Abstract
Background: The strong observational association between total homocysteine (tHcy) concentrations and risk of coronary artery disease (CAD) and the null associations in the homocysteine-lowering trials have prompted the need to identify genetic variants associated with homocysteine concentrations and risk of CAD.
Objective: We tested whether common genetic polymorphisms associated with variation in tHcy are also associated with CAD.
Design: We conducted a meta-analysis of genome-wide association studies (GWAS) on tHcy concentrations in 44,147 individuals of European descent. Polymorphisms associated with tHcy (P < 10−8) were tested for association with CAD in 31,400 cases and 92,927 controls.
Results: Common variants at 13 loci, explaining 5.9% of the variation in tHcy, were associated with tHcy concentrations, including 6 novel loci in or near MMACHC (2.1 × 10−9), SLC17A3 (1.0 × 10−8), GTPB10 (1.7 × 10−8), CUBN (7.5 × 10−10), HNF1A (1.2 × 10−12), and FUT2 (6.6 × 10−9), and variants previously reported at or near the MTHFR, MTR, CPS1, MUT, NOX4, DPEP1, and CBS genes. Individuals within the highest 10% of the genotype risk score (GRS) had 3-μmol/L higher mean tHcy concentrations than did those within the lowest 10% of the GRS (P = 1 × 10−36). The GRS was not associated with risk of CAD (OR: 1.01; 95% CI: 0.98, 1.04; P = 0.49).
Conclusions: We identified several novel loci that influence plasma tHcy concentrations. Overall, common genetic variants that influence plasma tHcy concentrations are not associated with risk of CAD in white populations, which further refutes the causal relevance of moderately elevated tHcy concentrations and tHcy-related pathways for CAD.
INTRODUCTION
Observational studies have consistently shown that elevated concentrations of total homocysteine (tHcy)6 are associated with an increased risk of coronary artery disease (CAD), stroke, and venous thrombosis in addition to other clinical conditions (1–6). However, it is uncertain whether these associations are causal, given the absence of protection against cardiovascular events in homocysteine-lowering trials (7). We therefore used a Mendelian randomization strategy to test whether long-term elevated concentrations of homocysteine are associated with an increased risk of CAD. Mendelian randomization is a method of using measured variation in genes of known function to examine the causal effect of a modifiable exposure on disease in nonexperimental studies. By examining whether common polymorphisms associated with variations in tHcy are also associated with CAD, we could explore the association between tHcy and CAD.
To date, 5 separate genome-wide association studies (GWAS) of blood tHcy concentrations have been published, including 4 in whites and 1 in Filipinos, which have collectively identified 7 independent loci definitively associated with tHcy concentrations (8–12). These include the MTHFR C667T variant (rs1801133), the most widely studied consistently associated polymorphism with tHcy, and variants at or near the CBS, MTR, DPEP1, NOX4, CPS1, and MUT genes. Of these genes, the effect of the MTHFR functional variant on CAD was studied previously. In a meta-analysis of published results based on 26,000 CAD cases and >31,000 controls, Lewis et al (13) reported a significant association between the C667T variant with CAD in Asian populations only; the association between this variant and CAD in whites was close to null and not statistically significant. In contrast, the MTHFR Studies Collaboration showed no association of MTHFR C677T with risk of CAD in unpublished studies, irrespective of folate status or ethnic group. The latter meta-analysis compared the associations of MTHFR C677T with CAD in a meta-analysis of 19 unpublished data sets involving 48,175 CAD cases with a meta-analysis of 86 published studies involving 28,617 CAD cases and showed that the discrepant results of the unpublished and published studies were explained by publication bias and other methodologic problems in the published studies of Asian populations (14).
To identify other common variants associated with risk of CAD through effects on tHcy concentrations, we extended previous efforts by performing a meta-analysis of 2.7 million genotyped and imputed single nucleotide polymorphisms (SNPs) from 10 GWAS, with data on a total of 44,147 individuals of European ancestry (including new data on 26,669 individuals) with measured blood tHcy concentrations, and tested whether common polymorphisms associated with variations in tHcy were also associated with CAD. We tested each individual tHcy-associated polymorphism and a weighted genotype risk score (GRS), composed of the top (highest P value) independent SNPs (as assessed by r2 < 0.2 between associated SNPs) consistently associated with tHcy concentrations, for their association with CAD in 31,400 cases of CAD and 92,927 controls.
SUBJECTS AND METHODS
Homocysteine meta-analysis study populations
The current meta-analysis included data from a total of 44,147 white individuals of European ancestry derived from 10 GWAS on tHcy concentrations (see supplementary materials under “Supplemental data” in the online issue for details of each cohort). Data from Invecchiare in Chianti (n = 1208), the Baltimore Longitudinal Study of Aging (BLSA) (n = 638), and the Nurses’ Health Study (NHS) (n = 1658) and part of the data (n = 13,974) from the Women's Genome Health Study (WGHS) were published previously (8–10). Thus, the current meta-analysis includes new data on 26,669 individuals from 6 new cohorts. Each study was approved by the institutional ethics review committees at the relevant organizations, and all subjects provided informed consent for the collection of DNA and its use for genetic analyses.
Coronary artery disease cohorts
We tested the association of the tHcy-associated variants with risk of CAD in 6 independent cohorts and in the Coronary ARtery DIsease Genome-wide Replication And Meta-Analysis (CARDIoGRAM) consortium (15) cohorts, which consisted of up to 31,400 cases of CAD and 92,927 controls. The 6 studies were as follows: 1) WGHS (9), 2) The London Life Sciences Prospective Population Study (LOLIPOP) (16), 3) Rotterdam I and II (17), 4) Precocious Coronary Artery Disease (PROCARDIS) (11), 5) Pennsylvania Catheterization Study (15), and 6) The Wellcome Trust Case Control Consortium (www.wtccc.org.uk/). All cases and controls were of European ancestry. The formal criteria used for the recruitment of cases and controls in each of these studies is summarized elsewhere (see Supplementary Table 1 under “Supplemental data” in the online issue).
Blood tHcy measurements
tHcy was measured in each cohort by using one of the following methods: isotope-dilution liquid chromatography–tandem mass spectrometry, gas chromatography–coupled mass spectrometry, HPLC, or enzymatic, immune, or chemiluminescence (see Supplementary Table 2 under “Supplemental data”).
Genotyping and quality control
The participants from each cohort were genotyped by using the Illumina Infinium HumanHap550, the Illumina Infinium HumanHap300, the Illumina HumanCNV370 BeadChips, or the Affymetrix 5.0 GeneChip 500K according to the manufacturers’ protocols and quality-control standards. The exclusion/filtering criteria for SNPs in each of the cohorts are described elsewhere (see Supplementary Table 2 under “Supplemental data” in the online issue).
Genotype imputation
Genotypes were imputed for all polymorphic SNPs based on HapMap CEU (haplotype map for people living in Utah with ancestry from northern and western Europe) phase II data (release 22, build 36) oriented to the positive strand. Hidden Markov model–based algorithms were used to infer unobserved genotypes probabilistically as implemented in either MACH (18), IMPUTE (19), or BimBam (20) Imputation quality-control metrics included the ratio of observed/expected variance of the allele dose ≤0.01 for MACH, proper information ≤0.40 for IMPUTE, or r2 ≥ 0.3 for BimBam. Detailed descriptions of the quality-control and imputation procedures are summarized for all studies elsewhere (see Supplementary Table 3 under “Supplemental data” in the online issue).
Genome-wide association analysis
All analyses were performed on natural log-transformed tHcy concentrations by using sex-specific, age-adjusted standardized residuals (z scores). Each study performed a genome-wide association analysis for tHcy concentrations by using either MACH2QTL (www.sph.umich.edu/csg/abecasis/MACH/index.html) or SNPTEST (www.stats.ox.ac.uk/˜marchini/software/gwas/snptest.html), which use the genotype dosage value as continuous additive predictors of tHcy concentrations in a linear regression framework or GenABEL (21) with the use of an additive genetic model while accounting for relatedness between the members of each family (see Supplementary Table 3 under “Supplemental data” in the online issue). Analysis of imputed genotype data accounted for uncertainty in each genotype prediction by using either the dosage information from MACH or BimBam or the genotype probabilities from IMPUTE. Genomic control was used to correct any inflation of test statistics due to population structure in each cohort before the meta-analysis.
Meta-analysis
The minor allele from HapMap CEU genotypes was used to define the coded allele in all analyses, regardless of frequency in individual cohorts. A total of 2,090,256 SNPs with a minor allele frequency ≥1% and directly genotyped or imputed with sufficient quality (see Supplementary Table 3 under “Supplemental data” in the online issue) were included in the meta-analysis. We performed an inverse-variance, weighted, meta-analysis of the summary data from the 10 studies to obtain a pooled estimate of the overall β coefficient and SE by using METAL (www.sph.umich.edu/csg/abecasis/metal/index.html). We considered the results to be genome-wide significant at α = 5 × 10−8.
Conditional analysis to discover independent signals
To test for the presence of additional, independent signals at each of the top loci we carried out secondary GWAS conditioning on the top SNPs—referred to as the lead SNP—that had reached genome-wide significance. We re-ran each GWAS under the same conditions used in the primary analysis but included the maximum likelihood allele dose of each of the top 18 SNPs as covariates in the model. Parameters from the conditional analyses were then meta-analyzed by using a fixed-effects model. The regions within one mega-base up- and downstream of the SNP conditioned on were examined for additional hits by using the significance criterion used for the original meta-analysis (α = 5 × 10−8). Any genome-wide significant SNP in these regions was considered as a second independent association in the region.
Risk of hyperhomocysteinemia
Using the method outlined by Horne et al (22), we generated a GRS equal to the sum of the expected number of risk alleles at each SNP weighted by their effect sizes (β coefficient from the tHcy meta-analysis). We used logistic regression to test the combined effect of the 18 loci associated with tHcy on the risk of hyperhomocysteinemia (defined as ≥15 μmol/L), with adjustments for age and sex, in the Rotterdam Study cohorts. In addition, we estimated the explained variance of the identified SNPs in the Rotterdam cohorts by calculating the R2 in a linear regression analysis, in which tHcy concentration was the dependent variant and the SNPs were the independent variable.
Association of tHcy loci with coronary artery disease
We sought to assess whether common polymorphisms associated with tHcy identified through this meta-analysis are also associated with CAD. The association between tHcy SNPs on CAD were tested in 2 ways:
1) Each of the 18 SNPs that were associated with tHcy concentrations was tested for association with risk of CAD in a case-control study with the use of data from the CARDIoGRAM consortium (22,233 CAD cases and 64,762 controls) and in 3 separate CAD cohorts: the WGHS (23) (758 CAD cases and 22130 controls), which was also used in the discovery stage; the LOLIPOP (24) study, which contributed 2793 CAD cases and 3756 controls from South Asia; and the PROCARDIS study (25), which contributed a total of 5616 CAD cases and 2279 controls.
2) We estimated the combined effect of the 18 loci associated with tHcy in 6 studies (WGHS, PROCARDIS, The Wellcome Trust Case Control Consortium, Pennsylvania Catheterization Study, LOLIPOP, and Rotterdam) by testing the association between an additive GRS for elevated tHcy concentrations and increased risk of CAD.
We conducted a logistic regression analysis treating either the SNPs or the GRSs as a continuous explanatory variable, including sex, age, and BMI as covariates. Output parameters were then meta-analyzed by using a fixed-effects model.
Mendelian randomization study of tHcy and CAD
To quantify the expected causal relation between tHcy variation and risk of CAD, we used the methods outlined by Minelli et al (26). Under the assumptions required for Mendelian randomization, the OR of CAD resulting from a unit change in tHcy (CADtHcy) is equivalent to the OR of CAD resulting from a unit change in the GRS (CADGRS) raised to the power of the inverse of the mean difference in tHcy per unit of the GRS (ΔtHcyGRS): OR(CADtHcy) = OR(CADGRS)1/ΔtHcyGRS. We used the CADGRS from the meta-analysis and the (ΔtHcyGRS) from the Rotterdam Study. We used the SE of the OR(CADGRS) to construct the 95% CIs for OR(CADGRS).
Expression quantitative trait loci analysis
For each of the 18 lead genome-wide significant tHcy SNPs, all proxy SNP identifiers were identified by using SNAP (27). All lead SNPs and their proxies (r2 > 0.7) were then searched against a collected database of expression SNP results including the following tissues: fresh lymphocytes (28), fresh leukocytes (29), leukocyte samples in individuals with celiac disease (30), lymphoblastoid cell lines (LCLs) derived from asthmatic children (31), HapMap LCLs from 3 populations (32), a separate study on HapMap CEU LCLs (33), peripheral blood monocytes (34), adipose and blood samples (35, 36), 2 studies on brain cortex (34, 37), and 3 large studies of brain regions, including the prefrontal cortex, visual cortex, and cerebellum (V Emilsson, personal communication, 2012); liver (38); osteoblasts (39); skin (40); and additional fibroblast, T cell, and LCL samples (41). The collected expression SNP results met criteria for the statistical significance for association with gene transcript levels as described in the original articles.
RESULTS
Homocysteine cohort characteristics
The mean age of each cohort varied from 47.5 to 74.4 y, and the cohort participants ranged in age from 17 to 79 y. Four cohorts—including TwinsUK, WGHS, and NHS—were composed predominantly of females, whereas the remainder of the cohorts had roughly the same number of males and females (women: 45–58%). Additional characteristics of the study populations are presented elsewhere (see Supplementary Table 2 under “Supplemental data” in the online issue).
Meta-analysis of genome-wide association scans of tHcy concentrations
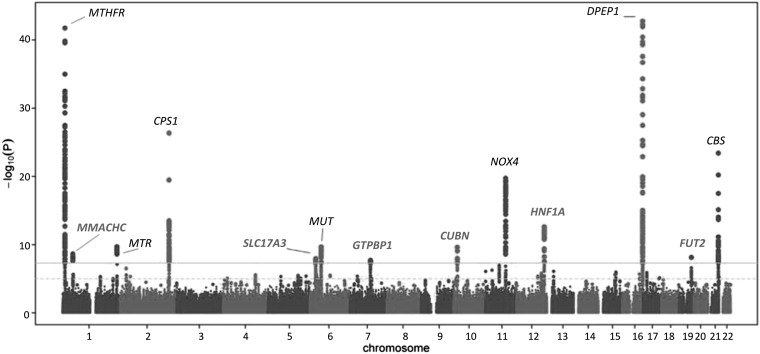
The meta-analysis identified SNPs from 13 independent genomic loci exceeding the GWAS threshold (P < 5 × 10−8) (see Supplementary Figure 1 under “Supplemental data” in the online issue, Figure 1, and Table 1). These included 6 previously unreported loci in or near MMACHC (P = 2.1 × 10−9), SLC17A3 (1.0 × 10−8), GTPB10 (1.7 × 10−8), CUBN (P = 7.5 × 10−10), HNF1A (1.2 × 10−12), and FUT2 (6.6 × 10−9). In addition, we confirmed 7 loci previously reported to be associated with tHcy concentrations at or near the MTHFR, MTR, CPS1, MUT, NOX4, DPEP1, and CBS genes. We failed to replicate previous associations with variants at or near the ZNF366 (rs7445013) and the PTPRD (rs973117) genes (11) (P = 0.29 and 0.39, respectively).
FIGURE 1.
Manhattan plot of a meta-analysis of 10 genome-wide association studies for total plasma total homocysteine concentrations. Genome-significant associations were defined as P = 5 × 10−8 and are depicted by a gray line. Novel total homocysteine genes/loci that are described for the first time in this article are shown in bold gray font.
TABLE 1.
SNPs associated with total plasma homocysteine concentrations1
| SNP | Nearest gene | Chromosome | Coded allele | Frequency | β (SD/allele) | SE | P(meta) | P(het) |
| Known loci2 | ||||||||
| rs1801133 | MTHFR | 1 | A | 0.34 | 0.1583 | 0.007 | 4.34 × 10minus104 | 6.90 × 10minus8 |
| rs2275565 | MTR | 1 | T | 0.21 | −0.0542 | 0.009 | 1.96 × 10minus10 | 0.05 |
| rs7422339 | CPS1 | 2 | A | 0.33 | 0.0864 | 0.008 | 4.58 × 10minus27 | 0.61 |
| rs9369898 | MUT | 6 | A | 0.62 | 0.0449 | 0.007 | 2.17 × 10minus10 | 0.66 |
| rs7130284 | NOX4 | 11 | T | 0.07 | −0.1242 | 0.013 | 1.88 × 10minus20 | 0.35 |
| rs154657 | DPEP1 | 16 | A | 0.47 | 0.0963 | 0.007 | 1.74 × 10minus43 | 0.09 |
| rs234709 | CBS | 21 | T | 0.45 | −0.0718 | 0.007 | 3.90 × 10minus24 | 0.44 |
| Novel loci | ||||||||
| rs4660306 | MMACHC | 1 | T | 0.33 | 0.0435 | 0.007 | 2.33 × 10minus9 | 0.18 |
| rs548987 | SLC17A3 | 6 | C | 0.13 | 0.0597 | 0.010 | 1.12 × 10minus8 | 0.45 |
| rs42648 | GTPB10 | 7 | A | 0.40 | −0.0395 | 0.007 | 1.97 × 10minus8 | 0.57 |
| rs1801222 | CUBN | 10 | A | 0.34 | 0.0453 | 0.007 | 8.43 × 10minus10 | 0.05 |
| rs2251468 | HNF1A | 12 | A | 0.65 | −0.0512 | 0.007 | 1.28 × 10minus12 | 0.43 |
| rs838133 | FUT2 | 19 | A | 0.45 | 0.0422 | 0.007 | 7.48 × 10minus9 | 0.31 |
| Additional SNPs associated after conditional analysis | ||||||||
| rs12134663 | MTHFR | 1 | A | 0.80 | −0.101 | 0.011 | 2.54 × 10minus21 | 0.43 |
| rs12780845 | CUBN | 10 | A | 0.65 | 0.0529 | 0.009 | 7.8 × 10minus10 | 0.73 |
| rs957140 | NOX4 | 11 | A | 0.45 | −0.045 | 0.008 | 2.43 × 10minus8 | 0.97 |
| rs12921383 | DPEP1/FANCA | 16 | T | 0.87 | −0.090 | 0.014 | 8.22 × 10minus11 | 0.96 |
| rs2851391 | CBS | 21 | T | 0.47 | 0.056 | 0.008 | 1.70 × 10minus12 | 0.10 |
β, β coefficient presented as the number of SD differences in homocysteine concentrations per allele; P(het), P value for the Cochran's heterogeneity statistic; P(meta), P value for the overall meta-analysis; SNP, single nucleotide polymorphism.
Conditional independence of variants associated with tHcy concentrations
Visual inspection of the regional association plots of the 13 loci suggested that, for a number of loci, there was evidence to support the presence of additional SNPs that showed an association with variation in tHcy concentrations independently of the lead variant. This was supported by the high level of significance despite the low correlation (r2 < 0.2) with the lead SNP and its position with respect to recombination hot spots.
Regional plots of the results from this conditional analysis are shown in Supplementary Figure 2. Our analysis identified ≥5 other SNPs—at or near MTHFR, CUBN, NOX4, DPEP1, and CBS—which showed a significant association with variation in tHcy concentrations (P < 5 × 10−8) that were conditionally independent of the lead variant at these loci. In addition, there was evidence of a secondary signal at the SLC17A3 locus on chromosome 6, although this did not reach genome-wide significance (results not shown). In summary, we identified a total of 18 independent SNPs from 13 loci that are significantly associated with tHcy concentrations (see Supplementary Table 4 under “Supplemental data” in the online issue).
Functional genomics of identified variants
We tested for an association of the 18 lead SNPs [or SNPs in high linkage disequilibrium (r2 > 0.8) with the lead SNP; ie, proxies] with cis-allelic expression of gene transcripts by using publically available databases (see Supplementary Table 5 under “Supplemental data” in the online issue). Associations were seen for transcripts of LOC126661, RASIP1, CDK10, and CBS. The most significant correlation with expression in several tissues was seen with CBS for rs2851391—the SNP that was identified after conditional analysis. Notably, the lead variant on HNF1A was not associated with differential expression of the gene in any of the tissues tested, including the liver.
Effect of tHcy loci and the tHcy risk score on hyperhomocysteinemia
We examined the combined effect of the top SNPs arising from the 13 associated tHcy loci on the risk of tHcy concentrations and hyperhomocysteinemia (defined as tHcy ≥15 μmol/L) in 5528 individuals from the prospective population-based Rotterdam Study I and II. Weighted risk allele scores were generated, and the population was divided into deciles according to risk score. The combined risk score explained 4.6% (Rotterdam Study I) to 5.9% (Rotterdam Study II) of the variance in tHcy concentrations. A highly significant linear increase in the mean tHcy concentrations of individuals with increasing values of GRS (P-trend: 5 × 10−57) is shown elsewhere (see Supplementary Figure 3 under “Supplemental data” in the online issue). Individuals within the highest 10% of the GRS (n = 552) had 3-μmol/L higher tHcy concentrations than did those in the lowest 10% of GRS (n = 552) (P = 1 × 10−36; ANCOVA). In addition, the GRS was significantly associated with the risk of hyperhomocysteinemia (OR: 1.40; 95% CI: 1.32, 1.49; P = 3 × 10−28 for each 1-SD increase in GRS).
Effect of tHcy loci and the tHcy risk score on CAD
Given the strong observational association between tHcy concentrations and risk of cardiovascular disease (42), we tested 1) each individual tHcy-associated SNP, and 2) the combined effect of the 18 SNPs from the 13 tHcy loci, with CAD risk in up to 31,400 CAD cases and 92,927 controls. We did not have Hcy concentrations available for most of the cases and controls who were used to assess the association between the GRS and CAD. The variants rs2251468 (OR: 0.93; 95% CI: 0.91, 0.95; P = 2.5 × 10−07) and rs7422339 (OR: 0.96; 95% CI: 0.93, 0.99; P = 0.01) (Table 2), located in the HNF1A and CPS1 genes, respectively, are associated with an increased risk of CAD, although the latter failed to pass the Bonferroni correction criteria of P < 0.004.
TABLE 2.
Association between total homocysteine–associated loci and risk of coronary artery disease in 31,400 cases and 92,927 controls1
| SNP | Nearest gene | Chromosome | Position | Allele2 | OR | OR (95L) | OR (95U) | P(meta) | Q | P(het) | I2 | No. of cases3 | No. of controls3 | Effects4 |
| rs154657 | DPEP1 | 16 | 88235597 | A | 1.01 | 0.99 | 1.04 | 0.40 | 2.14 | 0.54 | 0.00 | 29,314 | 91,599 | ++++ |
| rs1801133 | MTHFR | 1 | 11778965 | A | 1.01 | 0.98 | 1.04 | 0.50 | 9.21 | 0.03 | 0.67 | 15,974 | 78,259 | +–+ |
| rs1801222 | CUBN | 10 | 17196157 | A | 0.98 | 0.96 | 1.01 | 0.29 | 10.04 | 0.02 | 0.70 | 18,817 | 81,102 | –+– |
| rs2251468 | HNF1A | 12 | 119889509 | A | 0.94 | 0.91 | 0.96 | 2.5E-07 | 2.85 | 0.42 | 0.00 | 29,014 | 91,299 | – – – – |
| rs2275565 | MTR | 1 | 235115299 | G | 1.01 | 0.98 | 1.04 | 0.37 | 3.40 | 0.33 | 0.12 | 29,319 | 91,604 | +–++ |
| rs234709 | CBS | 21 | 43360033 | C | 1.00 | 0.97 | 1.02 | 0.81 | 1.43 | 0.49 | 0.00 | 23,110 | 84,432 | /–+– |
| rs42648 | GTPBP10 | 7 | 89815696 | A | 1.01 | 0.98 | 1.03 | 0.68 | 4.33 | 0.23 | 0.31 | 28,384 | 90,669 | –++ |
| rs4660306 | MMACHC | 1 | 45751262 | C | 1.00 | 0.98 | 1.03 | 0.84 | 2.96 | 0.40 | 0.00 | 29,591 | 91,876 | –+–+ |
| rs548987 | SLC17A3 | 6 | 25977350 | C | 1.00 | 0.96 | 1.04 | 0.92 | 0.92 | 0.82 | 0.00 | 18,612 | 80,897 | ++– |
| rs7130284 | NOX4 | 11 | 88788020 | C | 1.01 | 0.96 | 1.05 | 0.75 | 8.32 | 0.04 | 0.64 | 28,330 | 90,615 | –++ |
| rs7422339 | CPS1 | 2 | 211248752 | A | 0.96 | 0.93 | 0.99 | 0.01 | 1.34 | 0.72 | 0.00 | 20,900 | 83,185 | –+– |
| rs838133 | FUT2 | 19 | 53951341 | A | 1.02 | 0.98 | 1.05 | 0.31 | 3.79 | 0.28 | 0.21 | 14,060 | 76,345 | –+++ |
| rs9369898 | MUT | 6 | 49490152 | A | 1.02 | 0.99 | 1.04 | 0.22 | 8.65 | 0.03 | 0.65 | 28,492 | 90,777 | −+−+ |
I2, Higgins heterogeneity index; P(meta), P value for the overall meta-analysis; P(het), P value for the Cochran's heterogeneity statistic; Q, Cochran's heterogeneity statistic; SNP, single nucleotide polymorphism; 95L, lower 95th percentile; 95U, upper 95th percentile.
Allele denotes the modeled or coded allele.
Number with marker present.
Effects denotes the summary of effect directions of all the single studies used for the meta-analysis, where “+” indicates a positive (increased) effect of the coded allele on risk of disease, “−” indicates a negative (decreased disease risk) effect of the coded allele on risk of disease, and “/” indicates missing.
We failed to show an association between the tHcy GRS and an increased risk of CAD (OR: 1.01; 95% CI: 0.98, 1.04; P = 0.49). We quantified the expected causal relation between tHcy variation and risk of CAD with the use of a Mendelian randomization framework. Using the output from the GRS analysis for hyperhomocysteinemia and CAD—ie, OR(CADGRS) = 1.01 or β = 0.01, ΔtHcyGRS = 0.18, and SE of OR(CADGRS) = 0.016—the OR(CADtHcy) was calculated at 1.05 (95% CI: 0.85, 1.2).
DISCUSSION
Given the strong observational link between tHcy concentrations and risk of CVD (1–3, 6), we tested whether common polymorphisms associated with variation in tHcy are also associated with CAD. To this end, we carried out a meta-analysis of 10 independent genome-wide association studies comprising a total of 44,147 individuals and identified 6 novel loci in or near genes MMACHC, SLC17A3, GTPB10, CUBN, HNF1A, and FUT2 and replicating variants previously reported at or near the MTHFR, MTR, CPS1, MUT, NOX4, DPEP1, and CBS loci. Furthermore, we provide statistical evidence pointing to the fact that there are ≥2 significant, independent loci in MTHFR, CUBN, NOX4, DPEP1, CBS, and SLC17A3. We then tested for an association between each lead SNP at these loci and a GRS combining these variants with an increased risk of hyperhomocysteinemia and CAD. Our results showed that genetic variants at the HNF1A locus—which are associated with reduced concentrations of tHcy—and previously associated with Maturity Onset Diabetes of the Young increased the risk of type 2 diabetes, and plasma concentrations of C-reactive protein, total cholesterol, and low-density lipoprotein are also significantly associated with a reduced risk of CAD. It is therefore plausible that this association is not caused by Hcy concentrations but rather to the pleiotropic role of this DNA variant, possibly through lipid metabolism. Although the aggregate GRS was significantly associated with hyperhomocysteinemia (OR: 1.40), we failed to detect an association between the GRS and CAD.
We used tHcy genetic variants to examine whether Hcy concentrations are causally related to CAD. Previous meta-analyses of published studies of MTHFR C667T and CAD had reported associations between the MTHFR variant and CAD (13, 43, 44). However, the MTHFR Studies Collaboration reported no association of MTHFR C677T with CAD in unpublished studies and showed that the discrepant results of unpublished and published studies were a result of publication bias or other methodologic problems in the Asian studies. The current meta-analysis extends the findings for MTHFR C677T and CAD in European populations by studying the association of CAD with all available genetic variants that influence Hcy concentrations. With the possible exception of the HNF1A locus, our results did not provide any support for an association of the tHcy-associated variants with CAD, including the most widely studied MTHFR C677T variant. A formal assessment of the expected causal association under the assumptions of Mendelian randomization, with the GRS used as the instrumental variable, is consistent with the conclusion that tHcy does not cause CAD. Notably, our study 1) did not have the potential publication bias that a meta-analysis on published studies might have had, and 2) had sufficient power to pick up the previously reported effect size for MTHFR (30% increased risk) and the other SNPs. Power calculations showed that, for CAD, we had ∼90% power to detect an OR as low as 1.07 for SNPs with an allele frequency of ≥20%r (with an α = 0.004). Because the GRS was associated with a 3-μmol/L average elevation of tHcy, an 11% increased risk might plausibly have been anticipated from the observational studies if the association of Hcy with coronary artery disease seen in those studies was causal. With 12,138 such events among 45,705 individuals, this collaboration had >85% power to detect that difference. Therefore, these data indicate that a genetically raised concentration of Hcy itself is unlikely to be even a modest causal factor in CAD. Because the association of the MTHFR variant with tHcy concentrations varies by folate status, the effect of other variants for tHcy-related pathways on tHcy concentrations may also vary by folate status because of differences in national policies on folate fortification (14, 44, 46). Despite differences in the effect of MTHFR C677T on tHcy concentrations by folate status, a previous meta-analysis has shown that such differences do not explain the null associations of MTHFRC677T with risk of CAD (14), and there is no evidence that differences in folate status would influence the effects of other tHcy-related variants on risk of CAD.
It is important to highlight that our study had little power to detect associations with common variants of smaller effects or to rarer (2–5%) and/or rare (≤1%) variants. Collectively, the identified polymorphisms explained only a small proportion of the total variance in tHcy concentrations, and the effect sizes attributable to the associated variants were modest, ranging from SD differences of 0.09 to 0.19 in tHcy per allele. This suggests that either a significant proportion of the genetic variants are yet to be discovered or that the true heritability of tHcy is substantially lower than the often quoted 63%.
A second potential limitation pertained to the study design. We included data from 10 different GWAS studies on tHcy concentrations in our meta-analysis. Five of the 10 cohorts included participants from the United States, and 1 of these—BLSA—recruited subjects after mandatory food fortification. For the other 5 cohorts, participants were predominantly from northwestern European ancestry. Note that tHcy concentrations were qualitatively very similar across the US cohorts, although folate concentrations were notably different, ranging from 7.9 to 23.6 pg/mL across the 4 cohorts (FHS, CHS, BLSA, and NHS), which had folate concentrations available (see Supplementary Table 2 under “Supplemental data” in the online issue). Total Hcy concentrations seem to vary significantly across the European cohorts. These differences might be explained in part by differences in age, sex distribution, medical and lifestyle factors, and technical differences in the measurement techniques. However, because we used normalized tHcy values for each population separately, and did not compare crude mean concentrations, these differences should not have affected the overall outcome of the meta-analysis.
The other important limitations pertain to the sex and ethnic distribution of the cohorts studied. More than 82% of the individuals from the 10 cohorts were female, and participants from 9 of the cohorts had a predominantly white ancestry, which meant that we had insufficient power to assess differences in gene associations by sex and our conclusions are restricted to populations of northwestern European ancestry.
Finally, results from the conditional analyses based on different assumptions about the penetrance model of the lead (eg, MTHFR) loci may be warranted in future studies. Similarly, a thorough evaluation of the potential effect of other confounding factors (eg, lack of adjustment for B vitamin status), which can only be speculated about currently, needs to be tested in future studies.
Interestingly, 3 of our novel associations in genes MMACHC, CUBN, FUT2, and in the confirmed association with MUT, have either been previously associated with variation in constitutive vitamin B-12 concentrations—MUT, FUT2, and CUBN—or in the case of MMACHC, are excellent candidates through association with Mendelian inborn errors of vitamin B-12 transport. Data from TwinsUK show that tHcy and vitamin B-12 concentrations are correlated (r = −0.09, P < 10−8). Thus, the simplest explanations for this could be that, because vitamin B-12 is an important cofactor in Hcy metabolism, any genetic perturbation of vitamin B-12 concentrations could have a pleiotropic effect downstream on circulatory concentrations of Hcy.
Finally, previous studies have shown that tHcy concentrations increase with decreasing glomerular filtration rate, although the precise mechanism by which tHcy concentrations is related to glomerular filtration rate and consequently to CAD has not been established (46, 47). Our results support the idea that it is not tHcy itself, but something that tHcy is a marker for, that is a causal risk factor for CAD. One possible candidate is renal impairment or perhaps more likely, a particular molecule that, such as tHcy, is increased in the setting of renal impairment. Two of our associations with tHcy, namely with variants in or in the vicinity of DPEP1 and SLC17A3, were previously shown to be associated with renal function. The gene DPEP1, which encodes for an enzyme that is highly expressed and active in the proximal convoluted tubules, is implicated in the renal metabolism of glutathione and its conjugates. Because DPEP1 deficiency leads to increased urinary excretion of cysteine—a precursor of tHcy—it has been hypothesized that genetic variation at DPEP1 affects renal activity by altering the renal handling of amino acids. We used data from the Rotterdam Study cohorts and adjusted our analyses for creatinine concentrations to test whether our association was driven by individuals with altered renal function, but this did not affect the results of the GWAS (data not shown). However, because creatinine concentrations serve as only a rough proxy for kidney function, we cannot completely rule out that this association is driven by kidney function.
The results of the current study extend the findings of previous studies of MTHFR C677T and CAD (14) by assessing the relevance for CAD of all available genetic determinants of tHcy concentrations, alone and in combination, and provide further refutation of the causal relevance of moderately elevated tHcy concentrations to CAD in white populations.
Supplementary Material
Acknowledgments
Study-specific acknowledgments are provided as part of the Supplementary Material.
The authors’ responsibilities were as follows—JBJvM and KRA: conceived and designed the study; JBJvM, GP, SMS, AH, KRA: researched the data, contributed to the discussion, wrote the manuscript, and reviewed and edited the manuscript; JBJvM, TT, SHV, IC, XY, AM, CN, LMR, KRA: contributed to the analysis; SB, JCB, HB, MJB, CC, LF, A Hamsten, A Hofman, DJH, AG, ADJ, SK, EK, DPK, AALMK, JCC, PK, JL, BM, CN, CJO, BMP, PMR, FR, US, DSS, HS, JS, PMU, PV, GW, DMW, HW, JCMW, MdH, PJ, AGU, JSK, DJR, MPR, VM, DIC, and NJS: contributed material/data and contributed to discussions; and JBJvM and KRA: had full access to the data in this study and take complete responsibility for the integrity of the data and the accuracy of the data analysis. All authors reviewed and edited the manuscript, and the corresponding authors had full access to all of the data in the study and had final responsibility for the decision to submit the manuscript for publication. None of the authors had a conflict of interest. The sponsors of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Footnotes
Abbreviations used: BLSA, Baltimore Longitudinal Study of Aging; CAD, coronary artery disease; CARDIoGRAM, Coronary ARtery DIsease Genome-wide Replication And Meta-Analysis; CEU, people living in Utah of Northwest European ancestry; GRS, genotype risk score; GWAS, genome-wide association studies; HapMap, haplotype map; LCL, lymphoblastoid cell lines; LOLIPOP, The London Life Sciences Prospective Population Study; NHS, Nurses’ Health Study; PROCARDIS, Precocious Coronary Artery Disease; SNP, single nucleotide polymorphisms; tHcy, total homocysteine; WGHS, Women's Genome Health Study.
REFERENCES
- 1.Wald DS, Law M, Morris JK. Homocysteine and cardiovascular disease: evidence on causality from a meta-analysis. BMJ 2002;325:1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casas JP, Bautista LE, Smeeth L, Sharma P, Hingorani AD. Homocysteine and stroke: evidence on a causal link from Mendelian randomisation. Lancet 2005;365:224–32. [DOI] [PubMed] [Google Scholar]
- 3.Den Heijer M, Lewington S, Clarke R. Homocysteine, MTHFR and risk of venous thrombosis: a meta-analysis of published epidemiological studies. J Thromb Haemost 2005;3:292–9. [DOI] [PubMed] [Google Scholar]
- 4.Nelen WL. Hyperhomocysteinaemia and human reproduction. Clin Chem Lab Med 2001;39:758–63. [DOI] [PubMed] [Google Scholar]
- 5.Morris MS. Homocysteine and Alzheimer's disease. Lancet Neurol 2003;2:425–8. [DOI] [PubMed] [Google Scholar]
- 6.van Meurs JB, Dhonukshe-Rutten RA, Pluijm SM, van der Klift M, de Jonge R, Lindemans J, de Groot LC, Hofman A, Witteman JC, van Leeuwen JP, et al. Homocysteine levels and the risk of osteoporotic fracture. N Engl J Med 2004;350:2033–41. [DOI] [PubMed] [Google Scholar]
- 7.Clarke R, Halsey J, Lewington S, Lonn E, Armitage J, Manson JE, Bonaa KH, Spence JD, Nygard O, Jamison R, et al. Effects of lowering homocysteine levels with B vitamins on cardiovascular disease, cancer, and cause-specific mortality: meta-analysis of 8 randomized trials involving 37 485 individuals. Arch Intern Med 2010;170:1622–31. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka T, Scheet P, Giusti B, Bandinelli S, Piras MG, Usala G, Lai S, Mulas A, Corsi AM, Vestrini A, et al. Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am J Hum Genet 2009;84:477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paré G, Chasman DI, Parker AN, Zee RR, Malarstig A, Seedorf U, Collins R, Watkins H, Hamsten A, Miletich JP, et al. Novel associations of CPS1, MUT, NOX4, and DPEP1 with plasma homocysteine in a healthy population: a genome-wide evaluation of 13 974 participants in the Women's Genome Health Study. Circ Cardiovasc Genet 2009;2:142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazra A, Kraft P, Lazarus R, Chen C, Chanock SJ, Jacques P, Selhub J, Hunter DJ. Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum Mol Genet 2009;18:4677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mälarstig A, Buil A, Souto JC, Clarke R, Blanco-Vaca F, Fontcuberta J, Peden J, Andersen M, Silveira A, Barlera S, et al. Identification of ZNF366 and PTPRD as novel determinants of plasma homocysteine in a family-based genome-wide association study. Blood 2009;114:1417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange LA, Croteau-Chonka DC, Marvelle AF, Qin L, Gaulton KJ, Kuzawa CW, McDade TW, Wang Y, Li Y, Levy S, et al. Genome-wide association study of homocysteine levels in Filipinos provides evidence for CPS1 in women and a stronger MTHFR effect in young adults. Hum Mol Genet 2010;19:2050–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis SJ, Ebrahim S, Davey Smith G. Meta-analysis of MTHFR 677C->T polymorphism and coronary heart disease: does totality of evidence support causal role for homocysteine and preventive potential of folate? BMJ 2005;331:1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke R, Bennett DA, Parish S, Verhoef P, Dotsch-Klerk M, Lathrop M, Xu P, Nordestgaard BG, Holm H, Hopewell JC, et al. Homocysteine and coronary heart disease: meta-analysis of MTHFR case-control studies, avoiding publication bias. PLoS Med 2012;9:e1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preuss M, Konig IR, Thompson JR, Erdmann J, Absher D, Assimes TL, Blankenberg S, Boerwinkle E, Chen L, Cupples LA, et al. Design of the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Study: a genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet 2010;3:475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chambers JC, Zhao J, Terracciano CM, Bezzina CR, Zhang W, Kaba R, Navaratnarajah M, Lotlikar A, Sehmi JS, Kooner MK, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet 2010;42:149–52. [DOI] [PubMed] [Google Scholar]
- 17.Hofman A, van Duijn CM, Franco OH, Ikram MA, Janssen HL, Klaver CC, Kuipers EJ, Nijsten TE, Stricker BH, Tiemeier H, et al. The Rotterdam Study: 2012 objectives and design update. Eur J Epidemiol 2011;26:657–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Boehnke M, Abecasis GR. Efficient study designs for test of genetic association using sibship data and unrelated cases and controls. Am J Hum Genet 2006;78:778–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–13. [DOI] [PubMed] [Google Scholar]
- 20.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet 2007;3:e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics 2007;23:1294–6. [DOI] [PubMed] [Google Scholar]
- 22.Horne BD, Anderson JL, Carlquist JF, Muhlestein JB, Renlund DG, Bair TL, Pearson RR, Camp NJ. Generating genetic risk scores from intermediate phenotypes for use in association studies of clinically significant endpoints. Ann Hum Genet 2005;69:176–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridker PM, Chasman DI, Zee RY, Parker A, Rose L, Cook NR, Buring JE. Women's Genome Health Study Working G: Rationale, design, and methodology of the Women's Genome Health Study: a genome-wide association study of more than 25,000 initially healthy american women. Clin Chem 2008;54:249–55. [DOI] [PubMed] [Google Scholar]
- 24.Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, Froguel P, Balding D, Scott J, Kooner JS. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet 2008;40:716–8. [DOI] [PubMed] [Google Scholar]
- 25.Farrall M, Green FR, Peden JF, Olsson PG, Clarke R, Hellenius ML, Rust S, Lagercrantz J, Franzosi MG, Schulte H, et al. Genome-wide mapping of susceptibility to coronary artery disease identifies a novel replicated locus on chromosome 17. PLoS Genet 2006;2:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minelli C, Thompson JR, Tobin MD, Abrams KR. An integrated approach to the meta-analysis of genetic association studies using Mendelian randomization. Am J Epidemiol 2004;160:445–52. [DOI] [PubMed] [Google Scholar]
- 27.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008;24:2938–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Göring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet 2007;39:1208–16. [DOI] [PubMed] [Google Scholar]
- 29.Idaghdour Y, Czika W, Shianna KV, Lee SH, Visscher PM, Martin HC, Miclaus K, Jadallah SJ, Goldstein DB, Wolfinger RD, et al. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat Genet 2010;42:62–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heap GA, Trynka G, Jansen RC, Bruinenberg M, Swertz MA, Dinesen LC, Hunt KA, Wijmenga C, Vanheel DA, Franke L. Complex nature of SNP genotype effects on gene expression in primary human leucocytes. BMC Med Genomics 2009;2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, et al. A genome-wide association study of global gene expression. Nat Genet 2007;39:1202–7. [DOI] [PubMed] [Google Scholar]
- 32.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat Genet 2007;39:1217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwan T, Benovoy D, Dias C, Gurd S, Provencher C, Beaulieu P, Hudson TJ, Sladek R, Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat Genet 2008;40:225–31. [DOI] [PubMed] [Google Scholar]
- 34.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol 2008;6:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. Genetics of gene expression and its effect on disease. Nature 2008;452:423–8. [DOI] [PubMed] [Google Scholar]
- 36.Inouye M, Silander K, Hamalainen E, Salomaa V, Harald K, Jousilahti P, Mannisto S, Eriksson JG, Saarela J, Ripatti S, et al. An immune response network associated with blood lipid levels. PLoS Genet 2010;6:e1001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, Rohrer K, Zhao A, Marlowe L, Kaleem M, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet 2009;84:445–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol 2008;6:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grundberg E, Kwan T, Ge B, Lam KC, Koka V, Kindmark A, Mallmin H, Dias J, Verlaan DJ, Ouimet M, et al. Population genomics in a disease targeted primary cell model. Genome Res 2009;19:1942–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, Chen W, Weichenthal M, Ellinghaus E, Franke A, Cookson W, et al. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet 2010;87:779–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Gutierrez Arcelus M, Sekowska M, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 2009;325:1246–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brattström L, Wilcken DE. Homocysteine and cardiovascular disease: cause or effect? Am J Clin Nutr 2000;72:315–23. [DOI] [PubMed] [Google Scholar]
- 43.Wald DS, Morris JK, Wald NJ. Reconciling the evidence on serum homocysteine and ischaemic heart disease: a meta-analysis. PLoS ONE 2011;6:e16473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klerk M, Verhoef P, Clarke R, Blom HJ, Kok FJ, Schouten EG. MTHFR 677C→T polymorphism and risk of coronary heart disease: a meta-analysis. JAMA 2002;288:2023–31. [DOI] [PubMed] [Google Scholar]
- 45.Zee RY, Mora S, Cheng S, Erlich HA, Lindpaintner K, Rifai N, Buring JE, Ridker PM. Homocysteine, 5,10-methylenetetrahydrofolate reductase 677C>T polymorphism, nutrient intake, and incident cardiovascular disease in 24,968 initially healthy women. Clin Chem 2007;53:845–51. [DOI] [PubMed] [Google Scholar]
- 46.van Guldener C. Why is homocysteine elevated in renal failure and what can be expected from homocysteine-lowering? Nephrol Dial Transplant 2006;21:1161–6. [DOI] [PubMed] [Google Scholar]
- 47.Potter K, Hankey GJ, Green DJ, Eikelboom JW, Arnolda LF. Homocysteine or renal impairment: which is the real cardiovascular risk factor? Arterioscler Thromb Vasc Biol 2008;28:1158–64. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.