Abstract
Indels (insertions and deletions) are the second most common form of genetic variations in the eukaryotic genomes and are responsible for a multitude of genetic diseases. Despite its significance, detailed molecular mechanisms for indel generation are still unclear. Here we examined 2,656,597 small human and mouse germline indels, 16,742 human somatic indels, 10,599 large human insertions, and 5,822 large chimpanzee insertions and systematically analyzed the patterns of DNA cleavage intensities in the 200 base pair regions surrounding these indels. Our results show that DNA cleavage intensities close to the start and end points of indels are significantly lower than other regions, for both small human germline and somatic indels and also for mouse small indels. Compared to small indels, the patterns of DNA cleavage intensity around large indels are more complex, and there are two low intensity regions near each end of the indels that are approximately 13 bp apart from each other. Detailed analyses of a subset of indels show that there is slight difference in cleavage intensity distribution between insertion indels and deletion indels that could be contributed by their respective enrichment of different repetitive elements. These results will provide new insight into indel generation mechanisms.
As the second most abundant form of human genetic variations, indels (insertions and deletions) also emerge as a significant source of variation that accounts for the majority of differences between species1,2. The presence of indels also contributes to the pathogenesis of diseases3 and changes in gene expression and protein functionality4.
According to the Human Gene Mutation Database (HGMD)5, indels are associated with at least 22% human severe diseases such as cystic fibrosis, fragile X syndrome, Huntington disease, and as well as many types of cancer5,6. Indels in coding regions, even the ones that are in-frame, can lead to abnormal protein folding and protein degradation7. A well-known case of indel effects is cystic fibrosis, a genetic disease frequently caused by a 3-bp deletion within the coding region of CFTR8,9. Similarly, indels in noncoding regions can also cause human diseases due to expansion or shrinkage of repeats. A well-known case is fragile X syndrome caused by the expansion of short trinucleotide in the promoter region of the FMR1 gene10. This insertion changes the promoter methylation status and thus the gene expression pattern of FMR1.
With recent advances in next generation sequencing technology, many indel detection methods have been proposed11,12,13,14. All these studies yield encouraging results and play significant roles in understanding the origin of indels. These advances also provided a large amount of indel data and made it possible to analyze the genome-wide distribution of indels and their effects on humans15. However, there are still unanswered questions regarding how and where indel occurs.
DNA structural properties play important roles in many biological processes including protein-DNA interactions16, transcription initiation17, replication18, and meiotic recombination19, in which binding of proteins to DNA is influenced both by the sequence of nucleotides and by the shape of the DNA double helix20. DNA cleavage intensity is an effective index that can be used to predict the shape of the DNA backbone and the width of minor groove of genomic DNA at single-nucleotide resolution21,22. Since proposed by Tullius and Greenbaum23, it has been widely used to characterize structural features of DNA, such as functional noncoding regions24, nucleosomes25, replication origins18, and so on. However, no detailed systematic analysis of contribution of DNA structural property to the generation of indels has been performed.
As DNA cleavage intensity may affect DNA structure and exposure/accessibility to DNA binding enzymes and indels are thought to be generated by DNA amplification errors, we hypothesize that the formation of indels may correlate with DNA cleavage intensity. Therefore, in the present study, we conducted a computational analysis of indel distribution with respect to DNA cleavage intensity. We found that DNA cleavage intensity of the start and end points of indels was significantly lower than those in surrounding regions. This pattern not only holds in both human germline and somatic cells, but also holds in chimpanzee and mouse genomes, suggesting a model of indel formation in relation to DNA cleavage intensity. Our finding offers new clues to understand the mechanisms of indel formation and provides new direction for improvement of indel detection algorithms.
Results
Cleavage intensity profile surrounding the small indels
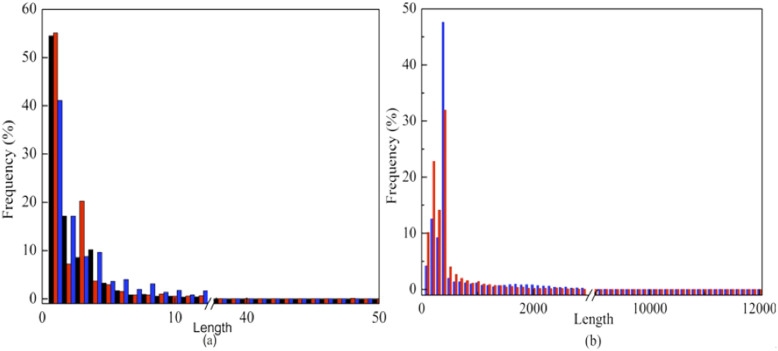
Altogether, we collected 2,656,597 small human and mouse indels (see Methods). Their detailed numbers on individual chromosomes are listed in Table 1, and their length distributions are shown in Figure 1a. Average lengths of human germline indels, human somatic indels, and mouse indels are 2 bps, 3 bps, and 4 bps, respectively.
Table 1. The number of indels on human, mouse, and chimpanzee chromosomes.
| Small indel | Large indel | ||||
|---|---|---|---|---|---|
| Human | |||||
| Chromosome | germline | somatic | Mouse | Human | Chimpanzee |
| 1 | 91,700 | 2,051 | 148,825 | 794 | 435 |
| 2 | 97,702 | 1,089 | 205,933 | 785 | a 228 |
| b 293 | |||||
| 3 | 82,332 | 1,089 | 39,977 | 677 | 439 |
| 4 | 84,756 | 737 | 59,073 | 744 | 425 |
| 5 | 75,478 | 944 | 38,930 | 725 | 341 |
| 6 | 76,750 | 942 | 36,397 | 696 | 374 |
| 7 | 68,170 | 656 | 184,170 | 564 | 339 |
| 8 | 60,078 | 681 | 42,752 | 654 | 324 |
| 9 | 47,942 | 714 | 61,826 | 414 | 230 |
| 10 | 56,568 | 590 | 54,165 | 485 | 304 |
| 11 | 56,102 | 964 | 54,835 | 470 | 271 |
| 12 | 56,306 | 981 | 26,924 | 537 | 241 |
| 13 | 43,956 | 214 | 31,788 | 496 | 215 |
| 14 | 38,940 | 603 | 21,557 | 355 | 219 |
| 15 | 34,408 | 574 | 65,727 | 315 | 143 |
| 16 | 31,024 | 521 | 84,238 | 322 | 177 |
| 17 | 32,410 | 958 | 35,663 | 262 | 131 |
| 18 | 33,732 | 346 | 104,192 | 315 | 183 |
| 19 | 27,078 | 787 | 61,572 | 195 | 103 |
| 20 | 24,700 | 319 | - | 210 | 157 |
| 21 | 17,016 | 205 | - | 163 | 89 |
| 22 | 15,364 | 322 | - | 112 | 57 |
| X | 47,555 | 455 | 81,244 | 309 | 104 |
| Total | 1,200,067 | 16,742 | 1,439,788 | 10,599 | 5,822 |
Figure 1. Length distribution of indels.
(a) The length distribution of human germline (black), human somatic (red) and mouse (blue) small indels. Their average lengths are 2 bps, 3 bps, and 4 bps, respectively. (b) The length distribution of large indels in the human (blue) and chimpanzee (red) genome. The average length of large indels is 840 bps for the human genome and 440 bps for the chimpanzee genome.
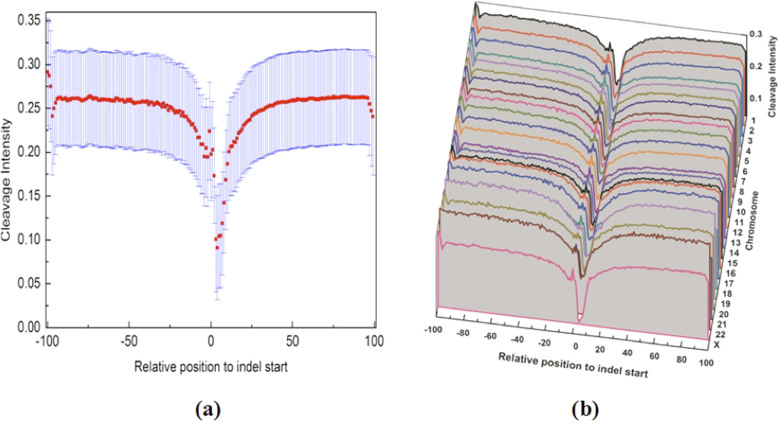
To investigate structural properties of the regions surrounding these indels, we calculated the DNA cleavage intensity of 200 bp sequences surrounding the indels, that is, −100 bp to +100 bp relative to the indel start sites (position 0) using ORChID220. The average cleavage intensity profile surrounding all the indels for the human genome is shown in Figure 2a and the one for individual chromosomes in Figure 2b (For clarity, individual chromosome's average cleavage intensity with 95% confidence interval is shown in Supplementary Figure S1). The pattern is amazingly consistent across all the chromosomes: cleavage intensity in the vicinity of indel start sites is significantly lower than other positions (Student's t-test, p<2.2 × 10−22). Similarly, the deep valley corresponding to very low cleavage intensity near indel start sites is also observed in the 16,742 human somatic indels (Figures 3a–b, Supplementary Figure S2) and the 1,439,788 mouse indels (Figures 4a–b, Supplementary Figure S3).
Figure 2. The average cleavage intensity profile of regions surrounding germline small indels in the human genome.
(a) for the entire genome. The average cleavage intensity for each position from −100 bp to +100 bp relative to indel start site was indicated by red rectangles. The blue bars represent the 95% confidence interval. (b) for individual chromosome. The average cleavage intensity profiles for the regions from −100 bp to +100 bp relative to indel start site on each human chromosome.
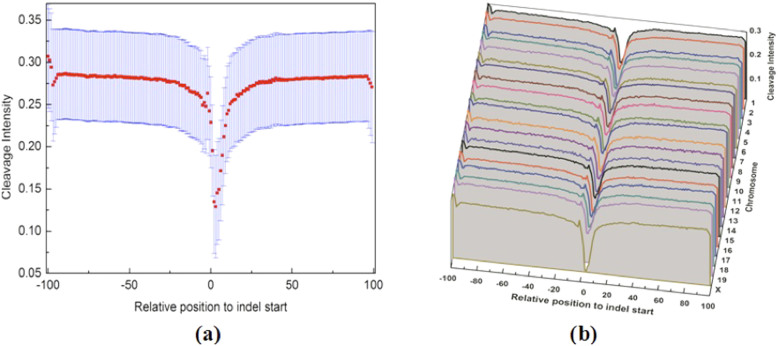
Figure 3. The average cleavage intensity profile of regions surrounding somatic small indels in the human genome.
(a) for the entire genome. The average cleavage intensity for each position from −100 bp to +100 bp relative to indel start site was indicated by red rectangles. The blue bars represent the 95% confidence interval. (b) for individual chromosome. The average cleavage intensity profiles for the regions from −100 bp to +100 bp relative to indel start site on each human chromosome.
Figure 4. The average cleavage intensity profile of regions surrounding small indels in the mouse genome.
(a) for the entire genome. The average cleavage intensity for each position from −100 bp to +100 bp relative to indel start site was indicated by red rectangles. The blue bars represent the 95% confidence interval. (b) for individual chromosome. The average cleavage intensity profiles for the regions from −100 bp to +100 bp relative to indel start site on each mouse chromosome.
As indels include insertion and deletion mutations, the observed pattern of cleavage intensity in and around indels could be the average effect of the two types of indels. An interesting question to ask is “do these two types of indels have the same distribution patterns with respect to cleavage intensity as the pooled indels”? To answer this question, we used the ancestral information provided by the 1000 Genomes project26 to infer the directionality of indels, and were able to annotate 185,234 insertions and 432,935 deletions for the human germline indels. The cleavage intensity profiles for the 200 positions from −100 bp to +100 bp relative to the start sites of these insertions and 432,935 deletions are shown in Supplementary Figures S4–S7. Overall, cleavage intensities around the start sites of both insertions and deletions are also significantly lower than their surrounding positions (Student's t-tests, p-value<1.6 × 10−22) and follow the same pattern as that of all small indels. However, compared to insertion indels, the contrast in cleavage intensity between indel vicinity and other surrounding regions is less pronounced for deletion indels (Figures S4 and S6).
Cleavage intensity profile surrounding large indels
Altogether, we obtained 10,599 and 5,822 large insertion indels in the human and chimpanzee genomes (see Methods), respectively. Detailed numbers for all the chromosomes are listed in Table 1, and length distributions are shown in Figure 1b. Average lengths of the large indels in the human and chimpanzee genomes are 840 bps and 440 bps, respectively.
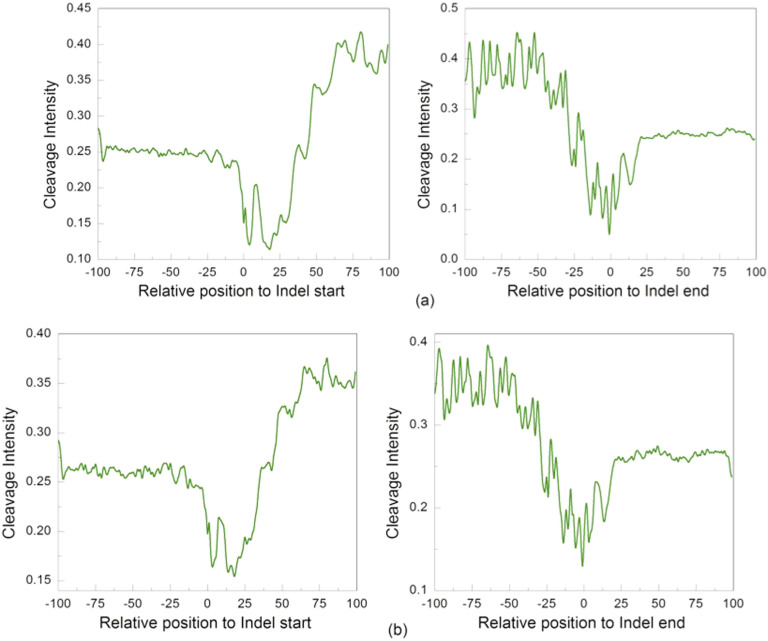
We next analyzed structural properties of the regions surrounding these large indels in both human and chimpanzee genomes by calculating DNA cleavage intensity. The average cleavage intensity profiles for the positions from −100 bp to +100 bp relative to the start and end sites of the large indels in both human and chimpanzee genomes are shown in Figure 5. Similar to the pattern shown by small indels, cleavage intensities near the start and end sites of large indels were also significantly lower than other positions (t-test, p<1.7 × 10−22). However, large indels have their own distinct pattern of cleavage intensity as compared to that of small indels. Two valleys located at about +3 bp and +18 bp downstream of the start site were observed. Moreover, two valleys located at about −14 bp and −1 bp upstream of the end site of the large indels were also observed in both human (Figure 5a) and chimpanzee genomes (Figure 5b).
Figure 5. Cleavage intensity for regions surrounding the start and end site of large indels.
(a) The average cleavage intensity profile for the positions from −100 bp to +100 bp relative to the start (top left panel) and end (top right panel) site of the large indels in the human genome. (b) The average cleavage intensity profile for the positions from −100 bp to +100 bp relative to the start (bottom left panel) and end (bottom right panel) site of the large indels in the chimpanzee genome.
Cleavage intensity profile surrounding SNPs
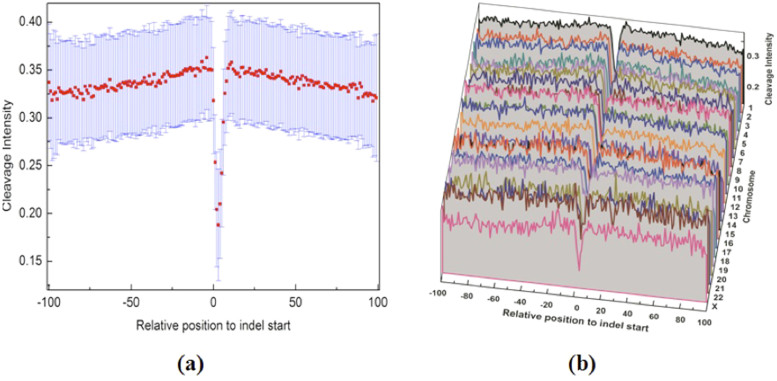
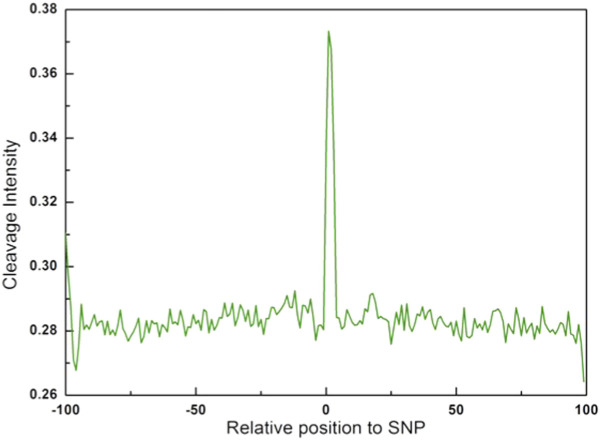
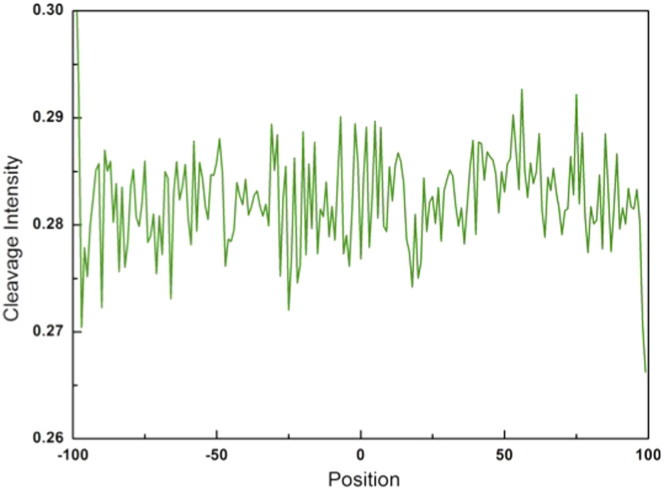
As a control analysis, we randomly sampled 17,000 human SNPs from UCSC genome database (hg19/snp138) and analyzed the cleavage intensity of surrounding sequences (from −100 to +100 bps). The average cleavage intensity profile of SNPs is shown in Figure 6. In contrast to indels, the cleavage intensity of SNP site is significantly higher than surrounding regions. Furthermore, we also randomly picked out 10,000 genomic positions and calculated the cleavage intensity for their surrounding sequences (from −100 to +100 bps). Figure 7 shows that the average cleavage intensity of random genomic regions exhibits random fluctuations and has no strong distribution pattern as compared to the selected sites, and therefore is dramatically different from that of indel regions and SNP regions (Figures 2,3,4,5). Taken together, these results ruled out the possibility that the observed lower cleavage intensity near start or end site of indels is due to sequence bias.
Figure 6. Cleavage intensity for regions surrounding SNPs in the human genome.
The average cleavage intensity profile for the positions from −100 bp to +100 bp relative to SNPs.
Figure 7. Cleavage intensity for random genomic positions in the human genome.
The average cleavage intensity profile for the positions from −100 bp to +100 bp relative to random genomic positions.
Discussion
In this work, we examined the cleavage intensity profile around 2,656,597 small indels and 16421 large indels in the human, chimpanzee, and mouse genomes. Small indels range from one to 50 bps and large indels from 80 to 12,000 bps. The indels obtained from the human 1000 Genomes projects26 and the mouse indels are expected to be enriched with germline indels, whereas the human somatic indels should be mostly somatic as the majority of them are identified through various cancer projects26.
For small indels, the cleave intensity profile shows a deep valley in the downstream of indel start sites (Figures 2,3,4 and Supplementary Figures S1–S3), and the cleavage intensity in the valley is significantly lower than other positions. The pattern holds for both insertions and deletions. Interestingly, insertions and deletions show two major differences. First, the contrast in cleavage intensity between indel vicinity and other surrounding regions is less pronounced for deletions than insertions (Figures S4 and S6). Second, the average cleavage intensities of insertions (Figures S4–S5) are a little higher than that of deletions (Figure S6–S7). To examine what may cause the differences, we ran RepeatMasker (http://www.repeatmasker.org) on the 200 bp (100 bp upstream and 100 bp downstream of indel occurrence sites) of the indel sites to identify repetitive sequences, and classified the insertions and deletions based on the types of repeats they have. If different types of repetitive sequences cause the different patterns seen in insertions and deletions, we expect that there will a nonrandom distribution of these repeat types. Indeed, the results of the hypothesis test27 as reported in Table 2 show that, compared with deletions, insertions are enriched in SINEs but short of LINEs, LTR retrotransposons, simple repeats, and DNA elements. Therefore, for the indels that we were able to identify insertions and deletions, the difference seen in their cleavage intensity seems to be caused by different repeat sequences.
Table 2. Results of the two-proportion z-test of repetitive elements in annotated insertions and deletions.
| Repetitive elements | Insertion | Deletion | p-valueb | |
|---|---|---|---|---|
| With repetitive elementa | LINE | 39,251 | 96,424 | 3e-4 |
| SINE | 37,440 | 68,514 | >0.9 | |
| LTR | 18,930 | 44,640 | 3e-4 | |
| Simple-repeat | 12,933 | 32,335 | 3e-4 | |
| DNA-element | 8,921 | 22,322 | 3e-4 | |
| Others | 2,685 | 6169 | - | |
| Without repetitive element | - | 65,074 | 162,531 | - |
| Total | - | 185,234 | 432,935 | - |
aThe numbers in each line indicate the number of insertions or deletions that contain repetitive elements.
bp-values of the two-proportion z-test.
Compared to that of small indels, the cleavage intensity profile of large indels shows a more complicated pattern: there are two valleys near the downstream of indel start sites, and also two valleys near the upstream of indel end sites (Figure 5). The patterns hold across chromosomes, species, and also regardless of whether the indels are somatic or germline. Therefore, our results suggest that indel distributions are strongly associated with DNA cleavage intensity and indels tend to occur in low cleavage intensity regions.
The observed distinct structural difference reflected by cleavage intensity between regions of close proximity to indels and those further away provides new insight into indel generation mechanisms. It has been demonstrated that small indels are generated due to strand slippage during DNA replication28,29. All the known DNA polymerases can generate indels30 due to DNA strand slippage in the process of DNA synthesis. Although DNA polymerases can monitor and correct mutations using the proofreading mechanism, efficiency of proofreading for indel mismatches varies with sequence context and structure28. It has been reported that many DNA polymerases monitor the correct base-pairing by hydrogen bonds with the minor groove and van der Wass contacts with bases30. However, abnormal geometry DNA sequences can result in steric clashes in and around the activate site that precludes efficient catalysis30. Therefore, the observed rigidity at the start site of small indels may facilitate template displacement involved in strand slippage initiation as demonstrated by a recent theoretical model29 and may also prevent polymerases from binding to this region and then lower down the proofreading efficiency of polymerases.
Besides strand slippage, other mechanisms of generating small indels require single-stranded or double-stranded breaks and repairs mechanisms such as break-induced replication, nonhomologous end joining, and microhomology-mediated end-joining31. All these processes require the action of different nucleases, primase, synthesis and the involvement of different nonreplicative, low fidelity repair polymerases with very different error rates of incorporating a wrong base31,32,33. Therefore, the cleavage intensity differences between regions of close proximity to indels and those further away may be helpful to the creation of single-stranded or double-stranded breaks and also may hinder the binding of nucleases, primase or polymerases to DNA, which is influenced by the shape of the DNA double helix.
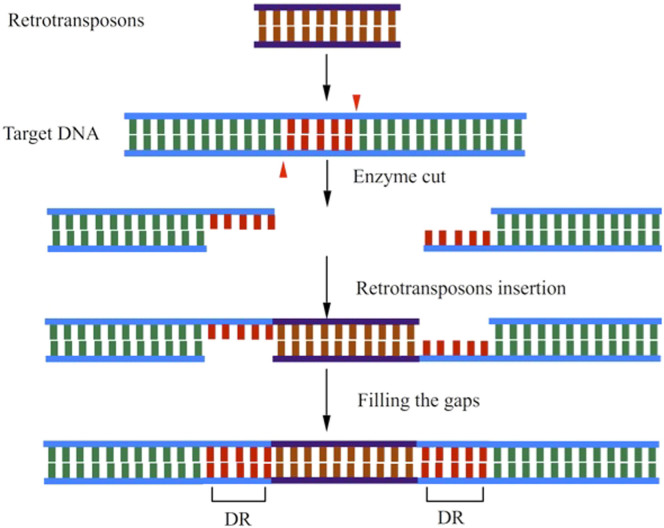
It also is interesting to consider why the cleavage intensity is significantly lower at both the start and end point of large indels (Figure 5). One mechanism of large indel generation is due to the proliferation and illegitimate recombination of transposable elements34,35, which is clearly different from that of small ones. Large indels considered in the present work are all associated with retrotransposons that move around by a "cut and paste" process in the genome36 (Polavarapu, et al. 2011). Shown in Figure 8, DNA at the target site is cut in an offset manner (like the "sticky ends" produced by some restriction enzymes) and after the transposon is ligated to the host DNA, gaps are filled in by the Watson-Crick base pairing rule. In this process, identical direct repeats (DR) will be generated at each end of the retrotransposon. The distance (about 13 bps) of the two pairs of valley observed at the both ends of large indels (Figure 5) is in accordance with the average length of the DR that is 13 bps37. Therefore, the observed rigidity at both ends of large indels may facilitate the endonuclease to recognize and cut the target DNA.
Figure 8. Mechanism of large indel generation through retrotransposition.
Two red angles indicate the enzyme cut site. DR is short for identical direct repeats indicated in red regions. The brown region at the bottom panel is the large indel generated due to the insertion of a retrotransposon.
Previous studies have shown that SNPs are preferentially distributed in nucleosome positioning regions, whereas indels seem to show different distribution patterns but it is unclear what DNA structural properties affect indel distribution38. Our current study provides insight into this problem, revealing the strong pattern that indels tend to locate in regions of the chromosome with low cleavage intensities, whereas SNPs tend to locate in regions with high cleavage intensities (Figure 6). Considering that genomic regions with high cleavage intensity are prone to form nucleosomes25, the observed distinct cleavage intensity patterns between indel and SNPs may be also attributable to their different distribution patterns relative to nucleosomes. We could also conjecture that DNA structural feature reflected by cleavage intensity boosts indel mutations in two ways regardless of indel generation mechanisms (i.e., strand slippage, unequal crossing over, retrotransposition, etc.). First, due to the low cleavage intensity in and near the regions where indels appear, errors resulting in indels are difficult to fix as the hydroxyl activity is low in the region and enzymes cannot easily find and fix the errors. Second, also because of the low cleavage intensity, the DNA in and near indels is rigid, fragile, and easy to break. For majority of the possible mechanisms of indel generation, DNA breaks, either one stranded or two stranded (e.g., the sticky double stranded breaks during retrotransposition), are involved during the process, and the low cleavage intensity is necessary and facilitate the break. The two valleys near both the start and end of large indels generated by retrotransposons (Figures 5) show strong support to our conjecture here.
Our current finding suggests that cleavage intensity can be used to assist the prediction and identification of indels. It is well known that indels pose great computational challenges to both short reads mapping and indel calling algorithms11 and there can be many false positives during indel calling39. With what is observed in our study, it is easily imaginable that cleavage intensity is an important DNA structural feature that one can consider when predicting or confirming the presence of indels, so indel calling tools can incorporate cleavage intensity as a main feature for training and classification of indels. In fact, cleavage intensity has already been incorporated into the prediction of a variety of biological properties, such as transcription factor binding sites40, eukaryotic core promoters17, and DNA replication origin18.
Methods
Human and mouse small indel data
The Ensembl variation database stores different types of variants including single nucleotide polymorphisms (SNPs), small indels (i.e., indel sizes are less than 50 bps), and structural variants from different species. However, information on indels is only limited to human and mouse genomes. From the Ensembl database, we extracted small indels of the mouse genome and small somatic indels of the human genome. From the 1000 Genomes Project website, http://www.1000genomes.org/, we also obtained the information of germline small indels in the human genome. To obtain a high quality dataset, indels were selected according to the following two criteria: (1) Indels with multiple annotations were discarded; (2) The selected indels are at least 100 bps apart from others. Finally, we obtained 1,200,067 germline and 16,742 somatic indels in the human genome, and 1,404,325 small indels in the mouse genome.
Based on the reference genome sequences of humans (hg19) and mice (mm10) obtained from the UCSC genome database (http://genome.ucsc.edu/), 200 bp sequences, 100 bps upstream and 100 bps downstream of the start position of each indel, were extracted from the two reference genomes.
The frequency of insertions and deletions and the frequency of frameshifting and non-frameshifting indels in human germline, human somatic and mouse small indels are shown in Supplementary Figures 8 and 9, respectively.
Human and Chimpanzee large indel data
The large indel (80 to 12,000 bps in length) data for human and chimpanzee genomes was obtained from Polavarapu, et al.40. Most of these indels were generated due to insertions that are associated with retrotransposons. Based on their data, we obtained 10,599 and 5,822 large insertion indels in the human and chimpanzee genomes, respectively. As these large indels were identified for different genome assemblies, to maintain the consistency, the same versions used by the original study, human hg17 and chimpanzee PanTro2, were obtained from the UCSC genome database (http://genome.ucsc.edu/) for downstream large indel analyses. Similarly, 200 bps, 100 bps upstream and 100 bps downstream of the start position of each indel, were extracted from the two reference genomes.
The frequency of frameshifting and non-frameshifting indels in Human and Chimpanzee large indels are shown in Supplementary Figure 10.
Calculation of cleavage intensity
Cleavage intensity indicates the likelihood of DNA cleavage by hydroxyl radicals and provides a map of local variation in the shape of DNA backbone. The lower the cleavage intensity is, the more rigid the DNA is. Cleavage intensity can be calculated from parameters for a set of tetranucleotides in a given DNA sequence. The parameters of the 44 ( = 256) tetranucleotides were derived from experiments in which DNA sequences were exposed to hydroxyl radicals21. Recently, Bishop et al.20 developed the ORChID2 algorithm (http://dna.bu.edu/orchid/) to calculate DNA cleavage intensity according to the following equation21,
 |
where Ci is the cleavage intensity at position i, Ti-j+1 the hydroxyl radical cleavage intensity of the tetramer starting at position i-j+1, and j the j-th nucleotide in the tetramer. The ends of the DNA are calculated similarly, except that cleavage data are retrieved from only one, two, or three tetramers, rather than four. Accordingly, we can compute the cleavage intensity for each nucleotide in a DNA sequence by using ORChID2. In this way, a DNA sequence is converted into a numerical sequence with each nucleotide represented by the DNA cleavage intensity.
Author Contributions
W.C. and L.Z. conceived and designed the experiments: W.C. and L.Z. performed the analysis and wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Supplementary_Figures
Acknowledgments
The authors would like to thank Prof. John McDonald for providing the data of large indels in Human and Chimpanzee genomes. This work was supported by the National Nature Scientific Foundation of China (No. 61100092) and the Nature Scientific Foundation of Hebei Province (No.C2013209105).
References
- Frazer K. A. et al. Genomic DNA insertions and deletions occur frequently between humans and nonhuman primates. Genome Res. 13, 341–346, 10.1101/gr.554603 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H. et al. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429, 382–388, 10.1038/nature02564 (2004). [DOI] [PubMed] [Google Scholar]
- Budde S. M. et al. Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem. Biophys. Res. Commun. 275, 63–68, 10.1006/bbrc.2000.3257 (2000). [DOI] [PubMed] [Google Scholar]
- Dayi S. U. et al. Influence of angiotensin converting enzyme insertion/deletion polymorphism on long-term total graft occlusion after coronary artery bypass surgery. Heart Surg. Forum 8, E373–377, 10.1532/HSF98.20051113 (2005). [DOI] [PubMed] [Google Scholar]
- Stenson P. D. et al. The Human Gene Mutation Database: 2008 update. Genome Medicine 1, 13, 10.1186/gm13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval A. & Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 62, 2447–2454 (2002). [PubMed] [Google Scholar]
- Hu J. & Ng P. C. SIFT Indel: predictions for the functional effects of amino acid insertions/deletions in proteins. PloS One 8, e77940, 10.1371/journal.pone.0077940 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F. S., Brooks L. D. & Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 8, 1229–1231 (1998). [DOI] [PubMed] [Google Scholar]
- Collins F. S. et al. Construction of a general human chromosome jumping library, with application to cystic fibrosis. Science 235, 1046–1049 (1987). [DOI] [PubMed] [Google Scholar]
- Warren S. T., Zhang F., Licameli G. R. & Peters J. F. The fragile X site in somatic cell hybrids: an approach for molecular cloning of fragile sites. Science 237, 420–423 (1987). [DOI] [PubMed] [Google Scholar]
- Albers C. A. et al. Dindel: accurate indel calls from short-read data. Genome Res. 21, 961–973, 10.1101/gr.112326.110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genet. 43, 491–498, 10.1038/ng.806 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303, 10.1101/gr.107524.110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman J. A., Isakov O. & Shomron N. Analysis of insertion-deletion from deep-sequencing data: software evaluation for optimal detection. Brief. Bioinform. 14, 46–55, 10.1093/bib/bbs013 (2013). [DOI] [PubMed] [Google Scholar]
- Liu M., Watson L. T. & Zhang L. Quantitative prediction of the effect of genetic variation using hidden Markov models. BMC Bioinformatics 15, 5, 10.1186/1471-2105-15-5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson W. K., Gorin A. A., Lu X. J., Hock L. M. & Zhurkin V. B. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. U. S. A. 95, 11163–11168 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeel T., Saeys Y., Bonnet E., Rouze P. & Van de Peer Y. Generic eukaryotic core promoter prediction using structural features of DNA. Genome Res. 18, 310–323, 10.1101/gr.6991408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Feng P. & Lin H. Prediction of replication origins by calculating DNA structural properties. FEBS Lett. 586, 934–938, 10.1016/j.febslet.2012.02.034 (2012). [DOI] [PubMed] [Google Scholar]
- Chen W., Feng P. M., Lin H. & Chou K. C. iRSpot-PseDNC: identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res. 41, e68, 10.1093/nar/gks1450 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop E. P. et al. A map of minor groove shape and electrostatic potential from hydroxyl radical cleavage patterns of DNA. ACS Chem. Biol. 6, 1314–1320, 10.1021/cb200155t (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J. A., Pang B. & Tullius T. D. Construction of a genome-scale structural map at single-nucleotide resolution. Genome Res. 17, 947–953, 10.1101/gr.6073107 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R. et al. The role of DNA shape in protein-DNA recognition. Nature 461, 1248–1253, 10.1038/nature08473 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D. & Greenbaum J. A. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr. Opin. Chem. Biol. 9, 127–134, 10.1016/j.cbpa.2005.02.009 (2005). [DOI] [PubMed] [Google Scholar]
- Parker S. C., Hansen L., Abaan H. O., Tullius T. D. & Margulies E. H. Local DNA topography correlates with functional noncoding regions of the human genome. Science 324, 389–392, 10.1126/science.1169050 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki T., Yachie N., Ogawa R., Saito R. & Tomita M. Computational analysis suggests a highly bendable, fragile structure for nucleosomal DNA. Gene 476, 10–14, 10.1016/j.gene.2011.02.004 (2011). [DOI] [PubMed] [Google Scholar]
- Genomes Project C. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073, 10.1038/nature09534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann E. L. & Romano J. P. Testing Statistical Hypotheses (3E ed.). (Springer, 2005). [Google Scholar]
- Garcia-Diaz M. & Kunkel T. A. Mechanism of a genetic glissando: structural biology of indel mutations. Trends Biochem. Sci. 31, 206–214, 10.1016/j.tibs.2006.02.004 (2006). [DOI] [PubMed] [Google Scholar]
- Montgomery S. B. et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 23, 749–761, 10.1101/gr.148718.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. DNA replication fidelity. J. Biol. Chem. 279, 16895–16898, 10.1074/jbc.R400006200 (2004). [DOI] [PubMed] [Google Scholar]
- De S. & Babu M. M. A time-invariant principle of genome evolution. Proc. Natl. Acad. Sci. U. S. A. 107, 13004–13009, 10.1073/pnas.0914454107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov Y. I., Shcherbakova P. V. & Rogozin I. B. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int. Rev. Cytol. 255, 41–132, 10.1016/S0074-7696(06)55002-8 (2006). [DOI] [PubMed] [Google Scholar]
- Rattray A. J. & Strathern J. N. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu. Rev. Genet. 37, 31–66, 10.1146/annurev.genet.37.042203.132748 (2003). [DOI] [PubMed] [Google Scholar]
- Devos K. M., Brown J. K. & Bennetzen J. L. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12, 1075–1079, 10.1101/gr.132102 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H. H. Jr Mobile elements: drivers of genome evolution. Science 303, 1626–1632, 10.1126/science.1089670 (2004). [DOI] [PubMed] [Google Scholar]
- Polavarapu N., Arora G., Mittal V. K. & McDonald J. F. Characterization and potential functional significance of human-chimpanzee large INDEL variation. Mobile DNA 2, 13, 10.1186/1759-8753-2-13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B. Gene IX. (Jones and Bartlett Publishers, 2007). [Google Scholar]
- Tolstorukov M. Y., Volfovsky N., Stephens R. M. & Park P. J. Impact of chromatin structure on sequence variability in the human genome. Nat. Struct. Mol. Biol. 18, 510–515, 10.1038/nsmb.2012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., Hagmann J., Koenig D., Weigel D. & Borgwardt K. Accurate indel prediction using paired-end short reads. BMC Genomics 14, 132, 10.1186/1471-2164-14-132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maienschein-Cline M., Dinner A. R., Hlavacek W. S. & Mu F. Improved predictions of transcription factor binding sites using physicochemical features of DNA. Nucleic Acids Res. 40, e175, 10.1093/nar/gks771 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_Figures