Abstract
Brain-derived neurotrophic factor (BDNF) is involved in the survival, development, and synaptic plasticity of neurons. BDNF is believed to be associated with the pathophysiology of psychiatric disorders. Several studies have suggested the relevance of DNA methylation in its promoter region with depression. Here, we report different methylation statuses in groups with different depressive scores or undergoing different levels of job-stress. DNA samples were extracted from the saliva of 774 Japanese workers, and the methylation status was determined using the Illumina HumanMethylation 450 K Microarray. Depressive symptoms were measured using the Kessler's K6 questionnaire. Job-stress scales were assessed via a self-administered questionnaire. Independent DNA pools were formed based on K6 and job-strain scores, and the methylation levels were compared among these pools. The average DNA methylation rate was significantly decreased in the highest K6 score group compared to the lowest group (methylated signals, 14.2% vs. 16.5%, P = 2 · 16 × 10−198). This difference remained for the CpG island in the promoter region (10.4% vs. 5.8%, P = 3 · 67 × 10−133). Regarding the job-strain score, there was a slight increase in the methylation level of the whole gene in the group with the highest score compared to that with the lowest score; however, these groups showed no difference in the promoter region. Our results revealed significant changes in the DNA methylation status of the complete human BDNF gene in persons with depression compared to normal individuals, especially in the promoter region of exon 1. This indicates that DNA methylation in this gene is a promising biomarker for diagnosing depression. © 2014 The Authors. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics Published by Wiley Periodicals, Inc.
Keywords: BDNF, epigenetic, K6 score, HumanMethylation 450K Microarray
INTRODUCTION
Brain-derived neurotrophic factor (BDNF) is a member of the neurotrophin family of growth factors and, has been considered to be an activity-dependent modulator of neuronal structure and function in the adult brain [Lu et al., 2008; Bibel and Barde, 2000]. BDNF promotes the proliferation, survival, and differentiation of neurons in the peripheral and central nervous systems [Lindsay et al., 1994; Acheson et al., 1995; Huang and Reichardt, 2001]. Its effects on synaptic plasticity in the brain have been described, and its signaling in the hippocampus is crucial for learning and memory [Tyler et al., 2002; Yamada and Nabeshima, 2003; Bekinschtein et al., 2007]. Cumulative evidence has demonstrated possible relations between BDNF and psychiatric disorders, such as depression [Brunoni et al., 2008; Sen et al., 2008], bipolar disorder [Grande et al., 2010], and dementia [Arancio and Chao, 2007]. Shimizu et al. suggested that decreased BDNF levels play a pivotal role in the pathophysiology of major depressive disorders [Shimizu et al., 2003].
The human BDNF gene is located on chromosome 11p14.1, spans about 70 kb [Liu et al., 2005], and contains 11 exons (Fig. 1). The last exon (exon 9) is the main protein-coding exon, and several distinct transcript classes are formed when transcription is initiated at various other exons [Pruunsild et al., 2007]. Variants of this gene are a priori candidates for involvement in memory and hippocampal function [Egan et al., 2003], as well as, in genetic predispositions to anxiety and depressive disorders [Chen et al., 2006].
Figure 1.
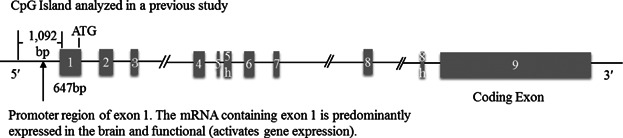
Schematic representation of human BDNF gene structure and position of the CpG-site-rich region that was analyzed in this study. The promoter of exon 1 was chosen for analysis because the first exon is generally considered as the transcription-relative region, and the promoter of the exon 1 of the BDNF gene has been described as a brain-specific inducible promoter. The target region of the current analyses is the same CpG island as that used in a previous study (Chr11: 27743473–27744564), with a length of 1,092 bp and including of 10 CpG sites. Exons are shown as boxes and the numbers of exons are shown below. Introns are shown as lines. The size of exon 1 was obtained from the database of Ensembl Genome Browser, and that of the CpG island of the exon 1 promoter was identified in a previous study. ATG indicates the position of the translational start codon, which leads to a prepro-BDNF protein.
Recently, epigenetic mechanisms, which exert lasting control over gene expression without altering the gene's nucleotide sequence, have been considered as mediators of stable changes in brain function. DNA methylation of cytosine residues resulting in the formation of 5-methylcytosyine, primarily at cytosine–guanine (CpG) dinucleotide-containing regulatory sequences, is chemically and biologically stable and is regarded as a reliable marker for prediction of relationships with various diseases, including affective disorders [Qureshi and Mehler, 2010]. CpG islands, which are genomic regions containing a high frequency of CpG sites and occur typically at, or near, the transcription start site of genes [Saxonov et al., 2006], attract more attention because they are important regulatory structures that function as genomic platforms for the regulation of transcription at their associated promoters [Deaton and Bird, 2011].
In the human genome, the first exon of genes is usually believed to be important in transcription [Delgado and León, 2006]; however, the BDNF gene is different from other genes because it contains multiple functional promoters that activate the expression of its transcripts [Pruunsild et al., 2007]. The promoter of exon 1 is considered as a brain-specific inducible promoter, and mRNAs containing exon 1 are expressed predominantly in the brain of rats [Timmusk et al., 1993] or humans, with a strong ability to activate gene expression [Pruunsild et al., 2007]. In addition, exon 1 contains an in-frame ATG codon that can be used as translation start site and lead to the prepro-BDNF protein [Pruunsild et al., 2007].
The methylation status of the BDNF gene, especially of its promoter region, represents one of the most remarkable parameters that can be evaluated regarding an association with mental diseases. DNA methylation alterations within promoters of exons are related with depression [Fuchikami et al., 2011; Kim et al., 2013], bipolar disorder [Perroud et al., 2008; D'Addario et al., 2012], and suicidal behavior [Keller et al., 2010]. In addition, epigenetic modifications of the BDNF gene in response to stress, fear memory, and early maltreatment have also been studied in rats [Roth et al., 2009; Fuchikami et al., 2010; Roth et al., 2011] and humans [Lubin et al., 2008; Unternaehrer et al., 2012], although the results were discordant.
In the current study, we examined the methylation levels of the entire BDNF gene, with a special focus on its promoter region, and aimed to detect its potential correlation with depression and stress scales assessed via a self-administered questionnaire in our occupational cohort.
MATERIALS AND METHODS
The present cross-sectional study is part of our occupational-cohort study of social class and health, which is supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan; the details of the cohort study can be found elsewhere [Miyaki et al., 2012]. Employees of a major Japanese manufacturing company, with offices located across the country, were recruited. Among the approximately 2,500 workers who were invited to participate in the study, 976 agreed to provide saliva samples, and, finally, 774 samples were obtained. The protocol and explanatory documents of our study were approved by the ethics committee of the National Center for Global Health and Medicine and that of Tokyo University Graduate School of Medicine. Written informed consent was obtained from each subject.
The participants were asked to provide approximately 2 ml of saliva. Genomic DNA was extracted from leukocytes using the Oragene® DNA Self-Collection Kit, OG-500 Tube Format (DNA Genotek, Ontario, Canada) [Birnboim, 2004]. All DNA samples were diluted to a working concentration of 20 ng/µL with TE buffer (10 mM Tris, 1 mM EDTA).
The 774 subjects were stratified into quartiles according to their depressive scales or job-strain scores. Depressive symptoms were assessed using the Japanese version of the Kessler 6 (K6) scale [Kessler et al., 2002]. K6 score, job demands, and job control were assessed using a self-administered questionnaire [Kessler et al., 2003]. The job strain score was defined as: job demands/job control × 2.
An independent pool was formed using 90 DNA samples that were selected randomly from each quartile group and consisted of a volume of 1 µL per sample; thus, there were a total of eight pools: four for the K6 score and four for the job-strain scale. This pooling method was validated in a previous study-demonstrating that a DNA pool including 89 individual samples can provide an accurate and reliable quantitative estimate of the average group DNA methylation [Docherty et al., 2009]. In each pool, the total DNA volume was 90 µL and the total amount was 1.8 µg. Genome-wide DNA methylation profiling was performed using Illumina Infinium HumanMethylation450 BeadChip arrays and kit (Illumina, San Diego, CA). Bisulfite-converted DNA (4 µL) was used for hybridization onto the HumanMethylation450 BeadChip, according to the Illumina Infinium HD Methylation protocol.
An Illumina iScan SQ scanner was used to create images of the single arrays, and the intensities of the images were extracted using the GenomeStudio (v.2011.1) Methylation module (v.1.9.0) software. The methylation score was calculated as a ratio of methylated-to-methylated plus unmethylated signal intensity.
The sequence of the whole BDNF gene was identified using the Ensembl Genome Browser (http://asia.ensembl.org/index.html, chr11:27676440-27743605). We calculated the methylation level of each CpG site covered by HumanMethylation450 BeadChip probes, and the average methylation levels of all merged CpG loci on the BDNF gene in all pools. Furthermore, the merged CpG loci on a CpG island located upstream of the exon 1 (Chr11: 27743473–27744564) region, which was identified by Perroud et al. [2008], were analyzed independently. Subsequently, the methylation levels of groups with the lowest (Q1) and highest (Q4) K6 score or job-strain quartiles were compared using Pearson's chi-squared test. The whole cohort was also divided into two subgroups according to depressive or stress scores: individuals with a K6 score greater than the median value (K6 = 10) were defined as “Depressive,” which corresponds to the total of the Q3 and Q4 groups, whereas those with a score lower than the median value were classified as “Normal,” including the Q1 and Q2 groups and subjects with a job-strain score greater than the median point (job-strain = 1.50) were defined as the “High stress group,” and those with a job strain score lower than 1.50 were classified as “Normal.” The average DNA methylation levels were compared between the Depressive and Normal groups using Pearson's chi-squared test. The IBM SPSS for Windows version 19.0J (IBM, Armonk, NY) software packages was used for all statistical analyses. Statistical significance for all analyses was defined as P < 0.05.
RESULTS
In the current study, a total of 2,664 workers were asked to provide their saliva and finally 774 DNA samples were obtained successfully. The participants were not different from those did not participate the study on age, sex ratio and other clinical characteristics (data not shown). From each K6 group ninety subjects were randomly selected to form the DNA pools, then a total of 360 subjects took part in this study and 32 were women. The basic clinical characteristics, job strain, and socioeconomic status factors (education level, household income, and job position) of these subjects are shown in Table1, and the mean (±standard deviation) values of these indices were compared among job strain groups. There was no significant difference on main indices.
Table I.
Comparison of Clinical Characteristics, Socioeconomic Status Scores, Job Stress, and Lifestyle Factors Among Subjects in Four K6 Groups
| Q1 | Q2 | Q3 | Q4 | P for trend | |
|---|---|---|---|---|---|
| Age (year) | 42.4 ± 9.9 | 43.5 ± 10.4 | 44.7 ± 10.1 | 42.6 ± 9.4 | 0.716 |
| Proportion of women (%) | 7.8 | 6.7 | 12.2 | 8.9 | 0.589 |
| Body mass index (kg/m2) | 23.5 ± 3.2 | 23.4 ± 3.2 | 23.0 ± 3.4 | 22.8 ± 2.6 | 0.143 |
| Years of education (year) | 14.8 ± 2.4 | 15.1 ± 2.5 | 14.6 ± 2.3 | 15.2 ± 2.4 | 0.596 |
| Annual household income (ten thousands yen/year) | 694.6 ± 271.5 | 654.4 ± 237.9 | 776.5 ± 352.9 | 716.9 ± 279.3 | 0.192 |
| Proportion of individuals in a management position (%) | 23.8 | 21.3 | 27.2 | 30.0 | 0.603 |
| Job strain score | 3.7 ± 2.8 | 3.9 ± 3.3 | 3.6 ± 2.9 | 3.4 ± 3.2 | 0.444 |
| Proportion of current smokers (%) | 23.8 | 26.3 | 25.9 | 28.8 | 0.422 |
| Proportion of subjects with habitual drinking (%) | 84.5 | 71.3 | 80.2 | 67.5 | 0.040* |
Values are shown as mean ± standard deviation or percentage. For continuous variables linear regression analysis was used, and for categorized variables Pearson's chi-square test was used to compare indices between the K6 groups.
P < 0.05.
The BDNF gene contains 97 CpG loci that can be detected by the Infinium HumanMethylation450 BeadChip. Of these five were locate in exons; 51, in introns, 21, in the 5′ upstream region; and 20, in the 3′ downstream region. Among the 21 loci located in the 5′ upstream region, 10 (Nos. 85–94 of the 97 loci) were located within the CpG island region on the promoter of exon 1. However, there was no CpG site on the promoter region of exon 4.
The DNA methylation status of each site was examined by calculating the rate of methylated-to-total signal intensity in all pools and was compared between the Q1 and Q4 groups. For K6 score groups, compared to the Q1 group, the Q4 group showed significant hypermethylation at 13 CpG sites, and hypomethylation at 43 CpG sites. Narrowing of the target region to the CpG island located on the promoter of exon 1 revealed that the methylation levels of three loci (Nos. 87, 92, and 94) were significantly decreased and those of two loci (Nos. 88 and 93) were increased in the Q4 group compared to the Q1 group. Locus No. 92 was the most striking locus because only 5.7% of the signals were methylated in the Q4 group, whereas 23.0% of the signals were methylated in the Q1 group (P = 3.63 × 10−300).
As shown in Figure 2, the average methylation level of the 97 loci included in the Q4 group was 14.2%, which was significantly lower than that observed in the Q1 group (16.5%, P = 2.16 × 10−198, chi-squared test) (Fig. 2). On the CpG island located in exon 1, the average methylation level for CpGs No. 85–94 of the Q4 group was only half of that for these CpGs of the Q1 group (5.8% and 10.4%, respectively); the difference between the two groups was significant (P = 3.67 × 10−133, chi-squared test).
Figure 2.
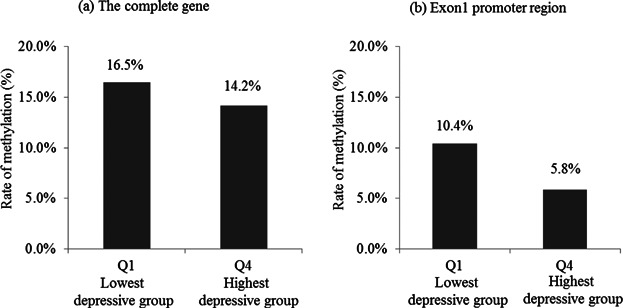
Comparison of the DNA methylation status between the groups with the lowest and highest depression levels in (a) the complete gene and (b) the promoter region of exon 1 of the human BDNF gene. The y-axis represents the rate of methylated signals. The group with the lowest depression level (Q1) had the lowest K6 score quartiles, and the group with the highest depression level (Q4) had the highest K6 quartiles.
Next, the subjects were divided into Depressive and Normal groups and the average DNA methylation levels were compared between them (Table2). Globally, the average methylation levels of the 97 loci in the Depressive and Normal groups were 14.1% and 16.9%, respectively (P < 0.001, chi-squared test). This difference remained significant for the promoter region of exon 1 (5.7% vs.10.8%, P value < 0.001) (Table2).
Table II.
Average DNA Methylation Rates for the CpG Sites on the Human BDNF Gene for the Normal Group and Depressive Groups
| CpG sites included | DNA methylation rate | P value | ||
|---|---|---|---|---|
| Merged from nos. 1–97 (whole gene) | Depressive group | Signal intensity | 65,969 | <0.001a |
| (K6 ≥10) | (%) | 14·1% | ||
| Normal group | Signal intensity | 68,831 | ||
| (K6 <10) | (%) | 16.9% | ||
| Merged from nos. 85–94 (exon 1 promoter) | Depressive group | Signal intensity | 2,531 | <0.001a |
| (K6 ≥10) | (%) | 5.7% | ||
| Normal group | Signal intensity | 4,414 | ||
| (K6 <10) | (%) | 10.8% |
The χ2 test was used to compare the differences in the percentage of methylated signals between the pools of the normal and depressive groups. The participants were divided into two groups at the median point of the K6 score (K6 = 10), and the group with the higher K6 score was defined as the “Depressive group.”
The methylation level was decreased in the depressive group compared with the normal group (P < 0.001).
Regarding the groups that were classified according to job strain, compared to the Q1 group, the Q4 group showed significantly higher methylation levels in 22 CpG sites and lower methylation levels in 28 sites. The average methylation level of all sites in the Q4 group was 14.4%, which was higher than that observed for the Q1 group (14.2%), with borderline significance (P = 0.045). However, the average methylation levels of the 10 sites located in the CpG island of exon 1 in the Q1 and Q4 groups were similar (5.6% and 5.7%, respectively), and only one locus showed a P value < 10−5 (No. 85). Thus, we observed no change in the level of methylation between the high-stress and normal groups, when subjects were stratified according to the median point of job strain (Table3).
Table III.
Average DNA Methylation Rates for the CpG Sites on the Human BDNF Gene for the Normal and High-Stress Groups (Negative Control)
| CpG sites included | DNA methylation rate | P value | ||
|---|---|---|---|---|
| Merged from nos. 1–97 (whole gene) | High-stress group | Signal intensity | 64,417 | 0.984 |
| (job strain ≥1.50) | (%) | 14.0% | ||
| Normal group | Signal intensity | 68,487 | ||
| (job strain <1.50) | (%) | 14.0% | ||
| Merged from nos. 85–94 (exon 1 promoter) | High-stress group | Signal intensity | 2,457 | 0.463 |
| (job strain ≥1.50) | (%) | 5.6% | ||
| Normal group | Signal intensity | 2,697 | ||
| (job strain <1.50) | (%) | 5.7% |
The χ2 test was used to compare the differences in percentage of methylated signals between the pools of the normal and high-stress groups. The participants were divided into two groups at the median point of job strain (job strain = 1.50), and the group with the higher job strain group was defined as the “high-stress group.”
DISCUSSION
In the current study, we investigated the methylation levels of the BDNF gene in groups with different depressive scores. To our knowledge, this is the first study that used the entire BDNF gene as the target region for investigation. We confirmed that the epigenetic alteration not only of the CpG sites within the CpG island located on the promoter region of exon 1, but also of those distributed throughout the gene, exhibit a relationship with depressive symptoms.
K6 is a self-report questionnaire developed by Kessler et al. [2002] and it takes only up to 3 min to complete this short screening questionnaire. The participants are asked to answer six questions about how frequently they experienced symptoms of depression and anxiety during the past 30 days. K6 has been thought as reasonably valid measure of depression among postpartum women and relatively cheap tools for estimating prevalence of postnatal depression [Baggaley et al., 2007]. The screening performance and acceptability of the Japanese versions of K6 has been validated [Sakurai et al., 2011]. Stratum-specific likelihood ratios (SSLRs), which express screening test characteristics and compare the usefulness of different tests were strikingly similar between the Japanese and the original versions [Furukawa et al., 2008].
Due to the difficulty of measuring the BDNF levels in the brain of living persons directly, it is important to develop new approach to determine the BDNF levels in order to explore the physiologic and pathological function of this neurotrophin. Although the source of circulating BDNF and its relationship with brain activity is unknown, it is evident that peripheral BDNF can serve as a marker of anti-depressant therapeutic efficacy [Sen et al., 2008] and peripheral and central BDNF levels are closely related [Karege et al., 2002]. Salivary BDNF was considered to be useful for stress-related research and human clinical investigations [Tirassa et al., 2012], in spite of the patterns of diurnal change of BDNF levels are different in serum and saliva. Previous epigenetic studies have suggested that changes on DNA methylation status are not limited to postmortem brain tissues, but can also be found in other peripheral tissues, including saliva [Rosa et al., 2008; Nohesara et al., 2011]. Saliva collection is considered as a safe and easy alternative to blood samples as a collection method of DNA for high-throughput genotyping [Abraham et al., 2012], in addition, DNA methylation patterns in saliva are relatively consistent with those in whole blood [Thompson et al., 2013].
In a previous study performed by another Japanese group, 11 CpG sites were found to be hypermethylated and 22 sites were hypomethylated in patients with major depression compared with healthy controls [Fuchikami et al., 2011]. Because of the limitation of the Illumina Infinium HumanMethylation450 BeadChip arrays, we were able to detect only four of these 33 sites. We observed a significant decrease in the methylation levels in the Q4 group compared to the Q1 K6 group at site No. 87, which is agreement with the results of Fuchikami et al. [2011], and, found no difference at two sites, nos. 85 and 89. However, site no. 94 yielded an inconsistent result: its methylation rate was decreased in the Q4 group (P = 1.96 × 10−5). Conversely, three sites, nos. 88, 92, and 93, that were not detected by Fuchikami et al. showed notable differences between the two groups, although the trends were opposite. Fuchikami et al. did not provide the average methylation rates of all sites; rather, the authors showed that there were more hypomethylated than hypermethylated sites in the Q4 group compared to the Q1 group, a trend that is consistent with the one detected here. Considering that the findings of this and the study by Fuchikami et al. are concordant on only four sites, our study provides new evidence in support of the importance of changes in DNA methylation. However, additional studies are needed to determine the exact role of epigenetic factors in the pathophysiology of depression.
Numerous studies have shown that reduced BDNF serum concentrations might be associated with depressive disorder [Sen et al., 2008; Satomura et al., 2011; Takebayashi et al., 2012], and that BDNF levels are increased significantly after antidepressant treatment [Brunoni et al., 2008], which indicates that BDNF is a protective factor against depression. Although it is generally believed that DNA methylation in the regulatory regions of genes plays a role in their silencing [Milutinovic et al., 2004], and several studies have reported higher levels of DNA methylation in bipolar disorder [Perroud et al., 2008; D'Addario et al., 2012], schizophrenia [Kordi-Tamandani et al., 2012] or poststroke depression [Kim et al., 2013], little evidence for the association of the methylation status of the exon 1 promoter region and risk of depressive symptoms has been found. Our study showed decreased methylated levels in the depressive group compared to the normal group; however, only 10 of the 81 CpG loci located in this CpG island of exon 1, could be evaluated in the current assay. Moreover, the promoter region of exon 4 could not be evaluated because of the absence of probes for this region. It is imperative to limit our efforts to identify all epigenetic alterations in this region and provide a general DNA methylation profile of the BDNF gene promoter region.
However, we detected several individual loci with significantly different methylation levels between the depressive and normal group, including loci nos. 87, 88, and 92–94. We think that these sites may be used as objective biomarkers for the diagnosing depression and assessing therapeutic interventions for depression. A previous study [Fuchikami et al., 2011] investigated all promoter CpG sites; however, their sample size was relatively small. Our observations are based on the screening data of four pooled samples comprising samples of 360 individuals; however, direct measurement of the DNA methylation status of these subjects should be performed in the future. The next step of our research group is to investigate whether the detected sites are associated with mental health in an independent, large-scale cohort.
Consistent with the results reported by Unternaehrer et al. [2012], our findings fail to reveal a significant difference in the DNA methylation levels of the promoter region among groups with different job-strain scores; however, a weak relationship was found for the whole gene. This result should be confirmed in further studies. Finally, CpG site No.85 can be considered as a potential marker for response to stress.
In summary, the present study provided new evidence for association between the DNA methylation status of the entire human BDNF gene and depressive symptoms. In particular, two-fold difference of methylation level was detected for the CpG island of the exon 1 promoter region between depressive and normal groups; this suggests that these loci are clinically significant and can be used as biomarkers for diagnosing depression and assessing antidepressant treatments.
Acknowledgments
The present study was supported by a Grant-in-Aid for Scientific Research (B) (No. 24390160) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (Chief: Dr. Koichi Miyaki), and by a Grant-in-Aid for Scientific Research on Innovative Areas (Research in a Proposed Research Area) (No. 21119002) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (Chief: Dr. Norito Kawakami).
REFERENCES
- Abraham JE, Maranian MJ, Spiteri I, Russell R, Ingle S, Luccarini C, Earl H, Pharoah P, Dunning A, Caldas C. Saliva samples are a viable alternative to blood samples as a source of DNA for high throughput genotyping. BMC Med Genomics. 2012;5:19. doi: 10.1186/1755-8794-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson A, Conover JC, Fandl JP, DeChiara TM, Russell M, Thadani A, Squinto SP, Yancopoulos GD, Lindsay RM. A BDNF autocrine loop in adult sensory neurons prevents cell death. Nature. 1995;374:450–453. doi: 10.1038/374450a0. [DOI] [PubMed] [Google Scholar]
- Arancio O, Chao MV. Neurotrophins, synaptic plasticity and dementia. Curr Opin Neurobiol. 2007;17:325–330. doi: 10.1016/j.conb.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Baggaley RF, Ganaba R, Filippi V, Kere M, Marshall T, Sombié I, Storeng KT, Patel V. Detecting depression after pregnancy: The validity of the K10 and K6 in Burkina Faso. Trop Med Int Health. 2007;12:1225–1229. doi: 10.1111/j.1365-3156.2007.01906.x. [DOI] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, Medina JH. Persistence of long-term memory storage requires a late protein synthesis- and BDNF-dependent phase in the hippocampus. Neuron. 2007;53:261–277. doi: 10.1016/j.neuron.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Bibel M, Barde YA. Neurotrophins: Key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Birnboim HC. 2004. DNA yield with Oragene™. Ottawa: DNA Genotek, Inc; [cited 2013 Aug 13]. Available from http://news.bio-medicine.org/?q=biology-technology/dna-yield-with-oragene-1232.
- Brunoni AR, Lopes M, Fregni F. A systematic review and meta-analysis of clinical studies on major depression and BDNF levels: Implications for the role of neuroplasticity in depression. Int J Neuropsychopharmacol. 2008;11:1169–1180. doi: 10.1017/S1461145708009309. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, Herrera DG, Toth M, Yang C, McEwen BS, Hempstead BL, Lee FS. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Addario C, Dell'Osso B, Palazzo MC, Benatti B, Lietti L, Cattaneo E, Galimberti D, Fenoglio C, Cortini F, Scarpini E, Arosio B, Di Francesco A, Di Benedetto M, Romualdi P, Candeletti S, Mari D, Bergamaschini L, Bresolin N, Maccarrone M, Altamura AC. Selective DNA methylation of BDNF promoter in bipolar disorder: Differences among patients with BDI and BDII. Neuropsychopharmacology. 2012;37:1647–1655. doi: 10.1038/npp.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado MD, León J. Gene expression regulation and cancer. Clin Transl Oncol. 2006;8:780–787. doi: 10.1007/s12094-006-0132-7. [DOI] [PubMed] [Google Scholar]
- Docherty SJ, Davis OS, Haworth CM, Plomin R, Mill J. Bisulfite-based epityping on pooled genomic DNA provides an accurate estimate of average group DNA methylation. Epigenetics Chromatin. 2009;2:3. doi: 10.1186/1756-8935-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Fuchikami M, Yamamoto S, Morinobu S, Takei S, Yamawaki S. Epigenetic regulation of BDNF gene in response to stress. Psychiatry Investig. 2010;7:251–256. doi: 10.4306/pi.2010.7.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchikami M, Morinobu S, Segawa M, Okamoto Y, Yamawaki S, Ozaki N, Inoue T, Kusumi I, Koyama T, Tsuchiyama K, Terao T. DNA methylation profiles of the brain-derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression. PLoS ONE. 2011;6:e23881. doi: 10.1371/journal.pone.0023881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa TA, Kawakami N, Saitoh M, Ono Y, Nakane Y, Nakamura Y, Tachimori H, Iwata N, Uda H, Nakane H, Watanabe M, Naganuma Y, Hata Y, Kobayashi M, Miyake Y, Takeshima T, Kikkawa T. The performance of the Japanese version of the K6 and K10 in the World Mental Health Survey Japan. Int J Methods Psychiatr Res. 2008;17:152–158. doi: 10.1002/mpr.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande I, Fries GR, Kunz M, Kapczinski F. The role of BDNF as a mediator of neuroplasticity in bipolar disorder. Psychiatry Investig. 2010;7:243–250. doi: 10.4306/pi.2010.7.4.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: Roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karege F, Schwald M, Cisse M. Postnatal developmental profile of brain-derived neurotrophic factor in rat brain and platelets. Neurosci Lett. 2002;328:261–264. doi: 10.1016/s0304-3940(02)00529-3. [DOI] [PubMed] [Google Scholar]
- Keller S, Sarchiapone M, Zarrilli F, Videtic A, Ferraro A, Carli V, Sacchetti S, Lembo F, Angiolillo A, Jovanovic N, Pisanti F, Tomaiuolo R, Monticelli A, Balazic J, Roy A, Marusic A, Cocozza S, Fusco A, Bruni CB, Castaldo G, Chiariotti L. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Andrews G, Colpe LJ, Hiripi E, Mroczek DK, Normand SL, Walters EE, Zaslavsky AM. Short screening scales to monitor population prevalences and trends in non-specific psychological distress. Psychol Med. 2002;32:959–976. doi: 10.1017/s0033291702006074. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Barker PR, Colpe LJ, Epstein JF, Gfroerer JC, Hiripi E, Howes MJ, Normand SL, Manderscheid RW, Walters EE, Zaslavsky AM. Screening for serious mental illness in the general population. Arch Gen Psychiatry. 2003;60:184–189. doi: 10.1001/archpsyc.60.2.184. [DOI] [PubMed] [Google Scholar]
- Kim JM, Stewart R, Kang HJ, Kim SY, Kim SW, Shin IS, Park MS, Kim HR, Shin MG, Cho KH, Yoon JS. A longitudinal study of BDNF promoter methylation and genotype with poststroke depression. J Affect Disord. 2013;149:93–99. doi: 10.1016/j.jad.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Kordi-Tamandani DM, Sahranavard R, Torkamanzehi A. DNA methylation and expression profiles of the brain-derived neurotrophic factor (BDNF) and dopamine transporter (DAT1) genes in patients with schizophrenia. Mol Biol Rep. 2012;39:10889–10893. doi: 10.1007/s11033-012-1986-0. [DOI] [PubMed] [Google Scholar]
- Lindsay RM, Wiegand SJ, Altar CA, DiStefano PS. Neurotrophic factors: From molecule to man. Trends Neurosci. 1994;17:182–190. doi: 10.1016/0166-2236(94)90099-x. [DOI] [PubMed] [Google Scholar]
- Liu QR, Walther D, Drgon T, Polesskaya O, Lesnick TG, Strain KJ, de Andrade M, Bower JH, Maraganore DM, Uhl GR. Human brain derived neurotrophic factor (BDNF) genes, splicing patterns, and assessments of associations with substance abuse and Parkinson's Disease. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:93–103. doi: 10.1002/ajmg.b.30109. [DOI] [PubMed] [Google Scholar]
- Lu Y, Christian K, Lu B. BDNF: A key regulator for protein synthesis-dependent LTP and long-term memory. Neurobiol Learn Mem. 2008;89:312–323. doi: 10.1016/j.nlm.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem. 2004;279:27915–27927. doi: 10.1074/jbc.M312823200. [DOI] [PubMed] [Google Scholar]
- Miyaki K, Song Y, Htun NC, Tsutsumi A, Hashimoto H, Kawakami N, Takahashi M, Shimazu A, Inoue A, Kurioka S, Shimbo T. Folate intake and depressive symptoms in Japanese workers considering SES and job stress factors: J-HOPE study. BMC Psychiatry. 2012;12:33. doi: 10.1186/1471-244X-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohesara S, Ghadirivasfi M, Mostafavi S, Eskandari MR, Ahmadkhaniha H, Thiagalingam S, Abdolmaleky HM. DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. J Psychiatr Res. 2011;45:1432–1438. doi: 10.1016/j.jpsychires.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Perroud N, Courtet P, Vincze I, Jaussent I, Jollant F, Bellivier F, Leboyer M, Baud P, Buresi C, Malafosse A. Interaction between BDNF Val66Met and childhood trauma on adult's violent suicide attempt. Genes Brain Behav. 2008;7:314–322. doi: 10.1111/j.1601-183X.2007.00354.x. [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: Bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Epigenetic mechanisms underlying human epileptic disorders and the process of epileptogenesis. Neurobiol Dis. 2010;39:53–60. doi: 10.1016/j.nbd.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Picchioni MM, Kalidindi S, Loat CS, Knight J, Toulopoulou T, Vonk R, van der Schot AC, Nolen W, Kahn RS, McGuffin P, Murray RM, Craig IW. Differential methylation of the X-chromosome is a possible source of discordance for bipolar disorder female monozygotic twins. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:459–462. doi: 10.1002/ajmg.b.30616. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. J Psychiatr Res. 2011;45:919–926. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Nishi A, Kondo K, Yanagida K, Kawakami N. Screening performance of K6/K10 and other screening instruments for mood and anxiety disorders in Japan. Psychiatry Clin Neurosci. 2011;65:434–441. doi: 10.1111/j.1440-1819.2011.02236.x. [DOI] [PubMed] [Google Scholar]
- Satomura E, Baba H, Nakano Y, Maeshima H, Suzuki T, Arai H. Correlations between brain-derived neurotrophic factor and clinical symptoms in medicated patients with major depression. J Affect Disord. 2011;135:332–335. doi: 10.1016/j.jad.2011.06.041. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Duman R, Sanacora G. Serum brain-derived neurotrophic factor, depression, and antidepressant medications: Meta-analyses and implications. Biol Psychiatry. 2008;64:527–532. doi: 10.1016/j.biopsych.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu E, Hashimoto K, Okamura N, Koike K, Komatsu N, Kumakiri C, Nakazato M, Watanabe H, Shinoda N, Okada S, Iyo M. Alterations of serum levels of brain-derived neurotrophic factor (BDNF) in depressed patients with or without antidepressants. Biol Psychiatry. 2003;54:70–75. doi: 10.1016/s0006-3223(03)00181-1. [DOI] [PubMed] [Google Scholar]
- Takebayashi N, Maeshima H, Baba H, Nakano Y, Satomura E, Kita Y, Namekawa Y, Nomoto H, Suzuki T, Arai H. Duration of last depressive episode may influence serum BDNF levels in remitted patients with major depression. Depress Anxiety. 2012;29:775–779. doi: 10.1002/da.21933. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Tirassa P, Iannitelli A, Sornelli F, Cirulli F, Mazza M, Calza A, Alleva E, Branchi I, Aloe L, Bersani G, Pacitti F. Daily serum and salivary BDNF levels correlate with morning-evening personality type in women and are affected by light therapy. Riv Psichiatr. 2012;47:527–534. doi: 10.1708/1178.13059. [DOI] [PubMed] [Google Scholar]
- Tyler WJ, Alonso M, Bramham CR, Pozzo-Miller LD. From acquisition to consolidation: On the role of brain-derived neurotrophic factor signaling in hippocampal-dependent learning. Learn Mem. 2002;9:224–237. doi: 10.1101/lm.51202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unternaehrer E, Luers P, Mill J, Dempster E, Meyer AH, Staehli S, Lieb R, Hellhammer DH, Meinlschmidt G. Dynamic changes in DNA methylation of stress-associated genes (OXTR, BDNF) after acute psychosocial stress. Transl Psychiatry. 2012;2:e150. doi: 10.1038/tp.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Nabeshima T. Brain-derived neurotrophic factor/TrkB signaling in memory processes. J Pharmacol Sci. 2003;91:267–270. doi: 10.1254/jphs.91.267. [DOI] [PubMed] [Google Scholar]


