Abstract
The circadian oscillator is astonishingly robust to changes in the environment but also to genomic changes that alter the copy number of its components through genome duplication, gene duplication, and homeologous gene loss. While studying the potential effect of aneuploidy on the Arabidopsis thaliana circadian clock, we discovered that a line thought to be trisomic for chromosome 3 also bears the gi-1 mutation, resulting in a short period and late flowering. With the help of whole-genome sequencing, we uncovered the unexpected complexity of this trisomic stock’s history, as its genome shows evidence of past outcrossing with another A. thaliana accession. Our study indicates that although historical aneuploidy lines exist and are available, it might be safer to generate new individuals and confirm their genomes and karyotypes by sequencing.
Keywords: trisomic line, circadian period, flowering time, GIGANTEA, deep-sequencing
The anticipation of daily transitions like dawn and dusk by a circadian clock provides a fitness advantage in cyanobacteria, plants, fungi, and animals (Dodd et al. 2005; Green et al. 2002; Xu et al. 2011; Yerushalmi et al. 2011). Although the circadian clock comprises a highly interconnected network of transcriptional activators and repressors, this adaptation to a rhythmic environment has been maintained while the expansion of gene families and whole-genome duplication or triplication events have altered the balance between different clock components. With the advent of genome-sequencing platforms, we are now in an ideal position to study the evolutionary trajectory of clock gene families, including gene copy number variation, and the birth and death of new copies. Because of its network properties, the circadian clock constitutes a great model system in which to test the gene balance hypothesis. Indeed, Albert Blakeslee observed already in the early 1920s that plants with three copies of a single chromosome are generally less vigorous and fertile than polyploid lines in which all chromosomes have been triplicated, leading to the formulation of genetic (or gene) balance (Blakeslee et al. 1920; Blakeslee 1921). In Arabidopsis thaliana, some circadian mutants like the toc1-1 allele were first thought to exhibit dosage sensitivity (Millar et al. 1995), but this was later attributed to a dominant-negative mutation in the repressor domain of the protein (Gendron et al. 2012). Most A. thaliana clock mutations have recessive effects, indicating that a single gene copy is sufficient to maintain period length close to 24 hr, and thus demonstrates some dosage compensation. However, increasing gene copy number for TOC1 (Mas et al. 2003) or ZTL (Somers et al. 2004) changes period length in a dose-dependent manner, suggesting that network stoichiometry is important. A related phenomenon has been observed in mice, where segmental trisomy 16 shortens the clock period in constant darkness, demonstrating that aneuploidy may lead to alterations in circadian rhythms (Ruby et al. 2010). Following whole-genome duplication, clock genes in Brassica rapa are more likely to be retained compared with their neighboring genes or random and control genes (Lou et al. 2012). Properly functioning circadian clocks confer fitness benefits (Dodd et al. 2005; Green et al. 2002), putting strong selective pressure on clock genes to be maintained in a stoichiometrically balanced fashion. We wished to determine what circadian phenotypes might be observed in trisomic lines generated by George Redeí and others in the 1960s and available from the Arabidopsis stock centers (Redeí 1962; Steinitz-Sears 1963). Although a stock presumed to be trisomic for chromosome 3, CS3227/N3227, had a shorter period, the cause was found to be a 5-bp deletion in the GIGANTEA gene found also in one of Redeí’s other stocks, gi-1 (Redeí 1962; Park et al. 1999). Whole-genome sequencing revealed that the supposedly trisomic stock is an introgression line, with one fifth of its genome coming from an Estland/Estonia (Est)-like accession. An accession from Est was one of ten Arabidopsis accessions widely studied by Friedrich Laibach (Laibach 1943) and later George Redeí (Redeí 1962), making it reasonable to deduce that this strain arose through inadvertent outcrossing.
Materials and Methods
Plant material and growth conditions
Seed stocks were ordered from stock centers: CS3227 [from Arabidopsis Biological Resource Center (ABRC)] and N3227 [from Nottingham Arabidopsis Stock Centre (NASC)], gi-1, gi-2, gi-3 (N3123, N3124, and N51 from NASC). The transfer DNA insertion allele gi-201 has been described (Martin-Tryon et al. 2007) and is presumed to be a null allele. Mutants ft-10 and fkf-1 came from laboratory stocks. The TOC1:LUC luciferase reporter line in the Columbia-2 (Col-2) background was described previously (Salomé and McClung 2005). All mutants and CS3227 were crossed to this line; mutant seedlings homozygous for the mutation in question (toc1-101, gi-1, gi-2, gi-201, lhy-20) were selected based on circadian or flowering time phenotypes. We did not observe any natural variation arising from crossing the Col-2 accession (harboring the luciferase (LUC) reporter) to the accessions Col-0 (background of toc1-101, lhy-20 and gi-201) or Col-1 (backgrounds for gi-1 and gi-2). Flowering time of all genotypes was scored as number of leaves (rosette and cauline) as well as days to flowering, in long days (16-hr light:8-hr dark, LD) at 23°, with a relative humidity of 65%; at least 20 plants were grown for each genotype.
Cotyledon movement
Seeds were surface-sterilized by the vapor-phase method (Clough and Bent 1998) and plated on Murashige and Skoog (MS) medium supplemented with 2% sucrose and 0.8% agar. After stratification for 3 d at 4° in the dark, plates were released and seedlings allowed to grow for 5 d under a photoperiod of 12-hr light: 12-hr dark, at 23°. On day 6, individual seedlings were transferred to 24-well cloning plates on a cube of growth medium and released into constant light at 23°. Cotyledon position was captured with surveillance cameras over 7 d. Postrun analysis was conducted with the help of the Kujata software package, and circadian parameters estimated by Fast-Fourier transform as previously described (Salomé and McClung 2005).
Circadian analysis of LUC reporters
Surface-sterilized seeds were plated on MS medium containing 1% sucrose and 0.5% agar and stratified for 2 d at 4° in the dark. After release in 23°, seedlings were entrained by light-dark cycles (16-hr light: 8-hr dark) for 7 d, and were then transferred to 96-well plates. Each well contained 200 µL of MS medium supplemented with 1% sucrose and 30 µL of 2.5 mM luciferin, potassium salt (Biosynth, Postfach, Switzerland). Plates were subjected to another entraining cycle and then moved into constant light and temperature for 5 d for luciferase activity measurements on a Perkin Elmer Topcount microplate luminescence counter. Circadian parameters were extracted from time-course data by Fast-Fourier transform as previously described (Salomé and McClung 2005). All experiments were performed at least three times (n = 12−24).
Hypocotyl length measurements
Surface-sterilized seeds were plated along the diagonal of Petri dishes containing full-strength MS medium supplemented with 1% sucrose and 0.8% agar. Plates were placed at 4° for 3 d in the dark before being released in short days for 6 d. Images of plates were acquired on a flat-bed scanner in transparent mode alongside a ruler. Hypocotyl length was measured with ImageJ (http://rsbweb.nih.gov/ij/).
Next-generation sequencing and data analysis
Genomic DNA from gi-1, gi-2, Est-1 from the Max Planck Institute (MPI; CS22683), Est from the Salk Institute (CS67485), and CS3227 was isolated with the QIAGEN Plant Mini kit. A total of 1 µg of genomic DNA was used as starting material for Illumina’s TruSeq PCR-free library preparation according to manufacturer’s instructions for 350-bp insert size (Illumina, San Diego, CA). Final libraries were quantified by quantitative polymerase chain reaction (qPCR Kapa Biosystems, Boston, MA) and confirmed by Bioanalyzer (Agilent Technologies, Santa Clara, CA) and sequenced on a HiSeq2000 instrument with 2 × 101-bp reads. Short reads were processed with the SHORE pipeline (Schneeberger et al. 2009); after filtering of reads that did not map to the Col-0 reference genome, genome-wide coverage was about 20x for gi-1, 28x for gi-2, and 24x for CS3227, and more than 30x for Est accessions. High-quality single-nucleotide polymorphisms (SNPs) and small deletions between Col-0 and other genotypes were derived from shore consensus by the use of a scoring matrix optimized for identifying homozygous positions that differ from the reference genome (scoring_matrix_hom.txt). The resulting SNP lists were compared in R (The R Foundation for Statistical Computing 2013) with the help of the merge and intersect functions.
Results
The genetic stock CS3227 has a short period
Seeds were obtained for a trisomic stock carrying three copies of chromosome 3 from the Arabidopsis Biological Resource Center. This stock, referred to herein as CS3227 (N3227 from NASC), displayed a short period when assayed by cotyledon movement (Figure 1A). This phenotype was observed in all seedlings from the original stock received from ABRC, as well as all subsequent generations derived from the original stock. This phenotype did not segregate in any of these populations (representing at least three generations of seed-to seed propagation). Plants trisomic for chromosome 3 are expected to exhibit yellow-green rosette leaves and reduced fertility (www.arabidopsis.org), but we did not select progeny based on these phenotypes at any point, suggesting that the short circadian period was not linked to a compromised dosage of clock genes located on chromosome 3 but rather to an independent genetic defect that was fixed in this stock.
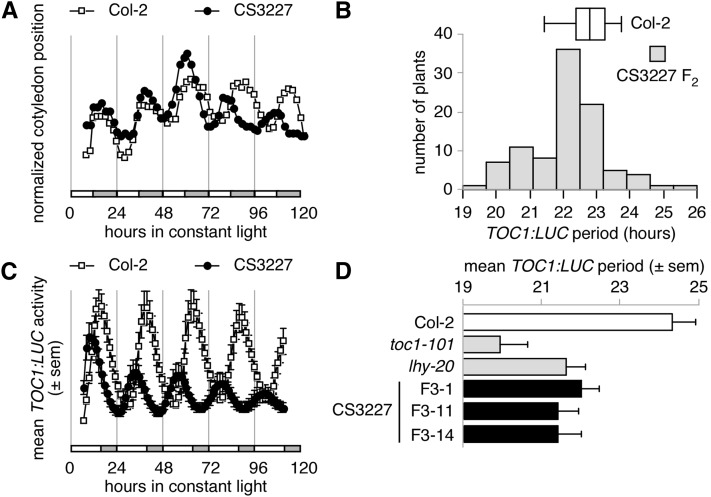
Figure 1.
CS3227 has a short period. Seedlings were grown under 12L:12D cycles for 5 (A) or 7 (B−D) d before release into constant light for recording of circadian parameters. (A) Cotyledon movement for Col-2 and CS3227 seedlings. (B) Segregation of the short period phenotype in an F2 population derived from a cross between CS3227 and a TOC1:LUC reporter in the Col-2 background. (C) Mean TOC1:LUC activity for Col-2 and CS3227 seedlings after release into constant light and temperature. (D) Mean TOC1:LUC period for Col-2, CS3227 F3 lines and the short period mutants lhy-20 and toc1-101, under the same conditions as (B) and (C). All data shown as means ± SEM (n = 12−24).
We introduced a TOC1:LUC luciferase reporter (in the Col-2 background; Salomé and McClung 2005) into CS3227 by genetic crossing. As shown in Figure 1 B and C, the short period phenotype segregated as a simple, recessive mendelian trait, further confirming that the circadian defect was not caused by aneuploidy. We selected several F2 seedlings with a short period and confirmed their phenotype in their progeny (Figure 1D and Table 1). Free-running period in CS3227 was 2 hr shorter than the stated parental line Col-2: 21.9 ± 0.1 hr for F3 lines (n = 12) vs. 24.2 ± 0.1 hr. CS3227 shortened period as much as lhy-20 (21.6 ± 0.1 hr, n = 12), a null allele of the LATE ELONGATED HYPOCOTYL gene, but remained longer than a strong allele of the TIMING of CAB2 1 gene (toc1-101: 19.7 ± 0.2 hr, n = 12; Figure 1D and Table 1).
Table 1. Circadian parameters of genotypes.
| Genotype | Period ± SEM | Amplitude ± SEM | RAEa ± SEM | n |
|---|---|---|---|---|
| Col-0 | 24.2 ± 0.1 | 19,435 ± 1653 | 0.17 ± 0.01 | 24 |
| gi-1 | 21.9 ± 0.2 | 6222 ± 770 | 0.41 ± 0.03 | 23 |
| gi-2 | 25.0 ± 0.4 | 3154 ± 404 | 0.31 ± 0.03 | 22 |
| gi-201 | 24.1 ± 0.1 | 1878 ± 179 | 0.16 ± 0.02 | 12 |
| CS3227 | 21.2 ± 0.2 | 6111 ± 945 | 0.27 ± 0.02 | 12 |
| Col-0 | 24.3 ± 0.1 | 15,580 ± 1785 | 0.14 ± 0.01 | 23 |
| CS3227 | 22.0 ± 0.1 | 4909 ± 1004 | 0.20 ± 0.02 | 12 |
| lhy-20 | 21.6 ± 0.1 | 3628 ± 282 | 0.12 ± 0.01 | 12 |
| toc1-101 | 19.9 ± 0.1 | 980 ± 115 | 0.20 ± 0.03 | 23 |
RAE refers to relative amplitude error, with an RAE of zero corresponding to a perfect sine wave, while RAEs closer to 1 denote weaker rhythms.
The genetic stock CS3227 is late flowering
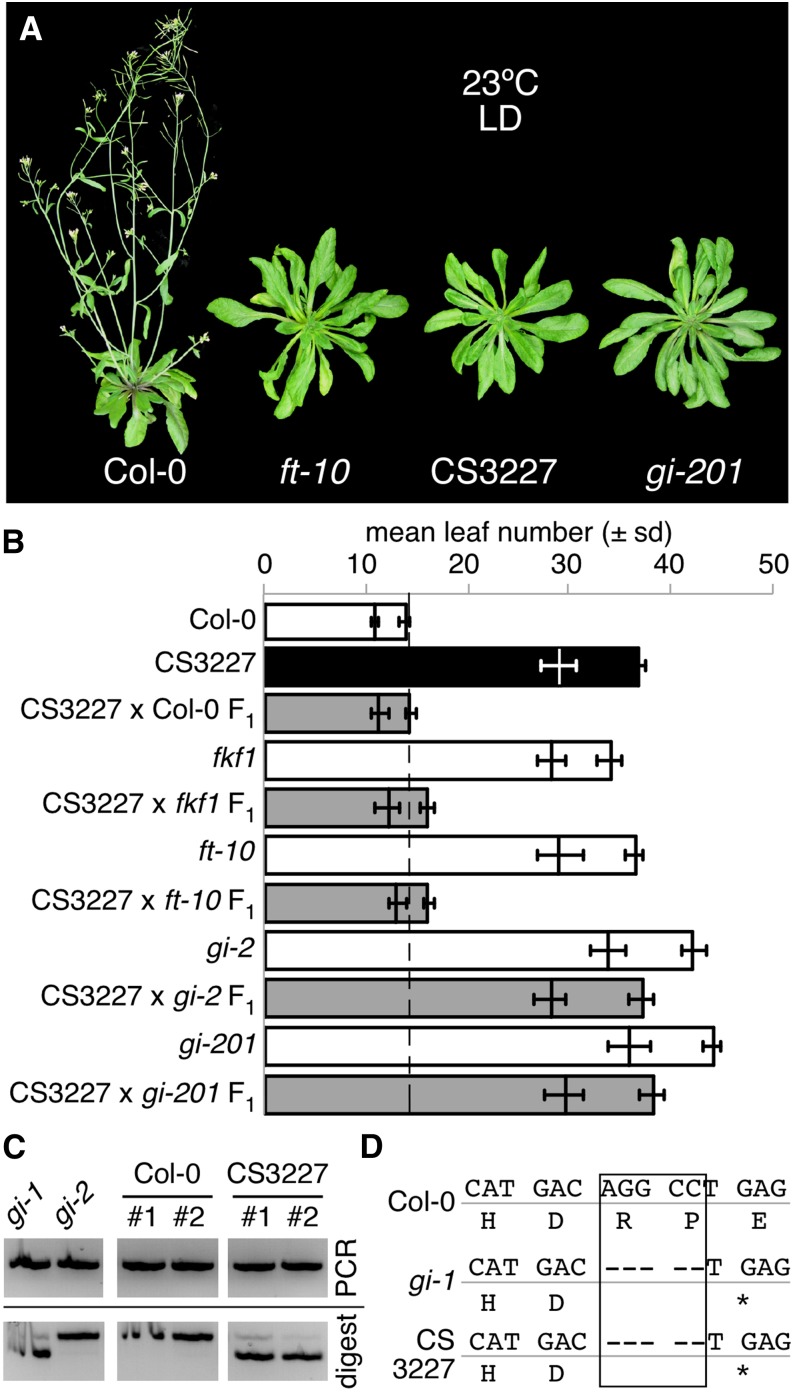
An additional phenotype, delayed flowering, was observed in CS3227 plants when grown in long days (16-hr light) at 23°. The delay in initiation of the reproductive phase was similar to the other late flowering mutants fkf1, ft, and gi (Figure 2 A and B). Late flowering and short period phenotypes of CS3227 always cosegregrated (Figure 2B), indicating that they were likely caused by the same genetic lesion. We tested whether CS3227 was allelic to known late flowering mutants by crossing CS3227 to fkf1, ft-10, gi-2, and gi-201. Only F1 plants from crosses between CS3227 and both gi alleles flowered late, suggesting that GIGANTEA might carry a lesion in the CS3227 stock.Mapping of the short period phenotype in an F2 population derived from a cross between CS3227 TOC1:LUC and the Ler accession independently confirmed strong linkage to the top arm of chromosome 1, where GI is located. Targeted sequencing of the GI locus in CS3227 revealed a 5-bp deletion near the 3′ end of the gene, resulting in the introduction of a premature stop after amino acid 1002. The deletion introduced a PsyI restriction site in GICS3227, and allowed an independent confirmation with a CAPS marker (Glazebrook et al. 1998). PCR amplification followed by PsyI digestion yielded two bands of the expected size for CS3227 (Figure 2C). Col-0 and gi-2 products did not carry the restriction site and were thus not cleaved by the enzyme. However, gi-1, which has exactly the same mutation (Park et al. 1999) as we found in CS3227, showed the same restriction pattern as CS3227 samples (Figure 2 C and D). We reordered the seed stock for CS3227 from NASC and found that all plants were homozygous for the gi-1 deletion (Table 2). We therefore concluded that the late flowering and short period phenotypes of CS3227 were caused by a small deletion in GI, which is identical to the gi-1 lesion.
Figure 2.
CS3227 is late flowering and carries the same deletion as gi-1. (A) Representative adult plants for Col-0, ft-10, CS3227, and gi-201 strains grown for 4 wk in long days (LD) at 23°. (B) Mean rosette and cauline leaf number in F1 plants from crosses between CS3227 and various late flowering mutants along with parental controls. (C) CAPS analysis of the genetic lesion found in the GIGANTEA gene in CS3227. Two independent DNA preparations were tested for Col-0 and CS3227 and are referred to as #1 and #2. PCR products were first precipitated to remove excess salts, as PsyI digest performed poorly when directly added to the PCR. Only the larger digest product is shown for gi-1 and CS3227 samples. PCR, polymerase chain reaction. (D) Alignment of genomic sequences for the GI locus in Col-0, gi-1, and CS3227, around the 5-bp deletion detected in CS327. The asterisk denotes the position of the premature stop codon introduced by the deletion.
Table 2. Frequencies of phenotypes and genotypes observed in a N3227 stock, obtained from NASC.
| Observed | Expecteda | % Observed | P Value | |
|---|---|---|---|---|
| Late flowering | 106 | 29−30 | 100 | 0 |
| gi-1 / gi-1 genotype | 106 | 29−30 | 100 | 0 |
NASC, Nottingham Arabidopsis Stock Centre.
Transmission of the trisomic copy of chromosome 3 is based on Steinitz-Sears, 1963 (5 trisomics from 18 offspring, frequency = 27.8%, Steinitz-Sears 1963).
CS3227 behaves like gi-1 for circadian phenotypes
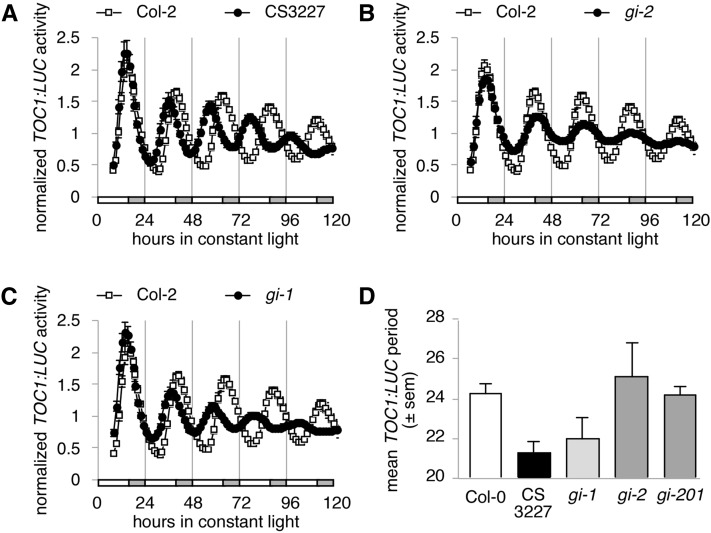
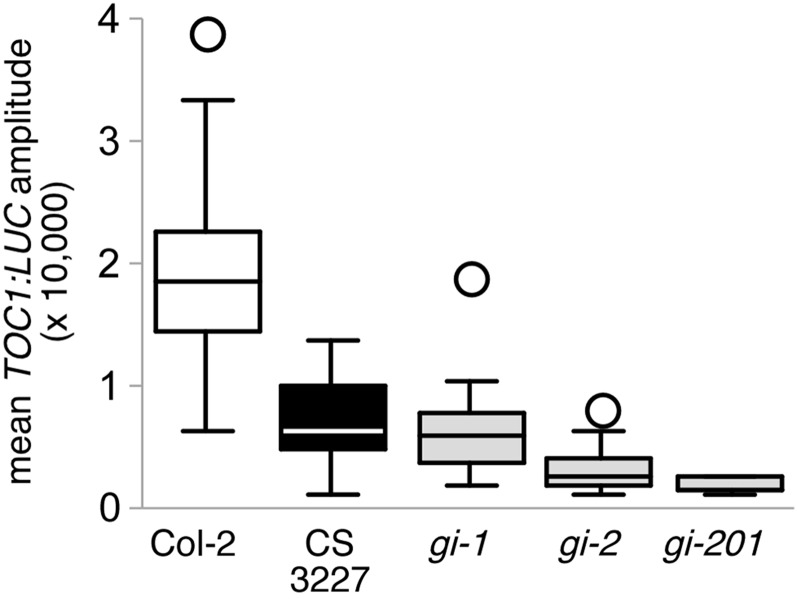
In light of the molecular lesion found in CS3227, we re-examined the circadian defects present in CS3227 alongside the weak gi allele gi-1 and the strong alleles gi-2 and gi-201. Both CS3227 and gi-1 displayed a similar short period, while the strong gi alleles did not affect period length significantly (Figure 3). Circadian amplitude of the reporter was markedly decreased in all gi mutants and in CS3227, with strong gi alleles having a stronger effect (with values about 10–14% of wild type, Figure 4. The same reporter was introgressed from Col-2 into all mutant backgrounds, allowing direct comparison of amplitudes). Again, gi-1 and CS3227 behaved in a similar fashion regarding circadian amplitude, which reached about 40% of wild-type values. The CS3227 genetic stock therefore exhibited all circadian defects known to occur in the gi-1 allele.
Figure 3.
Comparison of CS3227 with other gi alleles for circadian period. All seedlings were grown under 12L:12D cycles for 7 d before being released into constant light to record circadian parameters. Mean normalized TOC1:LUC activity for Col-2 and CS3227 (A), gi-2 (B), and gi-1 (C) after release into constant conditions. (D) Mean TOC1:LUC period for seedlings shown in (A−C). All data shown as means ± SEM (n = 12−24).
Figure 4.
Comparison of CS3227 with other gi alleles for circadian amplitude. Mean TOC1:LUC amplitude for Col-2 and several gi alleles. Data are shown as box and whiskers plot from at least 24 seedlings.
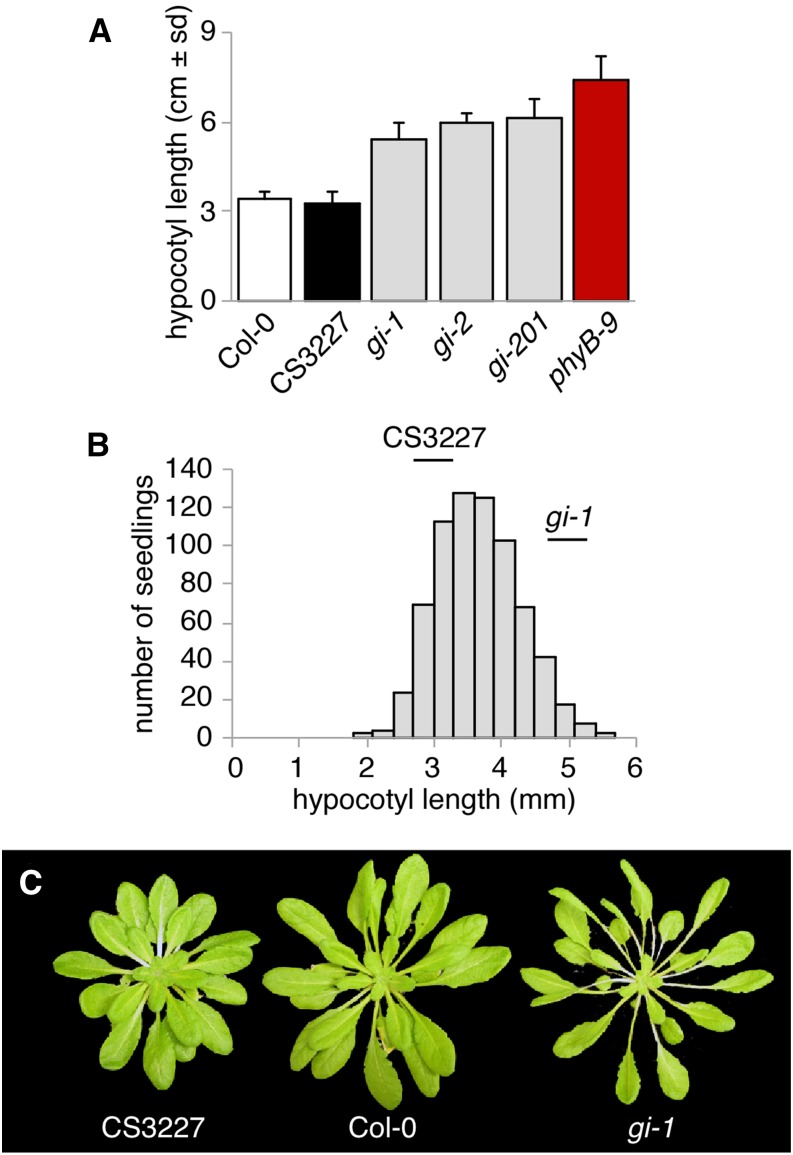
CS3227 differs from gi-1 for petiole and hypocotyl elongation
Not all aspects of CS3227 aligned with the phenotypes described for gi-1. GI loss of function alleles are characterized by elongated hypocotyls in red light and white light. We therefore measured hypocotyl length in Col-0, CS3227, and several gi alleles when grown in white light and short days. As expected, all gi alleles (gi-1, gi-2, and gi-201) had long hypocotyls under these conditions (mean length ~6 mm; with the Col-0 wild-type strain being about 3 mm in length) but remained slightly shorter than the red light photoreceptor mutant phyB-9 (Figure 5A). In contrast, CS3227 displayed a shortened hypocotyl that was comparable to Col-0. The long hypocotyl characteristic of a gi-1 mutant was recovered in an F2 population derived from a cross between CS3227 and Col-0, indicating that CS3227 also carried a modifier locus (or loci) that suppressed the long hypocotyl resulting from loss of GI function (Figure 5B). Petiole elongation is strongly promoted in gi mutants when grown in short days, as seen for gi-1 adult plants (Figure 5C). Petioles in CS3227 were much shorter, and were comparable to those seen in Col-0 (Figure 5C). We concluded that the CS3227 genetic stock carried both the gi-1 mutation (a 5-bp deletion) as well as a modifier locus (or loci) acting specifically in GI-dependent control of cell elongation. These results also alleviate potential concerns about seed contamination or simple mislabeling of CS3227, as it clearly displayed a number of phenotypes not seen in gi-1.
Figure 5.
CS3227 carries modifiers that suppress gi-induced petiole and hypocotyl elongation. (A) Mean hypocotyl length for Col-0, CS3227, and several gi alleles grown for 6 d in short days at 23°. (B) Distribution of hypocotyl length in an F2 population derived from a cross between CS3227 and Col-0. (C) Representative adult Col-0, gi-1, and CS3227 plants grown for 8 wk in short days at 23°.
Deep-sequencing of CS3227 reveals an unexpected pedigree
CS3227, gi-1, and gi-2 were isolated by George Redei in the early 1960s. That both CS3227 and gi-1 carry an identical 5-bp deletion raised the possibility of a shared genetic history of the two strains. We therefore sequenced the genomes of gi-1, gi-2, and CS3227 with the Illumina PCR-free DNA TruSeq protocol to identify polymorphisms relative to Col-0 in these genetic stocks and test for relatedness. SNP numbers relative to the Col-0 reference genome sequence are summarized in Table 3. As expected, gi-1 and gi-2 shared most of their SNPs, distributed along all 5 chromosomes. SNP numbers were low, consistent with the genetic background in which these two mutants were originally isolated (Col-1, which later gave rise to the commonly used Col-0 stock). A comparison with SNPs identified in CS3227 revealed a more complex picture. For much of the genome, CS3227 had 10−200 times more SNPs per chromosome than gi-1 or gi-2, but SNP density was distributed unevenly, with large-scale blocks of dense and sparse SNPs (Supporting Information, Figure S1, Figure S2, and Table 3). Chromosome 5 was an exception, with very few SNPs, many of which were shared with gi-1 and gi-2, presumably due to their common original background (Figure S1).
Table 3. Summary of SNP numbers identified in gi-1, gi-2, and CS3227 relative to the reference Col-0.
| Genotype | chr 1 | chr 2 | chr 3 | chr 4 | chr 5 |
|---|---|---|---|---|---|
| gi-1 | 391 | 210 | 251 | 775 | 335 |
| gi-2 | 366 | 206 | 248 | 793 | 346 |
| CS3227 | 78,415 | 2506 | 8857 | 55,318 | 488 |
SNP, single-nucleotide polymorphism.
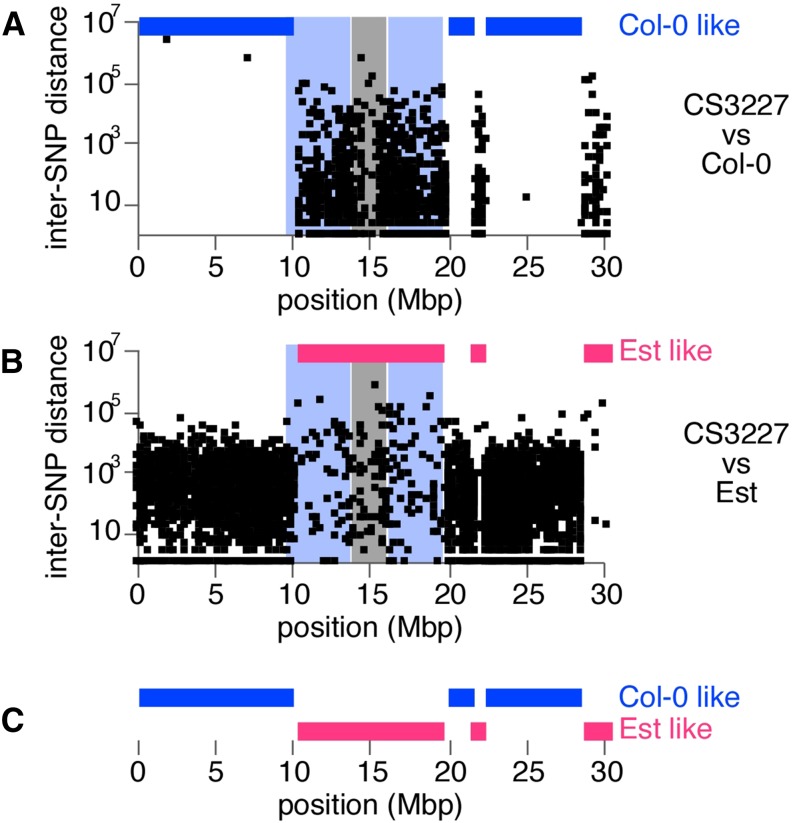
Although most Arabidopsis laboratories currently focus on one of the Col accessions or Ler, initial Arabidopsis research included 10 distinct accessions, all collected and distributed to the community by Friedrich Laibach (Laibach 1943). We therefore compared SNPs of a non−Col-0 portion of chromosome 1 from CS3227 with SNP datasets from seven of the nine non-Col accessions used by George Redeí that have been re-sequenced as part of the 1001 Genomes project (http://1001genomes.org) (Weigel and Mott 2009) and found a match with Est (from Estland/Estonia; Figure S3). Blocks of high similarity between Est and CS3227 coincided with regions with high SNP density relative to Col-0, and were found on all chromosomes except chromosome 5 (Figure 6 and Figure S2).
Figure 6.
Estland/Estonia (Est) is a likely candidate progenitor for the non−Col-0 genomic regions found in CS3227. The Est accession shared most of its SNPs with the non−Col-0 regions of CS3227, here shown for chromosome 1 but also for chromosomes 2−4. (A) Inter-SNP distance for all SNPs found along chromosome 1 in CS3227 when Col-0 is used as reference. (B) Inter-SNP distance for chromosome 1 of CS3227 with Est as reference genome. (C) Complementary pattern of Col-0−like and Est-like genomic regions along chromosome 1 of CS3227. SNP, single-nucleotide polymorphism.
Samples of the Est accessions have been sequenced at the MPI in Tübingen and at the Salk Institute, but only Est sequenced at the Salk Institute was a very good match to CS3227. We resequenced the Est accessions from Salk (CS67485) and from MPI (CS22683) with the PCR-free DNA TruSeq method used for our initial sequencing. Again, CS3227 was clearly related to Est from Salk, but not to Est-1 from MPI (Figure S3B). Both Est accessions displayed the lesioning/early senescence phenotype characteristic of accessions carrying a hyperactive allele of the ACCELERATED CELL DEATH 6 (ACD6) gene (Todesco et al. 2010). Of the two presumptive causal SNPs identified in the Est-1 allele of ACD6, only the polymorphism A566N was shared with the Est stock sequenced at the Salk Institute, suggesting that this SNP alone is sufficient to result in early onset senescence. CS3227 plants did not share this phenotype (Figure 2A); in agreement, the genomic location of ACD6 (about 8.3 Mb on chromosome 4) was of Col origin, although flanked by two Est regions (Figure S2).
Based on haplotype blocks, we identified clear breakpoints for all Est-like regions (Table 4). We counted several Est-like regions on chromosome 1 alone, indicative of at least eight independent crossover events resulting in the observed alternating pattern of Col-0-like and Est-like genomes. Because the average crossover rate for chromosome 1 is about 1.7 after a single meiosis (Salomé et al. 2012), this observation is consistent with an outcrossing event followed by many generations of selfing, leading to a pattern typical for recombinant inbred lines.
Table 4. Predicted Est-like intervals in CS3227.
| Chromosome | Left Border, bp | Right Border, bp |
|---|---|---|
| 1 | 10,180,611 | 19,875,136 |
| 21,543,989 | 22,196,062 | |
| 28,423,595 | 30,427,017 | |
| 2 | 0 | 302,692 |
| 3 | 1,732,545 | 4,274,768 |
| 4,289,162 | 4,622,728 | |
| 4 | 0 | 6,215,838 |
Est, Estland/Estonia.
Finally, we wished to confirm that our CS3227 stock was no longer carrying a third copy of chromosome 3. We therefore plotted normalized read support for all identified Est-like SNPs for CS3227. As shown in Figure 7, read counts for CS3227 were even and showed a pattern very close to that seen in Est from Salk. Importantly, normalized read count for all chromosomes was close to 1, including chromosome 3, indicating that any trisomic history in our stock had been lost prior to the generation we sequenced.
Figure 7.
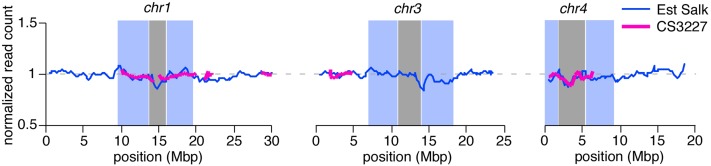
The sequenced CS3227 stock is not trisomic. Normalized read count (over all chromosomes) for Estland/Estonia (Est) Salk (blue line) and the Est-like genomic regions for CS3227 (magenta line).
Discussion
We initially set out to characterize what effect additional chromosomes might have on the A. thaliana circadian clock. When it became clear that we did not have trisomic individuals in our test population, we shifted our attention to line CS3227, initially described as being trisomic for chromosome 3, which exhibited a number of interesting phenotypes rarely seen in combinations in other A. thaliana mutants: a short circadian period (Figure 1 and Figure 3) and late flowering in long days (Figure 2A). We mapped the causal locus for these phenotypes and found that CS3227 harbored the gi-1 mutation, a 5-bp deletion in the GIGANTEA gene (Figure 2, B and C). Both gi-1 and CS3227 were isolated in the 1960s by George Redeí, anchoring the two stocks to a common origin.
Whole-genome sequencing of the CS3227 genome revealed a mosaic of Col-like and Est-like regions for all but chromosome 5, arguing against a simple seed contamination (Figure 6, Figure S2, and Figure S3). We did not observe greater read coverage for chromosome 3 (Figure 7), consistent with a lack of trisomy in our population. A delay in flowering time was not reported for newly synthesized chromosome 3 trisomics (Henry et al. 2010), leading us to conclude (i) that this phenotype is specific for CS3227 and the gi-1 deletion, and (ii) that chromosome 3 does not contain genes with major roles in the switch to flowering. Indeed, very few genes with major effects on flowering time have been mapped to chromosome 3.
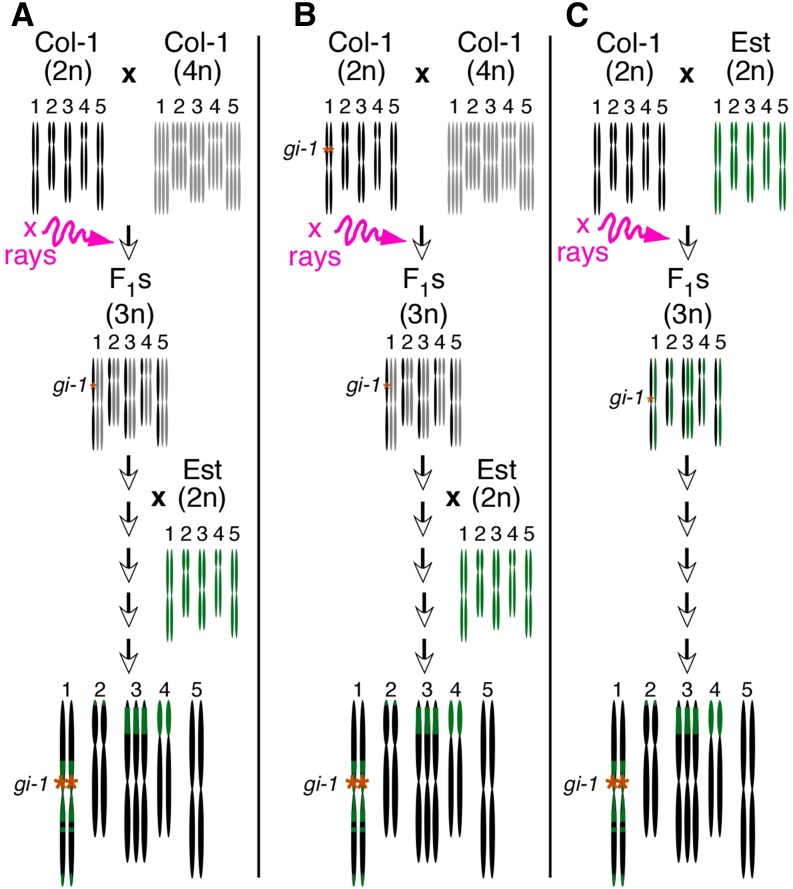
Even with full genome sequence from our CS3227 stock, it is difficult to reconstruct the exact chain of events that led to its genesis. Three major events must have taken place, the order of which is not clear: the isolation of a trisomic stock for chromosome 3; the introduction of the gi-1 deletion; and the introgression of an Est-like genome. Trisomic lines for other chromosomes do not share the gi-1 deletion (not shown), arguing against the model depicted in Figure 8B, in which the gi-1 mutant was used as diploid parent in the original diploid x tetraploid cross. gi-1 and the trisomic lines were both generated by irradiating A. thaliana with X rays (Redeí 1962; Steinitz-Sears 1963); it is therefore conceivable that gi-1 was isolated in the progeny of the diploid x tetraploid cross (Figure 8, A and C). Although we favor this scenario, we notice that gi-1 and CS3227 do not share more SNPs with each other than with gi-2. However, ionizing agents do not induce as many point mutations as EMS (Belfield et al. 2012) and may therefore not live an easily trackable footprint.
Figure 8.
Possible models to explain the genesis of CS3227. In models (A) and (B), the complement of primary trisomics was generated from a cross between a diploid and a tetraploid Col-1 parent. The nonequal segregation of chromosomes in the F1s was induced by X-ray irradiation, which may have caused the gi-1 deletion (A). Alternatively, gi-1 may have been the diploid parent (B). The introduction of the Est-like genome occurred after the isolation of trisomic individuals, either by targeted crossing or by outcrossing to a neighboring Est-like plant. In model (C), two diploid parents, Col-1 and Est-like, were crossed and the F1 plants X-ray irradiated. The extent of Est-like genome was subsequently largely lost following backcrosses to Col-1 or by single-seed decent. As in (A), model (C) posits the genesis of gi-1 as a consequence of X-ray irradiation of the F1 generation.
The introduction of the Est-like genome in the ancestral CS3227 stock is more difficult to date. Because of the number of apparent crossovers, most notably on chromosome 1, many generations must separate the initial cross and the current generation we characterized and sequenced. Other trisomic lines were negative for an Est-like genotype at a small number of tested markers, suggesting that the induction of trisomy may have predated the introduction of the Est-like genome. The model depicted in Figure 8A is therefore the most likely scenario to explain how CS3227 came about.
Although the study of changes in circadian parameters caused by aneuploidy merits further attention, it is clear that future efforts should begin with the generation, karyotyping, and phenotyping of aneuploid individuals from genetically defined parental lines. Chromosomal variants can now be easily identified by whole-genome sequencing (Henry et al. 2010), thus facilitating the precise identification of aneuploidy lines and the exact chromosome complement of their genomic make-up. It may also be informative to decrease clock gene copy number in a tetraploid background, to observe what possible circadian defects might arise from the loss of a clock gene following whole-genome duplication. Our study also highlights the power of whole genome sequencing to resolve issues arising from mixed genetic stocks, a non-negligible problem in Arabidopsis (Anastasio et al. 2011).
Supplementary Material
Acknowledgments
We thank Luz Rivero (Ohio State University, ABRC) for comments and discussion. We also thank Rob McClung (Dartmouth College) for sharing the CS3227 leaf movement data acquired during the PhD research of P.A.S. This work was supported by an EMBO Long Term Fellowship (P.A.S) and funds from the Max Planck Society (D.W.).
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.015156/-/DC1
Communicating editor: Z. Yang
Literature Cited
- Anastasio A. E., Platt A., Horton M., Grotewold E., Scholl R., et al. , 2011. Source verification of mis-identified Arabidopsis thaliana accessions. Plant J. 67: 554–566. [DOI] [PubMed] [Google Scholar]
- Belfield E. J., Gan X., Mithani A., Brown C., Jiang C., et al. , 2012. Genome-wide analysis of mutations in mutant lineages selected following fast-neutron irradiation mutagenesis of Arabidopsis thaliana. Genome Res. 22: 1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee A. F., 1921. Types of mutations and their possible significance in evolution. Am. Nat. 5: 254–267. [Google Scholar]
- Blakeslee A. F., Belling J., Farnham M. E., 1920. Chromosomal duplication and mendelian phenomena in datura mutants. Science 52: 388–390. [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dodd A. N., Salathia N., Hall A., Kevei E., Toth R., et al. , 2005. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Gendron J. M., Pruneda-Paz J. L., Doherty C. J., Gross A. M., Kang S. E., et al. , 2012. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 109: 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Drenkard E., Preuss D., Ausubel F. M., 1998. Use of cleaved amplified polymorphic sequences (CAPS) as genetic markers in Arabidopsis thaliana. Methods Mol. Biol. 82: 173–182. [DOI] [PubMed] [Google Scholar]
- Green R. M., Tingay S., Wang Z. Y., Tobin E. M., 2002. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129: 576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry I. M., Dilkes B. P., Miller E. S., Burkart-Waco D., Comai L., 2010. Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics 186: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laibach F., 1943. Arabidopsis thaliana (L.) Heynh. als Objekt für genetische und entwicklungsphysiologische Untersuchungen. Bot. Archiv 44: 439–455. [Google Scholar]
- Lou P., Wu J., Cheng F., Cressman L. G., Wang X., et al. , 2012. Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell 24: 2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon E. L., Kreps J. A., Harmer S. L., 2007. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143: 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P., Alabadi D., Yanovsky M. J., Oyama T., Kay S. A., 2003. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. J., Carre I. A., Strayer C. A., Chua N. H., Kay S. A., 1995. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163. [DOI] [PubMed] [Google Scholar]
- Park D. H., Somers D. E., Kim Y. S., Choy Y. H., Lim H. K., et al. , 1999. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582. [DOI] [PubMed] [Google Scholar]
- Redeí G. P., 1962. Supervital mutants of Arabidopsis. Genetics 47: 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The R Foundation for Statistical Computing 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Ruby N. F., Fernandez F., Zhang P., Klima J., Heller H. C., et al. , 2010. Circadian locomotor rhythms are normal in Ts65Dn “Down syndrome” mice and unaffected by pentylenetetrazole. J. Biol. Rhythms 25: 63–66. [DOI] [PubMed] [Google Scholar]
- Salomé P. A., McClung C. R., 2005. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P. A., Bomblies K., Fitz J., Laitinen R. A., Warthmann N., et al. , 2012. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity (Edinb) 108: 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A. H., et al. , 2009. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Somers D. E., Kim W. Y., Geng R., 2004. The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinitz-Sears L. M., 1963. Chromosome studies in Arabidopsis thaliana. Genetics 48: 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Balasubramanian S., Hu T. T., Traw M. B., Horton M., et al. , 2010. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465: 632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Mott R., 2009. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., DiAngelo J. R., Hughes M. E., Hogenesch J. B., Sehgal A., 2011. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 13: 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi S., Yakir E., Green R. M., 2011. Circadian clocks and adaptation in Arabidopsis. Mol. Ecol. 20: 1155–1165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.