Short abstract
Osteogenic lineage commitment is often evaluated by analyzing gene expression. However, many genes are transiently expressed during differentiation. The availability of genes for expression is influenced by epigenetic state, which affects the heterochromatin structure. DNA methylation, a form of epigenetic regulation, is stable and heritable. Therefore, analyzing methylation status may be less temporally dependent and more informative for evaluating lineage commitment. Here we analyzed the effect of mechanical stimulation on osteogenic differentiation by applying fluid shear stress for 24 hr to osteocytes and then applying the osteocyte-conditioned medium (CM) to progenitor cells. We analyzed gene expression and changes in DNA methylation after 24 hr of exposure to the CM using quantitative real-time polymerase chain reaction and bisulfite sequencing. With fluid shear stress stimulation, methylation decreased for both adipogenic and osteogenic markers, which typically increases availability of genes for expression. After only 24 hr of exposure to CM, we also observed increases in expression of later osteogenic markers that are typically observed to increase after seven days or more with biochemical induction. However, we observed a decrease or no change in early osteogenic markers and decreases in adipogenic gene expression. Treatment of a demethylating agent produced an increase in all genes. The results indicate that fluid shear stress stimulation rapidly promotes the availability of genes for expression, but also specifically increases gene expression of later osteogenic markers.
Keywords: epigenetics, osteogenesis, osteogenic differentiation, mechanotransduction
Introduction
Osteogenic lineage commitment of progenitor cells requires a sequence of concerted gene expressions. While the expression of early markers may be transient during the process of differentiation, a hallmark of maturity is the constant expression of late markers [1]. The epigenetic state of a cell, which affects heterochromatin architecture but does not change the sequence of deoxyribonucleic acid (DNA), can impact gene accessibility. Although many factors regulate gene expression, including biochemical signals and transcription factors, for a gene to be consistently expressed, it must first be accessible for binding by the transcription machinery.
Epigenetic regulation can include various modifications of histones, which can either increase or decrease binding to DNA, thereby influencing heterochromatin structure. Another form of epigenetic regulation is methylation of cytosines in CpG dinucleotides, where a cytosine is followed by a guanine on the same backbone. In contrast to histone modifications, methylation directly modifies DNA. Methylation of CpG dinucleotides can enhance binding to histones contributing to a more condensed chromatin, which is inhibitive toward transcription, and can also prevent binding of specific transcription factors, resulting in a general decrease in gene expression [2–6]. Methyl groups on CpG dinucleotides in the gene regulatory region have been documented to contribute to gene silencing [7].
Methylation is durable and can produce persistent changes in gene expression. It is also heritable, and the pattern is passed onto daughter cells during division. Differentiated cells have a unique phenotype compared to progenitor cells, and this is due to altered gene expression. Altered methylation facilitates the passing of the phenotypic state from a progenitor cell to its offspring as it enables the expression of genes characteristic of a lineage as well as prevents expression of genes for other lineages. Therefore, methylation may be more informative of the state of differentiation than gene expression alone, which may be transient [8–10].
Biophysical signals are a potent factor influencing osteogenic lineage commitment of mesenchymal stem cells (MSCs). Mechanical loading of bones is well known to result in increased osteogenesis. The role of stem cells in this process has been proposed to be proliferation and migration to the bone surface, followed by osteogenic differentiation [11,12]. Direct mechanical stimulation of MSCs in vitro demonstrates they are capable of sensing mechanical signals and committing to the osteogenic lineage [13,14]. Furthermore, mechanical stimulation in the form of fluid shear stress has been shown to upregulate both early [15] and late [16–18] osteogenesis.
Arnsdorf et al. previously demonstrated that mechanical stimulation resulted in a decrease in methylation at a single CpG site in the osteopontin (OPN) gene and a corresponding increase in gene expression [19]. Similar changes in methylation and gene expression were also observed with biochemical differentiation. In contrast, gene expression and methylation of osteocalcin (OCN) were not found to be responsive to mechanical stimulation, even though application of biochemical stimulation did promote a significant increase in gene expression. This suggests that biochemical and mechanical stimulation promote lineage commitment through different mechanisms, and these mechanisms may be gene-dependent. However, a limitation of that study is methylation was only evaluated at a single CpG site, and it is possible that methylation changes occurred at other sites.
Another mechanism by which stem cells may be induced to commit to the osteogenic lineage is through paracrine signaling. Osteocytes are embedded within the mineralized bone matrix and can signal other osteocytes and cells on the bone surface through dendrites. Osteocytes have also been implicated in sensing changes in mechanical strain, and transmitting signals to initiate mechanically induced modeling [20]. Mechanical loading may be detected by osteocytes, which then produce signals that are sensed by stem cells. These signals then promote stem cell migration to the bone surface and osteogenic differentiation. Indeed, direct mechanical stimulation has previously been demonstrated to be sensed by osteoblasts and osteocytes, resulting in changes in gene expression [21–24]. In a study by Hoey et al., when osteocytes were subjected to mechanical stimulation, paracrine signals were released that influenced stem cell osteogenic lineage commitment [25]. Osteocytes were subjected to fluid shear stress, and medium was then collected. This CM was applied to MSCs, and induced an increase in expression of Cox-2, an early osteogenic marker, and OPN. Surprisingly though, there was no change in Runx2 expression, which is an early osteogenic transcription factor. Thus, if genes regulated by Runx2 are changing expression levels without changes in Runx2, perhaps an altered methylation state is involved, and could provide a better indication of lineage commitment.
This CM model produced a robust induction of gene expression, and was used in this study to analyze the effect of mechanical stimulation on methylation. In this study, we used bisulfite sequencing to analyze methylation levels over the landscape of the regulatory regions of multiple genes. We found that methylation levels were indeed decreased for later osteogenic markers, indicating the potential for lineage commitment.
Methods and Materials
Cell Culture.
MLO-Y4 osteocyte-like murine cells (gift from Lynda Bonewald, University of Missouri) were maintained using Minimum Essential Medium α (Life Technologies, Grand Island, NY) supplemented with 5% fetal bovine serum (GE Healthcare Life Sciences, Logan, UT), 5% fetal calf serum (GE Healthcare Life Sciences) and 1% penicillin–streptomycin (Life Technologies). C3H10T1/2 mesenchymal progenitor cells were maintained in growth medium consisting of Dulbecco's Modified Eagle Medium low glucose (Life Technologies) supplemented with 10% fetal bovine serum (GE Healthcare Life Sciences) and 1% penicillin–streptomycin (Life Technologies).
MLO-Y4 cells were seeded at 4000 cells/cm2 on type I collagen-coated (BD Biosciences, San Jose, CA) flasks. Medium was changed 48 hr following seeding and cells remained static or were mechanically stimulated through application of fluid shear stress (described below) for 24 hr. Osteocyte CM was then collected by pooling media from five flasks for each condition. C3H10T1/2 murine cells (ATCC, Manassas, VA) were seeded at 1000 cells/cm2− on tissue culture polystyrene dishes and maintained in Dulbecco's Modified Eagle Medium low glucose, supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin for 48 hr. Six dishes of C3H10T1/2 cells were then treated with CM from either fluid shear stress stimulated or static MLO-Y4 cells for 24 hr. Ribonucleic acid (RNA) was then isolated separately from three dishes of C3H10T1/2 cells after 24 hr of CM treatment (n = 3) and DNA was isolated from the remaining three dishes (n = 1).
To investigate the effects of adding a demethylating agent, 5-Aza-2′-deoxycytidine (5-Aza-dC, Sigma, St. Louis, MO) was added to C3H10T1/2 growth medium to a final concentration of 10 μM. C3H10T1/2 cells seeded at 1000 cells/cm2 on tissue culture polystyrene dishes were treated with growth medium containing 5-Aza-dC for 72 hr before RNA isolation. Control cells were treated with growth medium for 72 hr before RNA isolation. Four dishes of cells were used for 5-Aza-dC treatment and for controls and RNA was isolated separately from each dish (n = 4). Cells used for analyzing the effects of 5-Aza-dC were not exposed to CM.
Application of Fluid Shear Stress to MLO-Y4 Osteocyte-Like Cells.
Rectangular flasks (82 × 92 mm; 10 ml media) containing MLO-Y4 cells at approximately 80% confluence were placed on a rocking platform. The platform oscillated at a frequency of 0.5 Hz with an amplitude of 1.5 cm for 24 hr. This system has previously been used to apply dynamic fluid flow to osteocytes [25], and has been shown to generate fluid flow induced shear stress across a layer of cells that may be similar to that experienced by osteocytes within the lacuna–canalicular network in bones [26]. Another benefit of this system was that soluble factors were not diluted, making this approach suitable for studying the effect of paracrine signaling from mechanically stimulated cells on the lineage commitment of progenitor cells.
RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction (PCR).
Tri Reagent (Sigma) was used to extract RNA from C3H10T1/2 cells. cDNA was then synthesized from extracted RNA using the Taqman reverse transcription kit (Life Technologies). Real-time quantitative PCR was then performed on cDNA samples using Taqman PCR Master Mix on an ABI Prism 7900HT sequence detection system (Life Technologies). Primers and probes for Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), Runx2, Dlx5, Osterix (OSX), OPN, OCN, PPARγ, fatty acid binding protein 4 (FABP4), and lipoprotein lipase (LPL) were obtained from Applied Biosystems (Life Technologies). Amplification curves for all genes investigated were recorded and relative gene levels between samples were quantified using the relative standard curve method. All samples were normalized to endogenous control GAPDH levels. All samples and standards were run in triplicate.
DNA Isolation and Bisulfite Treatment.
Cells were lifted and pelleted, and DNA was extracted using the DNeasy Tissue Kit (Qiagen, Germantown, MD). DNA was eluted in 100 μl of AE buffer (10 mM Tris-Cl, 0.5 mM EDTA, pH 9.0), and the concentration was measured using an ND-1000 Spectrometer (Thermo, Wilmington, DE). 1 μg of DNA was bisulfite treated using the EZ DNA Methylation kit (Zymo, Irvine, CA) to convert nonmethylated cytosines to uracils. 5 μl of M-dilution buffer was added to DNA. Water was added to adjust to 50 μl total volume, and the reaction was incubated at 42 °C for 25 min. After incubation, 100 μl of CT conversion reagent was added, and DNA samples were incubated at 50 °C for 15 hr protected from light. The resulting bisulfite-treated DNA was purified and eluted in 20 μl of M-elution buffer.
Primer Design and Bisulfite Specific PCR.
methprimer [27] was used to design primers (Table 1), which were specific for bisulfite converted DNA strands with no CpG dinucleotides in the original sequences to amplify each gene of interest. TaKaRa LA Taq Hot Start PCR Kit (Takara Bio Inc., Shiga, Japan) was used for bisulfite specific PCR amplifications with 30 ng of bisulfite treated DNA. The PCR regimen was as follows: 95 °C for 1 min, 35 cycles of denaturing (94 °C for 30 s), annealing (various temperatures for 30 s), and elongation (72 °C for 1 min), followed by 72 °C for 10 min. Annealing temperatures were varied depending on the target as follows: Runx2 at 60 °C, OSX at 63 °C, Dlx5 at 57 °C, OPN500 at 53, OPN-1000 at 57, OCN-500 at 58 °C, OCN-1500 at 57 °C, PPARγ at 53 °C, FABP4 and LPL at 57 °C. OPN500 was repeated for 38 cycles instead of 35 cycles. Specific amplification was confirmed by performing agarose gel electrophoresis and verifying a single band at 700 base pairs.
Table 1.
Primer sequences for bisulfite specific PCR
| Gene target | Forward primer | Reverse primer |
|---|---|---|
| Runx2 | 5′-GGA AGG AGA GAT AGA GGA ATA TTT ATA AGT-3′ | Reverse: 5′- ACC CCA AAA AAA ACT TTA CTA ACA C-3′ |
| Osterix | 5′-TGT TTT AGT TTT TTT GTG TGA GTG T-3′ | 5′-AAA ATC CAC CCT CTA ATT ACA ACT TTC C-3′ |
| Dlx5 | 5′-TAA TGG GGG ATG TTA TAG AAT TTA AAT TTA-3′ | 5′-CAA TCC CAA AAC CTA ACT CC-3′ |
| OPN (500 base pairs after transcription start site) | 5′-TTT TTA GAA AAT TGT TTT ATT TTA AAA GAG-3′ | 5′-AAC AAA TCA CTA CCA ATC TCA TAA TC-3′ |
| OPN (1000 base pairs prior to transcription start site) | 5′-GTT GTT TTA ATA GAG TAA TAA GGT TTA-3′ | 5′-CAT AAA ATT TTT ACC ACT ACC C-3′ |
| OCN (500 base pairs prior to transcription start site) | 5′-TTT TTT TGG GGT TTG GTT TT-3′ | 5′-TAA TTA ATT CTA TTT CCT CCC TAT TAT CTC-3′ |
| OCN (1500 base pairs prior to transcription start site) | 5′-AGA AAG AAA GAA TAT AAA TAA GTG AGA TGT-3′ | 5′-AAC CAA ACC CCA AAA AAA A-3′ |
| PPARγ | 5′-TTT AAG AAA AAT TTT GGT TAA ATA-3′ | 5′-AAA TTC TAA ATA CAT TTA TAA ATA ATC ACC-3′ |
| FABP4 | 5′-GTG TGA TGT TTT TGT GGG AAT TTG-3′ | 5′-TAC ATA CCC TAC TTT CCT TCT AAA TTA CTC-3′ |
| LPL | 5′-AAA TTT AGG ATA GTT TAA AAT GTT TGA TTA-3′ | 5′-CAA TTA CAA AAA ACA AAA TTC CTC-3′ |
Bisulfite Specific Sequencing.
The TOPO TA Cloning Kit for Sequencing (Life Technologies) was used to clone bisulfite specific PCR products into plasmid vectors (pCR4-TOPO) for sequencing. The TOPO cloning reaction was performed by adding 4 μl of fresh bisulfite specific PCR product to 1 μl of salt solution (200 mM NaCl, 10 mM MgCl2), and 1 μl of pCR4-TOPO vector and incubating for 10 min at room temperature. 2 μl of the TOPO cloning reaction was added to DH5α-T1 Competent E. coli (Life Technologies), and incubated on ice for 10 min. Cells were heat-shocked for 30 s at 42 °C and immediately transferred to ice. About 250 μl of super optimal broth with catabolite repression (Life Technologies) was added and the vial of cells was shaken horizontally (200 rpm) on an orbital shaker at 37 °C for 1 hr. 10–50 μl from each transformation was spread on selective Luria-Bertani agar (Life Technologies) plates containing 50 μg/ml kanamycin (Life Technologies) and 50 μg/ml ampicillin (Life Technologies) and incubated overnight at 37 °C. Plasmid DNA from at least 10 unique colonies per group was prepared and sequenced (GENEWIZ, Inc., South Plainfield, NJ) using T7 or T3 universal primers.
Data Analysis.
Data are expressed as mean ± standard error (SE). Gene expression levels were normalized against GAPDH mRNA assayed in the same tube. Unpaired t-tests with Welch's correction were used to compare cells receiving static or fluid flow CM (n = 3 each for static or fluid flow CM) and to compare control to 5-Aza-dC treated cells (n = 4 each for control or 5-Aza-dC treatment).
Sequences for at least 10 colonies for each gene target were analyzed. Uracils from bisulfite treatment were converted to thymines through bisulfite specific PCR, and thymine reads at CpG sites indicated nonmethylated cytosines, whereas cytosine reads at CpG sites indicated methylated cytosines. Methylation level for each CpG site was calculated as a percentage of reads that were cytosines. Individual values for methylation levels for each CpG site were then used to calculate the mean total methylation level for each gene. Paired t-tests were used to compare methylation levels with α = 0.05. Statistical tests were performed using graphpad prism (GraphPad Software, La Jolla, CA).
Results
Chemically Induced Demethylation Results in a General Increase in MSC Gene Expression.
After application of the demethylating agent, 5′-deoxyazacytidine (5-AZA-dC) for 72 hr, increase in expression of both osteogenic and adipogenic markers was observed (Fig. 1). Statistically significant increases in expression were measured for the osteogenic markers OPN (p < 0.01), OCN (p < 0.001), and Dlx5 (p < 0.01), as well as for adipogenic markers LPL (p < 0.01), FABP4 (p < 0.001), and PPARγ (p < 0.001). mRNA levels were increased by 3.0 ± 0.03-fold for OPN, 10.8 ± 1.4-fold for OCN, 1.7 ± 0.03-fold for Dlx5, 2.5 ± 0.07-fold for LPL, 2.1 ± 0.03-fold for FABP4, and 1.4 ± 0.006-fold for PPARγ.
Fig. 1.
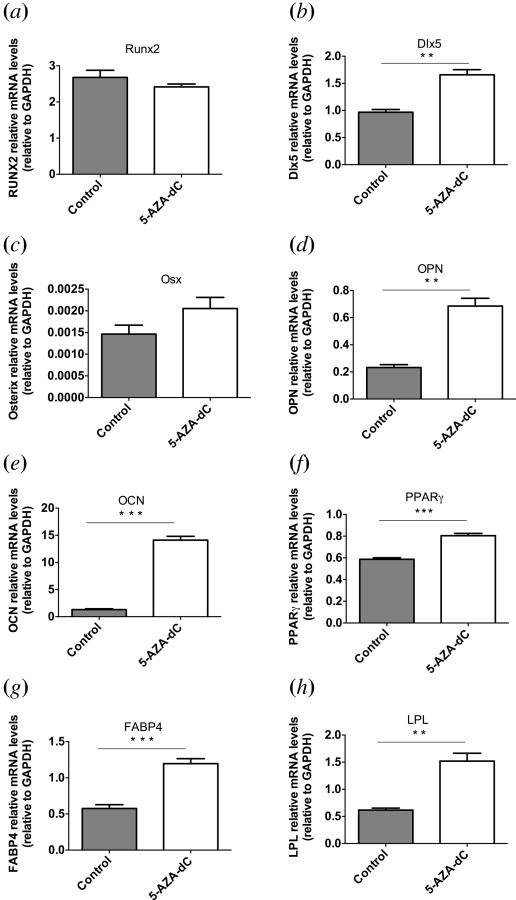
Gene expression changes after treatment with demethylating agent. mRNA levels for (a) Runx2, (b) Dlx5, (c) OSX, (d) OPN, (e) OCN, (f) PPARγ, (g) FABP4, and (h) LPL (bars indicate mean ± SE, n = 4, **p < 0.01, ***p < 0.001).
Fluid Shear Stress Stimulation Affects mRNA Levels of Osteogenic Markers in MSCs.
Gene expression was analyzed by measuring mRNA levels of multiple osteogenic markers. MSCs receiving CM from fluid shear stress stimulated osteocytes had different mRNA levels from MSCs receiving CM from static osteocytes (Fig. 2). A decrease in expression was observed with fluid shear stress stimulated medium for early osteogenic markers Runx2 (p < 0.05) and Dlx5 (p < 0.05). For later osteogenic markers, increases in expression for fluid shear stress stimulated CM compared to static CM were observed. Increases of 4.0 ± 0.02-fold for OSX, 2.7 ± 0.03-fold for OPN, and 28.2 ± 0.7-fold for OCN, statistically significant increases (p < 0.01, p < 0.001, and p < 0.01, respectively), were observed. In contrast, expression of adipogenic genes was decreased for PPARγ by 30 ± 0.1% (p < 0.01) and for FABP4 by 70 ± 0.05% (p < 0.0001). A decrease of 90 ± 0.5% was also measured for LPL. However, this was not statistically significant.
Fig. 2.
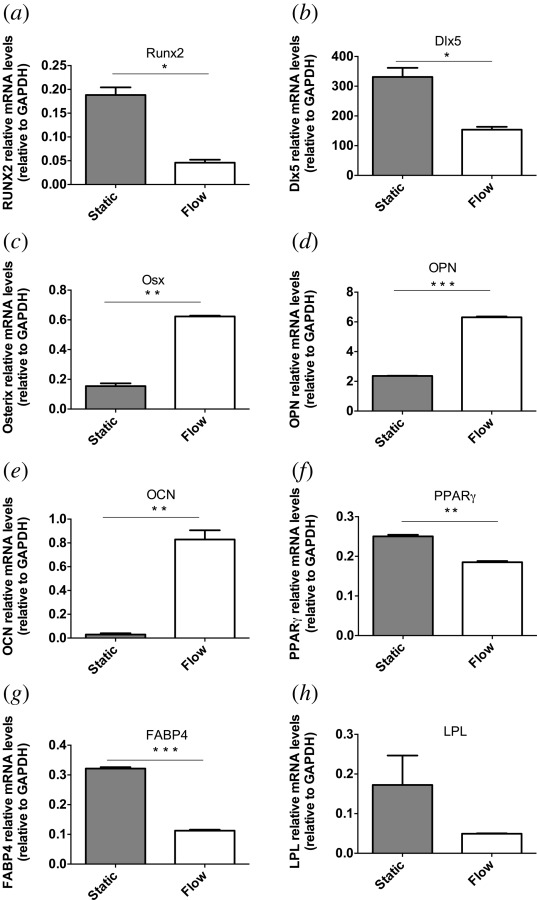
Gene expression changes after treatment with CM from fluid shear stress stimulated osteocytes. mRNA levels for (a) Runx2, (b) Dlx5, (c) OSX, (d) OPN, (e) OCN, (f) PPARγ, (g) FABP4, and (h) LPL (bars indicate mean ± SE, n = 3, *p < 0.05, **p < 0.01, ***p < 0.001).
Methylation Levels for Osteogenic Markers After Treatment With Static or Flow CM.
Methylation profiles of the osteogenic markers described above were analyzed. The percent methylation at CpG sites varied across the target area of the gene of interest, but in general cells receiving flow CM exhibited a decrease in methylation at a majority of sites for most of the markers evaluated compared to cells receiving static CM (Figs. 3(a)–3(e)). After receiving CM from osteocytes subjected to fluid flow, a statistically significant decrease in methylation in MSCs was observed for Dlx5 (Fig. 3(b)), Osx (Fig. 3(c)), OPN (Fig. 3(d)), and OCN (Fig. 3(e)) compared to MSCs receiving static CM. No change was measured for the early transcription factor Runx2.
Fig. 3.
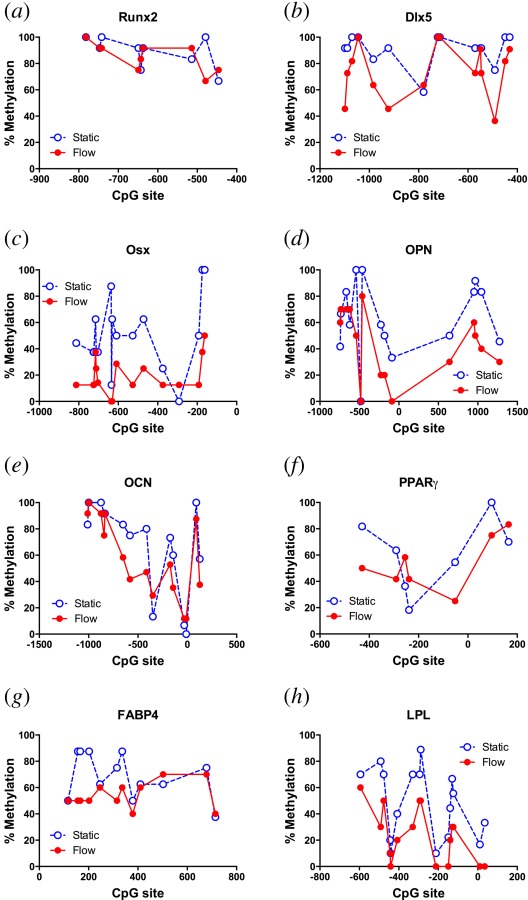
CpG methylation levels in cells treated with CM from static (open circles) or fluid shear stress stimulated (closed circles) osteocytes. Location of CpG site is shown as number of base pairs relative to transcription start site (0). Levels for (a) Runx2, (b) Dlx5, (c) OSX, (d) OPN, (e) OCN, (f) PPARγ, (g) FABP4, and (h) LPL (percentages were calculated from DNA from at least 10 colonies, n = 1).
CpG sites in the Runx2 promoter and gene were highly methylated with an average of 90 ± 4% methylation across all sites. After flow, despite a significant decrease in mRNA expression level, no significant difference in methylation was observed. Dlx5 mRNA expression decreased after treatment with flow CM compared to treatment with static CM. However, methylation levels showed a slight but statistically significant decrease of 17 ± 4% on average over all sites evaluated. In contrast, OSX methylation levels in cells treated with static CM averaged 50 ± 7% across all CpG sites evaluated in the promoter and gene. Treatment with flow CM resulted in a significant decrease in methylation levels with an average decrease of 32 ± 6% compared to cells receiving static CM. Nearly all CpG sites had a decrease after flow CM, with only one site showing a slight increase of 13%. In addition, 8 out of 16 sites with a decrease had a decrease of greater than 30%.
Methylation of extracellular matrix coding genes decreased with flow CM treatment. For OPN, cells treated with static CM had a methylation level of 63 ± 7% while cells treated with flow CM exhibited a decrease in methylation to 43 ± 7%. Methylation levels for individual CpG sites in cells treated with static CM also ranged widely from 0% to 100%. With flow CM treatment, 11 out of 15 sites evaluated showed a decrease in methylation, one had no change, while three had an increase. Furthermore, 5 out of the 11 sites with a decrease had a decrease of greater than 30%. For OCN, methylation levels were slightly decreased with flow CM (58 ± 8%) compared to static CM (68 ± 9%). Methylation levels of individual CpG sites also ranged widely from 0% to 100% for cells treated with static CM, with an average decrease of 10 ± 4% for most sites after flow CM treatment.
Methylation Levels for Adipogenic Markers After Treatment With Static or Flow CM.
The application of flow CM also induced changes in methylation levels for markers in the adipogenic lineage (Figs. 3(f)–3(h)). For the transcription factor PPARγ, despite exhibiting a decrease in gene expression with flow CM, no significant change in average methylation level was observed (Fig. 3(f)). For the later adipogenic markers FABP4 and LPL methylation decreased in response to flow CM compared to static CM from 69 ± 5% to 54 ± 3% and from 48 ± 7% to 24 ± 5%, respectively (Figs. 3(g) and 3(h)). For FABP4 the majority of CpG sites (9 out of 12) showed a decrease in methylation with 5 out of 12 sites exhibiting at least a 25% decrease. Similarly for LPL all CpG sites experienced a decrease in methylation with 11 out of 15 sites exhibiting at least a 20% decrease.
Discussion
We observed a decrease in methylation in response to treatment with CM from fluid shear stress stimulated osteocytes compared to CM from static osteocytes. Interestingly, although methylation decreased for both adipogenic and osteogenic genes, only osteogenic gene expression increased with flow CM. This is in contrast to treatment with a demethylating agent, where gene expression generally increased for the osteogenic and adipogenic markers tested. This implies that although a decrease in methylation can promote an increase in gene expression, other mechanisms must be involved in specifically inducing mechanically induced osteogenic lineage commitment.
Significant changes in methylation and gene expression were also detected within a short time of applying CM. Within 24 hr of application of flow CM, an increase in expression of later osteogenic markers was measured. In contrast, experiments involving biochemical induction typically probe for signs of osteogenic differentiation after at least 14 days of induction [28,29]. Furthermore, changes in gene expression were only observed after three days of application of the demethylating reagent. The rapid change in gene expression and methylation in response to physical stimulation indicates that physical stimulation may be more potent than biochemical stimulation for osteogenic differentiation.
In contrast to osteogenic markers, changes in methylation were not correlated with gene expression for adipogenic markers. A decrease in methylation may indicate exit from the proliferative state, and potential for differentiation. However, the methylation state of later osteogenic markers alone may not be sufficient for predicting lineage. Rather, in this case, methylation may act via regulation of late transcription factors. We found that for OSX, application of flow CM resulted in a significant decrease in methylation of 32% such that the final methylation level was only 18%, which was the lowest observed of the transcription factors investigated here. Methylation was also moderate for OSX after application of static CM at 50%. In comparison, methylation levels of the osteogenic transcription factors Runx2 and Dlx5 were around 90% with static CM, and decreased 4% and 17%, respectively, with flow CM.
Interestingly, the early transcription factor Runx2 was the only gene that did not show an increase in expression after treatment with the demethylating agent. In addition, gene expression decreased with flow CM while no change in methylation level occurred. Runx2 was also the most highly methylated gene investigated here after application of flow CM. Together, these findings indicate that Runx2 expression is insensitive to methylation. Other genes associated with early development have also been demonstrated to have similar relationships to methylation, with active gene expression despite high levels of methylation [30]. This suggests that while methylation may be correlated with gene accessibility for genes with medium or low levels of methylation, this relationship does not hold for high levels of methylation. Highly methylated genes important for early development may be more resistant to changes in methylation patterns and remain accessible for transcription.
We found that the relationship between methylation and expression was altered for adipogenic genes compared to osteogenic genes. Like the osteogenic markers, treatment with the demethylating agent resulted in increases in PPARγ, FABP4, and LPL gene expression. Interestingly though, after flow CM, decreases in mRNA levels of PPARγ, FABP4, and LPL were observed despite a decrease in methylation. Although a decrease in methylation may increase the accessibility of genes for transcription, the decrease in expression of PPARγ may have resulted in a subsequent decrease in the downstream targets FABP4 [31] and LPL [32,33].
Although we analyzed multiple sites for various lineage markers, one of the limitations of this study remains the number of CpG sites evaluated. We primarily analyzed the promoter regions near the transcription start sites as well as areas immediately following the transcription start sites. While these areas are often responsible for controlling gene expression [34], many other potential regions of gene expression control exist. Nonpromoter regions such as upstream regions (−1 to −20 kb from transcription start site), introns, exons, and intergenic regions have been demonstrated to bind Runx2 during osteogenic differentiation [35]. Finally, we were limited to evaluating a few target genes, but many more genes are expressed during the course of osteogenic and adipogenic differentiation. Future studies specifically analyzing the presence of hydroxymethylcytosine, which has been proposed to be an intermediate stage for demethylation, could also provide clarity on methylation state.
Predicting gene expression based on methylation state is complicated because methylation does not necessarily prevent access to the gene. In particular, some transcription factors or transcription binding proteins are enhanced by the presence of methylation. For example, MeCP2 preferentially binds to methylated regions [36]. Also, heterochromatin structure will ultimately influence gene accessibility, and while methylation may result in tight binding of DNA to histones, high levels of methylation may actually prevent binding. Furthermore, histone modifications are important in influencing heterochromatin structure. The scope of this study was limited to DNA methylation, but future studies involving the effect of mechanical stimuli on histone modifications could be informative.
In this study, we showed that mechanical signals potently stimulated osteogenic lineage commitment of progenitor cells. Mechanical signals promoted a general rapid decrease in DNA methylation for osteogenic markers, but increases in gene expression only occurred for later osteogenic markers despite the early time point. Lineage markers are transiently expressed during differentiation, and conclusions concerning differentiation state of MSCs are therefore temporally dependent. Analysis of DNA methylation combined with gene expression is therefore a more robust method for determining lineage commitment. The field of epigenetics has been advancing rapidly, creating an increasingly complex picture of gene regulation. Exciting new tools for analyzing the epigenetic state are constantly being developed, and are being applied to provide a more complete characterization of the epigenetic states of various cell types. Rigorous future analyses of epigenetic state have the potential to elucidate the varying mechanisms responsible for lineage commitment.
Acknowledgment
We thank K. Lee, A. Nguyen, and D. Hoey for technical assistance and discussions. This work was supported by the New York State Stem Cell Grant (NYSTEM) N089-210 and National Institutes of Health (NIH) grants AR054156, AR062177, and AR059038.
Glossary
Nomenclature
- CM =
conditioned medium
- DNA =
deoxyribonucleic acid
- FABP4 =
fatty acid binding protein 4
- GAPDH =
glyceraldehyde 3-phosphate dehydrogenase
- LPL =
lipoprotein lipase
- MSC =
mesenchymal stem cell
- OCN =
osteocalcin
- OPN =
osteopontin
- OSX =
osterix
- PCR =
polymerase chain reaction
- RNA =
ribonucleic acid
- 5-Aza-dC =
5′-deoxyazacytidine
Contributor Information
Julia C. Chen, Department of Biomedical Engineering, Columbia University, New York, NY 10027
Mardonn Chua, Department of Biotechnology, University of British Columbia, Vancouver, BC V6T 1Z4, Canada .
Raymond B. Bellon, Department of Chemical Engineering, Columbia University, New York, NY 10027
Christopher R. Jacobs, Mem. ASME, Department of Biomedical Engineering, Columbia University, New York, NY 10027.
References
- [1]. Kato, Y. , Windle, J. J. , and Koop, B. A. , 1997, “Establishment of an Osteocyte-Like Cell Line, MLO-Y4,” J. Bone Miner. Res., 12(12), pp. 2014–2023. [DOI] [PubMed] [Google Scholar]
- [2]. Antequera, F. , 2003, “Structure, Function and Evolution of CpG Island Promoters,” Cell. Mol. Life Sci., 60(8) pp. 1647–1658. 10.1007/s00018-003-3088-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Ballestar, E. , and Wolffe, A. P. , 2001, “Methyl-CpG-Binding Proteins. Targeting Specific Gene Repression,” Eur. J. Biochem., 268(1), pp. 1–6. 10.1046/j.1432-1327.2001.01869.x [DOI] [PubMed] [Google Scholar]
- [4]. El-Osta, A. , and Wolffe, A. P. , 2000, “DNA Methylation and Histone Deacetylation in the Control of Gene Expression: Basic Biochemistry to Human Development and Disease,” Gene Expression, 9(1–2) pp. 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Fuks, F. , Hurd, P. J. , and Wolf, D. , 2003, “The Methyl-CpG-Binding Protein MeCP2 Links DNA Methylation to Histone Methylation,” J. Biol. Chem., 278(6) pp. 4035–4040. 10.1074/jbc.M210256200 [DOI] [PubMed] [Google Scholar]
- [6]. Wolffe, A. P. , and Matzke, M. A. , 1999, “Epigenetics: Regulation Through Repression,” Science, 286(5439), pp. 481–486. 10.1126/science.286.5439.481 [DOI] [PubMed] [Google Scholar]
- [7]. Noer, A. , Sorensen, A. L. , and Boquest, A. C. , 2006, “Stable CpG Hypomethylation of Adipogenic Promoters in Freshly Isolated, Cultured, and Differentiated Mesenchymal Stem Cells From Adipose Tissue,” Mol. Biol. Cell, 17(8) pp. 3543–3556. 10.1091/mbc.E06-04-0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Friedl, G. , Schmidt, H. , and Rehak, I. , 2007, “Undifferentiated Human Mesenchymal Stem Cells (hMSCs) are Highly Sensitive to Mechanical Strain: Transcriptionally Controlled Early Osteo-Chondrogenic Response In Vitro,” Osteoarthritis Cartilage, 15(11) pp. 1293–1300. 10.1016/j.joca.2007.04.002 [DOI] [PubMed] [Google Scholar]
- [9]. Lachner, M. , 2002, “Epigenetics: SUPERMAN Dresses Up,” Curr. Biol., 12(12), pp. R434–R436. 10.1016/S0960-9822(02)00919-3 [DOI] [PubMed] [Google Scholar]
- [10]. Lachner, M. , and Jenuwein, T. , 2002, “The Many Faces of Histone Lysine Methylation,” Curr. Opin. Cell Biol., 14(3) pp. 286–298. 10.1016/S0955-0674(02)00335-6 [DOI] [PubMed] [Google Scholar]
- [11]. Turner, C. H. , Owan, I. , and Alvey, T. , 1998, “Recruitment and Proliferative Responses of Osteoblasts After Mechanical Loading In Vivo Determined Using Sustained-Release Bromodeoxyuridine,” Bone, 22(5), pp. 463–469. 10.1016/S8756-3282(98)00041-6 [DOI] [PubMed] [Google Scholar]
- [12]. David, V. , Martin, A. , and Lafage-Proust, M. H. , 2007, “Mechanical Loading Down-Regulates Peroxisome Proliferator-Activated Receptor Gamma in Bone Marrow Stromal Cells and Favors Osteoblastogenesis at the Expense of Adipogenesis,” Endocrinology, 148(5) pp. 2553–2562. 10.1210/en.2006-1704 [DOI] [PubMed] [Google Scholar]
- [13]. McBeath, R. , Pirone, D. M. , and Nelson, C. M. , 2004, “Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment,” Dev. Cell, 6(4), pp. 483–495. 10.1016/S1534-5807(04)00075-9 [DOI] [PubMed] [Google Scholar]
- [14]. Estes, B. T. , Gimble, J. M. , and Guilak, F. , 2004, “Mechanical Signals as Regulators of Stem Cell Fate,” Curr. Topics Dev. Biol., 60, pp. 91–126. 10.1016/S0070-2153(04)60004-4 [DOI] [PubMed] [Google Scholar]
- [15]. Hoey, D. A. , Tormey, S. , and Ramcharan, S. , 2012, “Primary Cilia-Mediated Mechanotransduction in Human Mesenchymal Stem Cells,” Stem Cells (Dayton, Ohio), 30(11), pp. 2561–2570. 10.1002/stem.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Li, Y. J. , Batra, N. N. , and You, L. , 2004, “Oscillatory Fluid Flow Affects Human Marrow Stromal Cell Proliferation and Differentiation,” J. Orthop. Res., 22(6), pp. 1283–1289. 10.1016/j.orthres.2004.04.002 [DOI] [PubMed] [Google Scholar]
- [17]. Kreke, M. R. , Huckle, W. R. , and Goldstein, A. S. , 2005, “Fluid Flow Stimulates Expression of Osteopontin and Bone Sialoprotein by Bone Marrow Stromal Cells in a Temporally Dependent Manner,” Bone, 36(6), pp. 1047–1055. 10.1016/j.bone.2005.03.008 [DOI] [PubMed] [Google Scholar]
- [18]. Kreke, M. R. , Sharp, L. A. , and Lee, Y. W. , 2008, “Effect of Intermittent Shear Stress on Mechanotransductive Signaling and Osteoblastic Differentiation of Bone Marrow Stromal Cells,” Tissue Eng. Part A, 14(4), pp. 529–537. 10.1089/tea.2007.0068 [DOI] [PubMed] [Google Scholar]
- [19]. Arnsdorf, E. J. , Tummala, P. , and Castillo, A. B. , 2010, “The Epigenetic Mechanism of Mechanically Induced Osteogenic Differentiation,” J. Biomech., 43(15), pp. 2881–2886. 10.1016/j.jbiomech.2010.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Tatsumi, S. , Ishii, K. , and Amizuka, N. , 2007, “Targeted Ablation of Osteocytes Induces Osteoporosis With Defective Mechanotransduction,” Cell Metab., 5(6), pp. 464–475. 10.1016/j.cmet.2007.05.001 [DOI] [PubMed] [Google Scholar]
- [21]. Batra, N. N. , Li, Y. J. , and Yellowley, C. E. , 2005, “Effects of Short-Term Recovery Periods on Fluid-Induced Signaling in Osteoblastic Cells,” J. Biomech., 38(9), pp. 1909–1917. 10.1016/j.jbiomech.2004.08.009 [DOI] [PubMed] [Google Scholar]
- [22]. Malone, A. M. , Anderson, C. T. , and Tummala, P. , 2007, “Primary Cilia Mediate Mechanosensing in Bone Cells by a Calcium-Independent Mechanism,” Proc. Natl. Acad. Sci. USA, 104(33), pp. 13325–13330. 10.1073/pnas.0700636104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. You, J. , Reilly, G. C. , and Zhen, X. , 2001, “Osteopontin Gene Regulation by Oscillatory Fluid Flow Via Intracellular Calcium Mobilization and Activation of Mitogen-Activated Protein Kinase in MC3T3-E1 Osteoblasts,” J. Biol. Chem., 276(16), pp. 13365–13371. 10.1074/jbc.M009846200 [DOI] [PubMed] [Google Scholar]
- [24]. You, J. , Yellowley, C. E. , and Donahue, H. J. , 2000, “Substrate Deformation Levels Associated With Routine Physical Activity are Less Stimulatory to Bone Cells Relative to Loading-Induced Oscillatory Fluid Flow,” ASME J. Biomech. Eng., 122(4), pp. 387–393. 10.1115/1.1287161 [DOI] [PubMed] [Google Scholar]
- [25]. Hoey, D. A. , Kelly, D. J. , and Jacobs, C. R. , 2011, “A Role for the Primary Cilium in Paracrine Signaling Between Mechanically Stimulated Osteocytes and Mesenchymal Stem Cells,” Biochem. Biophys. Res. Commun., 412(1), pp. 182–187. 10.1016/j.bbrc.2011.07.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Zhou, X. , Liu, D. , and You, L. , 2010, “Quantifying Fluid Shear Stress in a Rocking Culture Dish,” J. Biomech., 43(8), pp. 1598–1602. 10.1016/j.jbiomech.2009.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Li, L. C. , and Dahiya, R. , 2002, “MethPrimer: Designing Primers for Methylation PCRs,” Bioinformatics, 18(11), pp. 1427–1431. 10.1093/bioinformatics/18.11.1427 [DOI] [PubMed] [Google Scholar]
- [28]. Jaiswal, N. , Haynesworth, S. E. , and Caplan, A. I. , 1997, “Osteogenic Differentiation of Purified, Culture-Expanded Human Mesenchymal Stem Cells In Vitro,” J. Cell. Biochem., 64(2), pp. 295–312. [DOI] [PubMed] [Google Scholar]
- [29]. Bourne, S. , Polak, J. M. , and Hughes, S. P. , 2004, “Osteogenic Differentiation of Mouse Embryonic Stem Cells: Differential Gene Expression Analysis by cDNA Microarray and Purification of Osteoblasts by Cadherin-11 Magnetically Activated Cell Sorting,” Tissue Eng., 10(5–6), pp. 796–806. 10.1089/1076327041348293 [DOI] [PubMed] [Google Scholar]
- [30]. Sorensen, A. L. , Jacobsen, B. M. , and Reiner, A. H. , 2010, “Promoter DNA Methylation Patterns of Differentiated Cells are Largely Programmed at the Progenitor Stage,” Mol. Biol. Cell, 21(12), pp. 2066–2077. 10.1091/mbc.E10-01-0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Tontonoz, P. , Hu, E. , and Spiegelman, B. M. , 1994, “Stimulation of Adipogenesis in Fibroblasts by PPAR Gamma 2, a Lipid-Activated Transcription Factor,” Cell, 79(7), pp. 1147–1156. 10.1016/0092-8674(94)90006-X [DOI] [PubMed] [Google Scholar]
- [32]. Hill, M. R. , Young, M. D. , and McCurdy, C. M. , 1997, “Decreased Expression of Murine PPARgamma in Adipose Tissue During Endotoxemia,” Endocrinology, 138(7), pp. 3073–3076. [DOI] [PubMed] [Google Scholar]
- [33]. Schoonjans, K. , Peinado-Onsurbe, J. , and Lefebvre, A. M. , 1996, “PPARalpha and PPARgamma Activators Direct a Distinct Tissue-Specific Transcriptional Response Via a PPRE in the Lipoprotein Lipase Gene,” EMBO J., 15(19), pp. 5336–5348. [PMC free article] [PubMed] [Google Scholar]
- [34]. Meissner, A. , Mikkelsen, T. S. , and Gu, H. , 2008, “Genome-Scale DNA Methylation Maps of Pluripotent and Differentiated Cells,” Nature, 454(7205), pp. 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Wu, H. , Whitfield, T. W. , and Gordon, J. A. , 2014, “Genomic Occupancy of Runx2 With Global Expression Profiling Identifies a Novel Dimension to Control of Osteoblastogenesis,” Genome Biol., 15(3), p. R52. 10.1186/gb-2014-15-3-r52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Chahrour, M. , Jung, S. Y. , and Shaw, C. , 2008, “MeCP2, a Key Contributor to Neurological Disease, Activates and Represses Transcription,” Science, 320(5880), pp. 1224–1229. 10.1126/science.1153252 [DOI] [PMC free article] [PubMed] [Google Scholar]


