Abstract
Suberin is a lipid and phenolic cell wall heteropolymer found in the roots and other organs of all vascular plants. Suberin plays a critical role in plant water relations and in protecting plants from biotic and abiotic stresses. Here we describe a transcription factor, AtMYB41 (At4g28110), that can activate the steps necessary for aliphatic suberin synthesis and deposition of cell wall-associated suberin-like lamellae in both Arabidopsis thaliana and Nicotiana benthamiana. Overexpression of AtMYB41 increased the abundance of suberin biosynthetic gene transcripts by orders of magnitude and resulted in the accumulation of up to 22 times more suberin-type than cutin-type aliphatic monomers in leaves. Overexpression of AtMYB41 also resulted in elevated amounts of monolignols in leaves and an increase in the accumulation of phenylpropanoid and lignin biosynthetic gene transcripts. Surprisingly, ultrastructural data indicated that overexpression led to the formation of suberin-like lamellae in both epidermal and mesophyll cells of leaves. We further implicate AtMYB41 in the production of aliphatic suberin under abiotic stress conditions. These results provide insight into the molecular-genetic mechanisms of the biosynthesis and deposition of a ubiquitous cell wall-associated plant structure and will serve as a basis for discovering the transcriptional network behind one of the most abundant lipid-based polymers in nature.
Keywords: suberin, R2-R3 MYB, transcription factor, lignin, abiotic stress, Arabidopsis, Nicotiana benthamiana
Introduction
The colonization of land by plants required the acquisition of specialized modifications to the cell wall. Structures like lignified vasculature and suberized endodermal cells allowed plants to develop a homoiohydric lifestyle (the capacity to maintain a constant water status) required for terrestrial habitats. Suberin, an aliphatic and phenolic heteropolymer, is synthesized in many tissues of higher plants including seed coats, tree bark, periderms of mature roots and tubers, endodermal cells of young roots, and abscission scars (Pollard et al., 2008; Schreiber, 2010; Beisson et al., 2012; Franke et al., 2012). It is also deposited as a response to environmental stresses such as high salinity and wounding (Kolattukudy, 2001; Schreiber et al., 2005a,b; Franke et al., 2009). As such, suberin represents a structure vital to many physiological processes related to plant water status and protection from biotic and abiotic stressors.
Suberin is an acylglycerol polymer, generically termed a polyester, deposited on the inner surface of the cell wall of specific cell types such as endodermal and peridermal cells. Suberin structures have been partially inferred from chemical analyses of monomers or oligomers released by either total or partial depolymerization, respectively (Holloway, 1972; Franke et al., 2005; Graça and Santos, 2006; Santos and Graça, 2006). For example, root suberins are typically dominated by 18:1 ω-hydroxy fatty acids (ω-OH FAs) and 18:1 dicarboxylic fatty acids (DCA). Non-polymeric aliphatic waxes associated with suberized tissues have also been described (Espelie et al., 1980; Li et al., 2007a). Similar to suberin, plant cuticles comprise an aliphatic acylglycerol-based polyester matrix, cutin, with associated waxes. In contrast to suberin, the cuticle is deposited on the outermost surface of the epidermal cell wall of aerial plant organs. In Arabidopsis, cutin and suberin differ chemically in terms of degree of unsaturation, carbon chain length, and phenolic content. Arabidopsis cutin consists of approximately 50% 18:2 monomers. Conversely, 18:2 monomers only comprise about 2% of root suberin. Instead, Arabidopsis root suberin is dominated by 18:1 monomers (>55%). Arabidopsis cutin comprises almost exclusively 16- and 18-carbon monomers whereas suberin consists of a range of 16- to 24-carbon monomers. Suberin has a much higher phenolic content than cutin, primarily in the form of the phenylpropanoid ferulic acid.
The genetic resources of Arabidopsis and potato (Solanum tuberosum) have provided for the discovery of a number of suberin biosynthetic genes (Beisson et al., 2012; Li-Beisson et al., 2013). Despite this progress, no transcriptional regulators of suberin synthesis have been definitively identified. However, several transcription factors related to plant cuticle biosynthesis have been identified and characterized. These include members of the AP2/EREBP, the HD-ZIP IV, the WW domain-containing, and the R2-R3 MYB transcription factor or co-activator families (Yeats and Rose, 2013). Regulators of cuticle biosynthesis related to mRNA stability, RNA-mediated gene silencing, and post-translational modification have also been identified (Hooker et al., 2007; Lam et al., 2012; Lü et al., 2012). These studies indicate that the regulatory network underlying cuticle biosynthesis is quite intricate. In contrast, only two transcription factors have been implicated in suberin synthesis (Lasserre et al., 2008; Almeida et al., 2013). However, these speculations were based primarily on gene expression patterns (i.e. they are primarily expressed in suberizing tissues) – biochemical or ultrastructural evidence of their involvement in suberin biosynthesis is lacking.
Given that no loss-of-function mutants have thus far been identified, a gain-of-function strategy to induce ectopic suberin production offers an alternative approach to deciphering the transcriptional network underlying suberin synthesis. Knowledge of the transcriptional controls of the suberization process has the potential to increase our understanding of mechanisms that control suberin levels and the role of suberin in response to stress, and to enhance resistance to drought, salt, and pathogens in crop plants. Because suberin is rich in unsaturated DCAs and ω-OH FAs, increasing its production may also allow the development of renewable sources of bifunctional olefins with potential for replacing petrochemicals in the production of bioplastics and other specialty biomaterials (Gandini, 2008).
Here we present the characterization of a transcription factor, AtMYB41 (At4g28110), capable of activating the synthesis, export, assembly, and cell wall-localized deposition of a suberin-like structure. We further implicate AtMYB41 in the activation of aliphatic suberin synthesis under conditions of abiotic stress.
Results
Overexpression of AtMYB41 results in the ectopic production of aliphatic suberin and the deposition of suberin-like lamellae
Arabidopsis has more than 100 genes encoding R2-R3 MYB transcription factors known to regulate diverse plant-specific processes including the biosynthesis of anthocyanin, secondary cell walls, and lignin (Dubos et al., 2010). R2-R3 MYBs also regulate cell fate and identity, plant development, and responses to biotic and abiotic stresses (Martin and Paz-Ares, 1997; Stracke et al., 2001; Davies and Schwinn, 2003). AtMYB41 was previously described as a regulator of accumulation of cuticle lipid, based primarily on the observation that overexpression lines (AtMYB41 OE) exhibited phenotypes associated with a malformed cuticle (Cominelli et al., 2008). However, chemical and ultrastructural analyses were not performed on these lines. We extended these studies by further analyzing the published DNA microarray results from Arabidopsis plants overexpressing AtMYB41 (Cominelli et al., 2008). We found that many of the top 50 most up-regulated genes encoded enzymes known for their involvement in suberin synthesis [e.g. acyltransferases and reductases like ASFT (At5g41040), GPAT5 (At3g11430), and FAR4 (At3g44540)[ (Beisson et al., 2007; Molina et al., 2009; Domergue et al., 2010; Vishwanath et al., 2013) and included genes whose expression highly correlates with the expression of suberin biosynthetic genes.
Based on the above observations, we hypothesized that AtMYB41 functions as a regulator of suberin biosynthesis. Suberin is not normally produced in leaves. Instead, a biosynthetically related but distinct cuticle, comprising cutin that is impregnated with waxes, covers the epidermal surfaces of leaves and other aerial plant organs. We postulated that overexpression of AtMYB41 might lead to the ectopic accumulation of suberin in aerial organs such as leaves where a cuticle is normally produced. Similar to a previous report, we found that plants overexpressing AtMYB41 driven by the 35S promoter (AtMYB41 OE-9) (Cominelli et al., 2008) had phenotypes associated with surface defects including stunted growth, glossy leaf surfaces, elevated permeability to toluidine blue stain, and altered pavement cell shape (Figures S1–S3). Analysis of leaf cross sections of stably transformed AtMYB41 OE-9 Arabidopsis plants by transmission electron microscopy (TEM) revealed the presence of lamellar structures, alternating light and dark bands deposited on the internal surfaces of the primary cell walls of epidermal cells (Figures1 and S4). These lamellar structures strongly resemble the lamellae typical of suberized endodermal and peridermal root cells (Figure S5) (Enstone et al., 2002; Ma and Peterson, 2003; Franke and Schreiber, 2007; Molina et al., 2009).
Figure 1.
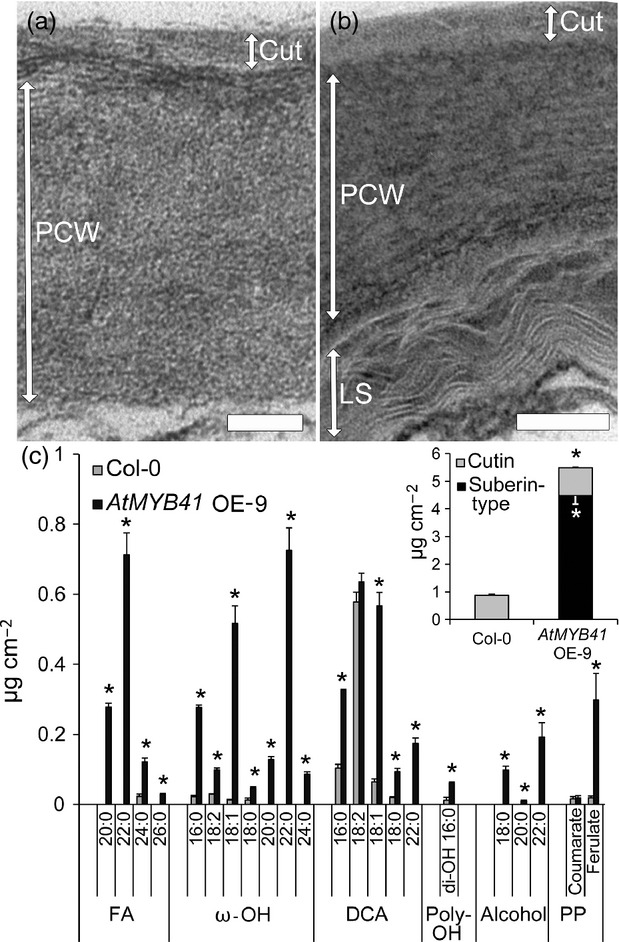
Overexpression of AtMYB41 in Arabidopsis leads to the ectopic deposition of lamellar structures in the cell walls of leaf epidermal cells and a leaf polyester monomer composition dominated by suberin-type monomers.(a) Transmission electron micrograph of a Col-0 (wild-type) leaf epidermal cell showing the intact cuticle and primary cell wall. Cut, cuticle; PCW, primary cell wall Scale bar = 100 nm.(b) Transmission electron micrograph of a leaf epidermal cell of AtMYB41 OE-9 showing a lamellar structure abutting the primary cell wall and an intact cuticle on the outer surface of the cell wall. Cut, cuticle; PCW, primary cell wall; LS, lamellar structure. Scale bar = 100 nm.(c) Leaf polyester monomer composition of wild type (Col-0) and AtMYB41 OE-9 from 6-week-old plants. Inset: total amounts of polyester grouped as cutin- and suberin-type monomers. All data are presented in micrograms per square centimeter of leaf area as mean values with SD (n = 4). FA, fatty acid; ω-OH, ω-hydroxy fatty acid; DCA, dicarboxylic fatty acid; di-OH, 10(9),16-dihydroxy fatty acid; alcohol, primary fatty alcohol; PP, phenylpropanoid. This experiment was performed twice with similar results. *Significant differences (P ≤ 0.01) as determined by Student's t-tests or Satterthwaite t-tests.
Analysis of the leaf polyester composition of AtMYB41 OE-9 plants revealed aliphatic monomers typical of Arabidopsis suberin in addition to the cutin monomers normally present (Figure1). Particularly diagnostic of suberin aliphatics were the 7-, 36-, and 15-fold increases in 18:1 DCA, 18:1 ω-OH FA, and ferulic acid content, respectively, accompanied by greater than twofold increases in 16:0 and 18:0 ω-OH FA and 16:0 and 18:0 DCA content (Figure1). The appearance of 18:0–22:0 primary fatty alcohols, 20:0–22:0 ω-OH FAs, 20:0–22:0 DCAs, and 20:0-26:0 fatty acids was also strongly indicative of suberin deposition. Inconsistent with the notion of AtMYB41 as a regulator of cutin synthesis, we observed no substantial difference in 18:2 DCA content (on a surface area basis). Collectively, stable overexpression of AtMYB41 in Arabidopsis led to the production of 4.5 times more suberin-type than cutin-type monomers. As such, this represents a very large flux of acyl-lipids and phenylpropanoid ferulic acid into suberin synthesis without apparent perturbation of cutin synthesis. Observation of young roots (endodermis), mature roots (periderm), and seed coat suberin aliphatics from AtMYB41 OE-9 plants revealed little difference in composition compared with the wild type (WT) (Figure S6).
To confirm the chemical phenotypes observed in the AtMYB41 OE-9 line, we generated additional, independent AtMYB41 overexpression lines (Appendices S1 and S2). Analysis of the leaves of T2 plants revealed polyesters comprising substantial amounts of suberin-type monomers (Figure S7). The aliphatic suberin content of these lines at 4 weeks of age ranged from roughly one to three times that of their cutin monomer content. Total cutin monomer amounts (normalized to dry mass) were similar across all lines with the exception of line #63 which had slightly less total cutin than the WT. Analysis of segregants with a WT visual appearance from two independent lines (negative controls) revealed no accumulation of suberin-type monomers.
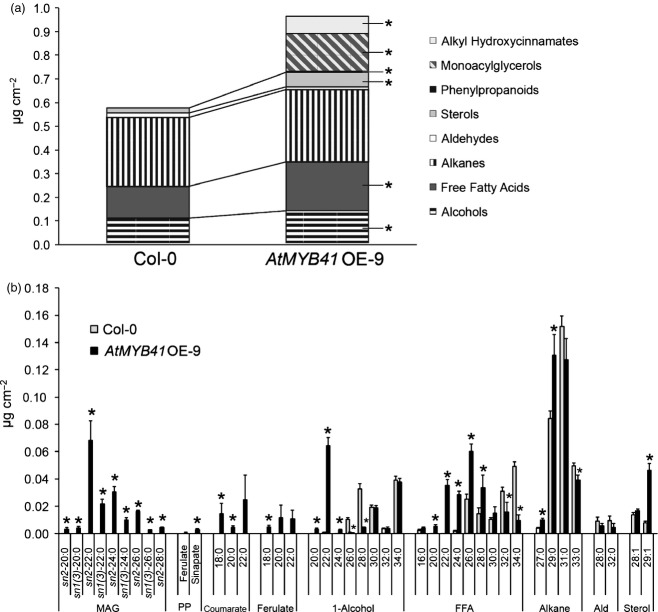
Overexpression of AtMYB41 in Arabidopsis also led to the production of atypical leaf wax components: alkyl hydroxycinnamates and monoacylglycerols (Figure2). Alkyl hydroxycinnamates and monoacylglycerols are waxes normally associated with the suberized periderm of Arabidopsis taproots (Li et al., 2007a; Kosma et al., 2012). Other notable changes in AtMYB41 OE-9 leaf waxes included substantial shifts in the chain-length distribution of primary fatty alcohols and free fatty acids as well as a large increase in β-sitosterol (C29:1 sterol) content; also consistent with the wax composition of suberized Arabidopsis taproots. Notably, AtMYB41 OE-9 plants had nearly double the amount of wax found in WT plants but without a compensatory reduction in alkanes, the dominant class of Arabidopsis cuticular waxes (Figure2). These results implicate AtMYB41 in the induction suberin-associated wax production.
Figure 2.
Overexpression of AtMYB41 leads to the accumulation of suberin-associated wax-type compounds in Arabidopsis leaf waxes.(a) Leaf wax class composition of Col-0 (wild-type) and AtMYB41 OE-9 Arabidopsis plants demonstrating the presence of atypical wax classes: alkyl hydroxycinnamates, monoacylglycerols (MAG), and phenylpropanoids (PP).(b) Leaf wax compositions. All data are presented in micrograms per square centimeter of leaf area as mean values with SD (n = 4). Coumarate, alkyl coumarate; Ferulate, alkyl ferulate; 1-Alcohol, primary fatty alcohol; FFA, free fatty acid; Ald, aldehyde. This experiment was performed twice with similar results. *Significant differences (P ≤ 0.01) as determined by Student's t-tests or Satterthwaite t-tests.
The chemical and ultrastructural phenotypes observed in AtMYB41-overexpressing Arabidopsis plants were also observed when AtMYB41 was transiently overexpressed in Nicotiana benthamiana leaves under the control of the 35S promoter (Figure3). In total, leaves possessed 22 times more suberin-type monomers than cutin-type monomers after 6 days of AtMYB41 expression (Figure3). This included a 49-fold increase in ferulic acid, 17-fold increases in 16:0 and 18:1 ω-OH FAs, >85-fold increases in 20:0–24:0 DCAs and ω-OH FAs, and 15- to 55-fold increases in 20:0–24:0 fatty acids (Figure3). Transmission electron microscopy (TEM) images of leaf cross sections revealed the presence of distinct lamellae abutting the internal surfaces of the primary cell walls of epidermal cells (Figure3) and mesophyll cells (Figure S8). These lamellar structures bore a striking resemblance to the lamellae normally found in the cell walls of suberized peridermis and endodermis cells from mature and young roots, respectively (Figure S5). Transient expression of other candidate R2-R3 MYB transcription factors identified from transcriptional co-expression analysis (Table S1), AtMYB45 (At3g48920) and AtMYB67 (At3g12720), in leaves of N. benthamiana did not result in the accumulation of suberin-type monomers (Figure S9). Similarly, the infiltration process did not appear to induce the accumulation of suberin-type monomers as a response to the wounding that could potentially occur during infiltration (Figure S9). Collectively, these results show that AtMYB41 overexpression is sufficient to induce a number of activities required to form suberin-like lamellae with a full complement of aliphatic suberin-type components. The lack of suberin-type monomer production via expression of other candidate MYBs indicates that AtMYB41 specifically induces aliphatic suberin production and that the production of suberin aliphatics in N. benthamiana leaves is not a general consequence of overexpressing Arabidopsis MYB transcription factors. Similarly, the observed induction of suberin-type monomers by transient overexpression of AtMYB41 is not a general consequence of wounding by the infiltration process.
Figure 3.
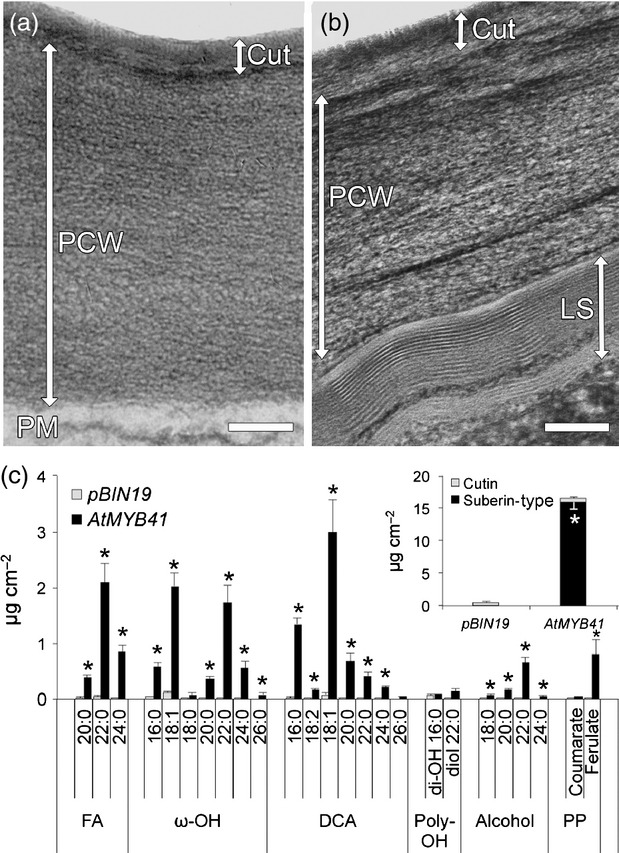
Transient expression of AtMYB41 in Nicotiana benthamiana leaves leads to ectopic deposition of suberin-like lamellae and a polyester monomer composition dominated by suberin-type monomers.(a) Transmission electron micrograph of a leaf epidermal cell of an infiltrated control (pBIN19, empty vector) showing an intact cuticle and the absence of a lamellar structure on the internal side of the primary cell wall. PM, plasma membrane; Cut, cuticle; PCW, primary cell wall. Scale bar = 100 nm.(b) Transmission electron micrograph of a leaf epidermal cell infiltrated with AtMYB41 showing a lamellar structure abutting the primary cell wall and an intact cuticle on the outer surface of the primary cell wall. PM, plasma membrane; Cut, cuticle; PCW, primary cell wall; LS, lamellar structure. Scale bar = 100 nm.(c) Leaf polyester monomer composition of N. benthamiana plants from infiltrated control plants (pBIN19 is the empty vector) and AtMYB41-infiltrated plants. Inset: total amounts of polyester grouped as cutin- or suberin-type monomers. All data are presented in micrograms per square centimeter of leaf surface area as mean values with SD (n = 3). Polyester determinations were made with leaves harvested 6 days after infiltration. FA, fatty acid; ω-OH, ω-hydroxy fatty acid; DCA, dicarboxylic fatty acid; di-OH, 10(9),16-dihydroxy; Alcohol, primary fatty alcohol; PP, phenylpropanoid. This experiment was performed three times with similar results. *Significant differences (P ≤ 0.01) as determined by Student's t-tests or Satterthwaite t-tests.
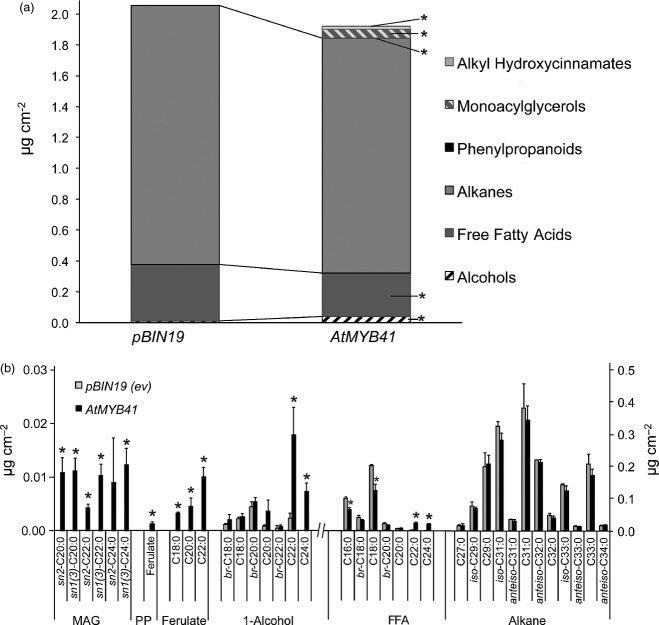
Transient expression of AtMYB41 in N. benthamiana also led to the production of atypical leaf surface waxes (Figures4 and S10). Similar to the results found with Arabidopsis plants overexpressing AtMYB41, alkyl hydroxycinnamates and monoacylglycerols were detected in extracts of leaf surface wax from N. benthamiana plants after 6 days of transiently expressing AtMYB41. Other notable changes in leaf waxes of N. benthamiana plants transiently expressing AtMYB41 included elevated amounts of straight-chain C22 and C24 fatty alcohols and free fatty acids, a reduced amount of C18 free fatty acid, and the appearance of free ferulic acid. The overall amount of fatty alcohols in N. benthamiana transiently expressing AtMYB41 was higher than in empty vector controls. Alkanes, the dominant class of N. benthamiana cuticular waxes, and glandular trichome-related diterpenes and acyl sugars (Slocombe et al., 2008) were generally unaffected by transient expression of AtMYB41 (Figures4 and S10). Transient expression of GPAT5 (At3g11430) led to the production of sn-2 and sn-1(3) monoacylglycerols (β and α MAGs, respectively), serving as a positive control for induction of suberin-associated MAG synthesis (Figure S10). These results further implicate AtMYB41 in induction of suberin-type wax production.
Figure 4.
Transient expression of AtMYB41 leads to the accumulation of suberin-associated wax-type compounds in Nicotiana benthamiana leaf waxes.(a) Leaf wax class composition of infiltrated control (pBIN19; ev, empty vector) and AtMYB41-infiltrated N. benthamiana plants demonstrating the presence of atypical wax classes: alkyl hydroxycinnamates, monoacylglycerols (MAG), and phenylpropanoids (PP).(b) Leaf wax constituent compositions. All data are presented in micrograms per square centimeter as mean values with SD (n = 4). Waxes were extracted 6 days after infiltration. MAG, monoacylglycerol; PP, phenylpropanoid; Ferulate, alkyl ferulate; Alcohol, primary fatty alcohol; FFA, free fatty acid; br, branched chain. This experiment was performed three times with similar results. *Significant differences (P ≤ 0.01) as determined by Student's t-tests or Satterthwaite t-tests.
Overexpression of AtMYB41 increases the abundance of suberin but not cuticle biosynthetic gene transcripts
The assembly of complex structures like suberin lamellae requires the coordinated expression of many biosynthetic genes encoding different enzyme activities. The accumulation of specific suberin-type monomers in Arabidopsis plants overexpressing AtMYB41 was consistent with the observed increases in suberin biosynthetic gene transcripts (Table1, Figure S11). For example, in agreement with the massive increase and appearance of specific chain lengths of ω-OH FAs and DCAs, CYP86A1 and CYP86B1 transcripts were elevated by more than 18 000- and 7000-fold, respectively. Cytochrome P450s CYP86A1 and CYP86B1 are responsible for the ω-hydroxylation (and possible further oxidation) of 16:0, 18:1 and 22:0, 24:0 fatty acids, respectively, and CYP86A1 is critical for the establishment of a proper lamellar structure in suberized periderm cells (Höfer et al., 2008; Compagnon et al., 2009; Molina et al., 2009). Similarly, the presence of suberin-type aliphatic monomers and associated MAG waxes would probably not be possible without the more than 8000-fold induction of GPAT5 transcript abundance. GPAT5 is critical for the proper synthesis of suberin and associated waxes via the synthesis of sn-2 MAG intermediates (Beisson et al., 2007; Li et al., 2007b; Molina et al., 2008; Yang et al., 2010). The 10-fold or greater elevation in FAR5, FAR4, and FAR1 transcript levels (Table1) was consistent with the appearance of specific chain lengths of primary fatty alcohols in both leaf polyesters and waxes (Figures1 and 2). FAR5, FAR4, and FAR1 are fatty acyl reductases responsible for the production of 18:0, 20:0, and 22:0 primary fatty alcohols, respectively, in suberized tissues (Domergue et al., 2010; Vishwanath et al., 2013). Furthermore, the elevated transcript abundance of the ferulate transferase-encoding ASFT and fatty acid elongase-encoding KCS2 correlated well with the observed presence of ferulic acid (Gou et al., 2009; Molina et al., 2009) and monomers with a chain length of >20 carbons (Franke et al., 2009; Lee et al., 2009), respectively.
Table 1.
Relative quantification of suberin, cuticle, and lignin biosynthetic and Casparian strip assembly gene expression in AtMYB41 OE-9 leaves. Data are presented as mean fold change values ± SEM from three biological replicates. Fold change was determined by the comparative CT (ΔΔCT) method. Fold change is defined as normalized gene transcript levels from AtMYB41 OE-9 mRNA relative to normalized gene transcript levels in wild-type (WT) samples. Target gene transcript levels were normalized to the transcript levels of the endogenous controls GAPC (At3g04120), EIF4A1 (At3g13920), and UBIQUITIN5 (At3g62250). The induction of suberin biosynthetic genes was verified by semi-quantitative RT-PCR analysis (Figure S11)
| Gene | AGI | Fold change relative to WT |
|---|---|---|
| AtMYB41 | At4g28110 | 743 ± 79 |
| Suberin biosynthetic genes | ||
| FAR1 | At5g22500 | 10 ± 2 |
| FAR4 | At3g44540 | 1762 ± 141 |
| FAR5 | At3g44550 | 399 ± 76 |
| GPAT5 | At3g11430 | 8230 ± 826 |
| ASFT | At5g41040 | 3207 ± 561 |
| KCS2/DAISY | At1g04220 | 167 ± 39 |
| FACT | At5g63560 | 2226 ± 697 |
| CYP86B1/RALPH | At5g23190 | 7224 ± 869 |
| CYP86A1/HORST | At5g58860 | 18 603 ± 4950 |
| Cuticle biosynthetic genes | ||
| CYP86A2/ATT1 | At4g00360 | 0.8 ± 0.1 |
| CER1 | At1g02205 | 0.6 ± 0.2 |
| FAR3/CER4 | At4g33790 | 0.1 ± 0.1 |
| KCS6/CER6 | At1g68530 | 1.1 ± 0.5 |
| HOTHEAD | At1g72970 | 0.2 ± 0.1 |
| GPAT4 | At1g01610 | 4.5 ± 1.7 |
| GPAT8 | At4g00400 | 0.4 ± 0.2 |
| Phenylpropanoid and lignin biosynthetic genes | ||
| PAL1 | At2g37040 | 8.6 ± 1.3 |
| CCoAMT1 | At4g34050 | 3.8 ± 0.7 |
| CAD5 | At4g34230 | 36.1 ± 7.7 |
| C4H/REF3 | At2g30490 | 5.3 ± 1.7 |
| PRXR9/PER30 | At3g21770 | 27.5 ± 6.8 |
| Casparian strip assembly genes | ||
| CASP1 | At2g36100 | 2 ± 0.2 |
| ESB1 | At2g28670 | 1 ± 0.3 |
The accumulation of cuticle biosynthetic gene transcripts was either unaffected or slightly lower in AtMYB41 OE-9 leaves (Table1). GPAT4 was the only exception, having transcripts that were four times more abundant in AtMYB41 OE-9 than in WT. This is not surprising, as GPAT4 is thought to participate in both suberin and cutin synthesis (Li-Beisson et al., 2013). Together, these data indicate that overexpression of AtMYB41 leads to increases in the accumulation of suberin-related but not cuticle biosynthetic gene transcripts.
Overexpression of AtMYB41 increases the abundance of phenylpropanoid and lignin biosynthetic gene transcripts and results in elevated lignin content
Suberin has been reported to possess both polyaliphatic and a polyphenolic domains (Bernards and Razem, 2001; Bernards, 2002). A number of studies on wound-induced periderm production in potato (Solanum tuberosum) tubers have indicated that the polyphenolic domain is similar to lignin but differs by containing a large proportion of hydroxycinnamic acids (especially ferulate) in addition to the hydroxycinnamyl alcohols normally found in lignin (Bernards et al., 1995; Yan and Stark, 2000). Recent work on understanding endodermis formation in Arabidopsis roots has proposed that Casparian strips in young root regions comprise exclusively the canonical lignin monomers (hydroxycinnamyl alcohols) typically associated with xylem vessel elements (Naseer et al., 2012; Geldner, 2013; Nawrath et al., 2013). Notwithstanding, it is clear that derivatives of the phenylpropanoid pathway, be they hydroxycinnamaic acids or the corresponding hydroxycinnamyl alcohols, are an important component of suberized tissues.
We sought to determine whether phenylpropanoid gene expression and metabolism were affected by overexpression of AtMYB41. We analyzed the abundance of transcripts encoded by phenylpropanoid and lignin biosynthetic genes and Casparian strip assembly genes in the leaves of Arabidopsis plants stably overexpressing AtMYB41. Nearly all gene transcripts analyzed showed a pattern of accumulation substantially higher than WT (Table1). This was particularly apparent with gene transcripts encoding phenylpropanoid and lignin metabolic genes. For example, PAL1 (At2g37040) transcripts were present at levels nearly nine times greater than wild-type levels. PAL1 encodes a phenylalanine ammonia lyase, which catalyzes the deamination of phenylalanine to cinnamic acid (Fraser and Chapple, 2011). CCoAMT1 (At4g34050) and C4H/REF3 (At2g30490) transcripts were present in amounts roughly four and five times greater than WT, respectively. CCoAMT1 encodes an O-methyltransferase (CCoAMT1) that catalyzes the methylation of the 3′ hydroxyl group of caffeoyl-CoA to form feruloyl-CoA (Humphreys and Chapple, 2002; Ibdah et al., 2003; Do et al., 2007). C4H/REF3 encodes a cytochrome P450 monooxygenase that catalyzes the hydroxylation of cinnamoyl-CoA at the 4′ carbon to form coumaroyl-CoA (Ruegger and Chapple, 2001; Schilmiller et al., 2009).
Overexpression of AtMYB41 increased the steady-state mRNA accumulation of two genes more specifically related to monolignol and lignin synthesis. CAD5 encodes a cinnamyl alcohol dehydrogenase involved in the reduction of cinnamaldehdyes to the corresponding cinnamyl alcohols in Arabidopsis (Sibout et al., 2003, 2005). CAD5 (At4g34230) levels were 36 times higher than in the WT. PRXR9/PER30 (At4g28110) encodes a class III peroxidase; class III peroxidases are thought to be involved in the activation of phenylpropanoids to phenoxy or phenyl radicals for oxidative coupling to form the lignin polymer (Boerjan et al., 2003; Passardi et al., 2004). In AtMYB41 OE-9 leaves, PRXR9/PER30 transcript levels were nearly 30 times higher than in WT leaves. The steady-state mRNA levels of one of the two genes tested related to Casparian strip assembly (Roppolo et al., 2011) was slightly elevated by overexpression of AtMYB41; CASP1 transcript levels were two times higher than WT in leaves of AtMYB41 OE-9. The transcript abundance of ESB1, which encodes a dirigent-domain-containing protein related to Casparian strip formation (Hosmani et al., 2013), was unaffected in AtMYB41 OE-9 leaves.
In order to correlate the increase in the abundance of phenylpropanoid-related gene transcripts imparted by overexpression of AtMYB41 with phenylpropanoid metabolites, we analyzed lignin by chromatographic analysis of the monomers released by thioacidolysis. Thioacidolysis, however, does present certain limitations; it is a degradative technique that only effectively releases monomers from 8-O-4′ ether linkages (Bernards, 2002). Nonetheless, it can serve as a proxy for estimating affected phenylpropanoid metabolism as it relates to lignin or lignin-like polymers. Analysis of the monomers released by thioacidolysis of AtMYB41 OE-9 leaf residues showed a doubling of syringyl (S) and guaiacyl (G) monomer amounts (Figure5). Similarly, analysis of N. benthamiana leaves transiently overexpressing AtMYB41 revealed a near doubling in S and G monomer amounts (Figure5). These results were also reflected in spectrophotometric determinations of the percentage of dry weight comprising of acetyl bromide-soluble lignin (%ABSL; Appendices S1 and S2). Leaf tissues from N. benthamiana plants transiently expressing AtMYB41 had roughly double the %ABSL compared with empty vector or uninfiltrated controls (Figure S12). Consistent with altered flux through the phenylpropanoid pathway, free sinapic acid was detected as a constituent of the leaf waxes of Arabidopsis plants stably overexpressing AtMYB41 (Figure2). Furthermore, ferulate dimers were identified in methanolysates (sodium-methoxide catalyzed transmethylation) from leaf tissues of Arabidopsis AtMYB41 OE-9 and N. benthamiana transiently expressing AtMYB41. A mass spectrum of one of the diferulates encountered is illustrated in Figure S13. These ferulate dimers are expected to be the products of the coupling of phenyl and/or phenoxy radicals, similar to the reactions proposed for the polymerization of lignin and the polyphenolic domain of suberin (Bernards, 2002; Razem and Bernards, 2002; Bernards et al., 2004).
Figure 5.
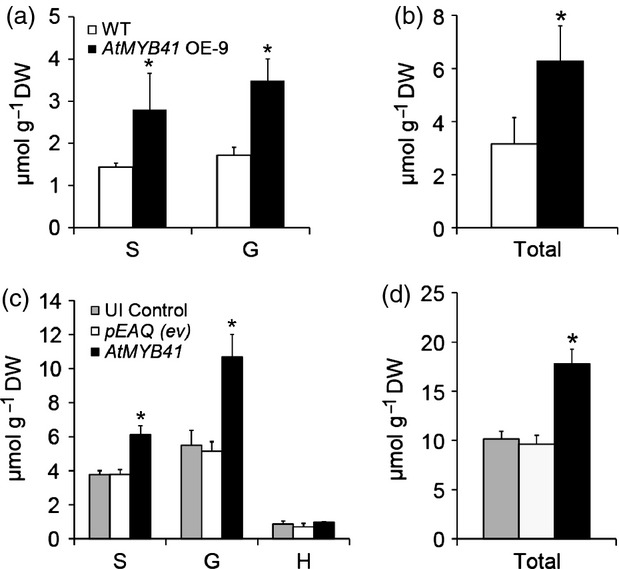
Overexpression in Arabidopsis or transient expression in Nicotiana benthamiana of AtMYB41 leads to elevated leaf lignin monomer content (presented as monomers released by thioacidolysis treatment of alcohol insoluble cell wall residue).(a, b) Lignin monomer composition and content, respectively, of the leaves from 6-week-old Col-0 (wild-type) and AtMYB41 OE-9 plants.(c, d) Lignin monomer composition and content, respectively, of uninfiltrated (UI) control, infiltration control (pEAQ; ev, empty vector), and AtMYB41-infiltrated N. benthamiana leaves. For N. benthamiana, leaves were harvested 6 days after infiltration. All data are presented in micromoles per gram dry weight with SD (n = 3–4). Monomers are: S, syringyl; G, guaiacyl; H, p-hydroxyphenyl. *Significant differences (P ≤ 0.01) when compared with pEAQ empty vector controls as determined by Student's t-tests or Satterthwaite t-tests.
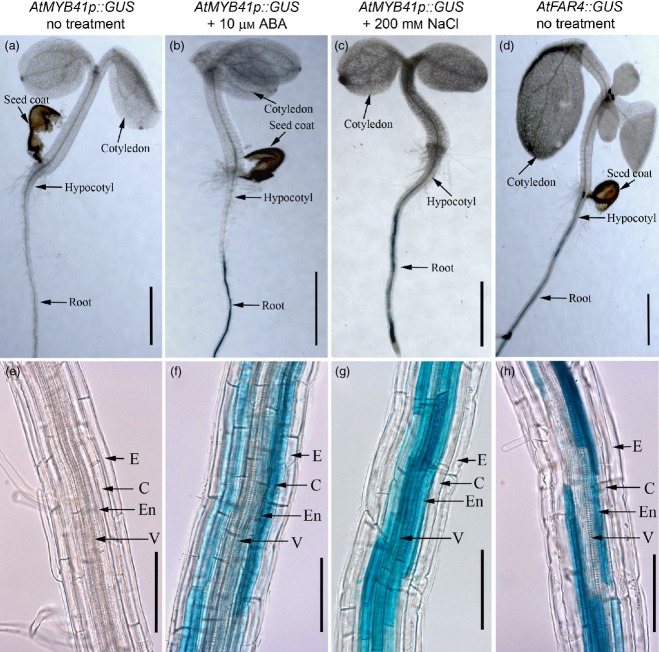
The AtMYB41 promoter is activated in endodermal cells by ABA and NaCl stress
AtMYB41 was previously implicated as playing a role in responses to abiotic stress in an abscisic acid (ABA)-dependent and phosphorylation-dependent manner (via mitogen-activated protein kinase activity) (Cominelli et al., 2008; Lippold et al., 2009; Hoang et al., 2012). Using AtMYB41promoter::GUS transcriptional fusions (AtMYB41p::GUS), we observed that the AtMYB41 promoter drove reporter gene expression in endodermal and surrounding cortical cells under ABA and sodium chloride (NaCl) treatment, but not under unstressed growth conditions (Figure6). Comparison with FAR4promoter::GUS (FAR4p::GUS) lines confirmed endodermal expression. These results demonstrate that the AtMYB41 promoter is active in the endodermis during periods of abiotic stress but not during normal growth.
Figure 6.
The GUS expression patterns in the primary root of transgenic, 5-day-old Arabidopsis seedlings harboring AtMYB41promoter::GUS (AtMYB41p::GUS) and AtFAR4promoter::GUS (AtFAR4p::GUS) fusions.(a, e) AtMYB41p::GUS untreated. (b, f) AtMYB41p::GUS in the presence of 10 μm abscisic acid (ABA) after 24 h. (c, g) AtMYB41p::GUS in the presence of 200 mm NaCl after 24 h. (d, h) AtFAR4p::GUS untreated. E, epidermis; C, cortex; En, endodermis; v, vascular cylinder. Scale bar: 0.5 cm in (a–d); 100 mm in (e–h).
Discussion
AtMYB41 activates the synthesis and deposition of suberin
Although suberin is one of the most abundant lipid polymers in nature, little is known about how its synthesis is regulated. Recent efforts have been made to understand the transcriptional control of periderm formation and have yielded lists of transcripts that are enriched specifically in the periderm of the cork oak (Quercus suber) or the periderm of potato (S. tuberosum) compared with other tissues (Soler et al., 2007, 2011). Recently, considerable progress has been made on the biosynthesis of suberin through the identification of a number of suberin-specific enzymes using the genetic resources of Arabidopsis and potato (Serra et al., 2009a,b, 2010; Ranathunge et al., 2011). However, to date, no transcription factors involved in regulating suberin synthesis have been identified.
AtMYB41 was originally described as a regulator of cuticle biosynthesis (Cominelli et al., 2008). However, our analysis of published DNA microarray data from AtMYB41 overexpression lines instead showed an extremely high accumulation of suberin biosynthetic gene transcripts. This prompted us to analyze the polyester compositions and cell wall ultrastructure of Arabidopsis and N. benthamiana cells overexpressing AtMYB41. The massive production of characteristic suberin-type monomers (18:1, 20:0–24:0 ω-OH FAs and DCAs) in both plant systems (Figures1 and 3) clearly indicated acyl flux into suberin-type monomer synthesis. Similarly, the presence of atypical waxes such as monoacylglycerols and alkyl hydroxycinnmates signifies the activation of suberin-associated wax biosynthesis. These experiments confirmed that overexpression of AtMYB41 activates the synthesis and deposition of both aliphatic suberin-type polyester and non-polymeric suberin-associated waxes. While the role of AtMYB41 in activating the synthesis and deposition of the polyphenolic domain of suberin (described by Bernards, 2002) was not definitively established, it was confirmed that overexpression of AtMYB41 has an effect on phenylpropanoid and lignin synthesis and related gene expression (Figure5, Table1). Overexpression of AtMYB41 is thus sufficient for the activation of both aliphatic and phenolic biosynthetic pathways and related gene expression. Collectively, these observations point to the dual chemical nature of suberin and highlight the significance of the phenylpropanoid pathway in suberin accumulation.
To date, only one report has described the transgenic production of suberin-type monomers in tissues normally covered with a cuticle (Li et al., 2007b). Co-overexpression of the suberin biosynthetic genes GPAT5 and CYP86A1 resulted in the production of about half as many suberin-like monomers as cutin without apparent modification of cutin content. In contrast, overexpression of AtMYB41 resulted in the production of 4.5 times more suberin-type than cutin-type aliphatic monomers in Arabidopsis (Figure1) and 22 times more suberin-type than cutin-type aliphatic monomers in N. benthamiana (Figure3). TEM analysis showed distinct lamellar structures abutting the inner surfaces of cell walls in the leaves of both Arabidopsis and N. benthamiana overexpressing AtMYB41 (Figures1 and 3), whereas ectopic co-overexpression of GPAT5 and CYP86A1 did not result in the production of lamellar structures abutting the primary cell wall nor a lamellate cuticle. This indicates that overproduction of suberin monomers via co-overexpression of multiple genes encoding activities for monomer synthesis alone is not sufficient to induce the ectopic assembly of suberin lamellae anywhere in or adjacent to the cell wall. It is striking that the overexpression of a single gene, AtMYB41, can activate and coordinate the large number of activities required to synthesize and assemble a complex macromolecular structure. Likewise, it is noteworthy that overexpression of AtMYB41 results in the assembly of lamellar structures at the subcellular site where suberin is normally deposited, adjoining the inner face of the primary cell wall.
The observation of leaf epidermal cells in AtMYB41-overexpressing plants that possess both a cuticle and suberin-like lamellae was intriguing. Few examples of plant cells producing both a cuticle and suberin-like lamellae exist in nature. The epidermal fiber cells from wild cotton species (Gossypium sp.) and the non-fibrous epidermal cells of cotton seeds (wild and domesticated) may represent the few cases in point, although it remains uncertain whether these cells possess a true cuticle or not (Ryser and Holloway, 1985). In Arabidopsis, suberin and cutin synthesis are spatiotemporally separated (e.g. cutin in the epidermis of expanding leaves and suberin in the endodermis of the root elongation zone, zone of maturation, and root periderm). In the case of AtMYB41 overexpression, this spatiotemporal separation has been uncoupled, resulting in the production of both aliphatic suberin and cutin in epidermal cells. Thus, a major question of biological import is brought to light. How do epidermal cells overexpressing AtMYB41 recognize and distinguish specific monomers and direct them to the external (for cutin) versus internal cell wall surfaces (for suberin)? This question is not limited to the specific situation of AtMYB41 overexpression but instead is an extension of a major unknown regarding normal cutin and suberin synthesis; how are suberin and cutin directed to different cell wall positions despite having similar compositions? Clearly, a multifaceted level of regulation is at play here.
Another striking observation was that production of suberin-like lamellae via overexpression of AtMYB41 was not limited to epidermal cells but was also evident in mesophyll cells (Figure S3). This is in stark contrast to the ectopic production of suberin-like monomers by co-overexpression of GPAT5 and CYP86A1 in which more than 90% of the resultant suberin-like aliphatic monomers were produced in epidermal cells; cells that are dedicated to the production of lipids for polyester synthesis in the form of cutin. Overexpression of AtMYB41 was sufficient to reprogram mesophyll cells to synthesize, assemble, and specifically deposit structures that they would not normally produce, namely extracellular lipids. In Arabidopsis and N. benthamiana, cutin, produced exclusively in epidermal cells, normally represents 0.3 and 0.7% of total leaf dry weight, respectively. Membrane fatty acids (produced in all leaf cells) on average comprise 4–5% of leaf dry weight (Yang and Ohlrogge, 2009). Overexpression of AtMYB41 resulted in the accumulation of lipid polyesters to comprise 1.1 and 2.4% of leaf dry weight in Arabidopsis and N. benthamiana leaves, respectively. This might be explained by the production of suberin aliphatics in both mesophyll and epidermal cells. Nonetheless, these are levels nearing those of membrane fatty acids, thereby representing a substantial flux of aliphatic acyl groups into suberin via AtMYB41 overexpression.
Although suberin monomers were produced in the leaves of Arabidopsis lines overexpressing AtMYB41, we did not observe alterations in the aliphatic suberin content or composition of roots or seed coats (Figure S6). It is possible that suberin deposition in these tissues is already at a maximal level because these are tissues where suberin is normally synthesized. Leaves represent a different situation, as they are starting from a point of nearly no suberin. Partner transcription factors could also be important for the elevated production of suberin by overexpression of AtMYB41. R2-R3 MYB transcription factors typically work in conjunction with bHLH, WD40, and other MYB transcription factors (Ramsay and Glover, 2005; Dubos et al., 2010) and these proteins could be differentially expressed or differentially modified at the post-translational level in the various tissues. Also, the 35S promoter has widely varying activities in different cell types; for example, the 35S promoter has low or patchy activity in seed coats (Young et al., 2008).
Biological function of AtMYB41
Collectively, our results demonstrate that overexpression of AtMYB41 is sufficient for the ectopic production of suberin-like lamellae, including a full complement of suberin-type aliphatics and elevated amounts of lignin monomers (Figures4, 6, S9, S10, and S12) with a concomitant increase in the accumulation of gene transcripts related to suberin, phenylpropanoid, and lignin biosynthesis (Table1). Furthermore, we found that the AtMYB41 promoter is active in the endodermis under conditions of abiotic stress but not under unstressed conditions (Figure6). This raises the question of the precise physiological role of AtMYB41. Like Cominelli et al. (2008), we were unable to obtain functional knock-out lines in Arabidopsis from publicly available T-DNA insertion mutant collections or by gene silencing approaches, possibly indicative of lethality or the involvement of genes with redundant function.
Based on our results, several possibilities exist for the biological function of AtMYB41. First, production of suberin-type monomers and lamellar structures by AtMYB41 overexpression could be a pleiotropic effect of strong overexpression. However, the high degree of coordinated gene expression encoding enzymatic, transporter, and polymerizing activities required to assemble a complex lamellar structure like suberin strongly argues against this. Further, the lack of induced suberin-type monomer production by transient expression of other candidate MYBs (Figure S9) corroborates a specific role for AtMYB41 in the synthesis of suberin aliphatics. Overall, our results indicate that AtMYB41 acts as a component of the regulatory network underlying stress-induced aliphatic suberin biosynthesis. Several lines of evidence support this hypothesis: (i) AtMYB41 induces the deposition of suberin-like materials when overexpressed; (ii) the AtMYB41 promoter is active in suberizing tissues under abiotic stress but not unstressed conditions; (iii) augmented root suberization is a recognized response to NaCl and ABA treatment (Reinhardt and Rost, 1995; Karahara et al., 2004; Schreiber et al., 2005a; Efetova et al., 2007; Franke et al., 2009); (iv) it is well-documented that AtMYB41 transcripts accumulate in roots and seedlings in response to various types of abiotic stress (Cominelli et al., 2008; Kosma et al., 2009; Lippold et al., 2009), and (v) stress-induced phosphorylation of AtMYB41 by a mitogen-activated protein kinase (MPK6) is required for the salt-tolerant phenotypes imparted by AtMYB41 overexpression (Hoang et al., 2012). Whether or not AtMYB41 acts via direct or indirect activation of suberin genes is not yet known. However, the coordination of multiple activities required for the deposition of such a lamellar structure (i.e. lipid and phenylpropanoid metabolism, transport to the cell wall, assembly, etc.) is reminiscent of the level of control exhibited by the transcriptional networks defined for secondary cell wall synthesis and seed maturation (Santos-Mendoza et al., 2008; Zhong et al., 2010). Specific MYB and NAC transcription factors act as master switches that regulate a downstream cascade of transcription factors more directly involved in activating the synthesis of lignin, cellulose, and hemicellulose components of secondary cell walls. Similarly, LEC2 (more globally involved in seed maturation processes) regulates seed oil biosynthesis by targeting WRI1, which is a direct regulator of fatty acid synthesis genes.
Here we have demonstrated that AtMYB41 is capable of activating the synthesis and deposition of suberin-like lamellae. While the exact biological function remains unclear, evidence suggests that this transcription factor plays a role in augmenting aliphatic suberization under conditions of abiotic stress. The demonstration that AtMYB41 can stimulate the ectopic production and deposition of suberin-type material provides a tool for generating plants with altered barrier properties. It also opens possibilities for the renewable production of bifunctional fatty acids in plants. Bifunctional fatty acids have potential as renewable chemical feedstocks for manufacturing bioplastics and other specialty chemicals normally synthesized from petrochemical products (Gandini, 2008).
Experimental Procedures
Plant materials
Arabidopsis thaliana Columbia-0 (Col-0) seeds were stratified at 4°C for 2–3 days and grown in a mixture of Promix PGX soil-less media (Premier Horticulture, http://www.pthorticulture.com/) and calcined clay granules (1:1, v/v; Profile Greens Grade). Nicotiana benthamiana seeds were grown in Promix MPV potting mixture (Premier Horticulture). All plants were grown in a growth chamber at 22°C, 40–60% humidity, a 16-h/8-h light/dark cycle, and a fluorescent light intensity of 100 (Arabidopsis) or 150 (N. benthamiana) μE m−2 sec−1.
Plasmid construction for transient overexpression
The AtMYB41 cDNA clone (PYAT4G28110) in the Gateway pENTR#x00AE;/D-TOPO#x00AE; vector was obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/). The AtMYB41 open reading frame was then recombined into the Gateway plant binary vector pMDC32 (Curtis and Grossniklaus, 2003). The 35S::AtMYB41 plasmid was transformed into Rhizobium radiobacter (Agrobacterium tumifaciens; GV3101::pMP90). Five-week-old N. benthamiana plants were used for all leaf infiltrations as previously described (Sparkes et al., 2006). Leaves were harvested for lipid and microscopic analyses 6 days after infiltration. pBIN19- or pEAQ (empty vector)-harboring R. radiobacter cultures were used for negative control plant infiltrations (Peyret and Lomonossoff, 2013).
Microscopy
For TEM analysis, plant tissue samples were prepared and analyzed as described by Molina et al. (2009). A modified staining procedure was used to enhance the contrast of suberin lamellae (Heumann, 1990). Images were processed with Adobe Photoshop CS6 (http://www.adobe.com/).
Quantitative (q)RT-PCR
Total RNA was isolated from 4-week-old Arabidopsis rosette leaves. RNA extraction, cDNA synthesis, and qRT-PCR were carried out as previously described (Vishwanath et al., 2013). The qRT-PCR reactions were performed with three biological replicates, each with three technical replicates. Transcripts were quantified using the comparative CT (ΔΔCT) method, and normalized using three endogenous control genes, eIF4A-1 (At3g13920), GAPC (At3g04120), and UBIQUITIN5 (At3g62250). The primers used for qRT-PCR are listed in Table S2.
Lipid analyses
Waxes were extracted from leaf materials by immersion in chloroform for 30 sec and derivatized and analyzed as previously described (Kosma et al., 2012). For polyester analysis, base-catalyzed depolymerization and GC-MS analysis were performed on ground delipidated leaves as previously described (Molina et al., 2006).
Lignin analysis
Lyophilized samples were ground into a fine powder with a ball mill and treated with solvents to remove pigments, proteins, lipids, and DNA from the material to create alcohol insoluble residue (AIR). Starch was removed from the residue with an amylase treatment. Thioacidolysis reactions were based on methodology described by Robinson and Mansfield (2009). Bisphenol E was used as an internal standard for quantification.
Promoter GUS construct design and analysis of GUS-expressing plants
To generate the AtMYB41promoter::GUS construct, the 1.35-kb 5′ upstream sequence of the AtMYB41 gene was amplified by PCR using genomic DNA as the template, cloned into pGEM#x00AE;-T Easy vector (Promega Corporation, http://www.promega.com/), and subcloned as a SalI–BamHI fragment into binary vector pBI101. The binary construct was introduced into R. radiobacter (GV3101::pMP90) for Arabidopsis (Col-0) transformation. Arabidopsis (Col-0) plants were transformed by floral dipping (Clough and Bent, 1998). GUS expression was analyzed as previously described (Domergue et al., 2010).
Acknowledgments
The authors would like to acknowledge Professors John Ohlrogge and Mike Pollard (Michigan State University) for space, material, guidance, and critical reading of the manuscript. The authors acknowledge Professor Chiara Tonelli and Eleonora Cominelli (Università degli Studi di Milano, Milano, Italy) for kindly donating AtMYB41 OE-9 seeds and Professor Wolfgang Werr (Universität zu Köln, Köln, Germany) for generously providing the pRT-Ω/Not1/Asc1 and pGPTV-KAN-Asc plasmids. The authors also extend thanks to Alicia Withrow from the Michigan State University Center for Advanced Microscopy for sample preparation and assistance with TEM analyses, Cliff Foster of the Great Lakes Bioenergy Research Center (GLBRC) Cell Wall Analysis Facility for assistance with lignin analyses, Adam Rice (Michigan State University) for assistance with plant care and lipid analyses, and Linda Danhof of the GLBRC Arabidopsis Service Center for plant transformation. Funding for this research was provided by the National Science Foundation grant MCB-0615563, the Great Lakes Bioenergy Research Center under Department of Energy contract DE-AC02-05CH11231, and by the Natural Sciences and Engineering Research Council of Canada.
Author Contributions
D.K.K, I.M., and O.R. designed the research; D.K.K., J.M., F.M.R., P.S., and R.B. performed the research; D.K.K., J.M., F.M.R., I.M., and O.R. analyzed the data; and D.K.K., J.M., I.M., and O.R. wrote the paper.
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Figure S1. Growth phenotype of AtMYB41 OE-9 plants.
Figure S2. Toluidine blue (TB) staining of AtMYB41 OE-9 leaves.
Figure S3. Scanning electron microscope images of wild-type (WT) and AtMYB41 OE-9 leaf surfaces.
Figure S4. Transmission electron microscope images of rosette leaf sections from AtMYB41 OE-9 plants.
Figure S5. Transmission electron microscope images of suberin lamellae in the cell wall of root endodermal and peridermal cells of wild-type (WT) Nicotiana benthamiana and WT (Col-0) root peridermal cells of Arabidopsis.
Figure S6. Root and seed suberin monomer compositions of AtMYB41 OE-9 and wild-type (WT) (Col-0) plants.
Figure S7. Amounts of leaf polyester, growth phenotypes, and leaf polyester compositions from independently generated AtMYB41 overexpression lines (OE).
Figure S8. Transient expression of AtMYB41 in Nicotiana benthamiana leads to the accumulation of lamellar structures deposited in the cell walls of mesophyll and epidermal cells 6 days after infiltration.
Figure S9. Transient expression of candidate MYB transcription factors AtMYB45 (At3g48920) and AtMYB67 (At3g12720) did not result in the ectopic accumulation of suberin monomers in N. benthamiana leaves and infiltration did not lead to wound-induced suberin production.
Figure S10. Transient expression of AtMYB41 leads to the accumulation of suberin-associated wax-like compounds in Nicotiana benthamiana leaf waxes.
Figure S11. Semi-quantitative RT-PCR analysis of suberin gene transcripts using cDNA prepared from leaves of wild-type (WT) and AtMYB41 OE-9 plants.
Figure S12. Transient expression of AtMYB41 results in elevated levels of lignin in Nicotiana benthamiana leaves.
Figure S13. Electron-impact (EI) mass spectrum of a trimethylsilyl-derivatized dimethyl diferulate released by sodium methoxide-catalyzed transmethylation of delipidated leaf tissue of AtMYB41 OE-9 Arabidopsis or AtMYB41-expressing Nicotiana benthamiana.
Table S1. Co-expression analysis of candidate suberin transcription factors.
Table S2. List of genes, loci, and primers used for quantitative RT-PCR and cloning.
Appendix S1. Materials and methods.
Appendix S2. References.
References
- Almeida T, Menendez E, Capote T, Ribeiro T, Santos C, Goncalves S. Molecular characterization of quercus suber MYB1, a transcription factor up-regulated in cork tissues. J. Plant Physiol. 2013;170:172–178. doi: 10.1016/j.jplph.2012.08.023. [DOI] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell. 2007;19:351–368. doi: 10.1105/tpc.106.048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M. Solving the puzzles of cutin and suberin polymer biosynthesis. Curr. Opin. Plant Biol. 2012;15:329–337. doi: 10.1016/j.pbi.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Bernards MA. Demystifying suberin. Can. J. Bot. 2002;80:227–240. [Google Scholar]
- Bernards MA, Razem FA. The poly(phenolic) domain of potato suberin: a non-lignin cell wall bio-polymer. Phytochemistry. 2001;57:1115–1122. doi: 10.1016/s0031-9422(01)00046-2. [DOI] [PubMed] [Google Scholar]
- Bernards MA, Lopez ML, Zajicek J, Lewis NG. Hydroxycinnamic acid-derived polymers constitute the polyaromatic domain of suberin. J. Biol. Chem. 1995;270:7382–7386. doi: 10.1074/jbc.270.13.7382. [DOI] [PubMed] [Google Scholar]
- Bernards MA, Summerhurst DK, Razem FA. Oxidases, peroxidases and hydrogen peroxide: the suberin connection. Phytochem. Rev. 2004;3:113–126. [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C. Over-expression of the Arabidopsis ATMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 2008;53:53–64. doi: 10.1111/j.1365-313X.2007.03310.x. [DOI] [PubMed] [Google Scholar]
- Compagnon V, Diehl P, Benveniste I, Meyer D, Schaller H, Schreiber L, Franke R, Pinot F. CYP86B1 is required for very long chain ω-hydroxyacid and α, ω -dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol. 2009;150:1831–1843. doi: 10.1104/pp.109.141408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Schwinn KE. Transcriptional regulation of secondary metabolism. Funct. Plant Biol. 2003;30:913–925. doi: 10.1071/FP03062. [DOI] [PubMed] [Google Scholar]
- Do CT, Pollet B, Thevenin J, Sibout R, Denoue D, Barriere Y, Lapierre C, Jouanin L. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta. 2007;226:1117–1129. doi: 10.1007/s00425-007-0558-3. [DOI] [PubMed] [Google Scholar]
- Domergue F, Vishwanath SJ, Joubes J, et al. Three Arabidopsis fatty acyl-Coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 2010;153:1539–1554. doi: 10.1104/pp.110.158238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Efetova M, Zeier J, Riederer M, Lee C-W, Stingl N, Mueller M, Hartung W, Hedrich R, Deeken R. A central role of abscisic acid in drought stress protection of agrobacterium-induced tumors on Arabidopsis. Plant Physiol. 2007;145:853–862. doi: 10.1104/pp.107.104851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enstone DE, Peterson CA, Ma FS. Root endodermis and exodermis: structure, function, and responses to the environment. J. Plant Growth Regul. 2002;21:335–351. [Google Scholar]
- Espelie KE, Sadek NZ, Kolattukudy PE. Composition of suberin-associated waxes from the subterranean storage organs of seven plants, parsnip, carrot, rutabaga, turnip, red beet, sweet potato and potato. Planta. 1980;148:468–476. doi: 10.1007/BF00552662. [DOI] [PubMed] [Google Scholar]
- Franke R, Schreiber L. Suberin - a biopolyester forming apoplastic plant interfaces. Curr. Opin. Plant Biol. 2007;10:252–259. doi: 10.1016/j.pbi.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L. Apoplastic polyesters in Arabidopsis surface tissues – a typical suberin and a particular cutin. Phytochemistry. 2005;66:2643–2658. doi: 10.1016/j.phytochem.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Franke R, Höfer R, Briesen I, Emsermann M, Efremova N, Yephremov A, Schreiber L. The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J. 2009;57:80–95. doi: 10.1111/j.1365-313X.2008.03674.x. [DOI] [PubMed] [Google Scholar]
- Franke RB, Dombrink I, Schreiber L. Suberin goes genomics: use of a short living plant to investigate a long lasting polymer. Front. Plant Sci. 2012;3:4. doi: 10.3389/fpls.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Chapple C. The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book. 2011;9:e0152. doi: 10.1199/tab.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandini A. Polymers from renewable resources: a challenge for the future of macromolecular materials. Macromolecules. 2008;41:9491–9504. [Google Scholar]
- Geldner N. The endodermis. Annu. Rev. Plant Biol. 2013;64:531–558. doi: 10.1146/annurev-arplant-050312-120050. [DOI] [PubMed] [Google Scholar]
- Gou JY, Yu XH, Liu CJ. A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis. Proc. Natl Acad. Sci. USA. 2009;106:18855–18860. doi: 10.1073/pnas.0905555106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graça J, Santos S. Glycerol-derived ester oligomers from cork suberin. Chem. Phys. Lipids. 2006;144:96–107. doi: 10.1016/j.chemphyslip.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Heumann HG. A simple method for improved visualization of the lamellated structure of cutinized and suberized plant cell walls by electron microscopy. Stain Technol. 1990;65:183–187. doi: 10.3109/10520299009108068. [DOI] [PubMed] [Google Scholar]
- Hoang MH, Nguyen XC, Lee K, Kwon YS, Pham HT, Park HC, Yun DJ, Lim CO, Chung WS. Phosphorylation by ATMPK6 is required for the biological function of AtMYB41 in Arabidopsis. Biochem. Biophys. Res. Commun. 2012;422:181–186. doi: 10.1016/j.bbrc.2012.04.137. [DOI] [PubMed] [Google Scholar]
- Höfer R, Briesen I, Beck M, Pinot F, Schreiber L, Franke R. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. J. Exp. Bot. 2008;59:2347–2360. doi: 10.1093/jxb/ern101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway PJ. Composition of suberin from corks of Quercus suber L. and Betula pendula Roth. Chem. Phys. Lipids. 1972;9:158–170. [Google Scholar]
- Hooker TS, Lam P, Zheng H, Kunst L. A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell. 2007;19:904–913. doi: 10.1105/tpc.106.049304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl Acad. Sci. USA. 2013;110:14498–14503. doi: 10.1073/pnas.1308412110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys JM, Chapple C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 2002;5:224–229. doi: 10.1016/s1369-5266(02)00257-1. [DOI] [PubMed] [Google Scholar]
- Ibdah M, Zhang XH, Schmidt J, Vogt T. A novel Mg2+-dependent O-methyltransferase in the phenylpropanoid metabolism of Mesembryanthemum crystallinum. J. Biol. Chem. 2003;278:43961–43972. doi: 10.1074/jbc.M304932200. [DOI] [PubMed] [Google Scholar]
- Karahara I, Ikeda A, Kondo T, Uetake Y. Development of the casparian strip in primary roots of maize under salt stress. Planta. 2004;219:41–47. doi: 10.1007/s00425-004-1208-7. [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE. Polyesters in higher plants. Adv. Biochem. Eng. Biotechnol. 2001;71:1–49. doi: 10.1007/3-540-40021-4_1. [DOI] [PubMed] [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lu S, Joubes J, Jenks MA. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 2009;151:1918–1929. doi: 10.1104/pp.109.141911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Molina I, Ohlrogge JB, Pollard M. Identification of an Arabidopsis fatty alcohol:caffeoyl-Coenzyme A acyltransferase required for the synthesis of alkyl hydroxycinnamates in root waxes. Plant Physiol. 2012;160:237–248. doi: 10.1104/pp.112.201822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Zhao LF, McFarlane HE, Aiga M, Lam V, Hooker TS, Kunst L. RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol. 2012;159:1385–1395. doi: 10.1104/pp.112.199646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasserre E, Jobet E, Llauro C, Delseny M. AtERF38At2g35700), an AP2/ERF family transcription factor gene from Arabidopsis thaliana, is expressed in specific cell types of roots, stems and seeds that undergo suberization. Plant Physiol. Biochem. 2008;46:1051–1061. doi: 10.1016/j.plaphy.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Lee S-B, Jung S-J, Go Y-S, Kim H-U, Kim J-K, Cho H-J, Park OK, Suh M-C. Two Arabidopsis 3-ketoacyl coa synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 2009;60:462–475. doi: 10.1111/j.1365-313X.2009.03973.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Ohlrogge J, Pollard M. Monoacylglycerols are components of root waxes and can be produced in the aerial cuticle by ectopic expression of a suberin-associated acyltransferase. Plant Physiol. 2007a;144:1267–1277. doi: 10.1104/pp.107.099432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge J. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc. Natl Acad. Sci. USA. 2007b;104:18339–18344. doi: 10.1073/pnas.0706984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, et al. Acyl-lipid metabolism. Arabidopsis Book. 2013;11:e0161. doi: 10.1199/tab.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK, Udvardi MK. AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 2009;149:1761–1772. doi: 10.1104/pp.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü S, Zhao H, Des Marais DL, et al. Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 2012;159:930–944. doi: 10.1104/pp.112.198697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Peterson CA. Curent insights into the development, structure, and chemistry of the endodermis and exodermis of roots. Can. J. Bot. 2003;81:405–421. [Google Scholar]
- Martin C, Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Molina I, Bonaventure G, Ohlrogge J, Pollard M. The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry. 2006;67:2597–2610. doi: 10.1016/j.phytochem.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Molina I, Ohlrogge JB, Pollard M. Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J. 2008;53:437–449. doi: 10.1111/j.1365-313X.2007.03348.x. [DOI] [PubMed] [Google Scholar]
- Molina I, Li-Beisson Y, Beisson F, Ohlrogge JB, Pollard M. Identification of an Arabidopsis feruloyl-Coenzyme A transferase required for suberin synthesis. Plant Physiol. 2009;151:1317–1328. doi: 10.1104/pp.109.144907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc. Natl Acad. Sci. USA. 2012;109:10101–10106. doi: 10.1073/pnas.1205726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Schreiber L, Franke RB, Geldner N, Reina-Pinto JJ, Kunst L. Apoplastic diffusion barriers in Arabidopsis. Arabidopsis Book. 2013;11:e0167. doi: 10.1199/tab.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dunand C. Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 2004;9:534–540. doi: 10.1016/j.tplants.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Peyret H, Lomonossoff G. The pEAQ vector series: the easy and quick way to produce recombinant proteins in plants. Plant Mol. Biol. 2013;83:51–58. doi: 10.1007/s11103-013-0036-1. [DOI] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci. 2008;13:236–246. doi: 10.1016/j.tplants.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Ranathunge K, Schreiber L, Franke R. Suberin research in the genomics era-new interest for an old polymer. Plant Sci. 2011;180:399–413. doi: 10.1016/j.plantsci.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Razem FA, Bernards MA. Hydrogen peroxide is required for poly(phenolic) domain formation during wound-induced suberization. J. Agric. Food Chem. 2002;50:1009–1015. doi: 10.1021/jf0110248. [DOI] [PubMed] [Google Scholar]
- Reinhardt DH, Rost TL. Salinity accelerates endodermal development and induces an exodermis in cotton seedling roots. Environ. Exp. Bot. 1995;35:563–574. [Google Scholar]
- Robinson AR, Mansfield SD. Rapid analysis of poplar lignin monomer composition by a streamlined thioacidolysis procedure and near-infrared reflectance-based prediction modeling. Plant J. 2009;58:706–714. doi: 10.1111/j.1365-313X.2009.03808.x. [DOI] [PubMed] [Google Scholar]
- Roppolo D, De Rybel B, Tendon VD, Pfister A, Alassimone J, Vermeer JEM, Yamazaki M, Stierhof YD, Beeckman T, Geldner N. A novel protein family mediates Casparian strip formation in the endodermis. Nature. 2011;473:380–383. doi: 10.1038/nature10070. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Chapple C. Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics. 2001;159:1741–1749. doi: 10.1093/genetics/159.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser U, Holloway PJ. Ultrastructure and chemistry of soluble and polymeric lipids in cell walls from seed coats and fibres of Gossypium species. Planta. 1985;163:151–163. doi: 10.1007/BF00393501. [DOI] [PubMed] [Google Scholar]
- Santos S, Graça J. Glycerol-ω-hydroxyacid-ferulic acid oligomers in cork suberin structure. Holzforschung. 2006;60:171–177. [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008;54:608–620. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C. Mutations in the CINNAMATE 4-HYDROXYLASE gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009;60:771–782. doi: 10.1111/j.1365-313X.2009.03996.x. [DOI] [PubMed] [Google Scholar]
- Schreiber L. Transport barriers made of cutin, suberin and associated waxes. Trends Plant Sci. 2010;15:546–553. doi: 10.1016/j.tplants.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Schreiber L, Franke R, Hartmann K. Effects of NO3 deficiency and NaCl stress on suberin deposition in rhizo- and hypodermal (rhcw) and endodermal cell walls (ecw) of castor bean (Ricinus communis L.) roots. Plant Soil. 2005a;269:333–339. [Google Scholar]
- Schreiber L, Franke R, Hartmann K. Wax and suberin development of native and wound periderm of potato (Solanum tuberosum L.) and its relation to peridermal transpiration. Planta. 2005b;220:520–530. doi: 10.1007/s00425-004-1364-9. [DOI] [PubMed] [Google Scholar]
- Serra O, Soler M, Hohn C, Franke R, Schreiber L, Prat S, Molinas M, Figueras M. Silencing of StKCS6 in potato periderm leads to reduced chain lengths of suberin and wax compounds and increased peridermal transpiration. J. Exp. Bot. 2009a;60:697–707. doi: 10.1093/jxb/ern314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra O, Soler M, Hohn C, Sauveplane V, Pinot F, Franke R, Schreiber L, Prat S, Molinas M, Figueras M. CYP86A33-targeted gene silencing in potato tuber alters suberin composition, distorts suberin lamellae, and impairs the periderm's water barrier function. Plant Physiol. 2009b;149:1050–1060. doi: 10.1104/pp.108.127183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra O, Hohn C, Franke R, Prat S, Molinas M, Figueras M. A feruloyl transferase involved in the biosynthesis of suberin and suberin-associated wax is required for maturation and sealing properties of potato periderm. Plant J. 2010;62:277–290. doi: 10.1111/j.1365-313X.2010.04144.x. [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Pollet B, Goujon T, Mila I, Granier F, Seguin A, Lapierre C, Jouanin L. Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiol. 2003;132:848–860. doi: 10.1104/pp.103.021048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Seguin A. CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell. 2005;17:2059–2076. doi: 10.1105/tpc.105.030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocombe SP, Schauvinhold I, McQuinn RP, Besser K, Welsby NA, Harper A, Aziz N, Li Y, Larson TR, Giovannoni J. Transcriptomic and reverse genetic analysesof branched-chain fatty acid and acyl sugar production in Solanum pennellii and Nicotiana benthamiana. Plant Physiol. 2008;148:1830–1846. doi: 10.1104/pp.108.129510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M, Serra O, Molinas M, Huguet G, Fluch S, Figueras M. A genomic approach to suberin biosynthesis and cork differentiation. Plant Physiol. 2007;144:419–431. doi: 10.1104/pp.106.094227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M, Serra O, Fluch S, Molinas M, Figueras M. A potato skin SSH library yields new candidate genes for suberin biosynthesis and periderm formation. Planta. 2011;233:933–945. doi: 10.1007/s00425-011-1350-y. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Vishwanath SJ, Kosma DK, Pulsifer IP, Scandola S, Pascal S, Joubès J, Dittrich-Domergue F, Lessire R, Rowland O, Domergue F. Suberin-associated fatty alcohols in Arabidopsis thaliana: distributions in roots and contributions to seed coat barrier properties. Plant Physiol. 2013;163:1118–1132. doi: 10.1104/pp.113.224410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Stark RE. Biosynthesis, molecular structure, and domain architecture of potato suberin: a 13C NMR study using isotopically labeled precursors. J. Agric. Food Chem. 2000;48:3298–3304. doi: 10.1021/jf000155q. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ohlrogge JB. Turnover of fatty acids during natural senescence of Arabidopsis, Brachypodium, and switchgrass and in Arabidopsis β-oxidation mutants. Plant Physiol. 2009;150:1981–1989. doi: 10.1104/pp.109.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Pollard M, Li-Beisson Y, Beisson F, Feig M, Ohlrogge J. A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proc. Natl Acad. Sci. USA. 2010;107:12040–12045. doi: 10.1073/pnas.0914149107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC. The formation and function of plant cuticles. Plant Physiol. 2013;163:5–20. doi: 10.1104/pp.113.222737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RE, McFarlane HE, Hahn MG, Western TL, Haughn GW, Samuels AL. Analysis of the golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell. 2008;20:1623–1638. doi: 10.1105/tpc.108.058842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye Z-H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010;15:625–632. doi: 10.1016/j.tplants.2010.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Growth phenotype of AtMYB41 OE-9 plants.
Figure S2. Toluidine blue (TB) staining of AtMYB41 OE-9 leaves.
Figure S3. Scanning electron microscope images of wild-type (WT) and AtMYB41 OE-9 leaf surfaces.
Figure S4. Transmission electron microscope images of rosette leaf sections from AtMYB41 OE-9 plants.
Figure S5. Transmission electron microscope images of suberin lamellae in the cell wall of root endodermal and peridermal cells of wild-type (WT) Nicotiana benthamiana and WT (Col-0) root peridermal cells of Arabidopsis.
Figure S6. Root and seed suberin monomer compositions of AtMYB41 OE-9 and wild-type (WT) (Col-0) plants.
Figure S7. Amounts of leaf polyester, growth phenotypes, and leaf polyester compositions from independently generated AtMYB41 overexpression lines (OE).
Figure S8. Transient expression of AtMYB41 in Nicotiana benthamiana leads to the accumulation of lamellar structures deposited in the cell walls of mesophyll and epidermal cells 6 days after infiltration.
Figure S9. Transient expression of candidate MYB transcription factors AtMYB45 (At3g48920) and AtMYB67 (At3g12720) did not result in the ectopic accumulation of suberin monomers in N. benthamiana leaves and infiltration did not lead to wound-induced suberin production.
Figure S10. Transient expression of AtMYB41 leads to the accumulation of suberin-associated wax-like compounds in Nicotiana benthamiana leaf waxes.
Figure S11. Semi-quantitative RT-PCR analysis of suberin gene transcripts using cDNA prepared from leaves of wild-type (WT) and AtMYB41 OE-9 plants.
Figure S12. Transient expression of AtMYB41 results in elevated levels of lignin in Nicotiana benthamiana leaves.
Figure S13. Electron-impact (EI) mass spectrum of a trimethylsilyl-derivatized dimethyl diferulate released by sodium methoxide-catalyzed transmethylation of delipidated leaf tissue of AtMYB41 OE-9 Arabidopsis or AtMYB41-expressing Nicotiana benthamiana.
Table S1. Co-expression analysis of candidate suberin transcription factors.
Table S2. List of genes, loci, and primers used for quantitative RT-PCR and cloning.
Appendix S1. Materials and methods.
Appendix S2. References.