Abstract
PIRIN (PRN) is a member of the functionally diverse cupin protein superfamily. There are four members of the Arabidopsis thaliana PRN family, but the roles of these proteins are largely unknown. Here we describe a function of the Arabidopsis PIRIN2 (PRN2) that is related to susceptibility to the bacterial plant pathogen Ralstonia solanacearum. Two prn2 mutant alleles displayed decreased disease development and bacterial growth in response to R. solanacearum infection. We elucidated the underlying molecular mechanism by analyzing PRN2 interactions with the papain-like cysteine proteases (PLCPs) XCP2, RD21A, and RD21B, all of which bound to PRN2 in yeast two-hybrid assays and in Arabidopsis protoplast co-immunoprecipitation assays. We show that XCP2 is stabilized by PRN2 through inhibition of its autolysis on the basis of PLCP activity profiling assays and enzymatic assays with recombinant protein. The stabilization of XCP2 by PRN2 was also confirmed in planta. Like prn2 mutants, an xcp2 single knockout mutant and xcp2 prn2 double knockout mutant displayed decreased susceptibility to R. solanacearum, suggesting that stabilization of XCP2 by PRN2 underlies susceptibility to R. solanacearum in Arabidopsis.
Keywords: PIRIN2, XCP2, papain-like cysteine protease, Arabidopsis thaliana, Ralstonia solanacearum, vascular pathogen
Introduction
PIRIN (PRN) proteins belong to the cupin domain-containing superfamily, which encompasses proteins with diverse functions (Dunwell et al., 2001, 2004). PRN proteins are highly conserved among mammals, plants, fungi and prokaryotic organisms. The single human PRN has been shown to interact with the nuclear factor I/CCAAT box transcription factor (Wendler et al., 1997) and the oncoprotein Bcl-3 (Dechend et al., 1999). It has been implicated in several processes, including transcriptional control, apoptosis, migration and cellular senescence of melanoma cells (Courtois and Gilmore, 2006; Ahmed and Milner, 2009; Miyazaki et al., 2010; Licciulli et al., 2011). Plants normally have small complements of PRN genes, most of which have poorly understood roles. However, Arabidopsis PRN1 is involved in blue light and ABA responses, seed germination and early seedling development (Lapik and Kaufman, 2003; Warpeha et al., 2007). TvPirin, a PRN gene in the parasitic plant Triphysaria versicolor, is transcriptionally up-regulated in roots after being exposed to the haustorium-inducing substance 2,6-dimethoxybenzoquinone (Bandaranayake et al., 2012). Transcription levels of Le-pirin, a tomato homolog of human PRN, dramatically increase during camptothecin-induced programmed cell death (PCD) (Orzaez et al., 2001) and, in barley, the transcription level of a PRN is reportedly induced by pathogen-derived trichothecenes (Boddu et al., 2007) that can induce cell death in plants (Nishiuchi et al., 2006; Boddu et al., 2007). Collectively, these results suggest that plant PRN genes participate in developmental and/or pathogen-related PCD.
Programmed cell death in plants as well as in many other organisms is controlled by the action of various types of proteases. A large part of the plant proteases accumulates in the central vacuole, where they are stored until the vacuole bursts with concomitant activation of their autolytic function after appropriate PCD-inducing stimuli (van der Hoorn, 2008; van Doorn et al., 2011; Bollhöner et al., 2013). Many of these vacuole-localized proteases belong to the C1A family of papain-like cysteine proteases (PLCPs), and several have been implicated in both developmental and pathogen-related PCD (van der Hoorn, 2008; Shindo and van Der Hoorn, 2008). For instance, the Arabidopsis PLCP Responsive to Dehydration 21A (RD21A) has been implicated in PCD on the basis of its interaction with Protein Disulfide Isomerase 5 (PDI5) which promotes PCD in endothelial cells of seeds (Andeme Ondzighi et al., 2008). RD21A was also recently shown to influence sensitivity to Botrytis cinerea infection (Shindo et al., 2012). The Arabidopsis PLCPs Xylem Cysteine Protease 1 (XCP1) and XCP2 are involved in cellular autolysis of xylem tracheary elements (TE) (Avci et al., 2008), while PLCP Responsive to Dehydration 19 (RD19) participates in Arabidopsis RRS1-R (RESISTANT TO RALSTONIA SOLANACEARUM 1-R)-mediated resistance to the bacterial pathogen Ralstonia solanacearum (Bernoux et al., 2008).
In a previous study, we identified several potential regulators of cell death in a differential gene expression analysis of a Zinnia elegans TE cell culture (Pesquet et al., 2013). Interestingly, expression of a Zinnia PRN gene coincided with TE PCD and was completely blocked when TE PCD was inhibited by silver thiosulfate (STS). Here, we provide evidence that the closest homolog of the Zinnia PRN in Arabidopsis, PRN2, can physically interact with three Arabidopsis PLCPs; XCP2, RD21A, and RD21B (Responsive to Dehydration 21B). To elucidate the underlying molecular mechanisms we investigated the nature of the interaction and obtained both in vitro and in planta evidence that PRN2 stabilizes XCP2 by inhibiting its autolysis. Interestingly, Arabidopsis null PRN2 and XCP2 mutants displayed increased resistance to the bacterial pathogen R. solanacearum. Genetic interaction was also observed between PRN2 and XCP2 during R. solanacearum infection. Collectively, these observations suggest that stabilization of XCP2 by PRN2 plays an important role in the compatible interaction between Arabidopsis and R. solanacearum.
Results
PRN2 null mutants have increased resistance to R. solanacearum
Analysis of the Arabidopsis genome indicated that the PRN gene At2g43120, denoted PRN2, encodes a protein which has the most similar sequence to a Zinnia protein (gi:219988534) (Figure S1a) that was previously identified as a potential regulator of PCD (Pesquet et al., 2013). PRN2 is expressed ubiquitously throughout the plant (Figure S1b). PRN2 has no known functional protein domains other than the typical cupin and pirin domains (gi:30689259). The protein lacks targeting signals, but observations of cells expressing fusion constructs using the native promoter showed that PRN2 is localized in both the cytosol and the nucleus (Figure S2). To investigate the function of the Arabidopsis PRN2 protein, we screened the NASC collection of T-DNA insertion lines (http://arabidopsis.info/) and identified two homozygous knockout lines, with T-DNA insertions at either position +77 (SM_3.15394) or +278 (SALK_079571) of the predicted open reading frame (Figure S1c). No full-length cDNA of PRN2 could be amplified by reverse transcription polymerase chain reaction (RT-PCR) from either of the two lines (Figure S1d), which we named prn2-1 (SM_3.15394) and prn2-2 (SALK_079571). We also generated two PRN2-overexpressing transgenic lines, PRN2OE6 and PRN2OE13, carrying constructs driven by the cauliflower mosaic virus 35S promoter. Overexpression of PRN2 in these lines was verified by quantitative (q)PCR (Figure S1e). Except for a slightly bushy growth pattern of one of the overexpressing lines (PRN2OE13), all the lines grew in a normal fashion (Figure S3).
To investigate a possible role of PRN2 in plant PCD, we first examined anatomical changes associated with PCD in the TEs of the prn2-1 mutant and the PRN2OE6 overexpressing line by transmission electron microscopy (TEM). No obvious changes between the two genotypes and Col-0 wild-type (WT) were observed in differentiation, occurrence of cell death or cellular autolysis of the xylem vessel elements (Figure S4). Next, we examined the mutants upon biotic stress induced by pathogens. Two prn2 mutants and two overexpressing lines were tested for resistance to five different pathogens, including Botrytis cinerea, Pseudomonas syringae pv. tomato (Pst), and three xylem-colonizing pathogens: Xanthomonas campestris pv. campestris (Xcc), Verticillium dahliae and Ralstonia solanacearum. No changes were observed in the prn2 mutants in response to infection with B. cinerea, V. dahliae, Pst or Xcc (Figures1a,b and S5). However, significant changes were observed in response to the bacterial wilt pathogen R. solanacearum. Four days post-inoculation (DPI) with R. solanacearum, all lines showed wilting symptoms, but the prn2 mutants had significantly fewer (P < 0.05) wilted leaves than WT (Figure1c). Conversely, symptoms developed more rapidly in the PRN2-overexpressing lines PRN2OE6 and PRN2OE13 than in WT (Figure1c). In accordance with the wilting symptoms, bacterial growth was significantly reduced (P < 0.001) in prn2-1 and prn2-2 plants and significantly increased (P < 0.001) in PRN2 overexpressors compared with WT plants at 3 DPI (Figure1d). The results demonstrate that PRN2 increases Arabidopsis susceptibility to R. solanacearum, thus the susceptibility of the Col-0 ecotype to R. solanacearum is at least partially mediated through PRN2.
Figure 1.
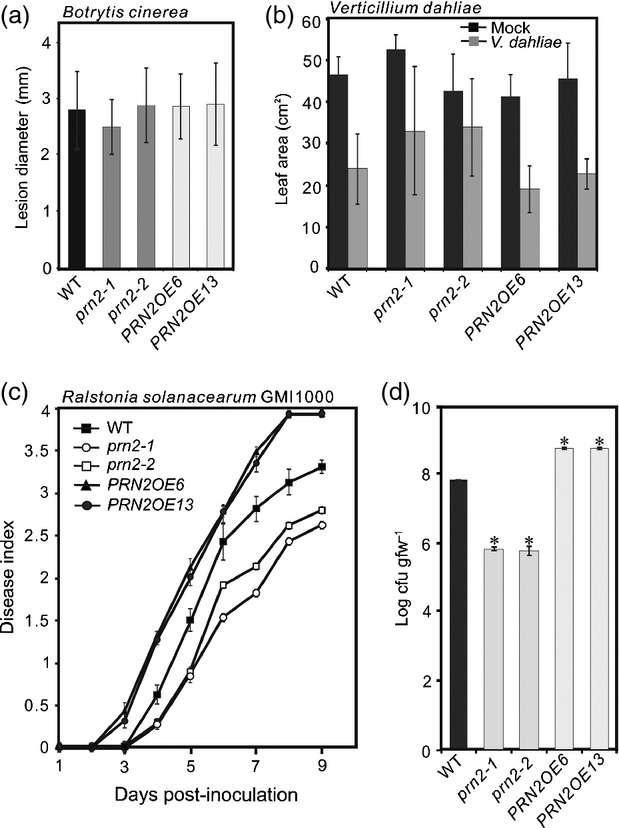
Arabidopsis PRN2 knockout mutants have enhanced resistance to Ralstonia solanacearum but not to Botrytis cinerea or Verticillium dahliae. (a) Plant resistance to B. cinerea. Diameter of lesions was observed 4 days post-inoculation (4 DPI) with B. cinerea. (b) Plant resistance to Verticillium dahliae. Foliar surface area was measured 21 DPI of V. dahliae- and mock-inoculated leaves. (c) Plant resistance to R. solanacearum GMI1000. Disease indices (DI) were scored 9 DPI of 30 plants per genotype (mean ± SD). DI 0 = no wilting, 1 = 25% of leaves per plant wilted, 2 = 50% of leaves per plant wilted, 3 = 75% of leaves per plant wilted, 4 = 100% of leaves per plant wilted. (d) Bacterial growth of R. solanacearum in the plant. Log transformed numbers of colony-forming units per gram fresh weight (cfu/gfw; means ± SD) are indicated for three biological replicates per genotype. Asterisks indicate significant differences (P < 0.001) between WT (Col-0) and prn2-1, prn2-2, PRN2OE6, and PRN2OE13 plants according to Student's t-test. All pathogen inoculation assays were repeated at least three times, and the R. solanacearum bacterial growth experiments twice, yielding similar results.
PRN2 physically interacts with XCP2, RD21A, and RD21B
To investigate the underlying molecular mechanism of PRN2 action in responses to R. solanacearum, we performed a yeast two-hybrid screen using the full-length Arabidopsis PRN2 as bait. The principal isolates included clones encoding the following three PLCPs: RD21A (At1g47128), RD21B (At5g43060), and XCP2 (At1g20850) (Figure2a and Table S1). The positive clones all encoded protein fragments encompassing both the pro-domain and the protease activity domain except for one XCP2 clone, which encoded a fragment containing only the protease activity domain (Figure2a).
Figure 2.
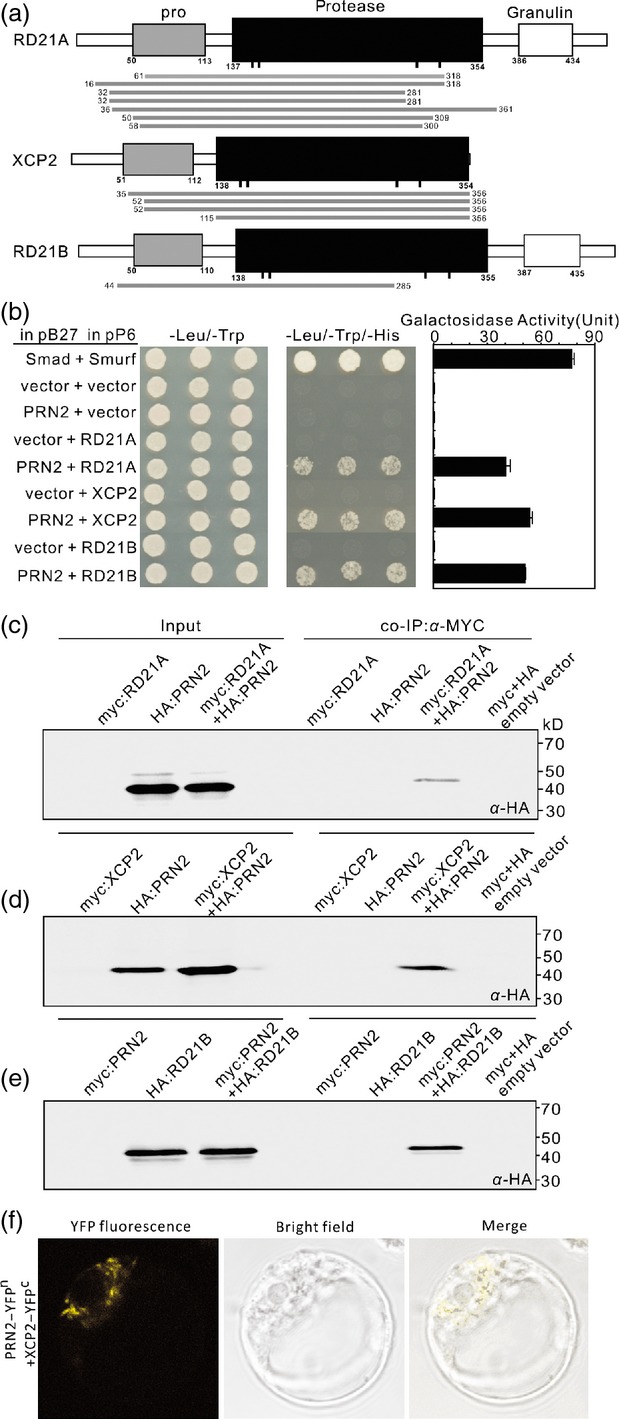
PRN2 interacts with RD21A, XCP2, and RD21B. (a) Identification of proteins interacting with PRN2 by yeast two-hybrid analysis. Alignment of putative amino acid sequences of the positive clones to those of full-length RD21A, XCP2, and RD21B proteins. The domain structure of the three papain-like cysteine proteases (PLCPs) consists of a pro-domain (pro, grey rectangles), a protease domain (black rectangles) with catalytic residues indicated by black bars, and a granulin domain (white rectangles). Putative amino acid sequences of the positive clones are shown with grey bars below each PLCPs. The numbers indicate start and stop positions of each positive clone. (b) One-by-one yeast two-hybrid assays. The results demonstrate interaction of PRN2 with RD21A, XCP2 and RD21B by activation of the reporter genes HIS3 and LacZ (β-galactosidase). Interaction between the human Smad and Smurf served as a positive control. Negative controls are represented by the vector combinations containing one or two empty vectors. The indicated combinations of plasmids were co-transformed into the yeast reporter strain, and interactions between the encoded proteins were assessed by growth on plates with yeast growth medium lacking Leu, Trp, and His (–Leu/–Trp/–His). Yeast growth on plates lacking Leu and Trp (–Leu/–Trp) shows the presence of the bait and prey vectors. The strength of activation of the second reporter gene (β-galactosidase) is shown in the chart to the right. (c–e) Co-immunoprecipitation (Co-IP) of PRN2 and XCP2. Co-IP with anti-Myc antibody is shown by western blot analysis, using anti-HA antibodies, for HA:PRN2 and myc:RD21A (c), HA:PRN2 and myc:XCP2 (d), and myc:PRN2 and HA:RD21B (e). Inputs are samples without Co-IP. The labeled bands in the Co-IP lanes indicate co-immunoprecipitated HA:PRN2 (c, d) and HA:RD21B (e). (f) BiFC assay of PRN2 and XCP2 interaction in Arabidopsis protoplasts. YFPN, N-terminal part of yellow fluorescent protein (YFP) (aa, 1–155); YFPC, C-terminal part of YFP (aa, 156–239). Further controls are shown in Figure S6 to prove specificity of the interaction.
Four tests verified the physical interactions. First, growth assays of yeast strains harboring the PRN2 bait vector and each of the three PLCP prey vectors on a histidine dropout medium showed that PRN2 interacted with the three proteases (Figure2b). Second, enzymatic assays using the β-galactosidase (β-gal) reporter gene constructs in the yeast strains revealed a high degree of interaction between PRN2 and each of the three PLCPs (Figure2b, right column). Third, co-immunoprecipitation (Co-IP) assays using Arabidopsis protoplasts confirmed the results of the yeast assays. Myc monoclonal antibody co-immunoprecipitated HA-tagged PRN2 protein together with myc-RD21A (Figure2c) or myc-XCP2 fusion protein (Figure2d) and HA-tagged RD21B protein together with myc-PRN2 (Figure2e). Fourth, bimolecular fluorescence complementation assays in Arabidopsis protoplasts confirmed the in vivo interaction between PRN2 and XCP2 in cytosolic compartments (Figures2f and S6). These findings demonstrate that PRN2 protein can physically interact with XCP2, RD21A, and RD21B both in vitro and in vivo.
PRN2 stabilizes XCP2 in vitro and in planta
To probe the nature of the interaction between PRN2 and the three PLCPs, we carried out protease activity profiling assays in Nicotiana benthamiana leaves transiently overexpressing the three PLCPs (XCP2, RD21A or RD21B) using an activity-based probe DCG-04, which is a biotinylated derivative of the irreversible PLCP inhibitor E-64 (van der Hoorn et al., 2004). Leaves transiently overexpressing XCP1 or Cathepsin B-like3 (CTB3) were used as controls. Protein extracts from the leaves were preincubated for 30 min with or without recombinant PRN2 (rPRN2) protein before adding DCG-04. After a 2-h incubation with DCG-04, one major band corresponding to each of the five protease was identified on blots using streptavidin-conjugated horseradish peroxidase (HRP) (Figure3a). The sizes of the proteins corresponded to the expected sizes of the active enzymes (Richau et al., 2012). Binding of DCG-04 was fully competed by treating with an excess of E-64 prior to labeling, indicating that DCG-04 specifically binds to PLCPs (Figure3a). Interestingly, in contrast to the expectation that PRN2 might bind and inhibit the proteases' activities, rPRN2 significantly enhanced DCG-04 labeling of protein extracts from leaves overexpressing XCP2 (Figure3a). A similar but smaller effect was observed for RD21A (Figure3a). These results suggest that PRN2 increases protease activities of XCP2 and RD21A.
Figure 3.
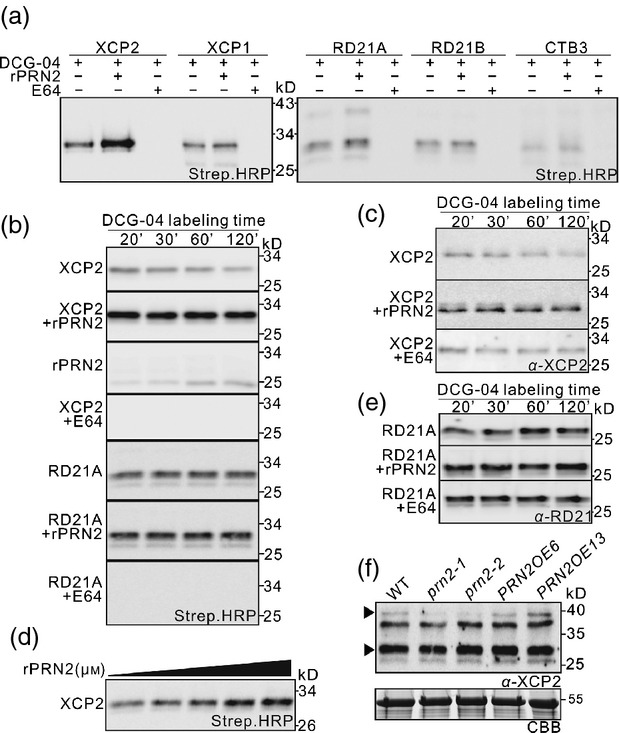
PRN2 stabilizes XCP2 in vitro and in planta. (a) Effect of PRN2 on the activities of five different papain-like cysteine proteases (PLCPs) in vitro. PLCP activity is shown in protein extracts of Nicotiana benthamiana leaves transiently overexpressing each Arabidopsis XCP2, RD21A, RD21B, XCP1, or CTB3 after labeling with DCG-04 in the presence or absence of recombinant PRN2 (rPRN2) or E64. Streptavidin–HRP detection shows one major band migrating around 30 kD for each of the five proteases. Enhanced signal in the second left lanes of XCP2 and RD21A indicates higher activity of these proteases in the presence of rPRN2. (b) Effect of PRN2 on PLCP activities over a 2-h time course. PLCP profiling of transiently overexpressed XCP2 or RD21A is shown during a 120-min time course of incubation in the presence or absence of rPRN2 or E64. The ‘rPRN2 panel’ is a negative control without any overexpressed proteases and reveals labeling of native PLCPs from N. benthamiana. (c) Effect of PRN2 on XCP2 stability in vitro over a 2-h time course. Western blot analysis with anti-XCP2 antibody (α-XCP2) is shown for protein extracts of N. benthamiana leaves transiently overexpressing XCP2 after incubation in the presence or absence of rPRN2 or E64. (d) Dosage effect of XCP2 stabilization by rPRN2. Western blot shows PLCP activity in protein extracts of N. benthamiana leaves transiently overexpressing XCP2 after labeling with DCG-04 in the presence of serial concentrations of rPRN2 (0, 0.0064, 0.064, 0.16, and 0.64 μm). (e) RD21 stability in vitro over a 2-h time course. Western blot analysis with anti-RD21A antibody (α-RD21) is shown in protein extracts of N. benthamiana leaves transiently overexpressing RD21A after incubation in the presence or absence of rPRN2 or E64. (f) PRN2 effect on XCP2 in planta. Western blot analysis with anti-XCP2 antibody (α-XCP2) is shown for total leaf protein extracts of Arabidopsis WT (Col-0), prn2-1, prn2-2, PRN2OE6, and PRN2OE13. The upper arrowhead indicates a specific band for XCP2 (as judged on the basis of the data in Figure S7a). Input proteins are visualized on a Coomassie Brilliant Blue (CBB)-stained protein gel.
Next, we examined effect of PRN2 on XCP2 and RD21A activities over time by incubating protein extracts of N. benthamiana leaves transiently overexpressing XCP2 or RD21A with DCG-04 and profiling the protease activities over a 2-h time course. The results revealed that XCP2 activity declined during the time course, but that the decline was suppressed in samples pretreated with rPRN2 (Figure3b). Western blot analysis using an antibody against XCP2 demonstrated that the decline in XCP2 activity resulted from gradual degradation of XCP2, which was prevented by preincubation with rPRN2 and E64 (Figure3c). The effect of PRN2 on XCP2 stability was shown to be dependent on the dosage of rPRN2 (Figure3d). The other protease, RD21A, did not show changes in stability or protease activity in response to PRN2 during the time course (Figure3b,e). These findings suggest that PRN2 stabilizes XCP2 by inhibiting XCP2 autolysis and led us to focus on PRN2-XCP2 interactions in further analysis.
To investigate effects of PRN2 on XCP2 in planta, XCP2 accumulation was analyzed in leaves of soil-grown prn2 knockout mutants and overexpressors. Labeling of four polypeptides (40, 37, 30, and 29 kD) with XCP2 antibody was observed in western blots of extracts of Col-0 (WT) leaves (Figure S7a). The 30-kD polypeptide (indicated by the lower arrowhead in the figure) is likely to be a mixture of polypeptides, including mature forms of XCP2 and RD21A (Yamada et al., 2001; van der Hoorn et al., 2004; Avci et al., 2008). Accordingly, it was less strongly labeled in extracts from xcp2, xcp1 xcp2, rd21a, and rd21ab leaves (Figure S7a). The 40-kD polypeptide (indicated by upper arrowhead) is likely to be the immature form of XCP2 (Avci et al., 2008), and was also absent in xcp2 and xcp1 xcp2 leaves (Figure S7a). Less of the 40-kD polypeptide was labeled by the XCP2 antibody in prn2-1 and prn2-2 than in WT leaf extracts (Figure3f), indicating that PRN2 stabilizes XCP2 in planta.
PRN2 competitively inhibits XCP2
A competitive DCG-04 labeling assay, in which DCG-04 probe was added to XCP2-overexpressing N. benthamiana leaf extracts simultaneously with rPRN2 provided indications of PRN2's action mechanism. In this assay, XCP2 activity was surprisingly suppressed in the presence of PRN2 at the start of the time course, but recovered to the same or somewhat higher activity level at the end of the time course experiment (Figure4a). This suggests that PRN2 inhibits XCP2 activity, but the inhibition is reversible and eventually results in stabilization of XCP2.
Figure 4.
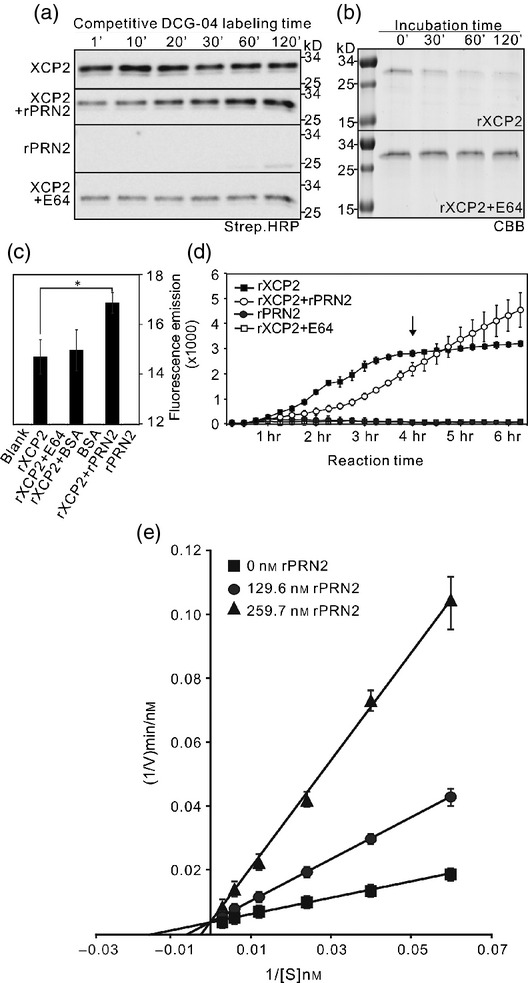
PRN2 stabilizes XCP2 by inhibiting its autolysis. (a) PRN2 effect on papain-like cysteine protease (PLCP) activities in a competitive assay. PLCP activity was profiled with DCG-04 over 120 min in protein extracts of N. benthamiana leaves transiently overexpressing XCP2 after incubation in the presence or absence of recombinant PRN2 (rPRN2) or E64. Streptavidin–HRP detection shows one major band of biotinylated PLCPs migrating around 30 kD for XCP2. The rPRN2 panel shows labeling of 25-kD polypeptides representing native PLCPs from N. benthamiana in negative controls with no overexpressed proteins. (b) The stability of recombinant XCP2 (rXCP2) during a 120-min time course in the presence or absence of E64, as detected by Coomassie Brilliant Blue staining (CBB). (c) Results of in vitro protease enzymatic assays of rXCP2 in the presence or absence of rPRN2, BSA or E64. Fluorescence emission indicates rXCP2 activity towards a casein substrate labeled with a fluorescent probe. Blank refers to a negative control without any added recombinant proteins. The asterisk indicates significant difference (P < 0.001) between the rXCP2 activities with and without rPRN2. (d) Enzyme progress curves of rXCP2 activity after incubation in the presence or absence of rPRN2 or E64. Fluorescence emission indicates rXCP2 activity towards the casein substrate labeled with a fluorescent probe. The arrow indicates the time point when rXCP2 activity was abolished without added rPRN2. (e) Lineweaver–Burk reciprocal plot of rXCP2 activity in the presence or absence of rPRN2. The reciprocal reaction velocity (1/V) is shown as a function of reciprocal substrate concentration [1/(S)] for rXCP2 activity when rXCP2 was incubated with serial concentrations of the casein substrate together with three different concentrations of PRN2, followed by protease enzymatic assay. The y-intercept is the same for the three rPRN2 concentrations, indicating that the Vmax (maximum reaction velocity) is not affected by rPRN2. The assays were performed three times yielding similar results.
To further investigate the inhibitory effect of PRN2 on XCP2 activity and the nature of the inhibitory mechanism, we conducted enzyme kinetic assays using a recombinant poly-His-tagged full-length XCP2 (rXCP2) protein together with rPRN2 (Figure S8a,b). The results indicated that rXCP2 autolysed with a similar time course to the plant-expressed XCP2 (Figure4b), and had a pH optimum of 4.5 for proteolysis of the standard casein substrate (Figure S8c,d). A protease activity assay using the fluorescence-based (BODiPY) EnzChek protease assay kit confirmed that rXCP2 has high activity, which was blocked by pretreatment with E64 and significantly enhanced (P < 0.01) by pretreatment with rPRN2, but not with Bovine Serum Albumin (BSA) used as a negative control (Figure4c). Enzyme progress-curve analysis (Duggleby, 1995) revealed that rXCP2 activity was indeed initially inhibited by rPRN2, but prolonged incubation with rPRN2 resulted in continued rXCP2 activity throughout the 6-h time course while incubation without rPRN2 abolished rXCP2 activity after just 4 h (Figure4d). The inhibition of rXCP2 activity by PRN2 was verified by a Lineweaver–Burk reciprocal plot of XCP2 activity in response to three different concentrations of rPRN2, which confirmed that XCP2 was inhibited by PRN2 in a competitive manner (Figure4e). The inhibition constant (Ki) was 2.226 × 10−8 for XCP2 with PRN2, as determined by non-linear regression (Leskovac, 2003). In these assays, it was also observed that rPRN2 was not degraded by rXCP2 (Figure S9).
PRN2 interacts with XCP2 during R. solanacearum infection
To investigate whether the PRN2-mediated stabilization of XCP2 contributes to the plant's response to R. solanacearum, we profiled PLCP activity and analyzed levels of the protein (by western blotting) in plants challenged with R. solanacearum. Leaves were harvested at day 0 (T0) post-inoculation. We also harvested wilted and unwil-ted leaves from plants scored as disease index1 (DI1; 25% of scored leaves per plant wilted), and designated them DI1(+) and DI1(−), respectively. The activity of PLCPs increased in WT DI1(−) and DI1(+) leaves in response to R. solanacearum infection (Figures5a,b and S10). The activities increased also in prn2-1 and prn2-2 leaves, but significantly less strongly (Figures5a,b and S10). Western blot analysis with the XCP2 antibody showed significant increases in the amounts of a 30-kD polypeptide in WT DI1(−) and DI1(+) leaves, but not in prn2-2 leaves (Figure5c). Also expression of XCP2 was increased in WT in response to R. solanacearum infection (Figure S11). Western blot analysis with the RD21 antibody showed similar induction of a 30-kD polypeptide in both WT and prn2-2 plants (Figure5d). Thus, the lower PLCP activity in prn2-2 mutants seems to be due to low level of XCP2, supporting the role of PRN2 in stabilizing XCP2 during R. solanacearum infection.
Figure 5.
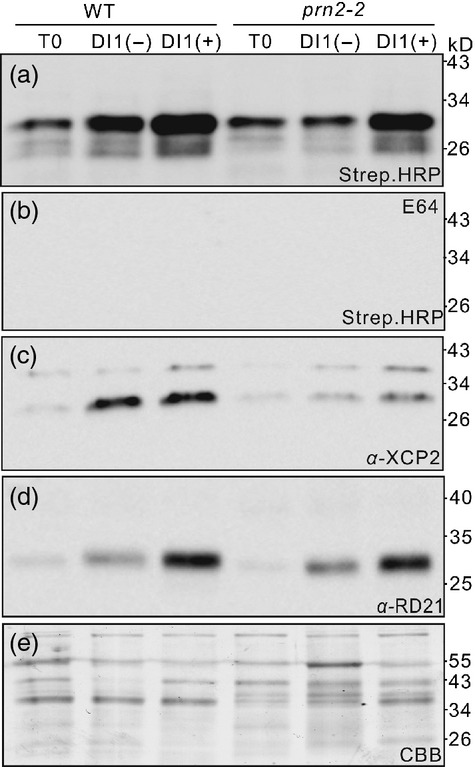
XCP2 induction by Ralstonia solanacearum is suppressed in the prn2-2 mutant. (a, b) Papain-like cysteine protease (PLCP) activity profiling with DCG-04 in protein extracts of leaves from WT (Col-0) and prn2-2 plants challenged by R. solanacearum. Active PLCPs are shown by streptavidin–HRP detection in the absence (a) or presence (b) of E64. (c) Western blot analysis with anti-XCP2 antibody (α-XCP2) of protein extracts of leaves from WT and prn2-2 plants challenged by R. solanacearum. (d) Western blot analysis with anti-RD21 antibody (α-RD21) of protein extracts of leaves from WT and prn2-2 plants challenged by R. solanacearum. (e) Input proteins are visualized by a Coomassie Brilliant Blue (CBB)-stained protein gel. T0, day 0 post-inoculation; DI1(−), unwilted leaves from DI1 plants; DI1(+), wilted leaves from DI1 plants. The assays were performed three times yielding similar results.
To further investigate this dependence, we inoculated an xcp2 knockout mutant (SALK_010938) (Avci et al., 2008), prn2-2 and the double mutant prn2-2 xcp2 with R. solanacearum, using Nd-1 (a resistant Arabidopsis accession) as a control. The prn2-2, xcp2, and prn2-2 xcp2 mutants were all more resistant to the pathogen than the WT plants and fewer disease symptoms appeared in their leaves (Figure6a,c). Growth rates of the bacteria were also slower in xcp2, prn2-2 and prn2-2 xcp2 leaf tissues (Figure6b). These results indicate that PRN2 and XCP2 both participate in a genetic pathway that increases the susceptibility to R. solanacearum, and thus support involvement of PRN2-mediated XCP2 accumulation in this pathway.
Figure 6.
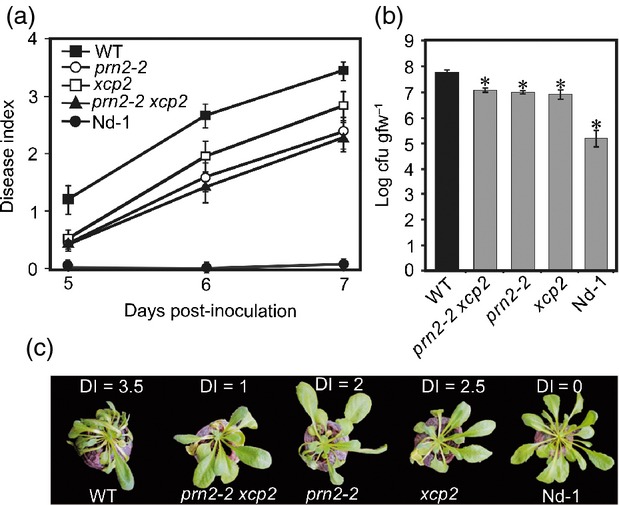
PRN2 interacts genetically with XCP2 during R. solanacearum infection. (a) Plant resistance to R. solanacearum GMI1000. Mean disease indices (see explanation in Figure1c legend) ± SD are shown for 30 plants per genotype 5, 6 and 7 days post-inoculation (DPI). (b) Growth of R. solanacearum bacteria in the plants. Log transformed mean numbers of colony-forming units per gram of fresh weight, cfu/gfw ± SD are shown for three biological replicates per genotype. The asterisks indicate significant differences (P < 0.001) between WT (Col-0) and the other genotypes according to Student's t-test. The pathogen inoculation assay was repeated three times, and the R. solanacearum bacterial growth experiments twice, yielding similar results. (c) Representative symptoms in each genotype at 7 DPI. Disease indices of these particular plants are indicated above each plant.
Discussion
The activity of proteolytic enzymes is commonly controlled by various kinds of protease inhibitors. Here, we report on the role of a PRN protein as a protease inhibitor in plants. We could show that the Arabidopsis PRN2 inhibits the enzymatic activity of the PLCP XCP2. This prevents autolytic degradation of XCP2, which in turn results in accumulation of XCP2 and increased overall XCP2 activity (Figure4). PRN2 does not possess any known protease inhibitor domains present in the previously identified PLCP inhibitors such as the serpins and cystatins. Therefore, PRN2 presumably acts in a different manner from the previously described cysteine protease inhibitors. The data we obtained from experiments with yeast and Arabidopsis protoplasts demonstrate that PRN2 interacts with three closely related PLCPs: XCP2, RD21A and RD21B (Figure2), suggesting that PRN2 could be involved in mechanisms involving several kinds of proteases. This is consistent with observations that different PLCPs can be present in the same protein complex (Andeme Ondzighi et al., 2008). Thus, we propose that PRN2 could function as a kind of scaffolding protein for PLCPs, allowing high local proteolytic activity whenever needed.
Several kinds of activities and protein interactions have been assigned to PRN proteins in various organisms, including human, bacteria, and several plant species. Human PRN has been associated with malignancies due to observed upregulation of its expression (inter alia) in breast cancer cells and in response to chronic cigarette smoking (Zhu et al., 2003; Gelbman et al., 2007), but the function has been shown only during the senescence and migration of melanoma cells (Miyazaki et al., 2010; Licciulli et al., 2011). The underlying molecular mechanism is believed to involve interactions with nuclear proteins, such as the transcriptional activator encoded by proto-oncogene B-cell lymphoma 3 (Bcl3) and various NF-κB proteins (Dechend et al., 1999; Liu et al., 2013). However, PRN proteins have functions also in other locations in addition to the nucleus. Notably, human PRN has been shown to dislocate in melanomas from the nucleus to the cytoplasm, where it seems to play a role in melanoma progression (Licciulli et al., 2010). In Arabidopsis, PRN1 is involved in ABA responses during germination and early seedling growth, possibly through the proven interaction with the plasma membrane-localized Gα subunit (Lapik and Kaufman, 2003). Our results show that even though the Arabidopsis PRN2 is localized to both the cytosol and the nucleus (Figure S2) the interaction between PRN2 and XCP2 takes place in the cytosol (Figures2f and S6). These results do not however exclude the possibility that plant PRN proteins, similar to the human counterpart, function also as transcriptional co-regulators in the nucleus.
We identified yet another role of the PRN proteins, as we showed that the Arabidopsis PRN2 is required for full susceptibility to the bacterial pathogen Ralstonia solanacearum (Figures1 and 6). Intriguingly, PRN2 shows specificity towards R. solanacearum and does not affect other xylem-colonizing pathogens, such as V. dahlia. However, whereas some defense components are found to target multiple xylem pathogens (Yadeta et al., 2014), others appear to be pathogen specific (Yadeta et al., 2011). Our data indicate further that the PRN2 function in the R. solanacearum response is mediated by interaction with PLCPs, and especially XCP2 (Figures5 and 6). No other evidence for a specific role of XCP2 in plant pathogenesis has been published, although it is implicated in pathogen resistance because heterologous expression of the Cladosporium fulvum virulence protein Avr2 suppresses XCP2 activity in Arabidopsis (van Esse et al., 2008). However, other PLCPs have well-documented functions in plant pathogenesis. For instance, in tomato the PLCP RCR3 (Required for Cladosporium resistance3) confers resistance to C. fulvum in the presence of the resistance gene Cf2 (Krüger et al., 2002), and contributes to resistance to Phytophthora infestans and a nematode (Song et al., 2009; Lozano-Torres et al., 2012). In Nicotiana benthamiana, silencing of a PLCP C14 reportedly causes hyper-susceptibility to P. infestans (Kaschani et al., 2010; Bozkurt et al., 2011). Arabidopsis plants with a mutant form of the PLCP RD19 are also more susceptible to Ralstonia in the presence of RRS1-R (Bernoux et al., 2008) and RD21A mutants are more susceptible to Botrytis (Shindo et al., 2012). Most importantly, in every previously reported case the PLCPs have been shown to increase the resistance to the invading pathogen. In sharp contrast, the xcp2 mutant has enhanced resistance to R. solanacearum, thus XCP2 reduces immunity to the bacterium (Figure6). This exceptional effect of PLCP function in pathogenesis suggests that the interaction between XCP2 and R. solanacearum does not involve a typical R-protein mediated resistance response, but is related to some other response that might be quite specific to R. solanacearum.
Resistance to R. solanacearum can be affected by specific structural features of the vasculature including pit membranes of vessel elements, contacts with neighbouring parenchymatic cells (Nakaho et al., 2000; Nakaho and Allen, 2009) and properties of the secondary cell walls (Hernandez-Blanco et al., 2007; Turner et al., 2009). XCP2 is normally expressed in the xylem vessel elements, through which R. solanacearum propagates and spreads throughout the plant (Digonnet et al., 2012). XCP2 is known to degrade cellular contents, especially after the developmental death of the vessel elements when the vacuolar contents have been mixed with the cytoplasm (Avci et al., 2008). Thus, impaired autolysis of the cellular remnants may inhibit propagation and/or dispersal of the bacteria in the vessels, thereby reducing symptom development in the xcp2 mutant. Another possible location of XCP2 action in R. solanacearum infection is the leaves, where R. solanacearum becomes a necrotrophic pathogen once it enters through the vasculature (Saile et al., 1997; Genin, 2010; Digonnet et al., 2012). Therefore, XCP2 activity may be required for full susceptibility to R. solanacearum by facilitating autolysis of the cellular contents in the leaves instead. Due to instability of the XCP2 protein, PRN2 may be then required to stabilize XCP2 and maintain its activity in cellular autolysis during R. solanacearum infection.
Experimental procedures
Plant material and growth conditions
Arabidopsis WTs Col-0 and Niedersenz (Nd-1; NASC accession number N1636), which are respectively fully susceptible and resistant to the R. solanacearum GMI1000 strain (Deslandes et al., 1998) were used in this study, in addition to prn2-1 (SM_3.15394), prn2-2 (SALK_079571) mutants, and the previously described knockout mutant xcp2 (SALK_010938) (Avci et al., 2008). To create PRN2 overexpression lines, full-length cDNA of PRN2 was amplified and cloned into pDONR207 vector by BP Clonase II (Invitrogen, www.lifetechnologies.com), followed by recombination into pK2GW7 destination vector (Karimi et al., 2002) and transformation into Arabidopsis by floral dipping. Plants were grown in soil, in growth chambers under short-day conditions (8 h light/16 h dark, 21/18°C, 70% relative humidity).
Plant inoculations
Ralstonia solanacearum GMI1000 inoculation by root dipping, disease rating and bacterial growth measurements were performed as previously described (Deslandes et al., 1998), using at least 30 plants of each genotype for the disease rating, and three per genotype for the bacterial growth measurement, in at least three independent experiments.
Botrytis cinerea inoculation was performed by placing two 4-μl drops of a conidial suspension (5 × 105 conidia/ml) in 12 g/l potato dextrose broth (Difco, www.bd.com) on each fully expanded leaf of tested plants. Plants were incubated at 20°C, 100% RH, under a 16 h/8 h light/dark regime. Disease progression was scored at 4 DPI by measuring the length and width of each lesion. At least four plants per genotype were analyzed in three independent experiments.
Verticillium dahliae inoculation was performed, essentially as previously described (Fradin et al., 2011), by dipping roots of 2-week-old soil-grown plants for 2 min in a conidial suspension (1 × 106 conidia/ml) in water. Mock inoculation was performed by dipping roots in water with no conidia. After replanting in soil, plants were incubated under standard greenhouse conditions (16 h/8 h light/dark regime, 70% RH). Symptoms were quantified at 21 DPI by determining the plants' foliar surface area using ImageJ version 1.47c (Reusche et al., 2012). At least four plants per genotypes were analyzed in two independent experiments, and eight per genotype in a final confirmatory experiment.
Pathogenicity assays with Pseudomonas syringae pv. tomato DC3000 and Xanthomonas campestris pv. campestris 8004 are described in Methods S1. The measured variables were expressed as mean ± SD and compared using Student's t-test. In the figures, significant P-values are shown as *(P < 0.001).
Yeast two-hybrid analysis
Yeast two-hybrid screening was performed by Hybrigenics, S.A., Paris, France (www.hybrigenics-services.com).
The full-length coding sequence (CDS) of AtPRN2 (At2 g43120) was amplified by PCR and cloned into pB27 as a C-terminal fusion to LexA (N-LexA-Pirin-C). The construct was checked by sequencing the entire insert and used as a bait to screen a random-primed Arabidopsis thaliana seedling cDNA library constructed using the plasmid pP6. pB27 and pP6 derive from the original pBTM116 (Vojtek and Hollenberg, 1995) and pGADGH (Bartel et al., 1993) plasmids, respectively.
In total, 63 million clones (six-fold the complexity of the library) were screened using a mating approach with Y187 (matα) and L40ΔGal4 (mata) yeast strains as previously described (Fromont-Racine et al., 1997). Overall, 354 His+ colonies were selected on a medium lacking tryptophan, leucine and histidine, and supplemented with 100 mm 3-aminotriazole to avoid bait autoactivation. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions. The resulting sequences were used to identify the corresponding interacting proteins in the GenBank database (NCBI) using a fully automated procedure.
One-by-one yeast two-hybrid interaction assays, including yeast growth on histidine dropout media (Kaiser et al., 1994) and β-galactosidase quantitative assays (Serebriiskii and Golemis, 2000), were performed as described in the Clontech Yeast Protocols Handbook (www.clontech.com/SE/Support/Product_Documents?sitex=10023:22372:US). The empty bait and prey vectors pB27, pP6, and two vectors carrying the fusion proteins LexA-Smad [pB27-Smad] and GAL4AD-Smurf [pP6-Smurf] that were used as positive controls, were obtained from Hybrigenics.
Transient expression in Arabidopsis protoplasts and co-immunoprecipitation assay
Full-length CDS of AtPRN2, AtXCP2, AtRD21A, and AtRD21B were cloned into the pRT104 3×HA or pRT104 3×myc vectors (Fulop et al., 2005) using BamHI and EcoRI restriction enzymes. Transfection in Arabidopsis protoplasts and co-immunoprecipitation were performed as previously described (Meskiene et al., 2003; Fulop et al., 2005). Myc-tagged proteins were co-immunoprecipitated from total protein extracts by incubating the extracts for 2 h at 4°C with 500 ng of 9E10C anti-myc monoclonal antibody (Covance, www.covance.com) and 10 μl of Protein G Sepharose (GE Healthcare, www.gehealthcare.com) Beads were then washed three times with washing buffer [1× phosphate-buffered saline (PBS) pH 7.4, 5% glycerol, 0.1% Igepal CA-630] and bound proteins were eluted with 50 μl of sodium dodecyl sulphate (SDS) loading buffer. Co-immunoprecipitation of the HA-tagged protein was detected by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by western blotting using the 16B12 anti-HA-POD monoclonal antibody (Roche, www.roche.com) and enhanced chemiluminescence detection (SuperSignal WestPico; Pierce, www.piercenet.com).
Microscopy
For bimolecular fluorescence complementation assay, full-length cDNAs of PRN2 and XCP2 were cloned into pUC-SPYNE or pUC-SPYCE vector (Walter et al., 2004) using BamHI and EcoRI restriction enzymes and transfected in Arabidopsis protoplasts. The two empty vectors, expressing YFPN and YFPC, were used as controls. Fluorescence of the protoplasts was visualized 24 h after transfection with a TCS SP2 (Leica, www.leica.com) confocal microscope and a 488 nm Ar/Kr laser line. YFPN/C fluorescence was detected with the excitation/emission combination of 514/525–535 nm.
TEM analysis and subcellular localization of PRN2:GFP fusion protein are described in Methods S1.
Protease activity profiling
XCP1, XCP2, RD21A, RD21B, and CTB3 were transiently overexpressed in Nicotiana benthamiana leaves by agroinfiltration as previously described (Richau et al., 2012). Protease activity profiling of N. benthamiana leaf extracts were analyzed, using a biotinylated DCG-04 probe, as previously described (Shindo et al., 2012). The protein extracts were labeled using 2 μM DCG-04 in a buffer containing 50 mm sodium acetate (pH 5.5) and 1 mm dithiothreitol (DTT) at room temperature. Inhibition assays were performed by DCG-04 labeling of the protein extracts with or without preincubation with 0.64 μm recombinant PRN2 protein or 20 μm E-64 for 30 min.
Competition assays were performed by adding the recombinant PRN2 protein (0.64 μm) or E-64 (20 μm) simultaneously with DCG-04 (2 μm) to the labeling reaction mixtures (100 μl). To monitor changes in activity over time 15 μl samples were taken at selected time points. The labeling reactions were stopped by adding 10 μl 5× SDS-PAGE sample buffer. Proteins in all samples were separated by SDS-PAGE, transferred onto nitrocellulose membrane (Bio-Rad www.biorad.com), and detected with streptavidin-HRP (1:3000; Ultrasensitive; Sigma, www.sigmaaldrich.com).
Papain-like cysteine proteases s activity profiling in protein extracts from Arabidopsis 4–6-week-old leaves challenged with R. solanacearum was performed as previously described (van der Hoorn et al., 2004).
Expression and purification of recombinant PRN2 and XCP2
Full-length CDSs of AtPRN2 and AtXCP2 were each cloned into pET28a(+) vector (Novagen, www.emdmillipore.com) using BamHI and XhoI restriction enzymes. The recombinant proteins were expressed in BL21 (DE3) Rosetta cells by growing cultures at 37°C with shaking at 250 rpm to OD600 0.5 and then inducing with 0.5 mm isopropyl-d-thiogalactoside (IPTG). Proteins were extracted and purified using Ni-NTA agarose beads as described by the manufacturer (Qiagen; www.qiagen.com). Recombinant PRN2 protein (rPRN2) was purified under native conditions. Recombinant XCP2 protein (rXCP2) was purified under denaturing conditions, followed by renaturation as previously described (Smith and Gottesman, 1989; Zhao et al., 2000). Briefly, purified XCP2 protein in elution buffer was added slowly (two drops per min) to 200 volumes of renaturation buffer (50 mm KH2PO4, 5 mm Ethylenediaminetetraacetic acid, 1 mm reduced glutathione,0.1 mm oxidized glutathione, 200 mm l-arginine pH 8.0) and stirred overnight at 4°C then concentrated at 4°C using Amicon Ultra-15 Centrifugal Filter Units (Millipore, www.millipore.com).
Protease enzymatic assays
In vitro protease enzymatic assays were performed using a fluorescence-based (BODiPY) EnzChek protease assay kit (Molecular Probes, http://products.invitrogen.com/ivgn/product/E6638). Autolysis of renatured XCP2 was examined by incubating it, with no other protein, in the presence or absence of E64 (10 μm) in a buffer containing 50 mm sodium citrate pH4.5 and 5 mm DTT (reducing agent). Inhibition assays were performed by preincubating the renatured rXCP2 protein (0.485 μM) with or without rPRN2 (0.75 μm) at 4°C for 2 h in a buffer containing 50 mm sodium citrate (pH4.5) and 5 mm DTT (reducing agent), then adding BODiPY-labeled casein substrate provided in the protease assay kit. The reaction was followed by monitoring changes in fluorescence (using a Tecan Infinite 200 PRO microplate reader with a 485 ± 12.5 nm excitation/530 ± 15 nm emission filter) indicating release of the probe from cleaved casein. Preincubation with BSA (0.75 μm) or E64 (5 μm) was used to provide negative and positive controls, respectively. Enzyme kinetic assays were performed by incubating rXCP2 (485 nm) with the substrate at serial concentrations (16.7, 25.0, 41.7, 83.3, 166.7 and 333.3 nm) together with various concentrations of rPRN2 (0, 129.6 and 259.7 nm). Samples were taken at several time points from each mixture to ensure that data points were representative of the initial velocity. The inhibition constant Ki was determined by non-linear regression, and Lineweaver–Burk plots was used to visualize the inhibition of rXCP2 by rPRN2. Each assay was performed at least three times yielding similar results and Student's t-test were applied to detect significant between-treatment differences in results.
Arabidopsis PRN2 peptide antibody and western blot analysis
The synthetic peptide (NH2−)MRAAINRANSLGGC(-CONH2) was used as an antigen for PRN2 antibody production in rabbit from Agrisera (www.agrisera.com).
For western blot analysis of plant extracts, Arabidopsis rosette leaves were homogenized in lysis buffer, including 25 mm Tris–HCl (pH 7.5–8.0), 10% glycerol, 75 mm NaCl (50–150 mm), 0.2% CA-630 (Igepal), 1 mm DTT and protease inhibitor (Roche, www.roche.com). The extracts were centrifuged at 18 000 g for 10 min at 4°C and the supernatant was taken. The protein concentration was determined by Bradford assay (Bio-rad, www.biorad.com). Protein samples were boiled in SDS-PAGE sample buffer and separated by SDS-PAGE, transferred onto nitrocellulose membrane (Bio-Rad, www.biorad.com), and detected with the SuperSignal western blotting system (ThermoScientific, www.thermoscientific.com).
Acknowledgments
We thank László Bakó (Umeå University) for technical help with transfection of Arabidopsis protoplasts and co-immunoprecipitation assays, Eric Beers (Virginia Polytechnic Institute and State University) for providing anti-XCP2 antibody and seeds of the xcp1 xcp2 double mutant, Ikuko Hara-Nishimura (Kyoto University) for providing anti-RD21 antibodies, Dr. Hermen Overkleeft for providing DCG-04, and Lenore Johansson (Umeå Core Facility Electron Microscopy, UCEM) for technical assistance with preparing TEM samples. We gratefully acknowledge financial support provided by the Kempe foundation, the Swedish Research Council FORMAS (Strong Research Environment BioImprove), the Swedish Research Council VR and the Swedish Governmental Agency for Innovation Systems VINNOVA (UPSC Berzelii Centre the Deutsche Forschungsgemeinschaft grant HO 3983/7-1, the Max Planck Society, COST CM1004 and Bio4Energy – a strategic research environment appointed by the Swedish government. The authors declare no conflict of interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Figure S1. Analysis of the Arabidopsis PRN2 and the genotypes used in the study.
Figure S2. Subcellular localization of Arabidopsis PRN2 protein.
Figure S3. Morphology of PRN2 knockout and overexpression lines.
Figure S4. Electron microscopy analysis of xylem vessel elements in the hypocotyls.
Figure S5. Results of pathogen inoculation assays.
Figure S6. BiFC assay of PRN2 and XCP2 interaction in Arabidopsis protoplasts.
Figure S7. Western blot analysis with XCP2 and RD21 antibody.
Figure S8. Expression and purification of recombinant PRN2 and XCP2.
Figure S9. The stability assay of rPRN2 in the presence of rXCP2.
Figure S10. Induction of PLCP activity in prn2-1 post-inoculation of R. solanacearum.
Figure S11. Expression of XCP2 after infection with a compatible R. solanacearum strain.
Table S1 Results of yeast two-hybrid screening using the full-length Arabidopsis PRN2 as a bait.
Table S2 All primer sequences used in this study.
Methods S1. Methods related to acquisition of the data in the supporting information.
References
- Ahmed SU, Milner J. Basal cancer cell survival involves JNK2 suppression of a novel JNK1/c-Jun/Bcl-3 apoptotic network. PLoS ONE. 2009;4:e7305. doi: 10.1371/journal.pone.0007305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andeme Ondzighi C, Christopher DA, Cho EJ, Chang SC, Staehelin LA. Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell. 2008;20:2205–2220. doi: 10.1105/tpc.108.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avci U, Petzold HE, Ismail IO, Beers EP, Haigler CH. Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J. 2008;56:303–315. doi: 10.1111/j.1365-313X.2008.03592.x. [DOI] [PubMed] [Google Scholar]
- Bandaranayake PC, Tomilov A, Tomilova NB, Ngo QA, Wickett N, dePamphilis CW, Yoder JI. The TvPirin gene is necessary for haustorium development in the parasitic plant Triphysaria versicolor. Plant Physiol. 2012;158:1046–1053. doi: 10.1104/pp.111.186858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P, Chien C, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley DA, editor. Cellular Interactions in Development: A practical Approach. Oxford: IRL Press; 1993. pp. 153–179. [Google Scholar]
- Bernoux M, Timmers T, Jauneau A, Briere C, de Wit PJ, Marco Y, Deslandes L. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell. 2008;20:2252–2264. doi: 10.1105/tpc.108.058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddu J, Cho S, Muehlbauer GJ. Transcriptome analysis of trichothecene-induced gene expression in barley. Mol. Plant Microbe Interact. 2007;20:1364–1375. doi: 10.1094/MPMI-20-11-1364. [DOI] [PubMed] [Google Scholar]
- Bollhöner B, Zhang B, Stael S, Denance N, Overmyer K, Goffner D, Van Breusegem F, Tuominen H. Post mortem function of AtMC9 in xylem vessel elements. New Phytol. 2013;200:498–510. doi: 10.1111/nph.12387. [DOI] [PubMed] [Google Scholar]
- Bozkurt TO, Schornack S, Win J, et al. Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl Acad. Sci. USA. 2011;108:20832–20837. doi: 10.1073/pnas.1112708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois G, Gilmore TD. Mutations in the NF-κB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- Dechend R, Hirano F, Lehmann K, Heissmeyer V, Ansieau S, Wulczyn FG, Scheidereit C, Leutz A. The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene. 1999;18:3316–3323. doi: 10.1038/sj.onc.1202717. [DOI] [PubMed] [Google Scholar]
- Deslandes L, Pileur F, Liaubet L, Camut S, Can C, Williams K, Holub E, Beynon J, Arlat M, Marco Y. Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum. Mol. Plant Microbe Interact. 1998;11:659–667. doi: 10.1094/MPMI.1998.11.7.659. [DOI] [PubMed] [Google Scholar]
- Digonnet C, Martinez Y, Denance N, Chasseray M, Dabos P, Ranocha P, Marco Y, Jauneau A, Goffner D. Deciphering the route of Ralstonia solanacearum colonization in Arabidopsis thaliana roots during a compatible interaction: focus at the plant cell wall. Planta. 2012;236:1419–1431. doi: 10.1007/s00425-012-1694-y. [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Beers EP, Dangl JL, et al. Morphological classification of plant cell deaths. Cell Death Differ. 2011;18:1241–1246. doi: 10.1038/cdd.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggleby RG. Analysis of enzyme progress curves by nonlinear regression. Methods Enzymol. 1995;249:61–90. doi: 10.1016/0076-6879(95)49031-0. [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Culham A, Carter CE, Sosa-Aguirre CR, Goodenough PW. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 2001;26:740–746. doi: 10.1016/s0968-0004(01)01981-8. [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- van Esse HP, Van ‘t Klooster JW, Bolton MD, Yadeta KA, van Baarlen P, Boeren S, Vervoort J, de Wit PJ, Thomma BP. The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell. 2008;20:1948–1963. doi: 10.1105/tpc.108.059394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GC, Joosten MH, Thomma BP. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 2011;156:2255–2265. doi: 10.1104/pp.111.180067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Fulop K, Pettko-Szandtner A, Magyar Z, Miskolczi P, Kondorosi E, Dudits D, Bako L. The Medicago CDKC;1-CYCLINT;1 kinase complex phosphorylates the carboxy-terminal domain of RNA polymerase II and promotes transcription. Plant J. 2005;42:810–820. doi: 10.1111/j.1365-313X.2005.02421.x. [DOI] [PubMed] [Google Scholar]
- Gelbman BD, Heguy A, O'Connor TP, Zabner J, Crystal RG. Upregulation of pirin expression by chronic cigarette smoking is associated with bronchial epithelial cell apoptosis. Respir. Res. 2007;8:10. doi: 10.1186/1465-9921-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin S. Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 2010;187:920–928. doi: 10.1111/j.1469-8137.2010.03397.x. [DOI] [PubMed] [Google Scholar]
- Hernandez-Blanco C, Feng DX, Hu J, et al. Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell. 2007;19:890–903. doi: 10.1105/tpc.106.048058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RA. Plant proteases: from phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA, Leeuwenburgh MA, Bogyo M, Joosten MH, Peck SC. Activity profiling of papain-like cysteine proteases in plants. Plant Physiol. 2004;135:1170–1178. doi: 10.1104/pp.104.041467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. New York: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kaschani F, Shabab M, Bozkurt T, Shindo T, Schornack S, Gu C, Ilyas M, Win J, Kamoun S, van der Hoorn RA. An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 2010;154:1794–1804. doi: 10.1104/pp.110.158030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JD. A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science. 2002;296:744–747. doi: 10.1126/science.1069288. [DOI] [PubMed] [Google Scholar]
- Lapik YR, Kaufman LS. The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein α-subunit GPA1 and regulates seed germination and early seedling development. Plant Cell. 2003;15:1578–1590. doi: 10.1105/tpc.011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskovac V. Comprehensive Enzyme Kinetics. New York: Kluwer Academic/Plenum Pub; 2003. [Google Scholar]
- Licciulli S, Luise C, Zanardi A, Giorgetti L, Viale G, Lanfrancone L, Carbone R, Alcalay M. Pirin delocalization in melanoma progression identified by high content immuno-detection based approaches. BMC Cell Biol. 2010;11:5. doi: 10.1186/1471-2121-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licciulli S, Luise C, Scafetta G, et al. Pirin inhibits cellular senescence in melanocytic cells. Am. J. Pathol. 2011;178:2397–2406. doi: 10.1016/j.ajpath.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Rehmani I, Esaki S, Fu R, Chen L, de Serrano V, Liu A. Pirin is an iron-dependent redox regulator of NF-κB. Proc. Natl Acad. Sci. USA. 2013;110:9722–9727. doi: 10.1073/pnas.1221743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Torres JL, Wilbers RH, Gawronski P, et al. Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl Acad. Sci. USA. 2012;109:10119–10124. doi: 10.1073/pnas.1202867109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I, Baudouin E, Schweighofer A, Liwosz A, Jonak C, Rodriguez PL, Jelinek H, Hirt H. Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J. Biol. Chem. 2003;278:18945–18952. doi: 10.1074/jbc.M300878200. [DOI] [PubMed] [Google Scholar]
- Miyazaki I, Simizu S, Okumura H, Takagi S, Osada H. A small-molecule inhibitor shows that pirin regulates migration of melanoma cells. Nat. Chem. Biol. 2010;6:667–673. doi: 10.1038/nchembio.423. [DOI] [PubMed] [Google Scholar]
- Nakaho K, Allen C. A pectinase-deficient Ralstonia solanacearum strain induces reduced and delayed structural defences in tomato xylem. J. Phytopathol. 2009;157:228–234. [Google Scholar]
- Nakaho K, Hibino H, Miyagawa H. Possible mechanisms limiting movement of Ralstonia solanacearum in resistant tomato tissues. J. Phytopathol. 2000;148:181–190. [Google Scholar]
- Nishiuchi T, Masuda D, Nakashita H, Ichimura K, Shinozaki K, Yoshida S, Kimura M, Yamaguchi I, Yamaguchi K. Fusarium phytotoxin trichothecenes have an elicitor-like activity in Arabidopsis thaliana, but the activity differed significantly among their molecular species. Mol. Plant Microbe Interact. 2006;19:512–520. doi: 10.1094/MPMI-19-0512. [DOI] [PubMed] [Google Scholar]
- Orzaez D, de Jong AJ, Woltering EJ. A tomato homologue of the human protein PIRIN is induced during programmed cell death. Plant Mol. Biol. 2001;46:459–468. doi: 10.1023/a:1010618515051. [DOI] [PubMed] [Google Scholar]
- Pesquet E, Zhang B, Gorzsas A, et al. Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. Plant Cell. 2013;25:1314–1328. doi: 10.1105/tpc.113.110593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusche M, Thole K, Janz D, Truskina J, Rindfleisch S, Drubert C, Polle A, Lipka V, Teichmann T. Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis. Plant Cell. 2012;24:3823–3837. doi: 10.1105/tpc.112.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richau KH, Kaschani F, Verdoes M, Pansuriya TC, Niessen S, Stuber K, Colby T, Overkleeft HS, Bogyo M, Van der Hoorn RAL. Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 2012;158:1583–1599. doi: 10.1104/pp.112.194001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saile E, McGarvey JA, Schell MA, Denny TP. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato pants by Ralstonia solanacearum. Phytopathology. 1997;87:1264–1271. doi: 10.1094/PHYTO.1997.87.12.1264. [DOI] [PubMed] [Google Scholar]
- Serebriiskii IG, Golemis EA. Uses of lacZ to study gene function: evaluation of beta-galactosidase assays employed in the yeast two-hybrid system. Anal. Biochem. 2000;285:1–15. doi: 10.1006/abio.2000.4672. [DOI] [PubMed] [Google Scholar]
- Shindo T, van Der Hoorn RAL. Papain-like cysteine proteases: key players at molecular battlefields employed by both plants and their invaders. Mol. Plant Pathol. 2008;9:119–125. doi: 10.1111/j.1364-3703.2007.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo T, Misas-Villamil JC, Horger AC, Song J, van der Hoorn RA. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS ONE. 2012;7:e29317. doi: 10.1371/journal.pone.0029317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Gottesman MM. Activity and deletion analysis of recombinant human cathepsin L expressed in Escherichia coli. J. Biol. Chem. 1989;264:20487–20495. [PubMed] [Google Scholar]
- Song J, Win J, Tian M, Schornack S, Kaschani F, Ilyas M, van der Hoorn RA, Kamoun S. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl Acad. Sci. USA. 2009;106:1654–1659. doi: 10.1073/pnas.0809201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, Jauneau A, Genin S, Tavella MJ, Vailleau F, Gentzbittel L, Jardinaud MF. Dissection of bacterial wilt on Medicago truncatula revealed two type III secretion system effectors acting on root infection process and disease development. Plant Physiol. 2009;150:1713–1722. doi: 10.1104/pp.109.141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Hollenberg SM. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 1995;255:331–342. doi: 10.1016/s0076-6879(95)55036-4. [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- Warpeha KM, Upadhyay S, Yeh J, Adamiak J, Hawkins SI, Lapik YR, Anderson MB, Kaufman LS. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 2007;143:1590–1600. doi: 10.1104/pp.106.089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler WM, Kremmer E, Forster R, Winnacker EL. Identification of pirin, a novel highly conserved nuclear protein. J. Biol. Chem. 1997;272:8482–8489. doi: 10.1074/jbc.272.13.8482. [DOI] [PubMed] [Google Scholar]
- Yadeta KA, Hanemian M, Smit P, Hiemstra JA, Pereira A, Marco Y, Thomma BP. The Arabidopsis thaliana DNA-binding protein AHL19 mediates verticillium wilt resistance. Mol. Plant Microbe Interact. 2011;24:1582–1591. doi: 10.1094/MPMI-04-11-0090. [DOI] [PubMed] [Google Scholar]
- Yadeta KA, Valkenburg DJ, Hanemian M, Marco Y, Thomma BP. The Brassicaceae-specific EWR1 gene provides resistance to vascular wilt pathogens. PLoS ONE. 2014;9:e88230. doi: 10.1371/journal.pone.0088230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Matsushima R, Nishimura M, Hara-Nishimura I. A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 2001;127:1626–1634. [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Johnson BJ, Kositsup B, Beers EP. Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol. 2000;123:1185–1196. doi: 10.1104/pp.123.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Reynolds L, Crnogorac-Jurcevic T, et al. Combination of microdissection and microarray analysis to identify gene expression changes between differentially located tumour cells in breast cancer. Oncogene. 2003;22:3742–3748. doi: 10.1038/sj.onc.1206428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Analysis of the Arabidopsis PRN2 and the genotypes used in the study.
Figure S2. Subcellular localization of Arabidopsis PRN2 protein.
Figure S3. Morphology of PRN2 knockout and overexpression lines.
Figure S4. Electron microscopy analysis of xylem vessel elements in the hypocotyls.
Figure S5. Results of pathogen inoculation assays.
Figure S6. BiFC assay of PRN2 and XCP2 interaction in Arabidopsis protoplasts.
Figure S7. Western blot analysis with XCP2 and RD21 antibody.
Figure S8. Expression and purification of recombinant PRN2 and XCP2.
Figure S9. The stability assay of rPRN2 in the presence of rXCP2.
Figure S10. Induction of PLCP activity in prn2-1 post-inoculation of R. solanacearum.
Figure S11. Expression of XCP2 after infection with a compatible R. solanacearum strain.
Table S1 Results of yeast two-hybrid screening using the full-length Arabidopsis PRN2 as a bait.
Table S2 All primer sequences used in this study.
Methods S1. Methods related to acquisition of the data in the supporting information.


