Abstract
Advances in molecular technologies make it possible to pinpoint genomic factors associated with complex human traits. For cognition and behaviour, identification of underlying genes provides new entry points for deciphering the key neurobiological pathways. In the past decade, the search for genetic correlates of musicality has gained traction. Reports have documented familial clustering for different extremes of ability, including amusia and absolute pitch (AP), with twin studies demonstrating high heritability for some music-related skills, such as pitch perception. Certain chromosomal regions have been linked to AP and musical aptitude, while individual candidate genes have been investigated in relation to aptitude and creativity. Most recently, researchers in this field started performing genome-wide association scans. Thus far, studies have been hampered by relatively small sample sizes and limitations in defining components of musicality, including an emphasis on skills that can only be assessed in trained musicians. With opportunities to administer standardized aptitude tests online, systematic large-scale assessment of musical abilities is now feasible, an important step towards high-powered genome-wide screens. Here, we offer a synthesis of existing literatures and outline concrete suggestions for the development of comprehensive operational tools for the analysis of musical phenotypes.
Keywords: musicality, individual differences, genetics, aptitude tests
1. Introduction
During the past few decades, our understanding of human biology has been transformed by advances in molecular methods. It has become routine to apply genetic techniques to studies of biomedical disorders, as well as to related traits that show individual variation in the general population. Genetic research has yielded novel mechanistic insights relevant both to understanding disease as well as normal function. In recent years, researchers have extended the reach of genetics and genomics beyond standard biomedical traits and have begun to tackle complex human-specific cognitive abilities, such as speech and language, with some success [1]. Genetic analysis of aspects of musical aptitude is a field that is still in its infancy [2]. In the current article, we discuss progress thus far and consider the promise that the post-genomic era holds for shedding light on the biological bases of human musicality, broadly defined here as the capacity to perceive (perceptual abilities), reproduce or create music (production abilities).
As for language, the enormous variability of musical expressions found around the world bears the hallmarks of culture. However, like language, an emerging consensus suggests that musicality may have deep biological foundations, and so warrants examination from a genetic perspective [3]. At the same time, if a trait is largely limited to our own species, this poses special challenges for deciphering the underlying biology [4,5]. When investigating these kinds of human capacities, it is important that we move beyond questions of species universals, also to recognize the value of studying variability [6]. In particular, major tools of genetics depend on assessing variability in observable aspects of anatomy, physiology, development, cognition, behaviour and so on (phenotypes), and then searching for correlations with variations at the genetic level (genotypes). Variability in musical aptitude is well documented within human populations and is not limited to exceptional cases of virtuoso musicians, or (at another extreme) people who are unable to appreciate or engage with music, despite adequate opportunity [7]. Clear evidence has emerged showing considerable individual variation in music-related skills throughout the general population [8], variation that is likely to have at least some basis in biology. Concomitantly, recent efforts to catalogue comprehensively the natural variability in modern human genomes have revealed a surprising degree of variation within populations, affecting virtually every genetic locus in some way [9,10]. Thus, human populations can be effectively treated as natural experiments for identifying biologically meaningful links between individual variation at different levels [6], allowing researchers to trace causal connections between particular genes and phenotypes of interest, in this case key features of musicality. Once relevant genes have been pinpointed, they can be used as entry points into the critical neurobiological pathways and can potentially complement other approaches to understanding musicality (as discussed elsewhere in this issue).
This should not be taken to imply that there exists a specific ‘gene for music’. Genes cannot directly specify behavioural or cognitive outcomes. They have highly indirect effects at best, encoding molecules (RNAs and proteins) that influence the ways in which neurons proliferate, migrate, differentiate and connect up with each other during brain development, and/or modulate the plasticity of circuits during learning (e.g. [11]). Moreover, musicality is a complex multifaceted phenotype, itself comprising many potentially distinct abilities [8,12], and an array of different genes may be involved. At this point, the genetic architecture underlying music-related skills is largely unknown. While extremes of musical ability might plausibly involve some rare monogenic effects, still to be discovered, it is likely that individual differences in the general population involve variants at multiple interacting genetic loci, the number of which has not yet been determined. In addition, we are not suggesting that environmental influences should be neglected. Socio-cultural variables, exposure to music and years of music training are well-known environmental factors that impact on aptitude [13–15]. Indeed, musicality may constitute an ideal system for studying interactions between genes and environment [12,16,17].
People harbour a diverse range of distinct types of genetic variants, which differ in frequency, size and functional impact (see the electronic supplementary material for an overview). In recent years, the technology for characterizing genomic variation has advanced at an astonishing pace, as the time and resources needed for genotyping and sequencing have been dramatically reduced. DNA chips allow for hundreds of thousands of known genetic variants to be simultaneously genotyped rapidly and at low cost, and can easily be scaled up to studies involving thousands of people. The advent of next-generation DNA sequencing means that already the entire genome of a person can be determined for a few thousand dollars in a matter of days, and the field continues to move forward [18]. Nonetheless, it is important to stress that success in genetic studies of any human trait of interest depends critically on a solid strategy for defining and characterizing the phenotype. Thus, advances in human genomics need to be matched by parallel advances in the area of phenomics.
In this article, we first review the available evidence concerning the links between genes or chromosomal regions that have been associated with ‘extreme’ musical phenotypes—that is, phenotypes that are only found in a small percentage of the general population and correspond to congenital impairments in musical ability on the one hand, or to rare faculties (such as absolute pitch (AP)) on the other hand. We will then move on to variability within the normal range of musical aptitudes of the general population, considering traits such as relative pitch (RP), music perception skills, and musical production and creativity. Finally, we will outline future research directions for the field and propose concrete suggestions for the development of comprehensive operational tools for the analysis of musical phenotypes.
2. Musicality at the extremes
(a). Disorders of music perception
Genetic investigations of neurodevelopmental disorders such as speech apraxia, specific language impairment and dyslexia have been crucial for uncovering the molecular bases of human speech and language skills [19]. Similar approaches can help to reveal the biological underpinnings of musicality (table 1) [20,34]. About 3% of the general population have difficulty detecting notes that are out-of-key in melodies, against a background of normal hearing, language and intelligence, and adequate environmental exposure [35]. The condition, often called tone-deafness, is now referred to as congenital amusia to distinguish this lifelong disorder from acquired forms of amusia that occur as the result of brain lesion [36,37]. Congenital amusia is not only characterized by a deficit in detecting mistuning in both melodic and acoustical contexts, but also by an inability to recognize familiar tunes without the help of the lyrics and difficulties to sing in tune. In both perception and production, rhythm is relatively spared [38]. The biological basis of the condition is further supported by the identification of brain abnormalities affecting grey and white matter in the right auditory and inferior frontal cortex [39], as well as reduced connectivity between these two regions [37].
Table 1.
Investigating the biological bases of musicality through extreme phenotypes and known genetic syndromes. Examples are given of the different types of approaches discussed in this article, along with key results from the relevant studies.
| focus | type of study | key findings | citations |
|---|---|---|---|
| congenital amusia | familial aggregation | in nine large families (n = 71) with an amusic proband, 39% of first-degree relatives were affected, while in 10 control families (n = 75) prevalence was only 3%. Sibling recurrence risk ratio was estimated at approximately 10.8 | [20] |
| AP | familial aggregation | different studies estimated sibling recurrence risk ratios of approximately 7.5–15.1. Prevalence was higher in people with early musical training, and also in families of East-Asian ethnicity; direction of causation unknown | [16,21–23] |
| twin study | concordance in identical twins (78.6%, 14 pairs) was significantly higher than that seen in non-identical twins (45.2%, 31 pairs) | [24] | |
| pharmacology | adult males taking valproate (a drug hypothesized to affect critical periods) learned to identify pitch better than those taking placebos | [25] | |
| linkage analysis | study of 45 European and 19 East-Asian families with multiple AP cases found suggestive linkage for multiple chromosomal regions, with inconsistent patterns in the two datasets. Strongest linkage for chromosome 8q24 in European families | [26] | |
| linkage analysis | investigation of 53 families (49 European and four Asian) failed to replicate top linkage peaks from prior AP work. High rates (20.1%) of self-reported synaesthesia in AP led the authors to run combined linkage of 53 AP families with 36 synaesthesia families. Strongest joint linkage on chromosomes 6q14–q16 and 2q22-q24 | [27] | |
| musicality in known genetic syndromes | phenotyping | it has been suggested that children with WBS (due to 7q11.23 microdeletion) have increased auditory sensitivity, musical interest, creativity and expressivity. Other studies argue that these children show a wide range of musicality profiles, and some may even have elevated risk of amusia | [28–30] |
| phenotyping | rare mutations of the FOXP2 transcription factor gene cause a severe speech and language disorder. One study of musical ability in a particularly large family with a FOXP2 disruption suggested that mutation carriers had selective problems with perception and production of rhythms, while pitch-related abilities were normal | [31–33] |
Congenital amusia tends to show clustering within families (familial aggregation). That is, the condition is present at higher rates in relatives of affected people than expected on the basis of prevalence in the general population. In 2007, Peretz et al. [20] studied 71 members of nine large families with an amusic proband, and 75 members of 10 control families, using an online battery to assess amusia via an anomalous pitch detection task, a control time asynchrony detection task and a detailed questionnaire. The results confirmed that congenital amusia involves deficits in processing musical pitch but not musical time, and also showed strong evidence of familial aggregation. In amusic families, 39% of first-degree relatives were affected, compared with only 3% in control families [20]. The sibling recurrence risk ratio was estimated as approximately 10.8, meaning that if you are a sibling of someone with congenital amusia then you have an almost 11-fold increased risk of being amusic yourself.
Observations of familial aggregation are supportive of genetic involvement, but might also be (partly or wholly) explained by shared family environment. As explained in box 1, twin studies can be used to pull apart these effects and obtain a robust estimate of heritability. To our knowledge, no formal twin study of congenital amusia has yet been reported, but there has been a broader study showing strong heritability for pitch perception [40], as discussed in §3. Nonetheless, by collecting families in which multiple relatives show congenital amusia [20], it becomes possible to try mapping the locations of potential susceptibility genes. Such work is currently underway and will benefit from the recent advances in genomic technologies.
Box 1. Do genes contribute?
Even without molecular data, it is possible to investigate contributions of genetic factors to phenotypes of interest. For a qualitatively defined trait, such as presence or absence of a particular disorder, researchers can ask whether cases tend to cluster within families and assess whether inheritance is consistent with simple single-gene patterns of transmission, or more likely to involve multiple factors. Increased incidence of a trait in relatives of a proband is often taken as evidence of genetic involvement, but could also be due to environmental factors that are shared by family members. Twin studies allow these types of contributing factors to be teased apart. In its simplest form, this approach assesses concordance of a phenotype in pairs of identical twins (who have almost identical genomes) and compares it to the concordance seen for pairs of non-identical twins (who share around 50% of their genetic variations, just like non-twin siblings). Elevated concordance in the identical twins provides evidence of genetic involvement. In fact, twin designs typically go further by directly incorporating quantitative trait data and using the twin–twin correlation structure to partition the phenotypic variation into that due to additive genetic factors, common environment (shared by twins) and unique environment (unshared by twins). The proportion of phenotypic variance that is accounted for by genetics gives a formal estimate of heritability. Statistical tools have become more sophisticated over the years, and it is now routine to apply structural equation modelling and maximum-likelihood methods to large twin datasets, asking questions that extend far beyond heritability estimation. What is the contribution of genetic factors at different ages, and is this due to the same or different sets of genes? How much of the covariance between two correlated traits involves common genetic and/or environmental contributions? Are sex differences likely to play a role? Is there evidence of gene–environment interaction or correlation underlying a trait? Quantitative methods can also be used in multigenerational families for partitioning the observed phenotypic variance and estimating heritability (variance component models). Quantitative genetic methods depend on certain assumptions (outside the scope of the current article), some of which have been challenged. More importantly, the concept of heritability itself is very often misunderstood by non-specialists. Heritability is a useful statistic that describes variance in a given population at a specific time with a particular set of genetic variations and environmental factors. It is not an intrinsic fixed property of a phenotype, and it does not reveal anything about the biology of an individual, nor of how malleable a trait might be. For example, heritability estimates of certain features (including general intelligence) are well known to increase with age. Changes in environment (such as many of the developments of modern medicine) can radically alter the heritability of a trait, either diminishing or exaggerating the relative contributions of genetic variation.
In recent years, another form of congenital amusia that affects rhythm but not pitch has been discovered [41,42]. So far the number of cases that have been described is very small. Little is known about the prevalence of such disorders, and whether they show familial aggregation. This represents a potentially interesting area for future investigation.
(b). Rare faculties
AP, the ability to identify or produce a musical tone (e.g. middle C or concert A) without reference to an external standard [43], is an unusual skill found only in a small percentage of people. AP involves at least two separate cognitive skills: memory for pitch, which seems to be widespread among humans [44] and non-human animals [45], and the ability to attach labels to stimuli (e.g. classifying tones with different spectral characteristics, such as piano or voice, and consequently labelling their pitch class), which appears to be rarer [43]. In early reports, the prevalence of AP in the general population was estimated to be 1 in 10 000 [46], but more recent studies suggest that it may be found in as many as 1 in 1500 people [21]. It has been proposed that this is a dichotomous trait, with a clear phenotypic separation between AP possessors and non-AP possessors [47]. As a discrete and easily quantifiable cognitive phenotype, AP may be particularly feasible for use in genetic studies [47,48] (but see [49]). However, its relevance to musicality remains questionable, especially given that most professional musicians do not possess AP [48].
In 1988, Profita & Bidder [21] were among the first to explore the hypothesis of a genetic basis for the condition, in a study of 35 people with AP, across 19 families. Subsequent familial aggregation studies reported sibling recurrence risk ratios between 7.5 and 15.1 [16,22,23], consistent with a role for genetic factors. Further evidence of a significant genetic contribution has been found in studies of twins with AP; the concordance of the condition in 14 pairs of identical twins was 78.6%, as compared with a concordance of 45.2% in 31 pairs of non-identical twins [24].
Environmental factors are also strongly implicated in AP, albeit in a complex manner. A robust link between AP and early music training has been uncovered [16,23], with a significantly higher prevalence of the condition in people who began their musical training at a very young age. Thus, early music training could potentially be a crucial environmental factor contributing to AP. On the other hand, this same pattern of data could be explained by assuming that a genetic predisposition to AP increases the likelihood that a child receives early music training. Hence, the direction of causation is difficult to establish [16]. In any case, it seems likely that both early musical training and genetic predisposition contribute together to the development of AP. Another unexplained observation is that there are higher rates of AP for people of East-Asian ethnicity [23]. Again, there are several alternative hypotheses that could account for this well-documented effect; certain cultural groups may respond to early signs of AP with more intensive parental efforts at music education, the increased AP prevalence may be a consequence of culture-specific educational systems that are more effective at fostering this ability, or the findings may have a genetic explanation, reflecting ethnic differences in frequencies of susceptibility alleles [23].
A recent intriguing observation comes from studies of valproate, an inhibitor of the histone deacetylase enzyme, which can act to put a ‘brake’ on critical-period learning [50]. Administration of this enzyme to adult males apparently reopens the critical-period learning of AP [25]. Neuroimaging studies have also been revealing. Relative to non-AP possessors, AP possessors exhibit anatomical differences in the temporal lobe and other areas [51,52], as well as differences in the cortical processing of pitch information [53,54].
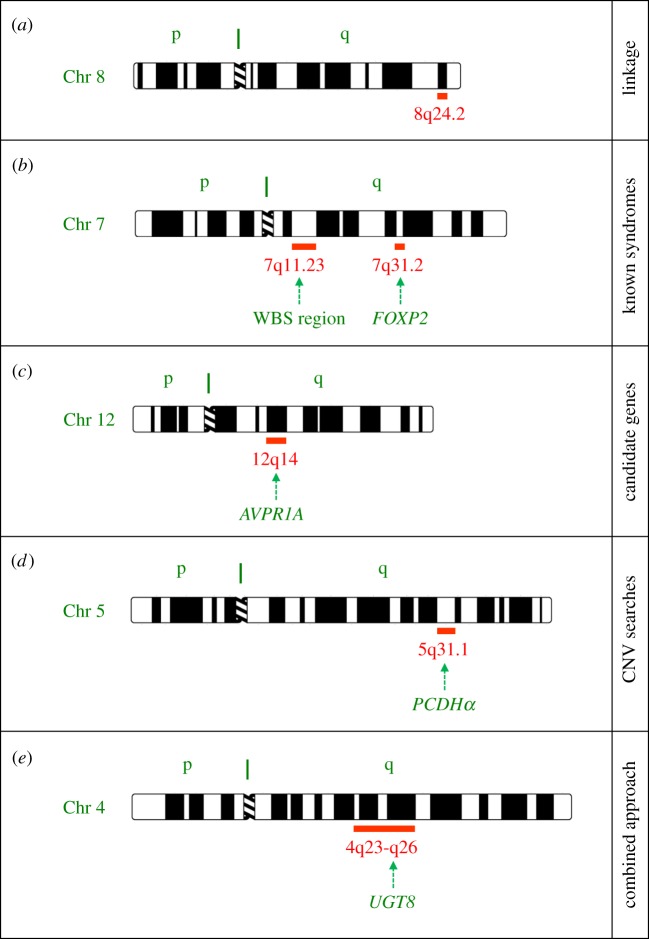
Researchers studying AP have used linkage analyses in families (box 2) to search for chromosomal regions that may harbour genes involved in the condition [26,27]. In a 2009 study, Theusch et al. [26] investigated 73 families with multiple AP-possessors including 45 families of European descent and 19 families with East-Asian ancestry. They found suggestive evidence for linkage to several different chromosomal regions, with strongest evidence on chromosomal band 8q24.21 in the European families (figure 1a). There was little consistency between the pattern of findings in the European and East-Asian datasets [26]; this genetic heterogeneity is interesting in light of the population differences in AP prevalence that have been documented.
Box 2. Tracing connections between genotypes and phenotypes.
Familial clustering and twin studies may provide support for genetic involvement in a human trait. How do we pinpoint the critical genes? In the early days of gene mapping, linkage analysis came to the fore. In this approach, researchers treat polymorphic genetic markers like signposts marking different chromosomal regions. They track how such genetic markers are transmitted to different members of a family, asking whether any particular chromosomal interval is linked to inheritance of the trait of interest. Robust statistical methods are used to ensure that an observed co-segregation between a genetic marker and the phenotype is not a chance finding. Linkage analysis is equally applicable to qualitative (i.e. dichotomous or ‘yes/no’) and quantitative traits, and can involve a pre-specified genetic model or be model-free. Data from different families may be combined; if the same genetic factors influence the phenotype (even if the precise mutation differs in each family), then this may help localize the gene(s) responsible. Nonetheless, linkage has low resolution, implicating large regions (loci) containing multiple genes, and is not well suited for detecting genetic effects that account for only a small proportion of phenotypic variance. Association analysis, a complementary method with different strengths and weaknesses, tests for correlations between particular gene variants and a trait at the population level. It has greater power than linkage to uncover small effect sizes and allows for higher resolution mapping. Still, due to linkage disequilibrium, a polymorphism that shows significant association is often not causal, but could be indexing an (as yet undiscovered) causal variant nearby. The first association studies typically focused on testing small numbers of polymorphisms from selected candidate genes, either based on hypotheses about the biology of the trait, or targeting regions highlighted by linkage. In recent years, it became quick and inexpensive to carry out systematic genome-wide genotyping capturing much of the polymorphic content of a phenotyped sample, allowing researchers to perform hypothesis-free association screening at high density across the genome. These screens involve an enormous amount of multiple testing (hundreds of thousands of polymorphisms in each individual), so rigorously adjusted thresholds for statistical significance are required to avoid false positives. Together with the fact that most complex traits are likely to involve many genes with very small effect sizes, such studies require sample sizes of thousands of individuals to achieve adequate power. In the post-genomic era, scientists also now make use of copy number variation (CNV) data and rare variations emerging from next-generation sequencing studies. Again, the key to success is use of robust statistics and replication in independent samples, to discount spurious genotype–phenotype relationships. Ultimately, studies of gene function in model systems are needed to demonstrate true causal connections.
Figure 1.
Connecting genes to musicality—some selected examples from the literature. Ideograms of chromosomes are shown with the cytogenetic bands of interest indicated. Each chromosome has a short (p) arm and a long (q) arm, separated by a structure called a centromere. When treated with certain stains, chromosomes display consistent banding patterns that are used to denote specific locations with respect to the centromere. (a) Linkage analysis of extreme phenotypes. The first linkage screen of families with AP highlighted a peak on chromosome 8q24.2 [26]. Subsequent AP studies have pointed instead to other regions elsewhere in the genome, some of which overlap with linkages to synaesthesia [27]. No specific AP-related genes have yet been identified. Linkage analysis has also been used to investigate musical aptitudes using quantitative phenotypes, as detailed in the main text. (b) Studies of musicality in known genetic disorders. WBS [55] and FOXP2-associated speech/language disorder [56], both involving chromosome 7, have been investigated in relation to musicality [28–30,57] and rhythm [33]. (c) Candidate genes. In some cases, particular candidate genes have been targeted based on hypotheses about their biological effects, and polymorphisms have been tested for association with music-related phenotypes. The AVPR1A gene is one well-studied example [58–61]. However, recent genome-wide screens failed to find significant effects for any prior-studied candidates, including AVPR1A [62,63]. (d) CNVs. A recent study searched for CNVs in people with low or high musical aptitude or musical creativity [64]. A number of interesting regions were reported, such as the PCDHα cluster on chromosome 5, found to be deleted in some individuals with low music. Nonetheless, as discussed in main text, for rare CNVs observed in only a few individuals it can be difficult to show causality, and so these findings await confirmation in independent samples. (e) Combined approach, using linkage, association, CNV analyses and sequencing. Park et al. [63] studied pitch-production accuracy in a multi-stage approach. They began with a linkage screen, identifying a broad linkage peak on chromosome 4q23, and followed up with association analyses of the surrounding region, eventually zooming in on the UGT8 gene in 4q26 as a candidate. Further independent evidence to support UGT8 came from identification of a CNV spanning that region as well as variants identified by large-scale sequencing. (Ideograms are adapted from http://www.pathology.washington.edu/research/cytopages/idiograms/human/.) (Online version in colour.)
In 2013, Gregersen et al. [27] studied an independent set of 53 AP families (49 European, four Asian) and identified modest evidence of linkage implicating different chromosomal regions from the prior work. More intriguingly, this later study also uncovered evidence suggesting phenotypic and genetic overlaps between AP and synaesthesia, another rare condition. For people with synaesthesia, a stimulus in one sensory modality automatically evokes a perceptual experience in another modality; for instance, particular pitches, keys or timbres may evoke specific sensations of colour. Like AP, synaesthesia is thought to involve genetic contributions, with some clues as to chromosomal regions of interest, but no definitive genes yet identified [65]. Gregersen et al. [27] uncovered unusually high rates (20.1%) of self-reported synaesthesia in people with AP, which motivated them to do a joint linkage study, combining their set of 53 AP families together with 36 families from a prior screen of synaesthesia [65]. Joint evidence of linkage was seen on chromosomes 6 and 2, but since the regions implicated are large, containing many genes (box 2), further studies are needed to pinpoint potential causal variants.
A possible drawback of most AP studies published so far is that they rely on the explicit labelling of pitches and are therefore limited to people with musical training. However, methods have been developed for detecting AP without requiring explicit labelling [66–68]. Thus, musical training may not be necessary for AP [69], underlining the need to test for the presence of this condition in non-musicians.
(c). Altered musicality in known genetic syndromes
The above discussions concern identification of rare music-specific conditions, followed by a search for genetic correlates. A complementary approach is to target existing syndromes, where the causative gene or genes are already known, and to investigate whether there are any consequences for musicality of affected people (table 1). This is an area that has been little explored, but could prove fruitful. Here, we will briefly mention two examples from the literature, both of which (by coincidence) involve genes on chromosome 7 (figure 1b). Williams–Beuren syndrome (WBS) is a well-characterized microdeletion syndrome with a prevalence of approximately 1 in 7500 people, in which as many as 28 neighbouring genes in 7q11.23 may be deleted [55]. People with WBS often show a distinctive cognitive/behavioural profile, which has been much studied by researchers interested in tracing connections between genes and brain functions. The typical WBS phenotype includes mild-to-moderate cognitive impairments, disparity between verbal and spatial skills, with receptive language being a relative strength compared with other abilities, as well as hypersociability, increased empathy, anxiety and attention deficits [55,57,70]. It has been argued that people with WBS show increased auditory sensitivity, heightened emotional responses to music and relative strengths in musical interest, creativity and expressivity, in contrast to other neurodevelopmental disorders [28]. On the other hand, close examinations of particular music perception, production and learning skills associated with WBS have revealed a more complex story, with considerable phenotypic variability from one affected person to another [29]. These issues are beyond the scope of the present article, but have been discussed in detail by Lense et al. [30], who found that incidence of amusia in WBS was probably higher than that seen in the general population.
Elsewhere on chromosome 7 lies FOXP2, a regulatory gene that modulates the expression of other genes [56]. Rare mutations that disrupt FOXP2 cause a severe speech and language disorder. Affected people have problems coordinating sequences of orofacial movements during speech (known as developmental verbal dyspraxia or childhood apraxia of speech), as well as spoken and written impairments in many aspects of expressive and receptive language. A number of different FOXP2 mutations have been identified thus far [56]. One of these has been particularly intensively studied, as it was found in 15 affected relatives of a large multigenerational pedigree, known as the KE family [31,32]. A study of musical ability in affected members of this family reported reduced performance in tasks involving perception and production of vocal and manual rhythms, while pitch-related abilities appeared to be preserved [33]. These findings are interesting in light of functional evidence implicating FOXP2 in sensorimotor integration and motor-skill learning [56]. Further studies of rhythmic abilities in the KE family and other independent cases of FOXP2 disruption are needed to shed further light on this area.
3. Genetic contributions to individual differences in the general population
We now turn our attention to the normal spectrum of musical abilities and survey findings linking individual phenotypic differences to genetic variation (summarized in table 2).
Table 2.
Investigating the biological bases of musicality through individual differences in the general population.
| focus | type of study | key findings | citations |
|---|---|---|---|
| pitch perception | twin study | performance on the Distorted Tunes Test [71] in 136 identical and 148 non-identical twin pairs from general population yielded heritability estimates of approximately 71–80% | [40] |
| musical memory | candidate genes | targeted study in 82 students reported provisional association of musical memory with an epistatic interaction between common promoter variants of the genes AVPR1A and SLC6A4 | [72] |
| pharmacology | arginine vasopressin was administered to 25 males, yielding effects on musical memory, mood and attentiveness, without affecting digit span | [73] | |
| battery of music perception tasks (Karma Test, Seashore Pitch and Rhythm Subtests) | genome-wide linkage scan | phenotypic scores in 15 families (n = 234) had heritabilities of 42% (Karma), 57% (Seashore Pitch), 21% (Seashore Rhythm) and 48% (composite score). Linkage screening revealed a significant peak on chromosome 4q22 and suggestive evidence at 8q13–21. A linkage region on 18q overlapped with one seen in prior studies of dyslexia | [74] |
| candidate genes | nineteen families (n = 343) were genotyped for polymorphisms of AVPR1A, SLC6A4, COMT, DRD2 and TPH1. Some haplotypes of AVPR1A were associated with aptitude on the music perception tasks. The other candidate genes showed no significant associations after multiple-testing correction | [60] | |
| screen for CNVs | study of five families (n = 170) and 172 unrelated subjects. Nine people with low perception scores carried a 5q31 deletion spanning PCDHα; deletion frequency did not significantly differ in people with high scores. Duplication of 8q24 (cf. AP) seen in one person with low scores, but absent in low-scoring relatives. Genome-wide CNV burden did not differ between people with high/low scores | [64] | |
| genome-wide linkage and association scan | linkage scan in 76 families (n = 767) identified strongest evidence at 4p14–13 and 4p12–q12, other peaks at 16q21–22, 18q12–21, 22q11. Locations of most linkages differed from prior music-related studies. Genome-wide association scan in same dataset found strongest evidence at 3q21 near to GATA2. Some association seen for PCDH7 in 4p15. No association for usual candidate genes (e.g. AVPR1A) | [62] | |
| musical creativity self-report | candidate genes | analysis of phenotypic scores in 19 families (n = 343) yielded heritabilities of 40% (composing), 46% (arranging), 62% (improvising) and 84% (composite score). Testing of genes AVPR1A, SLC6A4, COMT, DRD2 and TPH1 showed no significant associations after correction for multiple-testing | [60] |
| screen for CNVs | study of five families (n = 170) and 172 unrelated subjects. In two families, some people with high creativity carried a 2p22 duplication, spanning GALM. Genome-wide CNV burden did not differ between people with high/low creativity | [64] | |
| pitch production | genome-wide linkage scan and targeted association | seventy-three families (n = 1008) completed pitch-production task. Linkage screen in 70 families (n = 862) found a significant peak on 4q23. Genotyping of SNPs from the region in 53 families (n = 630) revealed significant association near to UGT8. Authors subsequently identified a non-synonymous SNP in UGT8, and a CNV deletion in the region, each showing association | [63] |
Given that pitch perception is a central component of musicality, and perhaps one of the most amenable for large-scale testing, it is not surprising that this facet has been examined more thoroughly than others. In one of the earliest twin studies conducted on music perception abilities in the general population, 136 identical and 148 non-identical twin pairs were administered the Distorted Tunes Test [71], in which they judged whether familiar melodies contained ‘wrong notes' [40]. The scores on this test, considered a proxy for the participants' ability to judge successive pitch intervals, were estimated to have a heritability of 71–80%, with no significant effect of shared environment.
Genetic contributions to AP have been studied more extensively, most likely because it can be treated as a dichotomous trait, but RP abilities are probably more relevant to everyday music listening [75]. Indeed, RP allows a listener to identify a familiar tune by means of its interval structure (or contour), instead of its constituent pitches (or absolute frequencies), and allows the detection of ‘wrong notes'. Importantly, AP and RP appear to correspond to two different pitch-processing abilities [76], and the RP performance of AP possessors is fairly variable [77–79]. A 2010 study by Hove et al. [80] shows that, as with AP, individuals of East-Asian ethnicity tend to display better RP abilities than Caucasian subjects. Interestingly, this East-Asian advantage did not extend to a rhythm perception task and was not modulated by tone language experience.
Few studies have examined the genetic correlates of musical memory, and so far these have focused on testing particular candidate genes for association (box 2 and figure 1c). The choice of candidate genes has been motivated by prior studies outside the music domain; for example, some studies of musical memory have targeted arginine vasopressin receptor 1a (AVPR1A) and serotonin transporter (SLC6A4) genes, because common polymorphisms of those genes had been previously reported to be associated with creative dance performance [58]. A study of musical and phonological memory in 82 students found provisional evidence that these skills were associated with a gene × gene epistatic interaction between promoter region polymorphisms of the two candidate genes [72]. In a follow-up to this work, intranasal administration of the arginine vasopressin hormone in 25 males was reported to affect musical working memory as well as mood and attentiveness levels, without influencing digit span test scores, suggesting a complex interaction between this hormone, musical memory and affective states [73]. Arginine vasopressin and its receptor have been broadly implicated in social behaviours in rodents and humans [81].
A series of studies investigated genetic contributions to musical aptitudes (at multiple levels from heritability to linkage mapping and association analyses) in an expanding sample of extended Finnish families (table 2) [59,60,62,64,74]. In the first of these studies [74], 15 families (234 people) were tested on a battery of music perception tests comprising the Karma Music Test [82], which measures participants' ability to detect structural changes in abstract sound patterns, and Seashore's Pitch and Rhythm Subtests, which are based on pairwise comparisons. By analysing the quantitative phenotype data using a variance component model (box 1), the authors obtained heritability estimates of 42% for the Karma Music Test, 57% for Seashore's Pitch Subtest, and 21% for Seashore's Rhythm Subtest. Linkage mapping using the quantitative traits revealed significant linkage on chromosome 4q22, as well as suggestive evidence on chromosome 8q13–21 [74]. The latter shows some overlap with a region of suggestive linkage identified in one of the AP studies [26], thus implying a potential link between general music perception aptitudes and rare faculties. Interestingly, there was also some evidence of linkage to a region on 18q that had previously been implicated in developmental dyslexia [83]. As noted in box 2, linkage regions are typically large and contain many different genes, so findings of overlapping linkages with distinct phenotypes need further investigation to establish whether there is indeed a shared genetic basis.
Later work by the Finnish group [60] tested for association of selected candidate genes (based on biological hypotheses from previous literature) with musical aptitudes, as measured by the Karma and Seashore tests, in an expanded dataset of 19 Finnish families (343 individuals). Participants were also probed about their musical creativity using a questionnaire; the resulting scores were highly heritable and correlated with performance on the music perception tests. The authors reported that certain haplotypes of AVPR1A (figure 1c) were associated with music perception aptitudes, while there was little support for the variants of the other candidate genes that they tested (serotonin transporter SLC6A4, catechol-O-methyltranferase COMT, dopamine receptor D2 DRD2 and tyrosine hydroxylase 1 TPH1). In a follow-up study involving AVPR1A and SCL6A4 polymorphisms [59], the music listening activities of 31 Finnish families (437 members) were surveyed, suggesting associations between AVPR1A haplotypes, but not SCL6A4 haplotypes, and active music listening.
The same research team also performed a preliminary investigation of genome-wide CNVs in five extended families and in 172 unrelated participants [64]. They used the quantitative scores on the Karma and Seashore tests to define cases of low musical aptitude in their sample. A deletion at 5q31.1 (figure 1d) was found in 54% of ‘low’ cases in two of the extended families, although the frequencies in the other members of these families were not reported, so the strength of the genotype–phenotype correlation remains unclear. In the set of unrelated participants, deletion of 5q31.1 was observed in 2 of 28 ‘low’ cases (7%), as compared to 0 of 40 cases of ‘high’ musical aptitude, but this difference in frequency is not statistically significant. Nonetheless, as the deletion spans the protocadherin alpha (PCDHα) gene cluster (figure 1d), which encodes cell adhesion proteins that are important for brain development, the observations warrant further investigation in samples with adequate power. One case of low musical aptitude in one of the large families carried a duplication of 8q24.22, overlapping with the top linkage region from an early study of AP [26], but this CNV did not segregate with the phenotype in the family, making the finding difficult to interpret. The authors also performed CNV analyses in relation to self-reports of musical creativity [64]. For example, they highlighted a duplication of 2p22.1 found in 27% of ‘creative’ relatives within two families; this CNV spanned glucose mutarotase (GALM), a gene that is linked to serotonin metabolism. There was no evidence that high/low musical aptitude or musical creativity were associated with an overall increase in CNVs or with an excess of large CNVs [64].
Most recently, the Finnish group conducted a genome-wide study of 767 individuals from 76 families, phenotyped with the music perception tests described above [62]. They screened hundreds of thousands of single-nucleotide polymorphisms (SNPs) across the genome, using these data to test not only for linkage, but also for association (box 2). The best evidence for linkage was found on chromosome 4, with strongest peaks at 4p14–13 and 4p12–q12. In this study, there were also weaker regions of linkage at other genomic locations on chromosome 4, including one that showed some overlap with the 4q22 interval implicated in the prior linkage screen on a smaller subset of the families [74]. Additional regions elsewhere in the genome showed evidence of linkage in the set of 76 families, including 16q21–22.1, 18q12.3–21.1 and 22q11.1–.21, but they did not replicate any findings from prior studies of music-related phenotypes. Moreover, none of the top linkage regions contained SNPs that showed robust evidence of association with the traits. Although linkage and association are different types of tests (box 2), it is unusual that there were no genetic markers showing convergent evidence from both methods [62]. Neighbouring the 4p14 linkage peak, but outside the region of linkage evidence, the authors identified association with SNPs that were next to protocadherin 7 (PCDH7), a gene known to be expressed in the cochlea and the amygdala. The strongest associations in the genome were observed for SNPs in 3q21.3, in the vicinity of the GATA2 (GATA-binding protein 2) gene. This gene encodes a transcription factor that determines identity of GABAergic neurons in the midbrain and has been implicated in development of several organs, including cochlear hair cells and the inferior colliculus. Overall, the study suggested interesting connections to known molecular pathways implicated in auditory processing, but did not support the findings from prior targeted studies on candidate genes such as AVPR1A [62].
No large-scale twin studies have focused specifically on music production abilities, although in 1989 Coon and Carey analysed music-related data obtained from an earlier survey containing a battery of personality and interest questionnaires [84]. Heritability estimates were higher for participation in singing activities than for self-reported music abilities. Heritability was higher for males than for females. A more recent study used self-reported data from 1685 twin pairs (12–24 years old) to estimate the heritability of aptitude and exceptional talent across different domains such as language, mathematics, sports, as well as music [85]. Heritability estimates for music aptitudes were again higher for males (66%) than for females (30%). However, in both studies, no objective assessment of musical abilities was obtained.
Research exploring genetic contributions to music production abilities has largely focused on singing abilities, which is probably the most widespread such behaviour in the general population. Morley and colleagues investigated the AVPR1A and SLC6A4 polymorphisms that were previously associated with musical abilities [60,72] (but see [62] for non-replication) and creative dancing [58], testing for their association with choir participation in 523 subjects [61]. Significant association was detected for a SLC6A4 polymorphism but not found for AVPR1A haplotypes proposed to be connected with musical skills in other studies.
Park et al. [63] invited 1008 individuals from 73 Mongolian families to participate in a pitch-production accuracy test. Family-based linkage analyses using over a thousand genetic markers across the genome identified a peak on 4q23 (figure 1e), in an interval that shows some overlap with regions of interest in studies of music perception [62,74] (note, however, that the genomic positions of the peak regions of chromosome 4 linkage in the most recent Finnish study [62] were somewhat different from earlier work on smaller samples [74]). The authors went on to investigate the linked region in detail, using data obtained from SNP genotyping in 53 of the families, and testing for association. They were eventually able to zoom in on a SNP near to the gene UGT8 (figure 1e) showing highly significant association with performance on the production task. Further analyses uncovered a non-synonymous SNP as well as a CNV in this region that provided further support for a relationship between UGT8 variations and musical phenotypes [63]. UDP glycosyltransferase 8 catalyses the transfer of galactose to ceramide, a key step in the biosynthesis of galactocerebrosides, which are important components of myelin membranes in the nervous system.
4. Phenomics of musicality in the post-genomic era
Dramatic advances in molecular technologies, particularly the development of next-generation DNA sequencing, are set to make a major impact on gene mapping studies of families with music-related disorders or exceptional skills. As for other cognitive traits, the road ahead will still be challenging, as it remains difficult to pinpoint aetiological gene variants against a genomic background containing many potential candidates, but developments in analyses of gene function will help to resolve this [1]. Moreover, the advent of high-throughput large-scale genotyping and sequencing now raise the potential to reliably detect complex genetic effects on musical abilities in the general population. Crucially, investigations of other complex human phenotypes indicate that thousands of participants are needed to achieve adequate power for genome-wide association scans (GWAS; box 2). The largest genetic association studies of musical skills reported thus far (e.g. [62,63]; table 2) have involved sample sizes that are small when compared with GWAS studies in other complex genetic traits, and so have been relatively underpowered. Studies with low power may fail to detect effects that are biologically real and, at the same time, are more susceptible to false-positive results, in which spurious genotype–phenotype correlations are observed [86]. The lack of replication of linkage and association findings in music-related studies thus far may stem in part from this issue of low power, especially given that the underlying genetic architecture (number of genes involved, effect sizes, etc.) is still unknown. Indeed, this is a problem that has broadly affected studies across human genetics as a whole, including investigations of many standard biomedical traits. These difficulties are now being overcome by improved study designs with high power to accommodate small genetic effect sizes and/or substantial degrees of heterogeneity.
The success of genetic studies of musical ability also depends critically on a robust, objective and reliable measure of the phenotype. Yet many of the studies discussed so far have used self-reports (e.g. musical creativity studies [60,64], twin studies on music production aptitudes [84,85]). Furthermore, as pointed out by Levitin [12], scores obtained on traditional assessments of musical aptitude, like the Seashore test, are not highly correlated with real-world musical achievement. The great majority of earlier tests were designed for specific music education purposes [87], and consequently tend to overlook other, more general, musical skills such as the abilities to verbally communicate about music and to use music to modulate emotional states efficiently [8,88].
Thus, there is a need for objective, validated, measures that correlate with expressed musicality and that can be used to assess large numbers of people systematically. Ideally, a test battery would have the following characteristics:
(1) Capture a broad array of musical abilities including the perception, memory and production of pitch and rhythm;
(2) Be designed to be administered to individuals with limited or no formal musical training, in order to obtain measures that are widely applicable to the general population;
(3) Have a version appropriate for preschool children, to investigate phenotypic differences before formal musical training;
(4) Cover a wide range of difficulty so that there is power to detect differences at both the low and high ends of ability, which may be most informative;
(5) Be culture-independent, or at least have culture-independent components, thus allowing comparisons between people from different cultures and reducing confounding factors when assessing potential genetic predispositions associated with specific phenotypes;
(6) Include covariates such as amount of musical training;
(7) Be designed to be administered robustly online to enable rapid large-scale phenotyping and
(8) Be of sufficiently short duration that large numbers of people will agree to participate.
A test battery that met these criteria could be administered to existing population cohorts that have already received genome-wide genotyping for studies unrelated to musical abilities. This kind of phenotype-driven approach could potentially be applied across multiple cohorts, and meta-analyses of the resulting GWAS datasets would yield suitably large sample sizes to achieve high power for detecting subtle genotype–phenotype connections. Other potential practical applications include fractionating musical ability by examining which aspects of musical ability correlate specifically with other cognitive traits or genetic characteristics [12].
While there have been critiques on fundamental issues of method and control in Web-delivered experiments [89,90], this type of data collection has great potential for music perception and cognition research, especially in domains where versatility and ecological validity are at stake [91,92]. Probing music perceptual skills can now be done reliably, due to recent technological advances in presenting audio over the Internet, for example by using file formats such as MPEG4 that guarantee optimal sound quality on different computer platforms at different transmission rates. However, when it comes to collecting and uploading individual sound files, there remains a lack of standardization, most notably with respect to timing. Therefore, music production experiments (such as, for instance, tapping or singing along with a stimulus) are still unreliable. Hence, at this point in time, it is most realistic to focus on phenotypes related to music perception abilities while also collecting information on other aspects of the phenotype through survey-style questionnaires.
There are several candidate components of musicality suggested in the literature [93]. With regard to perceptual abilities, RP [94,95], tonal encoding of pitch [96], beat or pulse perception [97,98] and metrical encoding of rhythm [98] are a good starting point for a phenomics of musicality. For example, the following specific tests could, in principle, be implemented in an Internet-based survey that could be administered to a broad population in less than 30 min:
(1) RP ability [99].
(2) Melodic memory [8].
(3) Beat perception: identifying the tempo of a musical excerpt, either by comparing two excerpts in different tempi and judging whether they are different or not, or by judging whether an isochronous rhythm is on or off the beat with respect to the underlying music (task based on [100,101]; cf. [102]).
(4) Metre perception: judging whether two excerpts are rhythmically (dis)similar using classes of rhythms in simple and compound metres (classification task based on [103]; cf. [104]).
One test battery covering most of these aspects, the Goldsmiths Musical Sophistication Index (Gold-MSI) questionnaire and test battery [8], has been validated on a large population. The Gold-MSI, which can be completed in 20 min, includes a melodic memory task based on a comparison paradigm [105], a beat-tracking task (based on [101]), and a self-report questionnaire covering a broad spectrum of musical behaviours. Furthermore, data from the Gold-MSI have been correlated and validated with other test batteries and with personality measures such as the Ten Item Personality Inventory (TIPI) [106].
Of course, for a fuller understanding of genetic contributions to musicality, there are many aspects of phenotypic variation beyond what is proposed above that could prove to be important. Certain of these aspects could potentially be probed in a less objective manner in questionnaires, or some of them might be administered to subsets of the thousands participating in the core 30 min test. For example, sensitivity to expressive timing nuances [107] or musical timbre [108] might be connected with consistent genetic variation. Psychophysical tasks measuring auditory streaming abilities [109] or the sensitivity to acoustical features such as roughness and harmonicity [110,111] could also prove informative, although the sound fidelity they require could be difficult to ensure in an online setting. Despite these difficulties with administration, such tasks are relatively culture-free and could form the basis for a test of musicality that could be administered across cultures. Musical production abilities, such as pitch reproduction accuracy (e.g. [63]) and metre tapping accuracy, are undoubtedly critical components of the musical phenotype, but as noted in §2 can be evaluated more reliably in the laboratory than via Internet-based experiments. Finally, it would be of great benefit to obtain indices of social and emotional responses to music, as well as musical behaviour in the sense of attendance at and participation in musical events. It would be possible to get at least crude estimates of these attributes through online questionnaires (cf. [112]).
5. Broader perspectives
A primary focus of this article has concerned the potential biological bases of individual differences in musical abilities. We note that the phenomics of musicality can also be investigated at the level of populations, although such studies typically involve comparing musical cultures and genetic relationships rather than assessing musical aptitudes. For example, one study has described a relationship between genetic distance and similarity in the folk music styles across 31 Eurasian nations [113]. A more recent report obtained significant correlations between folk song structure and mitochondrial DNA variation among nine indigenous Taiwanese populations [114]. The magnitude of these correlations was similar to that of the correlations between linguistic distance (based on lexical cognates) and genetic distance for the same populations. Interestingly, although musical and linguistic distances were both correlated with genetic distance, musical and linguistic distances were not significantly correlated with one another.
Crucially, genetic studies of individual differences in humans should be seen as one part of a broader framework for identifying the underpinnings of musicality. This might include comparative work assessing relevant skills in non-human animals [4]. Moreover, new possibilities are opened up once key genes have been identified; their evolutionary history can be traced, molecular networks can be teased apart in human neurons and ancestral functions can be studied in animal models. At the same time, the evolutionary history of cultural markers, including music, can be informed by phylogenetic studies comparing human populations, and possibly non-human animals. New technologies offer promising prospects in both respects. On the one hand, the fields of molecular and developmental neurobiology provide an ever-growing toolkit of sophisticated methods that can be used to decipher how particular genes of interest contribute to the development and plasticity of neural circuits in model systems and humans themselves. On the other hand, the implementation of online-based testing procedures enables a systematic assessment of musical aptitudes on an unprecedented scale. Together, these developments will likely result in a paradigmatic shift in this research field, ushering in a new era for the exploration of the biological bases of musicality.
Supplementary Material
Acknowledegments
The authors wish to thank David Huron for his suggestions and comments on an earlier version of the manuscript.
Glossary
- Allele
Alternative forms of the same gene or genomic position
- Association analysis
Statistical test of whether there are non-random correlations between a certain phenotypic trait (either a qualitatively defined affection status or a quantitative measure) and specific allelic variants
- Chromosomal band
Each human chromosome has a short arm (‘p’) and long arm (‘q’), separated by a centromere. Each chromosome arm is divided into regions, or cytogenetic bands, that can be seen using a microscope and special stains. These bands are labelled p1, p2, p3, q1, q2, q3, etc. counting from the centromere outwards. At higher resolutions, sub-bands can be seen within the bands, also numbered from centromere outwards
- Copy number variation
Structural alteration of a chromosome giving an abnormal number of copies of a particular section of DNA, due to a region of the genome being deleted or duplicated. Copy number variations (CNVs) may occur in an array of sizes, from hundreds to several million nucleotides
- Epistasis
When a single phenotype involves interactions between two or more genes
- Genotype
The genetic constitution of an individual. Can refer to the entire complement of genetic material, a specific gene or a set of genes
- Genome-wide association scans
Systematic hypothesis-free testing of association at hundreds of thousands (perhaps millions) of different genetic markers across the entire genome. Involves a huge amount of multiple testing, requiring appropriate adjustments when evaluating significance of results in order to avoid false-positive findings
- Haplotype
A cluster of several neighbouring polymorphisms on a chromosome that tend to be inherited together
- Heritability
The proportion of variability in a characteristic that can be attributed to genetic influences. A statistical description that applies to a specific population, it can vary if the environment changes
- Linkage analysis
Enables mapping of the rough genomic location of a gene implicated in a given trait. This method tracks the inheritance of polymorphic genetic markers as they are transmitted in families, assessing whether they co-segregate with a trait of interest, in a way that is unlikely to be due to chance
- Non-synonymous
A nucleotide change in a gene that alters the amino acid sequence of the encoded protein. Contrasts with synonymous substitutions which preserve the usual amino acid sequence
- Next-generation DNA sequencing
Newly emerged high-throughput technologies that allow DNA sequences to be determined at dramatically lower costs and much more rapidly than the standard approaches that were previously available
- Phenomics
The robust measurement of physical, biochemical, physiological and behavioural traits of organisms, and how they alter due to changes in genes and environment
- Phenotype
The appearance of an individual in terms of a particular characteristic; physical, biochemical, physiological, etc. resulting from interactions between genotype, environment and random factors
- Polymorphism
A position in the genome that contains variation in the population and therefore has more than one possible allele. At present, the most commonly studied of these are SNPs involving a single nucleotide at a specific point in the genome
- Proband
The index case who triggers investigation of a particular family to isolate the potential genetic factors involved in a given trait
- Promoter region
A region at the beginning of each gene that is responsible for its regulation, allowing it to be switched on or off in distinct cell types and developmental stages
- Recurrence risk
The likelihood that a trait will be observed elsewhere in a pedigree, given that at least one family member exhibits the trait. Can be defined for specific types of relationships, such as siblings
Author contributions
B.G. and S.E.F. wrote the majority of the initial draft of the paper. H.H. and L.T. contributed to the section ‘Phenomics of musicality’. I.P. contributed to the section ‘Musicality at the extremes'. This article is based on ideas exchanged during the ‘What Makes Us Musical Animals? Cognition, Biology and the Origins of Musicality’ workshop which took place at the Lorentz Center (Leiden, The Netherlands) in April 2014. All authors contributed to the ideas presented in this article and edited the manuscript.
References
- 1.Deriziotis P, Fisher SE. 2013. Neurogenomics of speech and language disorders: the road ahead. Genome Biol. 14, 204 ( 10.1186/gb-2013-14-4-204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan YT, McPherson GE, Peretz I, Berkovic SF, Wilson SJ. 2014. The genetic basis of music ability. Front. Psychol. 5, 658 ( 10.3389/fpsyg.2014.00658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peretz I. 2006. The nature of music from a biological perspective. Cognition 100, 1–32. ( 10.1016/j.cognition.2005.11.004) [DOI] [PubMed] [Google Scholar]
- 4.Hoeschele M, Merchant H, Kikuchi Y, Hattori Y, ten Cate C. 2015. Searching for the origins of musicality across species. Phil. Trans. R. Soc. B 370, 20140094 ( 10.1098/rstb.2014.0094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohrmeier M, Zuidema W, Wiggins GA, Scharff C. 2015. Principles of structure building in music, language and animal song. Phil. Trans. R. Soc. B 370, 20140097 ( 10.1098/rstb.2014.0097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vernes SC, Fisher SE. 2015. Genetics and the language sciences. Annu. Rev. Linguist. 1, 6.1–6.22. ( 10.1146/annurev-linguist-030514-125024) [DOI] [Google Scholar]
- 7.Peretz I. 2013. The biological foundations of music: insights from congenital amusia. In Psychology of music (ed. Deutsch D.), pp. 551–564. San Diego, CA: Elsevier. [Google Scholar]
- 8.Mullensiefen D, Gingras B, Musil J, Stewart L. 2014. The musicality of non-musicians: an index for assessing musical sophistication in the general population. PLoS ONE 9, e89642 ( 10.1371/journal.pone.0089642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65. ( 10.1038/nature11632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lappalainen T, et al. 2013. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511. ( 10.1038/nature12531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher SE. 2006. Tangled webs: tracing the connections between genes and cognition. Cognition 101, 270–297. ( 10.1016/j.cognition.2006.04.004) [DOI] [PubMed] [Google Scholar]
- 12.Levitin DJ. 2012. What does it mean to be musical? Neuron 73, 633–637. ( 10.1016/j.neuron.2012.01.017) [DOI] [PubMed] [Google Scholar]
- 13.Hargreaves DJ, Zimmerman MP. 1992. Developmental theories of music learning. In Handbook of research on music teaching and learning: a project of the music educators national conference (ed. Colwell R.), pp. 377–391. New York, NY: Schirmer. [Google Scholar]
- 14.Hannon EE, Trainor LJ. 2007. Music acquisition: effects of enculturation and formal training on development. Trends Cogn. Sci. 11, 466–472. ( 10.1016/J.Tics.2007.08.008) [DOI] [PubMed] [Google Scholar]
- 15.Trainor LJ, Unrau A. 2012. Development of pitch and music perception. Springer Handb. Audit. 42, 223–254. ( 10.1007/978-1-4614-1421-6_8) [DOI] [Google Scholar]
- 16.Baharloo S, Johnston PA, Service SK, Gitschier J, Freimer NB. 1998. Absolute pitch: an approach for identification of genetic and nongenetic components. Am. J. Hum. Genet. 62, 224–231. ( 10.1086/301704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hambrick DZ, Tucker-Drob EM. In press. The genetics of music accomplishment: evidence for gene-environment correlation and interaction. Psychon. Bull. Rev. ( 10.3758/s13423-014-0671-9) [DOI] [PubMed] [Google Scholar]
- 18.Gilad Y, Pritchard JK, Thornton K. 2009. Characterizing natural variation using next-generation sequencing technologies. Trends Genet. 25, 463–471. ( 10.1016/j.tig.2009.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham SA, Fisher SE. 2013. Decoding the genetics of speech and language. Curr. Opin. Neurobiol. 23, 43–51. ( 10.1016/j.conb.2012.11.006) [DOI] [PubMed] [Google Scholar]
- 20.Peretz I, Cummings S, Dube MP. 2007. The genetics of congenital amusia (tone deafness): a family-aggregation study. Am. J. Hum. Genet. 81, 582–588. ( 10.1086/521337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Profita J, Bidder TG. 1988. Perfect pitch. Am. J. Med. Genet. 29, 763–771. ( 10.1002/ajmg.1320290405) [DOI] [PubMed] [Google Scholar]
- 22.Baharloo S, Service SK, Risch N, Gitschier J, Freimer NB. 2000. Familial aggregation of absolute pitch. Am. J. Hum. Genet. 67, 755–758. ( 10.1086/303057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregersen PK, Kowalsky E, Kohn N, Marvin EW. 1999. Absolute pitch: prevalence, ethnic variation, and estimation of the genetic component. Am. J. Hum. Genet. 65, 911–913. ( 10.1086/302541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theusch E, Gitschier J. 2011. Absolute pitch twin study and segregation analysis. Twin Res. Hum. Genet. 14, 173–178. ( 10.1375/twin.14.2.173) [DOI] [PubMed] [Google Scholar]
- 25.Gervain J, Vines BW, Chen LM, Seo RJ, Hensch TK, Werker JF, Young AH. 2013. Valproate reopens critical-period learning of absolute pitch. Front. Syst. Neurosci. 7, 102 ( 10.3389/fnsys.2013.00102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theusch E, Basu A, Gitschier J. 2009. Genome-wide study of families with absolute pitch reveals linkage to 8q24.21 and locus heterogeneity. Am. J. Hum. Genet. 85, 112–119. ( 10.1016/j.ajhg.2009.06.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregersen PK, Kowalsky E, Lee A, Baron-Cohen S, Fisher SE, Asher JE, Ballard D, Freudenberg J, Li W. 2013. Absolute pitch exhibits phenotypic and genetic overlap with synesthesia. Hum. Mol. Genet. 22, 2097–2104. ( 10.1093/hmg/ddt059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levitin DJ. 2005. Musical behavior in a neurogenetic developmental disorder: evidence from Williams Syndrome. Ann. NY Acad. Sci. 1060, 325–334. ( 10.1196/annals.1360.027) [DOI] [PubMed] [Google Scholar]
- 29.Lense M, Dykens E. 2013. Musical learning in children and adults with Williams syndrome. J. Intellect. Disabil. Res. 57, 850–860. ( 10.1111/j.1365-2788.2012.01611.x) [DOI] [PubMed] [Google Scholar]
- 30.Lense MD, Shivers CM, Dykens EM. 2013. (A)musicality in Williams syndrome: examining relationships among auditory perception, musical skill, and emotional responsiveness to music. Front. Psychol. 4, 525 ( 10.3389/fpsyg.2013.00525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher SE, Vargha-Khadem F, Watkins KE, Monaco AP, Pembrey ME. 1998. Localisation of a gene implicated in a severe speech and language disorder. Nat. Genet. 18, 168–170. ( 10.1038/ng0298-168) [DOI] [PubMed] [Google Scholar]
- 32.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. 2001. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413, 519–523. ( 10.1038/35097076) [DOI] [PubMed] [Google Scholar]
- 33.Alcock KJ, Passingham RE, Watkins K, Vargha-Khadem F. 2000. Pitch and timing abilities in inherited speech and language impairment. Brain Lang. 75, 34–46. ( 10.1006/brln.2000.2323) [DOI] [PubMed] [Google Scholar]
- 34.Stewart L. 2008. Fractionating the musical mind: insights from congenital amusia. Curr. Opin. Neurobiol. 18, 127–130. ( 10.1016/j.conb.2008.07.008) [DOI] [PubMed] [Google Scholar]
- 35.Peretz I, Hyde KL. 2003. What is specific to music processing? Insights from congenital amusia. Trends Cogn. Sci. 7, 362–367. ( 10.1016/S1364-6613(03)00150-5) [DOI] [PubMed] [Google Scholar]
- 36.Peretz I, Ayotte J, Zatorre RJ, Mehler J, Ahad P, Penhune VB, Jutras B. 2002. Congenital amusia: a disorder of fine-grained pitch discrimination. Neuron 33, 185–191. ( 10.1016/S0896-6273(01)00580-3) [DOI] [PubMed] [Google Scholar]
- 37.Hyde KL, Zatorre RJ, Peretz I. 2011. Functional MRI evidence of an abnormal neural network for pitch processing in congenital amusia. Cereb. Cortex 21, 292–299. ( 10.1093/cercor/bhq094) [DOI] [PubMed] [Google Scholar]
- 38.Hyde KL, Peretz I. 2004. Brains that are out of tune but in time. Psychol. Sci. 15, 356–360. ( 10.1111/j.0956-7976.2004.00683.x) [DOI] [PubMed] [Google Scholar]
- 39.Hyde KL, Lerch JP, Zatorre RJ, Griffiths TD, Evans AC, Peretz I. 2007. Cortical thickness in congenital amusia: when less is better than more. J. Neurosci. 27, 13 028–13 032. ( 10.1523/JNEUROSCI.3039-07.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drayna D, Manichaikul A, de Lange M, Snieder H, Spector T. 2001. Genetic correlates of musical pitch recognition in humans. Science 291, 1969–1972. ( 10.1126/science.291.5510.1969) [DOI] [PubMed] [Google Scholar]
- 41.Phillips-Silver J, Toiviainen P, Gosselin N, Piche O, Nozaradan S, Palmer C, Peretz I. 2011. Born to dance but beat deaf: a new form of congenital amusia. Neuropsychologia 49, 961–969. ( 10.1016/j.neuropsychologia.2011.02.002) [DOI] [PubMed] [Google Scholar]
- 42.Launay J, Grube M, Stewart L. 2014. Dysrhythmia: a specific congenital rhythm perception deficit. Front. Psychol. 5, 18 ( 10.3389/fpsyg.2014.00018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deutsch D. 2013. Absolute pitch. In Psychology of music, 3rd edn, pp. 141–182. San Diego, CA: Elsevier. [Google Scholar]
- 44.Schellenberg EG, Trehub SE. 2003. Good pitch memory is widespread. Psychol. Sci. 14, 262–266. ( 10.1111/1467-9280.03432) [DOI] [PubMed] [Google Scholar]
- 45.Weisman RG, Mewhort DJK, Hoeschele M, Sturdy CB. 2012. New perspectives on absolute pitch in birds and mammals. In Handbook of comparative cognition (eds Wasserman EA, Zentall TR.), pp. 67–82. New York, NY: Oxford University Press. [Google Scholar]
- 46.Bachem A. 1955. Absolute pitch. J. Acoust. Soc. Am. 27, 1180–1185. ( 10.1121/1.1908155) [DOI] [Google Scholar]
- 47.Athos EA, Levinson B, Kistler A, Zemansky J, Bostrom A, Freimer N, Gitschier J. 2007. Dichotomy and perceptual distortions in absolute pitch ability. Proc. Natl Acad. Sci. USA 104, 14 795–14 800. ( 10.1073/pnas.0703868104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregersen PK. 1998. Instant recognition: the genetics of pitch perception. Am. J. Hum. Genet. 62, 221–223. ( 10.1086/301734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitouch O. 2003. Absolutist models of absolute pitch are absolutely misleading. Music Percept. 21, 111–117. ( 10.1525/mp.2003.21.1.111) [DOI] [Google Scholar]
- 50.Morishita H, Hensch TK. 2008. Critical period revisited: impact on vision. Curr. Opin. Neurobiol. 18, 101–107. ( 10.1016/j.conb.2008.05.009) [DOI] [PubMed] [Google Scholar]
- 51.Loui P, Li HC, Hohmann A, Schlaug G. 2011. Enhanced cortical connectivity in absolute pitch musicians: a model for local hyperconnectivity. J. Cogn. Neurosci. 23, 1015–1026. ( 10.1162/jocn.2010.21500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bermudez P, Lerch JP, Evans AC, Zatorre RJ. 2009. Neuroanatomical correlates of musicianship as revealed by cortical thickness and voxel-based morphometry. Cereb. Cortex 19, 1583–1596. ( 10.1093/cercor/bhn196) [DOI] [PubMed] [Google Scholar]
- 53.Loui P, Zamm A, Schlaug G. 2012. Enhanced functional networks in absolute pitch. Neuroimage 63, 632–640. ( 10.1016/j.neuroimage.2012.07.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zatorre RJ, Perry DW, Beckett CA, Westbury CF, Evans AC. 1998. Functional anatomy of musical processing in listeners with absolute pitch and relative pitch. Proc. Natl Acad. Sci. USA 95, 3172–3177. ( 10.1073/pnas.95.6.3172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martens MA, Wilson SJ, Reutens DC. 2008. Williams syndrome: a critical review of the cognitive, behavioral, and neuroanatomical phenotype. J. Child Psychol. Psychiatry 49, 576–608. ( 10.1111/j.1469-7610.2008.01887.x) [DOI] [PubMed] [Google Scholar]
- 56.Fisher SE, Scharff C. 2009. FOXP2 as a molecular window into speech and language. Trends Genet. 25, 166–177. ( 10.1016/j.tig.2009.03.002) [DOI] [PubMed] [Google Scholar]
- 57.Levitin DJ, Cole K, Chiles M, Lai Z, Lincoln A, Bellugi U. 2004. Characterizing the musical phenotype in individuals with Williams Syndrome. Child Neuropsychol. 10, 223–247. ( 10.1080/09297040490909288) [DOI] [PubMed] [Google Scholar]
- 58.Bachner-Melman R, et al. 2005. AVPR1a and SLC6A4 gene polymorphisms are associated with creative dance performance. PLoS Genet. 1, e42 ( 10.1371/journal.pgen.0010042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ukkola-Vuoti L, Oikkonen J, Onkamo P, Karma K, Raijas P, Jarvela I. 2011. Association of the arginine vasopressin receptor 1A (AVPR1A) haplotypes with listening to music. J. Hum. Genet. 56, 324–329. ( 10.1038/jhg.2011.13) [DOI] [PubMed] [Google Scholar]
- 60.Ukkola LT, Onkamo P, Raijas P, Karma K, Jarvela I. 2009. Musical aptitude is associated with AVPR1A-haplotypes. PLoS ONE 4, e5534 ( 10.1371/journal.pone.0005534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morley AP, Narayanan M, Mines R, Molokhia A, Baxter S, Craig G, Lewis CM, Craig I. 2012. AVPR1A and SLC6A4 polymorphisms in choral singers and non-musicians: a gene association study. PLoS ONE 7, e31763 ( 10.1371/journal.pone.0031763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oikkonen J, Huang Y, Onkamo P, Ukkola-Vuoti L, Raijas P, Karma K, Vieland VJ, Jarvela I. In press. A genome-wide linkage and association study of musical aptitude identifies loci containing genes related to inner ear development and neurocognitive functions. Mol. Psychiatry. ( 10.1038/mp.2014.8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park H, et al. 2012. Comprehensive genomic analyses associate UGT8 variants with musical ability in a Mongolian population. J. Med. Genet. 49, 747–752. ( 10.1136/jmedgenet-2012-101209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ukkola-Vuoti L, Kanduri C, Oikkonen J, Buck G, Blancher C, Raijas P, Karma K, Lahdesmaki H, Jarvela I. 2013. Genome-wide copy number variation analysis in extended families and unrelated individuals characterized for musical aptitude and creativity in music. PLoS ONE 8, e56356 ( 10.1371/journal.pone.0056356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asher JE, Lamb JA, Brocklebank D, Cazier JB, Maestrini E, Addis L, Sen M, Baron-Cohen S, Monaco AP. 2009. A whole-genome scan and fine-mapping linkage study of auditory-visual synesthesia reveals evidence of linkage to chromosomes 2q24, 5q33, 6p12, and 12p12. Am. J. Hum. Genet. 84, 279–285. ( 10.1016/j.ajhg.2009.01.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ross DA, Olson IR, Marks LE, Gore JC. 2004. A nonmusical paradigm for identifying absolute pitch possessors. J. Acoust. Soc. Am. 116, 1793–1799. ( 10.1121/1.1758973) [DOI] [PubMed] [Google Scholar]
- 67.Ross DA, Marks LE. 2009. Absolute pitch in children prior to the beginning of musical training. Ann. NY Acad. Sci. 1169, 199–204. ( 10.1111/j.1749-6632.2009.04847.x) [DOI] [PubMed] [Google Scholar]
- 68.Plantinga J, Trainor LJ. 2008. Infants’ memory for isolated tones and the effects of interference. Music Percept. 26, 121–127. ( 10.1525/Mp.2008.26.2.121). [DOI] [Google Scholar]
- 69.Ross DA, Olson IR, Gore JC. 2003. Absolute pitch does not depend on early musical training. Ann. NY Acad. Sci. 999, 522–526. ( 10.1196/annals.1284.065) [DOI] [PubMed] [Google Scholar]
- 70.Ng R, Lai P, Levitin DJ, Bellugi U. 2013. Musicality correlates with sociability and emotionality in Williams syndrome. J. Ment. Health. Res. Intellect. Disabil. 6, 268–279. ( 10.1080/19315864.2012.683932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalmus H, Fry DB. 1980. On tune deafness (dysmelodia): frequency, development, genetics and musical background. Ann. Hum. Genet. 43, 369–382. ( 10.1111/j.1469-1809.1980.tb01571.x) [DOI] [PubMed] [Google Scholar]
- 72.Granot RY, Frankel Y, Gritsenko V, Lerer E, Gritsenko I, Bachner-Melman R, Israel S, Ebstein RP. 2007. Provisional evidence that the arginine vasopressin 1a receptor gene is associated with musical memory. Evol. Hum. Behav. 28, 313–318. ( 10.1016/j.evolhumbehav.2007.05.003) [DOI] [Google Scholar]
- 73.Granot RY, Uzefovsky F, Bogopolsky H, Ebstein RP. 2013. Effects of arginine vasopressin on musical working memory. Front. Psychol. 4, 712 ( 10.3389/fpsyg.2013.00712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pulli K, Karma K, Norio R, Sistonen P, Goring HH, Jarvela I. 2008. Genome-wide linkage scan for loci of musical aptitude in Finnish families: evidence for a major locus at 4q22. J. Med. Genet. 45, 451–456. ( 10.1136/jmg.2007.056366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miyazaki K. 2004. How well do we understand absolute pitch? Acoust. Sci. Technol. 25, 426–432. ( 10.1250/ast.25.426) [DOI] [Google Scholar]
- 76.Ziv N, Radin S. 2014. Absolute and relative pitch: global versus local processing of chords. Adv. Cogn. Psychol. 10, 15–25. ( 10.2478/v10053-008-0152-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miyazaki K, Rakowski A. 2002. Recognition of notated melodies by possessors and nonpossessors of absolute pitch. Percept. Psychophys. 64, 1337–1345. ( 10.3758/BF03194776) [DOI] [PubMed] [Google Scholar]
- 78.Renninger LB, Granot RI, Donchin E. 2003. Absolute pitch and the P300 component of the event-related potential: and exploration of variables that may account for individual differences. Music Percept. 20, 357–382. ( 10.1525/mp.2003.20.4.357) [DOI] [Google Scholar]
- 79.Miyazaki K. 1993. Absolute pitch as an inability: identification of musical intervals in a tonal context. Music Percept. 11, 55–72. ( 10.2307/40285599) [DOI] [Google Scholar]
- 80.Hove MJ, Sutherland ME, Krumhansl CL. 2010. Ethnicity effects in relative pitch. Psychon. Bull. Rev. 17, 310–316. ( 10.3758/PBR.17.3.310) [DOI] [PubMed] [Google Scholar]
- 81.Insel TR. 2010. The challenge of translation in social neuroscience: a review of oxytocin, vasopressin, and affiliative behavior. Neuron 65, 768–779. ( 10.1016/j.neuron.2010.03.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karma K. 2007. Musical aptitude definition and measure validation: ecological validity can endanger the construct validity of musical aptitude tests. Psychomusicology 19, 79–90. ( 10.1037/h0094033) [DOI] [Google Scholar]
- 83.Fisher SE, et al. 2002. Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat. Genet. 30, 86–91. ( 10.1038/ng792) [DOI] [PubMed] [Google Scholar]
- 84.Coon H, Carey G. 1989. Genetic and environmental determinants of musical ability in twins. Behav. Genet. 19, 183–193. ( 10.1007/BF01065903) [DOI] [PubMed] [Google Scholar]
- 85.Vinkhuyzen AA, van der Sluis S, Posthuma D, Boomsma DI. 2009. The heritability of aptitude and exceptional talent across different domains in adolescents and young adults. Behav. Genet. 39, 380–392. ( 10.1007/s10519-009-9260-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, Munafo MR. 2013. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 14, 365–376. ( 10.1038/nrn3475) [DOI] [PubMed] [Google Scholar]
- 87.Boyle JD, Radocy RE. 1987. Measurement and evaluation of musical experiences. New York, NY: Schirmer Books. [Google Scholar]
- 88.Murphy C. 1999. How far do tests of musical ability shed light on the nature of musical intelligence? Br. J. Music Edu. 16, 39–50. ( 10.1017/S0265051799000133) [DOI] [Google Scholar]
- 89.Mehler J. 1999. Experiments carried out over the Web. Cognition 71, 187–189. ( 10.1016/S0010-0277(99)00029-3) [DOI] [PubMed] [Google Scholar]
- 90.McGraw KO, Tew MD, Williams JE. 2000. The integrity of Web-delivered experiments: can you trust the data? Psychol. Sci. 11, 502–506. [DOI] [PubMed] [Google Scholar]
- 91.Honing H, Reips U. 2008. Web-based versus lab-based studies: a response to Kendall. Emp. Musicol. Rev. 3, 73–77. [Google Scholar]
- 92.Honing H, Ladinig O. 2008. The potential of the internet for music perception research: a comment on lab-based versus Web-based studies. Emp. Musicol. Rev. 3, 4–7. [Google Scholar]
- 93.Honing H, ten Cate C, Peretz I, Trehub SE. 2015. Without it no music: cognition, biology and evolution of musicality. Phil. Trans. R. Soc. B 370, 20140088 ( 10.1098/rstb.2014.0088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trehub SE. 2003. The developmental origins of musicality. Nat. Neurosci. 6, 669–673. ( 10.1038/Nn1084) [DOI] [PubMed] [Google Scholar]
- 95.Justus T, Hutsler JJ. 2005. Fundamental issues in the evolutionary psychology of music: assessing innateness and domain specificity. Music Percept. 23, 1–27. ( 10.1525/Mp.2005.23.1.1) [DOI] [Google Scholar]
- 96.Peretz I, Coltheart M. 2003. Modularity of music processing. Nat. Neurosci. 6, 688–691. ( 10.1038/Nn1083) [DOI] [PubMed] [Google Scholar]
- 97.Honing H. 2012. Without it no music: beat induction as a fundamental musical trait. Ann. NY Acad. Sci. 1252, 85–91. ( 10.1111/J.1749-6632.2011.06402.X) [DOI] [PubMed] [Google Scholar]
- 98.Fitch WT. 2013. Rhythmic cognition in humans and animals: distinguishing meter and pulse perception. Front. Syst. Neurosci. 7, 68 ( 10.3389/fnsys.2013.00068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mullensiefen D, Halpern AR. 2014. The role of features and context in recognition of novel melodies. Music Percept. 31, 418–435. ( 10.1525/Mp.2014.31.5.418) [DOI] [Google Scholar]
- 100.Iversen JR, Patel AD, Ohgushi K. 2008. Perception of rhythmic grouping depends on auditory experience. J. Acoust. Soc. Am. 124, 2263–2271. ( 10.1121/1.2973189) [DOI] [PubMed] [Google Scholar]
- 101.Iversen JR, Patel AD. 2008. The Beat Alignment Test (BAT): surveying beat processing abilities in the general population. In 10th Int. Conf. on Music Perception and Cognition ICMPC10, Sapporo, Japan, 25–29 August 2008, pp. 465–468. Sapporo, Japan: Hokkaido Printing Planning. [Google Scholar]
- 102.Honing H. 2013. Structure and interpretation of rhythm in music. In Psychology of music, 3rd edn (ed. Deutsch D.), pp. 369–404. London, UK: Academic Press. [Google Scholar]
- 103.Hannon EE, Trehub SE. 2005. Metrical categories in infancy and adulthood. Psychol. Sci. 16, 48–55. ( 10.1111/j.0956-7976.2005.00779.x) [DOI] [PubMed] [Google Scholar]
- 104.Cao E, Lotstein M, Johnson-Laird P. 2014. Similarity and families of musical rhythms. Music Percept. 31, 444–469. ( 10.1525/mp.2014.31.5.444) [DOI] [Google Scholar]
- 105.Bartlett JC, Dowling WJ. 1980. Recognition of transposed melodies: a key-distance effect in developmental perspective. J. Exp. Psychol. Hum. Percept. Perform. 6, 501–515. ( 10.1037/0096-1523.6.3.501) [DOI] [PubMed] [Google Scholar]
- 106.Gosling SD, Rentfrow PJ, Swann WB., Jr 2003. A very brief measure of the Big-Five personality domains. J. Res. Pers. 37, 504–528. ( 10.1016/S0092-6566(03)00046-1) [DOI] [Google Scholar]
- 107.Honing H, Ladinig O. 2009. Exposure influences expressive timing judgments in music. J. Exp. Psychol. Hum. Percept. Perform. 35, 281–288. ( 10.1037/a0012732) [DOI] [PubMed] [Google Scholar]
- 108.Law LN, Zentner M. 2012. Assessing musical abilities objectively: construction and validation of the profile of music perception skills. PLoS ONE 7, e52508 ( 10.1371/journal.pone.0052508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huron D. 1989. Voice denumerability in polyphonic music of homogeneous timbres. Music Percept. 6, 361–382. ( 10.2307/40285438) [DOI] [Google Scholar]
- 110.McDermott JH, Lehr AJ, Oxenham AJ. 2010. Individual differences reveal the basis of consonance. Curr. Biol. 20, 1035–1041. ( 10.1016/j.cub.2010.04.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cousineau M, McDermott JH, Peretz I. 2012. The basis of musical consonance as revealed by congenital amusia. Proc. Natl Acad. Sci. USA 109, 19 858–19 863. ( 10.1073/pnas.1207989109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mas-Herrero E, Zatorre RJ, Rodriguez-Fornells A, Marco-Pallares J. 2014. Dissociation between musical and monetary reward responses in specific musical anhedonia. Curr. Biol. 24, 699–704. ( 10.1016/J.Cub.2014.01.068) [DOI] [PubMed] [Google Scholar]
- 113.Pamjav H, Juhasz Z, Zalan A, Nemeth E, Damdin B. 2012. A comparative phylogenetic study of genetics and folk music. Mol. Genet. Genomics 287, 337–349. ( 10.1007/s00438-012-0683-y) [DOI] [PubMed] [Google Scholar]
- 114.Brown S, Savage PE, Ko AM, Stoneking M, Ko YC, Loo JH, Trejaut JA. 2014. Correlations in the population structure of music, genes and language. Proc. R. Soc. B 281, 20132072 ( 10.1098/rspb.2013.2072) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.