. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Stock D. W., Jeffery W. R., 2004. Hedgehog signalling controls eye degeneration in blind cavefish. Nature 431: 844–847. [DOI] [PubMed] [Google Scholar]
- Yoshizawa M., Yamamoto Y., O’Quin K. E., Jeffery W. R., 2012. Evolution of an adaptive behavior and its sensory receptors promotes eye regression in blind cavefish. BMC Biol. 10: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.