Abstract
Ribonucleic acid-mediated transcriptional gene silencing (known as RNA-directed DNA methylation, or RdDM, in Arabidopsis thaliana) is important for influencing gene expression and the inhibition of transposons by the deposition of repressive chromatin marks such as histone modifications and DNA methylation. A key event in de novo methylation of DNA by RdDM is the production of long non-coding RNA (lncRNA) by RNA polymerase V (Pol V). Little is known about the events that connect Pol V transcription to the establishment of repressive chromatin modifications. Using RNA immunoprecipitation, we elucidated the order of events downstream of lncRNA production and discovered interdependency between lncRNA-associated proteins. We found that the effector protein ARGONAUTE4 (AGO4) binds lncRNA independent of the RNA-binding protein INVOLVED IN DE NOVO2 (IDN2). In contrast, IDN2 binds lncRNA in an AGO4-dependent manner. We further found that the de novo DNA methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) also associates with lncRNA produced by Pol V and that this event depends on AGO4 and IDN2. We propose a model where the silencing proteins AGO4, IDN2 and DRM2 bind to lncRNA in a stepwise manner, resulting in DNA methylation of RdDM target loci.
Keywords: non-coding RNA, DNA methyltransferase, Argonaute, INVOLVED IN DE NOVO2, Arabidopsis thaliana
Introduction
Eukaryotic genomes contain potentially mobile genomic elements called transposons. As new transposition events can damage genomes, for instance by causing insertion mutations (Belancio et al., 2010; Hancks and Kazazian, 2012), transcriptional gene silencing keeps transposons silent. Moreover, this process is important for gene expression, presumably by targeting transposons embedded in the promoters of genes (Zheng et al., 2013; Zhong et al., 2012; Le Thomas et al., 2013; Taliaferro et al., 2013). Transcriptional gene silencing may control targets by establishing DNA methylation and other repressive chromatin modifications (Bernstein and Hake, 2006; Grewal and Elgin, 2007; Jiang and Pugh, 2009; Hargreaves and Crabtree, 2011). In Arabidopsis thaliana, methylated cytosines are present in three different sequence contexts: symmetrical (CG and CHG, where H stands for any base except G) and asymmetrical (CHH) (Chan et al., 2005). After initial methylation of DNA, CG and CHG methylation can be maintained by copying the information from the parental strand after DNA replication. In contrast, CHH methylation needs to be deposited de novo after each round of DNA replication in a process called RNA-directed DNA methylation (RdDM). In RdDM, the DNA-dependent RNA polymerase IV (Pol IV) produces a transcript that is converted into double-stranded RNA (dsRNA) by the RNA-dependent RNA Polymerase RDR2 (Xie et al., 2004; Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005; Haag and Pikaard, 2011; Law et al., 2011; Wierzbicki, 2012). The resulting dsRNA is processed into 24 nucleotide (nt) long fragments by DICER-LIKE 3 (DCL3) (Xie et al., 2004; Kasschau et al., 2007) and one strand of this small interfering RNA (siRNA) is bound by the protein ARGONAUTE4 (AGO4) (Qi et al., 2006). The siRNA is thought to mediate association of AGO4 via base pairing with a long non-coding RNA (lncRNA), produced by another DNA-dependent RNA Polymerase called Pol V, at the silencing targets (Wierzbicki et al., 2008, 2009, 2012; Zheng et al., 2013). In addition to AGO4, the lncRNA-binding proteins SPT5L and IDN2 are also important for de novo DNA methylation (Ausin et al., 2009; He et al., 2009; Zheng et al., 2010; Zhu et al., 2013). Finally, the de novo DNA methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2) is recruited and deposits DNA methyl marks (Cao and Jacobsen, 2002; Naumann et al., 2011).
While production of siRNA is generally well understood (Haag and Pikaard, 2011; Wierzbicki, 2012), the order of events downstream of the synthesis of Pol V transcripts is less well studied. So far, AGO4 has been suggested to bind lncRNA in an siRNA-dependent manner, though limited AGO4 stability in rdr2, a mutant in siRNA synthesis, weakens this hypothesis (Li et al., 2006; Wierzbicki et al., 2009). Moreover, IDN2 was shown to act downstream of Pol V and to be important for RdDM, though its exact role remains a mystery. IDN2 was reported to interact with 5′-overhangs of dsRNA in vitro (Ausin et al., 2009) and, more recently, that it binds Pol V transcripts in vivo (Zhu et al., 2013). In addition to IDN2, the genome of Arabidopsis encodes IDN2-like (IDNL) 1 and 2 (also called FDM1 and FDM2, respectively) that are both involved in RdDM (Ausin et al., 2012; Xie et al., 2012a). They were proposed to work with IDN2 in a complex (Ausin et al., 2012; Xie et al., 2012b) and FDM1 was shown to bind 5′-overhangs of dsRNA in vitro similarly to IDN2 as well as unmethylated DNA in vitro (Ausin et al., 2009; Xie et al., 2012a,b). A complex consisting of IDN2 and FDM1 was proposed to simultaneously bind lncRNA and DNA at RdDM targets (Ausin et al., 2012; Xie et al., 2012b), thereby bringing the Pol V-transcript and the target DNA into closer proximity.
Even though many factors in RdDM have been described (reviewed in Haag and Pikaard, 2011; Wierzbicki, 2012), the order of events downstream of Pol V transcript production and any potential interdependences of lncRNA-binding proteins remain unclear. We discovered that there are two classes of AGO4-bound RdDM targets differing in their dependency on IDN2 or IDN2-like proteins. At loci requiring IDN2, RdDM components bind lncRNA in a certain order downstream of Pol V transcript production. While AGO4 binds to lncRNA independently of IDN2, IDN2 requires AGO4 for its association with Pol V transcripts. These events result in direct or indirect association of DRM2 with lncRNA and methylation of the target DNA.
Results
RdDM loci dependent on IDN2 are a distinct set of silencing targets
We previously demonstrated that Pol V-mediated binding of AGO4 to chromatin is a general feature of RdDM targets (Zheng et al., 2013). To test if CHH DNA methylation on AGO4-bound loci requires other proteins expected to work downstream of Pol V, we analysed published whole-genome bisulfite sequencing datasets (Stroud et al., 2013). In agreement with the previously published results (Stroud et al., 2013), 90% of the AGO4-bound regions which lost CHH methylation in the nrpe1 mutant also lost CHH methylation in the drm1/drm2 mutant (Figure1a). This is consistent with DRM1 and DRM2 being the main de novo DNA methyltransferases in RdDM (Cao and Jacobsen, 2002; Cao et al., 2003). In contrast, only 32% of these loci also lost CHH methylation in the idn2 mutant (Figure1a), suggesting that IDN2 works only on a subset of RdDM targets.
Figure 1.
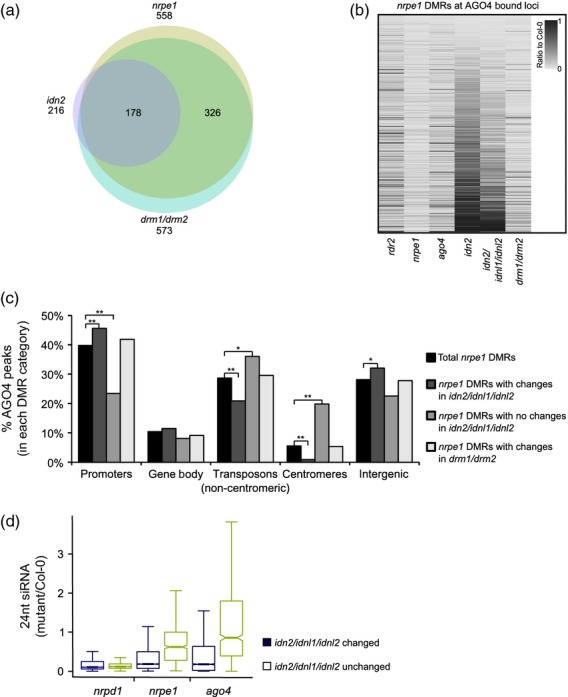
A subset of AGO4-bound RNA-directed DNA methylation (RdDM) targets requires IDN2. (a) Overlaps of AGO4-bound regions of differential CHH methylation (DMRs) between Col-0 and specified mutants. Most nrpe1 DMRs (beige) also have reduced CHH methylation in drm1/drm2 (blue). These overlap with idn2 (purple) DMRs. Total DMRs are 558 nrpe1, 573 drm1/drm2 or 216 idn2 out of 820 AGO4-bound loci. (b) Comparison of methylation levels on AGO4-bound nrpe1 DMRs. Most nrpe1 DMRs have reduced CHH methylation in rdr2, ago4 and drm1/drm2. A subset of nrpe1 DMRs has reduction in idn2 or in the idn2/idnl1/idnl2 triple mutant, but most have no or intermediate changes. nrpe1 DMRs were defined as having at least a four-fold reduction in CHH methylation. All values are represented as a ratio to Col-0. (c) Classification of AGO4-bound nrpe1 DMRs. Any DMRs overlapping promoters of protein-coding genes, gene bodies, transposons, centromeric regions or intergenic regions were counted. Total AGO4-bound nrpe1 DMRs were plotted as a baseline (black). nrpe1 DMRs were then divided into regions that have losses in CHH methylation in idn2/idnl1/idnl2 (dark grey) and regions that have no change (grey). nrpe1 DMRs with CHH methylation losses in drm1/drm2 were also plotted (light grey). Bars represent the percentage of each category that overlapped a particular genomic feature. *P < 0.05, **P < 0.001 (chi-square test). (d) Production of small interfering RNA (siRNA) on IDN2/IDNL1/IDNL2-dependent regions requires Pol IV, Pol V and AGO4. Total nrpe1 DMRs were classified as either idn2/idnl1/idnl2 changed (blue, meCHH ≤25% of Col-0) or unchanged (green, meCHH ≥75% of Col-0). The 24 nt siRNAs in either nrpd1, nrpe1 or ago4 overlapping each category were counted and plotted as a ratio to Col-0 with reads per million normalization.
To test if the IDN2 homologs IDNL1 and IDNL2 compensate for IDN2 in the idn2 mutant, we compared levels of CHH methylation on AGO4-bound nrpe1 differentially methylated regions (DMRs). The triple mutant idn2/idnl1/idnl2 had lower levels of CHH methylation than the single idn2 mutant. However, there was still a substantial number of loci with DNA methylation in contrast to the nrpe1, ago4 and drm1/drm2 mutants (Figure1b). This indicates that only a subset of RdDM targets requires IDN2, IDNL1 or IDNL2 for CHH methylation. Remaining RdDM targets could be silenced independently of IDN2 and its homologs. Alternatively, yet untested IDN2 homologs (Xie et al., 2012a) may work at these loci.
To better understand the difference between RdDM targets with IDN2/IDNL1/IDNL2-dependent or -independent CHH methylation, we performed a classification analysis of these regions. The IDN2/IDNL1/IDNL2-dependent loci were more likely to overlap gene promoters, while IDN2/IDNL1/IDNL2-independent targets were more likely to overlap transposons or centromeric regions (Figure1c). Additionally, we compared siRNA production on IDN1/IDNL1/IDNL2-dependent and -independent regions. While only Pol IV was required for siRNA biogenesis at IDN2/IDNL1/IDNL2-independent regions (Figure1d), 24 nt siRNA levels were low in the Pol IV mutant nrpd1, the Pol V mutant nrpe1 and the ago4 mutant at IDN2/IDNL1/IDNL2-dependent targets, suggesting that siRNA production requires Pol IV, Pol V and AGO4 at these loci (Figure1d). This observation might indicate that IDN2 could be involved in the production of Pol V-dependent siRNAs (Mosher et al., 2008; Zheng et al., 2010; Lee et al., 2012) and that this event requires AGO4.
Together, these results suggest that AGO4-binding sites with IDN2/IDNL1/IDNL2-dependent CHH methylation represent RdDM targets, where all tested silencing factors are involved in agreement with the accepted model of this process (Haag and Pikaard, 2011). Therefore, we selected this category of loci for further analysis of interdependences between proteins involved in RdDM.
Binding of AGO4 to lncRNA does not require IDN2
To resolve the order of events occurring on Pol V transcripts, we performed RNA immunoprecipitation (RIP) experiments with polyclonal antibodies against AGO4, IDN2 and DRM2. This method allows us to detect RNA bound to the pulled-down proteins after immunoprecipitation. By using appropriate mutant plants, we can determine if an interacting RNA is produced by Pol V and if other proteins are also required for this association. For our assays, we selected four RdDM targets with IDN2/IDNL1/IDNL2-dependent CHH methylation.
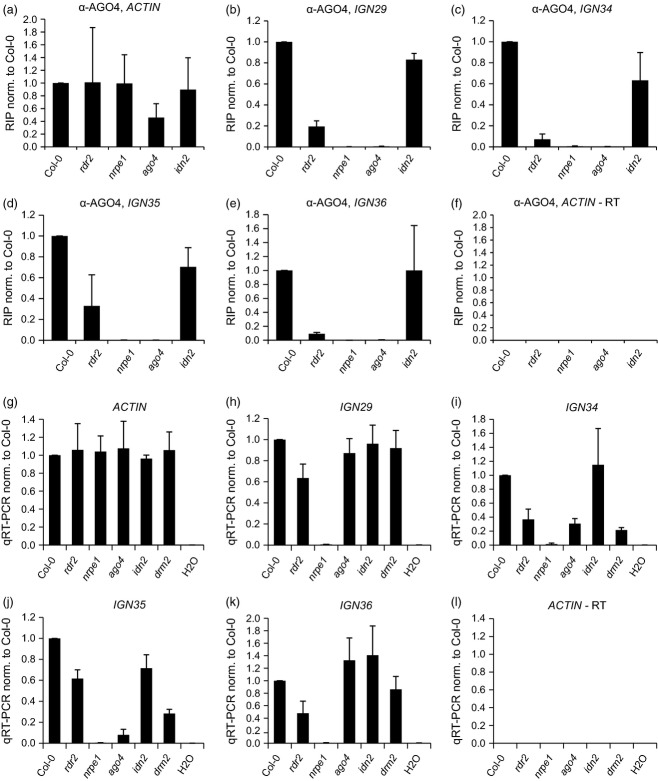
To determine if the binding of AGO4 to lncRNA depends on IDN2, we performed RIP experiments with an α-AGO4 antibody using seedlings of Col-0 wild type, the idn2 mutant as well as nrpe1, ago4 and rdr2 mutants. Non-specifically immunoprecipitated ACTIN2 RNA was used to monitor for comparable levels of total RNA in the samples (Figure2a). Quantitative (q)RT-PCR reactions without reverse transcriptase allowed detection of potential contamination with genomic DNA (Figure2f). RNA immunoprecipitation using Col-0 wild type pulled down RNA from all four of the tested loci, while we observed a loss of this signal in the ago4 mutant. This result indicates that AGO4 associates with RNA at these loci (Figure2b–e). The signal was not detectable in the nrpe1 mutant either (Figure2b–e), indicating that the transcripts that AGO4 interacts with are produced by Pol V. Recovery of RNA was strongly reduced in the rdr2 mutant, which suggests that siRNA is required for the association of AGO4 with Pol V transcripts. This result is consistent with data obtained at other loci (Wierzbicki et al., 2009); however, reduced stability of AGO4 in the rdr2 mutant (Li et al., 2006; Wierzbicki et al., 2009) makes this result difficult to interpret. In contrast, levels of immunoprecipitated Pol V transcripts were not significantly changed in idn2 compared with the wild type (Figure2b–e), indicating that IDN2 is not important for the interaction of AGO4 with lncRNA.
Figure 2.
Binding of AGO4 to long non-coding RNA (lncRNA) does not require IDN2. (a)–(f) RNA immunoprecipitation (RIP) experiments using an α-AGO4 antibody and nuclei isolated from wild type and mutant seedlings. Mean values and standard deviations of at least two biological repetitions are shown. Experiments were performed on the following loci: IGN29 (b), IGN34 (c), IGN35 (d), and IGN36 (e). Pol II-transcribed ACTIN2 served as a control for equal total RNA levels (a) and for potential contamination with genomic DNA (f). (g–l) Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) with total RNA isolated from seedlings. Mean values and standard deviations of three biological repetitions are shown. Four Pol V-transcribed loci with reduced DNA methylation in idn2 were analysed: IGN29 (h), IGN34 (i), IGN35 (j), and IGN36 (k). Pol II-transcribed ACTIN2 served as a control for equal total RNA levels (g) and potential contamination with genomic DNA (l).
To test if altered Pol V transcript levels in any of the tested mutants are affecting the results of the RIP assay, we performed qRT-PCR on total RNA. Loss of qRT-PCR signal in nrpe1 demonstrated that all four loci are indeed transcribed by Pol V (Figure2h–k). Levels of Pol V transcripts were not significantly changed in idn2 at any of the tested loci, while they were slightly reduced in rdr2. For ago4 and drm2 mutants, the amounts of Pol V transcripts were not substantially altered at IGN29 nor at IGN36 (Figure2h,k), while they were partially reduced at two other loci (Figure2i,j). These results indicate that despite some limited locus-specific variability, differences in the accumulation of Pol V transcripts between tested mutants do not account for the effects observed in the RIP experiments.
In summary, the association of AGO4 with Pol V transcripts at the tested loci does not require IDN2. This suggests that IDN2 works downstream of or in parallel to the binding of AGO4 to lncRNA.
Binding of IDN2 to lncRNA requires siRNA and AGO4
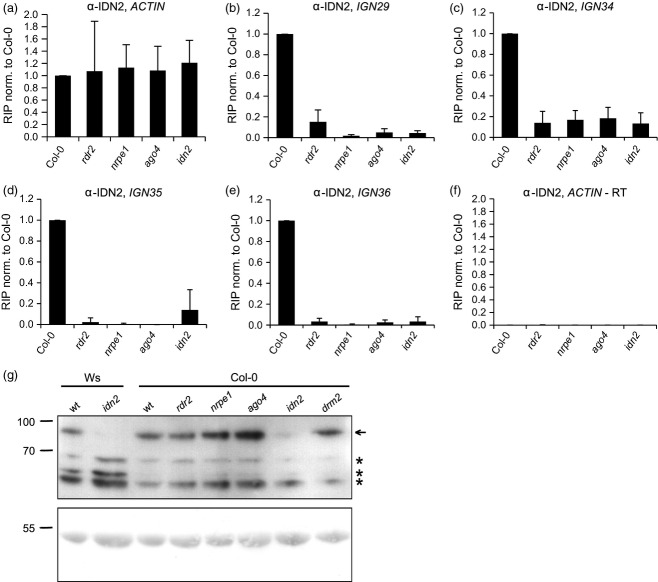
Even though binding of lncRNA by AGO4 does not seem to require IDN2, the association of IDN2 with Pol V transcripts might need AGO4. Alternatively, both proteins could bind lncRNA independently of each other. To distinguish between these two scenarios and to determine if IDN2 actually binds Pol V transcripts at studied loci, we performed RIP experiments with an α-IDN2 antibody in the Col-0 wild type, rdr2, nrpe1, ago4 and idn2. While immunoprecipitated RNA was detectable in Col-0, qRT-PCR signals were strongly reduced in the idn2 mutant (Figure3b–e), indicating that IDN2 associates with the tested RNAs. The signal was also lost in the nrpe1 mutant (Figure3b–e), which shows that IDN2 binds Pol V transcripts. No RNA above the idn2 background level was recovered from rdr2 or ago4 mutants (Figure3b–e). This result indicates that binding of IDN2 to Pol V transcripts requires AGO4 and potentially siRNAs, although the lower RIP signal in rdr2 may be explained by reduced protein levels of AGO4 in rdr2 as well.
Figure 3.
Binding of long non-coding RNA (lncRNA) by IDN2 depends on small interfering RNA (siRNA) and AGO4. (a–f) RNA immunoprecipitation (RIP) experiments with an α-IDN2 antibody and nuclei isolated from wild type and RNA-directed DNA methylation (RdDM) mutant seedlings. Mean values and standard deviations of at least three biological replicates are shown. Experiments were performed on the following loci: IGN29 (b), IGN34 (c), IGN35 (d), and IGN36 (e). Pol II-transcribed ACTIN2 served as a control for equal total RNA levels (a) and for potential contamination with genomic DNA (f). (g) Western blot with an α-IDN2 antibody and proteins isolated from wild type (wt) and RdDM mutant seedlings. Ponceau staining of the membrane shows comparable loading of protein extracts. The experiment was performed twice and one representative biological replicate is depicted.
To test if levels of the IDN2 protein are comparable between studied mutants, we performed Western blots with the α-IDN2 antibody. Specific signal was not detectable in the knock-out idn2-2 mutant (Ws background) and strongly reduced in the idn2-1 mutant (Col-0 background), confirming antibody specificity. The IDN2 protein levels were not significantly changed in any of the other RdDM mutants tested when compared with the wild type (Figure3g), indicating that results of RIP are not caused by differences in IDN2 protein accumulation. Therefore, we conclude that binding of IDN2 to lncRNA is dependent on siRNA and AGO4. This suggests that IDN2 works downstream of AGO4.
The DRM2 protein associates with lncRNA produced by Pol V
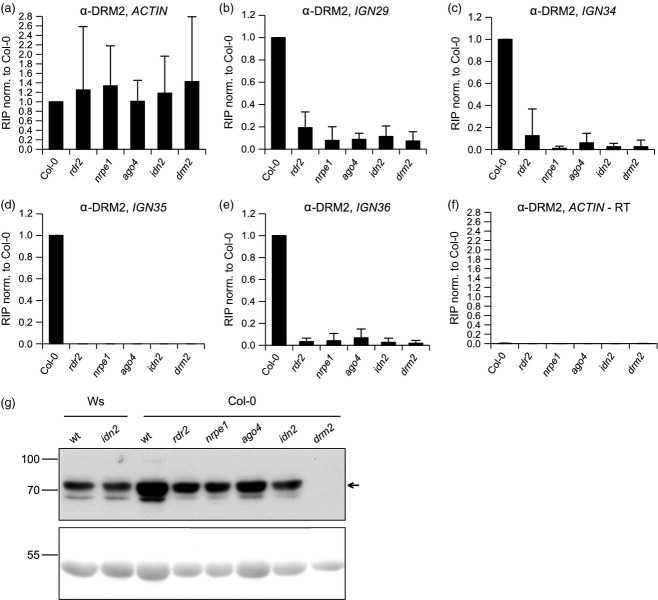
Experiments described above as well as previously published work (He et al., 2009; Wierzbicki et al., 2009; Rowley et al., 2011; Zhu et al., 2013) have demonstrated that three proteins (AGO4, SPT5L and IDN2) found to work downstream of Pol V actually associate with Pol V transcripts. Therefore, we hypothesized that DRM2 may also bind lncRNA produced by Pol V. To test this hypothesis, we performed RIP experiments with an α-DRM2 antibody. We were able to amplify RNA from the Col-0 wild type at all four tested loci, while the signal was undetectable or was strongly reduced in the drm2 mutant (Figure4b–e), indicating that DRM2 associates with RNA at these loci. The signal was also undetectable or strongly reduced in the nrpe1 mutant (Figure4b–e). Therefore, we concluded that that DRM2 associates specifically with Pol V transcripts at these loci.
Figure 4.
Association of DRM2 with Pol V transcripts requires small interfering RNA (siRNA), AGO4 and IDN2. (a–f) RNA immunoprecipitation (RIP) experiments with an α-DRM2 antibody and nuclei isolated from wild type and RNA-directed DNA methylation (RdDM) mutant seedlings. Mean values and standard deviations of at least two biological replicates are shown. Experiments were performed on the following loci: IGN29 (b), IGN34 (c), IGN35 (d), and IGN36 (e). Pol II-transcribed ACTIN2 served as a control for equal total RNA levels (a) and for potential contamination with genomic DNA (f). (g) Western blot with an α-DRM2 antibody and proteins isolated from wild type (wt) and RdDM mutant seedlings. Ponceau staining of the membrane shows comparable loading of protein extracts. The experiment was performed twice and one representative biological replicate is depicted.
The DRM2 protein requires AGO4 and IDN2 to bind to lncRNA
The association of DRM2 with lncRNA may be independent of the two other lncRNA-binding proteins IDN2 and AGO4. Alternatively, DRM2 might require prior association of these two proteins as well as the presence of siRNA. To test these possibilities, we included the rdr2, ago4 and idn2 mutants in our analysis. The α-DRM2 RIP experiments showed a similar loss of qRT-PCR signal in the rdr2, ago4 and idn2 mutants compared with drm2 and nrpe1 (Figure4b–e), suggesting that binding of DRM2 to Pol V transcripts requires AGO4, IDN2 and siRNA. To test if the observed effects were caused by reduced stability of DRM2 in the studied mutants, we performed a Western blot with the α-DRM2 antibody on total protein extracts. A specific signal was not detectable in the drm2 mutant, confirming antibody specificity. In the other tested mutants, levels of DRM2 protein were comparable to those in the Col-0 wild type (Figure4g), indicating that the results from the RIP assays are not caused by altered protein levels in RdDM mutants. Together, these results suggest that the association of DRM2 with Pol V transcripts occurs downstream of AGO4 and IDN2.
Association of AGO4, IDN2 and DRM2 with Pol V transcripts is important for CHH DNA methylation
Our RIP experiments showed that AGO4, IDN2 and DRM2 bind lncRNA at AGO4-bound DMRs. If this event is important for CHH methylation, DNA methylation levels should be reduced in all the mutants at these loci. To test this hypothesis, we performed Chop-PCR experiments in all the mutants as well as in the wild type. In a Chop-PCR assay, genomic DNA is digested with methylation-sensitive restriction enzymes and subsequently amplified by PCR. If DNA methylation is present, the enzyme cannot cut and a PCR product can be amplified. If DNA methylation is lost, then the enzyme digests the DNA and no PCR product can be observed. At all tested loci DNA methylation levels were reduced to a similar extent in the rdr2, nrpe1, ago4, idn2 and drm2 mutants (Figure5a), indicating that siRNAs produced by RDR2, Pol V transcripts, AGO4, IDN2 as well as DRM2 are all required for CHH methylation at these regions.
Figure 5.
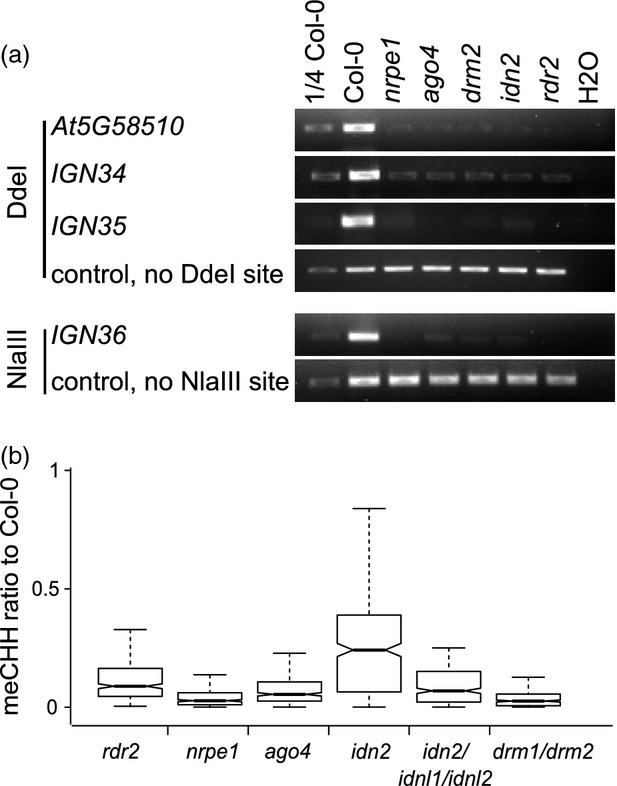
Pol V transcripts, small interfering RNA (siRNA), AGO4, IDN2 and DRM2 are necessary for CHH methylation at IDN2-dependent loci. (a) Chop-PCR with genomic DNA isolated from wild type and RNA-directed DNA methylation (RdDM) mutant seedlings. The DNA was digested with methylation-sensitive restriction enzymes and amplified by PCR. Three Pol V-transcribed loci with reduced DNA methylation levels in idn2 were analysed in addition to a control locus (At5G58510) that has been previously shown to have Pol V- and AGO4-dependent CHH methylation (Zheng et al., 2013). Regions lacking a restriction site were used as a loading control. The experiment was performed three times and one representative biological replicate is shown. (b) Methylation status of AGO4-bound nrpe1 differentially methylated regions (DMRs) as classified by the idn2/idnl1/idnl2 triple mutant. Those DMRs with a dependency on idn2/idnl1/idnl2 were selected and methylation levels in rdr2, ago4, idn2 and drm1/drm2 relative to the wild type were plotted. Dependency was defined as at least a four-fold reduction in CHH methylation compared with Col-0.
We also tested the relationship between DNA methylation levels in tested mutants throughout the genome. To do so, we analysed published datasets (Stroud et al., 2013) and compared all genomic regions where DNA methylation was reduced in the idn2/idnl1/idnl2 triple mutant. These regions had consistently strong reductions of CHH methylation levels in rdr2, nrpe1, ago4 and drm1/drm2 mutants (Figure5b). Together, these results suggest that all of the RdDM components included in the RIP experiments are important for DNA methylation at the studied category of loci. This is consistent with our results suggesting that AGO4, IDN2 and DRM2 are working together in a stepwise fashion to establish CHH DNA methylation.
Discussion
We have previously shown that association of AGO4 with Pol V-produced lncRNA is necessary for binding of AGO4 to chromatin and subsequent methylation of DNA throughout the genome (Wierzbicki et al., 2009; Zheng et al., 2013). To test if other RdDM components acting downstream of Pol V (i.e. IDN2 and DRM2) work in a more locus-specific way, we analysed published DNA methylation datasets (Stroud et al., 2013), focusing on AGO4-bound genomic regions. While DRM1/DRM2 (as expected) is required for CHH methylation at these loci, IDN2 and IDN2-related proteins are not always required for RdDM. In fact, there are two distinct categories of RdDM targets differing not only in their dependency on IDN2/IDNL1/IDNL2 but also in their location (Figure1c) and in the involvement of other RdDM factors in siRNA biogenesis (Figure1d). In particular, IDN2-dependent targets are enriched in promoters and intergenic regions while they are depleted in transposons and centromeres. It is possible that other IDN2-related proteins (Xie et al., 2012a; Butt et al., 2014) with a different preference for target loci might compensate for IDN2 at these loci. Alternatively, the function of IDN2 or IDN2-related proteins might not be required at these targets. For instance, the repetitive nature of transposons and centromeric sequences might already give rise to dsRNA that could be further processed by a dicer protein. In contrast, at non-repetitive regions such as promoters and intergenic sequences, IDN2 might be required for second-strand synthesis of non-coding RNA produced by Pol V, as indicated by our analysis of 24 nt siRNA on IDN2-dependent and independent regions (Figure1d).
As we wanted to understand the order of events downstream of Pol V production, we focused our study on IDN2/IDNL1/IDNL2-dependent loci and asked if lncRNA produced by Pol V at representative loci is bound by AGO4, IDN2 and DRM2. We also determined if these proteins bind to lncRNA in a particular order. We first tested if IDN2 is required for recruiting AGO4 to RdDM targets using RIP with an α-AGO4 antibody. While the experiments revealed that IDN2 is not essential for binding of lncRNA by AGO4, we confirmed that siRNA produced by RDR2 seems to be important for this event, as previously suggested (Wierzbicki et al., 2009; Zheng et al., 2013). Therefore, AGO4 binds to lncRNA independently of IDN2 (Figure6).
Figure 6.
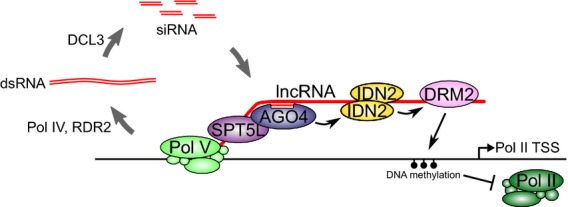
Model of stepwise association of silencing proteins with long non-coding RNA (lncRNA) produced by Pol V to mediate CHH methylation. At IDN2/IDNL1/IDNL2-dependent loci, AGO4 interacts with lncRNA produced by Pol V in a manner at least partially dependent on small interfering RNA (siRNA). In turn, IDN2 binds to lncRNA possibly recognizing a 5′-overhang of a double-stranded RNA consisting of siRNA and lncRNA. Finally, DRM2 directly or indirectly associates with the Pol V transcript in a way dependent on IDN2 and AGO4, and the DNA at the locus is methylated.
As IDN2 is not required for the association of AGO4 with lncRNA, it might work either downstream or in parallel to AGO4. To distinguish between these possibilities, we performed α-IDN2 RIP experiments. The results suggest that binding of IDN2 to Pol V transcripts requires AGO4 and siRNA (Figure6). As IDN2 protein levels are not changed in any of the mutants tested besides idn2, this result could be explained by siRNA being primarily important for the association of AGO4 with lncRNA via base pairing. In turn, AGO4 could interact with IDN2, thereby recruiting it to Pol V transcripts. So far, no direct interaction has been observed between AGO4 and IDN2 in purified protein complexes (Ausin et al., 2012). However, an interaction between FDM1 and AGO4 mediated by RNA has been reported (Xie et al., 2012a), suggesting simultaneous association of AGO4 and an IDN2 complex containing FDM1 with RNA. As IDN2 was reported to bind 5′-overhangs of dsRNA (Ausin et al., 2009), AGO4 might deposit the siRNA on the lncRNA. Then, IDN2 could bind to the resulting dsRNA now containing a 5′-overhang, as was proposed previously (Ausin et al., 2012).
As IDN2 and AGO4, two proteins working downstream of Pol V production, were shown to bind lncRNA, we wanted to test if DRM2 also associates with Pol V transcripts. Our RIP experiments demonstrated that DRM2 binds Pol V transcripts in an AGO4- and IDN2-dependent manner. We therefore propose that AGO4 binds a Pol V transcript in an siRNA-guided manner followed by association of IDN2 with subsequent recruitment of DRM2 (Figure6).
Our experiments demonstrating the binding of DRM2 to lncRNA involved formaldehyde crosslinking, which may induce covalent bond formation not only between proteins and nucleic acids but also between proteins and proteins. Therefore, we cannot distinguish between DRM2 directly binding to Pol V transcripts or requiring a lncRNA-binding protein that mediates an indirect association of DRM2 to lncRNA. Future studies will be necessary to address this question as we were unsuccessful in our preliminary attempts to replace formaldehyde with UV crosslinking, which specifically fixes protein–nucleic acid interactions. Similarly, tests for potential protein–protein interactions between IDN2 and DRM2 by yeast-two-hybrid and transient expression in tobacco were inconclusive. Moreover, no direct protein–protein interaction between FDM1 and DRM2 has been reported (Xie et al., 2012b). So far only an indirect association between AGO4 and DRM2 mediated by RDM1 has been proposed (Gao et al., 2010). However, Pol V transcripts are reduced in the rdm1 mutant (Law et al., 2010), which makes in vivo interactions of this protein with Pol V transcripts hard to study.
It is possible that DRM2 interacts directly with Pol V transcripts, which would be consistent with reports from other organisms (Mohammad et al., 2010; Schmitz et al., 2010; Holz-Schietinger and Reich, 2012; Di Ruscio et al., 2013). In murine cells, Dnmt3b was suggested to specifically recognize DNA:RNA triplexes (Schmitz et al., 2010), while human DNMT3A and DNMT1a were reported to interact directly with a specific RNA (Holz-Schietinger and Reich, 2012; Di Ruscio et al., 2013). This binding event can depend on specific elements of a RNA, as murine Dnmt1 was observed to bind a certain region of a particular lncRNA in order to be recruited to DNA methylation targets (Mohammad et al., 2010). Similarly, DRM2 might recognize and bind distinct regions or structural features of lncRNA. It is therefore feasible that AGO4 binds to Pol V transcripts and deposits siRNAs, which in turn establish 5′-overhangs that are recognized by IDN2. Binding of IDN2 could introduce a conformational shift in the RNA that could allow association of DRM2. Alternatively, this interaction could require IDN2 to somehow stabilize Pol V transcripts, similarly to SGS3 stabilizing precursors for siRNA (Yoshikawa et al., 2005; Fukunaga and Doudna, 2009). However, we did not observe any major changes in Pol V transcript levels in the idn2 mutant (Figure2h–k), suggesting that IDN2 probably does not affect the stability of lncRNA. As a third possibility, IDN2 could be necessary to prime second-strand synthesis of Pol V transcripts by an RNA-dependent RNA polymerase. This would be consistent with our analysis of Pol V-dependent siRNA, the partial reduction of siRNA levels on some RdDM targets in the idn2 mutant (Zheng et al., 2010) and the observation that FDM1/FDM2 is important for the synthesis of Pol V-dependent siRNAs (Xie et al., 2012a). Hence, DRM2 could hypothetically recognize a dsRNA primed by an IDN2–FDM1 complex. Future experiments elucidating what happens on Pol V transcripts could provide further insights into these open questions.
Experimental procedures
Plant material
nrpe1 (nrpd1b-11) and ago4 [ago4-1 (Zilberman et al., 2003) introgressed into the Col-0 background] have been described previously (Onodera et al., 2005; Wierzbicki et al., 2009). idn2-1 (Ausin et al., 2009) was kindly provided by Steven Jacobsen (UCLA/MCDB, Los Angeles, CA, USA). idn2-2 in the Ws background (FLAG_550B05) was obtained from INRA. drm2-2 (SAIL_70_E12) was described previously (Wierzbicki et al., 2008).
Bioinformatics analysis of DMRs
The genome-wide bisulfite sequencing data used were previously reported by Stroud et al. (2013) (GEO accession number GSE39901). Differentially methylated regions (DMR) were defined as having <25% methylation in the mutant versus wild type on AGO4 peaks (Zheng et al., 2013). The weighted Venn diagram was created using the Venneuler package in R. A list of AGO4 peaks with DNA methylation data is provided in Table S1 in Supporting Information.
To create a heatmap, AGO4 peaks with a CHH methylation signal present in Col-0 and nrpe1 to Col-0 CHH methylation ratio of <0.25 were kept. Relative methylation signals on these peaks were plotted in R as a mutant/Col-0 ratio with Col-0 as the maximum signal.
AGO4 peaks with a nrpe1 to Col-0 CHH methylation ratio of <0.25 were further divided into idn2/idnl1/idnl2 changed (<25% of Col-0) and unchanged (>75% of Col-0) DMR categories. These categories were intersected with TAIR10 annotated genomic features, counted and plotted.
Small RNA with 24 nt that overlapped these nrpe1 DMR categories (idn2/idnl1/idnl2 changed and unchanged) were counted and plotted as boxplots with the ratio of siRNA of each mutant to Col-0. The mutants examined for siRNA were nrpd1, nrpe1 (SRA accession number SRA054962; Wierzbicki et al., 2012) and ago4 (GEO accession number GSE16545; Havecker et al., 2010).
Quantitative RT-PCR and RIPs
The RNA immunoprecipitation experiments were performed with 3 g of approximately 3-week-old seedlings fixed with 0.5% formaldehyde as described (Rowley et al., 2013). Values were normalized to the wild type. Experiments were performed in at least two biological replicates.
For qRT-PCR, total RNA was isolated from approximately 3-week-old seedlings using the SV Total RNA Isolation kit from Promega (http://www.promega.com/) according to manufacturer's recommendations. In addition to the on-column DNase I digestion, the RNA was digested with Turbo DNase as described (Rowley et al., 2013). Three micrograms of total RNA was used for cDNA synthesis with random primers (Invitrogen, http://www.invitrogen.com/). The means and standard deviations were obtained from three biological replicates. The number of test loci was determined by the amount of RNA recovered in RIP and the requirement to perform all assays in multiple biological repeats. The following primer pairs were used: IGN29 JA227 CGTTTGTTTATGTAGGGCGAAAG and JA228 TAAAACTTTTCCCGCCAACCA (Zhu et al., 2013), IGN34 GB396 ATGAATAACAAATTTGGAGTCGTC and GB397 CCCTTTCATCGACACTGACA, IGN35 GB588 GACGGACCAAACGATTTCAT and GB589 TTCCTCTTTGAGCTTGACCA, IGN36 GB646 CAGTTTTGGGTGCGGTTTAT and GB647 GACAAAAATTGCTTTAGACCATGA.
Antibody production
A DRM2 fragment (amino acids 29–423, amplified with the following primers: DRM2F1 CACCATGCAGTGTAGGGTCGAAAATCTAGCT and DRM2R1 CTAATGCTTAGGCGGTTCTGGTTCTTC) was used for production of a polyclonal antibody in rabbits. The antibody was purified as described in Zhu et al. (2013). α-IDN2 and α-AGO4 antibodies were described previously (Wierzbicki et al., 2009; Zhu et al., 2013).
Western blots
Total protein extracts were isolated from 100 mg of approximately 3-week-old Arabidopsis seedlings and separated on 12% polyacrylamide gels. Western blots were performed using α-IDN2 (1:333) and α-DRM2 (1:500) antibodies. Donkey anti-rabbit secondary antibody (1:2000) conjugated with horseradish peroxidase was used (Santa Cruz Biotechnology, http://www.scbt.com/).
Chop-PCR
Genomic DNA was isolated from 100 mg of approximately 3-week-old seedlings using a DNeasy Plant kit (Qiagen, http://www.qiagen.com/) according to the manufacturer's recommendations. One hundred nanograms of DNA was digested with 10 U of NlaIII or DdeI, respectively, and PCRs were performed as described in Zheng et al. (2013). One representative experiment is shown from three biological replicates. The following primers were used: AT5G58510 JR529 AGAGATCCGCTTCGGGAAAG and JR530 AGAAACCATTGATAGAGATGGTCTTAG, IGN34 JR883 ATTGCCCGACGACTCCAAAT and JR884 AACTTTTGTAAGTCATGGTGTGTGTT, IGN35 JR885 GGGCCGGGCTTAGAGGATAG and JR886 CACATCTTCTACACGTGTCTTTAGGC, DdeI control JR535 TCCAAGATTGAGGCCAAATTA and JR536 AAAAGGAGTGGCCAAGTTGGAA, IGN36 JR887 GATTTTGATATTGTTACAGCATTGTT and JR888 TCCATATTCAGTACTTTTTAACCTACC, NlaIII control JR889 ACCGTTTGTTTATGTAGGGCGAAA and JR890 AAGATAACAGAAAAGACGATGATGACG.
Acknowledgments
This work was supported by the National Science Foundation grant MCB 1120271 to ATW and the Austrian Science Fund (FWF) fellowship J3199-B09 to GB.
Authors’ Contributions
Quantitative RT-PCRs and RIP experiments were done by GB, bioinformatic analysis was performed by MJR, Western blots were done by JK, DRM2 antigen production and subsequent antibody purification were performed by YZ and Chop-PCRs were done by IA. The manuscript was written by GB and ATW.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Table S1. Changes in CHH DNA methylation AGO4-bound genomic regions.
References
- Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat. Struct. Mol. Biol. 2009;16:1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Greenberg MVC, Simanshu DK, et al. INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc. Natl Acad. Sci. USA. 2012;109:8374–8381. doi: 10.1073/pnas.1206638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Roy-Engel AM, Deininger PL. All y'all need to know ‘bout retroelements in cancer. Semin. Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Hake SB. The nucleosome: a little variation goes a long way. Biochem. Cell Biol. 2006;84:505–517. doi: 10.1139/o06-085. [DOI] [PubMed] [Google Scholar]
- Butt H, Graner S, Luschnig C. Expression analysis of Arabidopsis XH/XS-domain proteins indicates overlapping and distinct functions for members of this gene family. J. Exp. Bot. 2014;65:1217–1227. doi: 10.1093/jxb/ert480. Available at: [Accessed April 1, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- Chan SW-L, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Doudna JA. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009;28:545–555. doi: 10.1038/emboj.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Liu H-L, Daxinger L, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SIS, Elgin SCR. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X-J, Hsu Y-F, Zhu S, et al. An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Holz-Schietinger C, Reich NO. RNA modulation of the human DNA methyltransferase 3A. Nucleic Acids Res. 2012;40:8550–8557. doi: 10.1093/nar/gks537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJM. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu J-K, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr. Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Tóth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Gurazada SGR, Zhai J, Li S, Simon SA, Matzke MA, Chen X, Meyers BC. RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics. 2012;7:781–795. doi: 10.4161/epi.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW-L, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl Acad. Sci. USA. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann U, Daxinger L, Kanno T, Eun C, Long Q, Lorkovic ZJ, Matzke M, Matzke AJM. Genetic evidence that DNA methyltransferase DRM2 has a direct catalytic role in RNA-directed DNA methylation in Arabidopsis thaliana. Genetics. 2011;187:977–979. doi: 10.1534/genetics.110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Qi Y, He X, Wang X-J, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet. 2011;7:e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Böhmdorfer G, Wierzbicki AT. Analysis of long non-coding RNAs produced by a specialized RNA polymerase in Arabidopsis thaliana. Methods. 2013;63:160–169. doi: 10.1016/j.ymeth.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K-M, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Aspden JL, Bradley T, Marwha D, Blanchette M, Rio DC. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: alternative pre-mRNA splicing and transcriptional repression. Genes Dev. 2013;27:378–389. doi: 10.1101/gad.210708.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT. The role of long non-coding RNA in transcriptional gene silencing. Curr. Opin. Plant Biol. 2012;15:517–522. doi: 10.1016/j.pbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012;26:1825–1836. doi: 10.1101/gad.197772.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Ren G, Costa-Nunes P, Pontes O, Yu B. A subgroup of SGS3-like proteins act redundantly in RNA-directed DNA methylation. Nucleic Acids Res. 2012a;40:4422–4431. doi: 10.1093/nar/gks034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Ren G, Zhang C, Yu B. The DNA- and RNA-binding protein FACTOR of DNA METHYLATION 1 requires XH domain-mediated complex formation for its function in RNA-directed DNA methylation. Plant J. 2012b;72:491–500. doi: 10.1111/j.1365-313X.2012.05092.x. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Xing Y, He X-J, Li W, Hu Y, Yadav SK, Oh J, Zhu J-K. An SGS3-like protein functions in RNA-directed DNA methylation and transcriptional gene silencing in Arabidopsis. Plant J. 2010;62:92–99. doi: 10.1111/j.1365-313X.2010.04130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Rowley MJ, Böhmdorfer G, Sandhu D, Gregory BD, Wierzbicki AT. RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J. 2013;73:179–189. doi: 10.1111/tpj.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat. Struct. Mol. Biol. 2012;19:870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Rowley MJ, Böhmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Changes in CHH DNA methylation AGO4-bound genomic regions.