Significance
The αβ T-cell receptor (TCR) on mammalian T lymphocytes recognizes intracellular pathogens to afford protective immunity. Detection of various foreign peptides bound to MHC molecules as TCR ligands occurs during immune surveillance where mechanical forces are generated through cell movement. Using single-molecule optical tweezer assays, we show with isolated and complete receptors on single T cells that both sensitivity and specificity of the biological T-lymphocyte response is dependent upon force-based interactions. Our work demonstrates a catch-and-release αβTCR structural conversion correlating with ligand potency wherein a strongly binding/compact state transitions to a weakly binding/extended state. An allosteric mechanism controls bond strength and lifetime, supporting a model in which quaternary αβTCR subunit associations regulate TCR recognition under load.
Keywords: mechanosensor, T-cell receptor, peptide discrimination, optical tweezers, catch bond
Abstract
The αβ T-cell receptor (TCR) on each T lymphocyte mediates exquisite specificity for a particular foreign peptide bound to a major histocompatibility complex molecule (pMHC) displayed on the surface of altered cells. This recognition stimulates protection in the mammalian host against intracellular pathogens, including viruses, and involves piconewton forces that accompany pMHC ligation. Physical forces are generated by T-lymphocyte movement during immune surveillance as well as by cytoskeletal rearrangements at the immunological synapse following cessation of cell migration. The mechanistic explanation for how TCRs distinguish between foreign and self-peptides bound to a given MHC molecule is unclear: peptide residues themselves comprise few of the TCR contacts on the pMHC, and pathogen-derived peptides are scant among myriad self-peptides bound to the same MHC class arrayed on infected cells. Using optical tweezers and DNA tether spacer technology that permit piconewton force application and nanometer scale precision, we have determined how bioforces relate to self versus nonself discrimination. Single-molecule analyses involving isolated αβ-heterodimers as well as complete TCR complexes on T lymphocytes reveal that the FG loop in the β-subunit constant domain allosterically controls both the variable domain module’s catch bond lifetime and peptide discrimination via force-driven conformational transition. In contrast to integrins, the TCR interrogates its ligand via a strong force-loaded state with release through a weakened, extended state. Our work defines a key element of TCR mechanotransduction, explaining why the FG loop structure evolved for adaptive immunity in αβ but not γδTCRs or immunoglobulins.
Antigen recognition by T lymphocytes is a crucial feature of adaptive immunity. This process requires the interaction of a clone-specific T-cell receptor (TCR) via its membrane distal variable module with a cognate peptide bound to a major histocompatibility complex molecule (pMHC) (refs. 1 and 2 and references therein). “Foreign” peptide antigens derived from infectious or other cell-altering processes including oncogenic transformation are presented either on a surface of the perturbed cell directly or indirectly via cross-presentation on antigen-presenting cells (APC). In either case, ligation of the relevant TCRαβ heterodimer initiates a cascade of T-cell signaling events following exposure of the immunoreceptor tyrosine-based activation motif (ITAM) elements in the cytoplasmic tail of the noncovalently associated subunits (CD3εγ, CD3εδ, and CD3ζζ) composing the TCR complex in 1:1:1:1 dimer stoichiometry. This accessibility allows the active kinase, Lck, to bind and phosphorylate ITAMs followed by recruitment and activation of a second tyrosine kinase, ZAP-70 (3–6). In turn, multiple downstream pathways are engaged, including transcriptional regulators controlling activation and differentiation of T cells (7, 8). Thymocyte development is also regulated by the TCR–pMHC interaction as it relates to repertoire selection, generating millions of useful TCRs with diverse specifications (reviewed in ref. 9).
The MHC-bound peptides recognized by T cells are typically 8–11 amino acids in length with single amino acid substitutions readily sensed and discriminated by a TCR. During immune surveillance, an individual high-avidity T cell has the capacity to detect several copies of a specific pMHC on the APC that expresses 100,000 chemically similar pMHC molecules. Discrimination of foreign vs. myriad self-peptides is manifest with precision; if this were not the case, then either autoimmunity or immunodeficiency would result. How TCRs are capable of mediating such exquisite specificity and sensitivity has remained a mystery, especially in view of the fact that, unlike with B-cell receptors, there is no somatic hypermutation of TCR genes and monomeric TCR–pMHC affinities are orders of magnitude weaker than high-affinity antibody–antigen interactions (10).
Very recently, it has become clear that the TCR functions as a mechanosensor. Kim et al. provided the first, to our knowledge, direct evidence of the influence of mechanical force in TCR activation (11). Using an optically trapped bead coated with pMHC or anti-CD3 monoclonal antibody for engaging the TCR, T cells were mechanically triggered by applying an oscillating tangential force to the cell surface while monitoring their activation via intracellular calcium flux. Importantly, piconewton (pN) force application with cognate pMHC but not irrelevant pMHC triggered activation. Additional mechanosensor evidence was provided in subsequent studies by Li using a micropipette to demonstrate shear force associated with activation (12) and by Husson using a biomembrane force probe (BFP) to reveal pushing and pulling associated with T-cell triggering (13). Triggering was also shown to depend on substrate stiffness (14). Prediction that a nonlinear mechanical response such as catch bond formation might facilitate TCR-based recognition (11) was elegantly confirmed by Zhu and coworkers using a BFP (15). Given the complexities of surface-antigen display on an APC and the physical forces associated with cellular migration, specific structural adaptations must exist for the TCR to work in such a chemically and physically “noisy” environment. Here we use single-molecule (SM) techniques to search for such relevant mechanotransduction features, identifying the TCRαβ heterodimer as a finely tuned allosteric regulator of force-driven peptide discrimination.
Results
Structural Features of the Cβ FG Loop as a Focus of Single-Molecule Analysis.
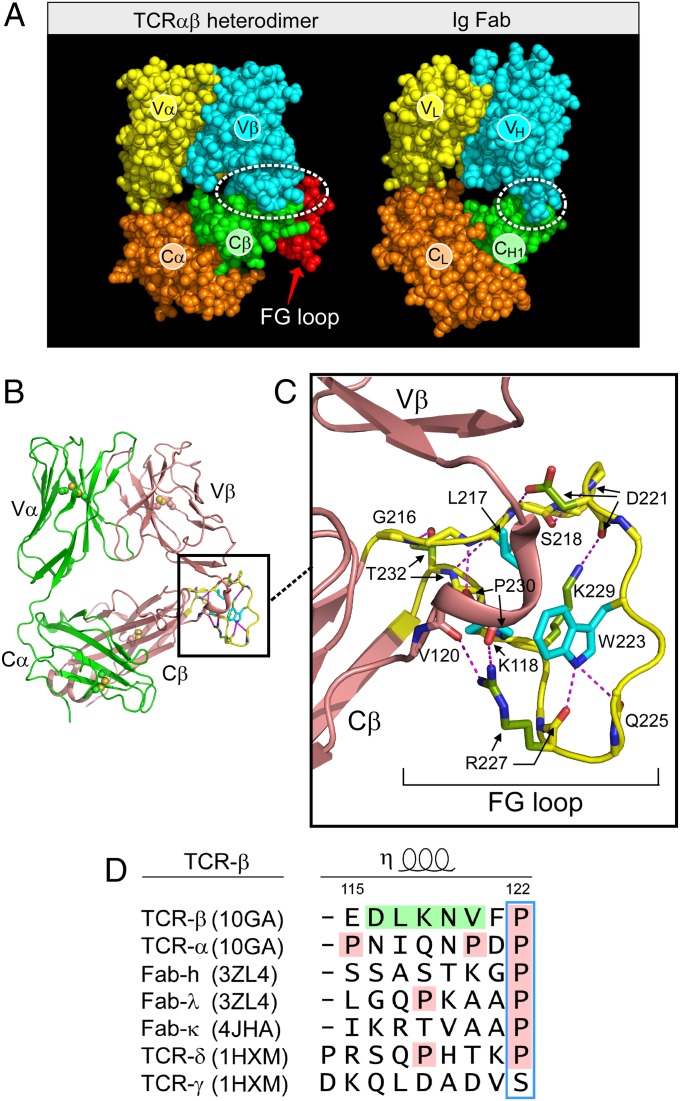
As shown in Fig. 1A, the TCRαβ heterodimer is very similar in overall topology to an Ig Fab fragment, consisting of a paired variable module (VαVβ vs. VLVH) linked to a constant domain module (CαCβ vs. CLCH1). Moreover, the overall height and width of these structures are similar (16). Closer inspection, however, reveals two striking differences. First, the CαCβ module manifests an obvious asymmetry, exposing residues on the ABD β-sheet of the Cβ domain that are buried in the symmetrical Fab (Fig. 1 B and A, green). Second, the size of the VβCβ interface is very different from that of the VHCH1 interface. In particular, the buried surface area between Vβ and Cβ domains is ∼350 Å2 on each side whereas that between VH and CH1 is only ∼150 Å2. This more robust VβCβ interface is contributed to by the CβFG loop (Fig. 1A, red), a 12-residue-long insertion unique to mammalian αβTCRs (17) and accounting for almost one-third of the buried surface area (SI Appendix, Fig. S3). As shown in Fig. 1C, the CβFG loop is well structured. At the middle of the loop is W223, which uses the NH group at its indole ring to form two hydrogen bonds to the main chain carbonyl oxygen atoms from Q225 and R227. This fixes the W223 residue in a position to serve as the center of a mini-hydrophobic core joined by L217 and P230 (cyan) (16). In addition, a 310 helix connector between the Vβ and Cβ domains is spatially adjacent to this CβFG loop. This helix is unique to TCRβ (Fig. 1D). A 310 helix is an overwound secondary structure and might be induced to extend when subject to a pulling force, as could the CβFG loop, given the absence of disulfide bonds therein to preclude force-driven extension. Because deletion of the CβFG loop attenuated antigen-triggered T-cell function (18, 19), we focused on this αβTCR feature to determine if it is associated with mechanotransduction. Note that throughout the designation “αβTCR” will be used to refer to the complete TCR complex (i.e., including the associated CD3 subunits) whereas the designation “TCRαβ” refers exclusively to the clonotypic heterodimeric component of the complex.
Fig. 1.
The CβFG loop region includes a structured element expanding the β-subunit interface between its variable and constant domains and an adjacent Vβ-Cβ connector. (A) Comparison of TCRαβ heterodimer (1OGA) and Ig Fab (4JFX) fragments with comparable domains delineated in their respectively colored CPK format. (B) Ribbon diagram of TCRαβ heterodimer with FG loop highlighted in yellow. (C) Magnified view of the CβFG loop region with three conserved hydrophobic loop residues indicated in blue and hydrogen bonds in dashed red. Note the 310 helix in the connector linking Vβ and Cβ domains in brown. (D) Connector sequences of different V-C domains indicated with numbering of sequence according to TCRβ and relevant structural alignment. PDB numbering for each structure is indicated.
Single-Molecule Assay Isolating the TCRαβ–pMHC Complex.
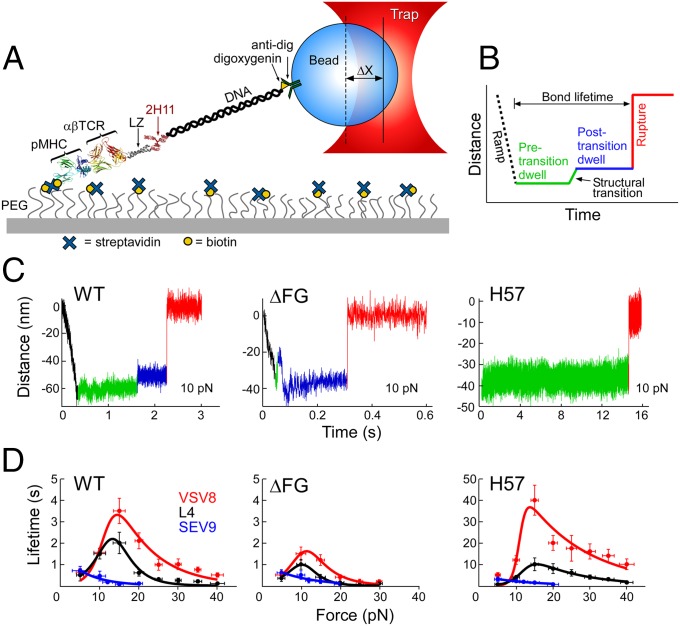
Two single-molecule assays were developed for probing the TCRαβ–pMHC complex under load. The first isolates the N15TCRαβ–pMHC interaction to a coverslip surface (Fig. 2A) in a tethered bead configuration. This TCR is specific for the vesicular stomatitis virus octapeptide (VSV8) bound to murine H-2Kb. Biotinylated pMHC molecules (VSV8/Kb) were bound through streptavidin to a PEG-pacified surface containing sparse biotin-PEG molecules. The TCRαβ heterodimer is fused to an acid-base leucine zipper (LZ) at the C terminus and tethered to an ∼300-nm-long 1,010-bp DNA molecule covalently bound to the zipper-specific antibody 2H11 and functionalized with digoxygenin on opposite 5′ ends. Force is applied to the TCRαβ–pMHC complex through an optically trapped anti-digoxigenin–coated bead. To measure the TCRαβ–pMHC bond strength in the isolated SM assay, tethered beads were visualized, centered in the detector laser, and calibrated for position sensing using automated procedures. Lifetime measurements were performed by translating the sample relative to the fixed trap and holding at a fixed position/force until bond rupture, identified as an abrupt snap back of the bead position within the trap. Conformational extensions are seen as upward displacements of the bead within the trap (Fig. 2 B and C).
Fig. 2.
SM studies. (A) Cartoon showing single-molecule tether assay for probing the lifetime of αβTCR–pMHC bonds. “ΔX” denotes displacement of bead out of the center of the trap. (B) Loading profile for measuring bond lifetime. Increase in distance represents greater separation along the system path. An initial ramp phase, indicated by a black dashed line, loads the tether to a fixed force, “Pre-transition dwell,” indicated by a green line. Frequently, a signature structural transition is observed, indicated by green to blue line, followed by a “Post-transition dwell,” in blue. “Rupture,” indicated by red line, is seen as an abrupt upward step. (C) Representative traces at 10 pN for WT, ΔFG, and H57 Fab. The WT trace shows a typical transition and rupture. ΔFG traces typically transition early, here occurring during the initial ramp phase. H57 Fab traces typically show no transition and dwell for longer periods before rupture. Low-amplitude motions in the green baseline (see WT) were also observed. (D) Force-bond lifetime plots for WT, ΔFG, and H57 Fab showing catch bonds for both VSV8 and L4 and slip bond character for SEV9. Catch bonds peak at ∼15 pN for WT and shift to lower force for ΔFG with significant reduction in bond lifetime. H57 stabilized the FG loop exhibiting dramatic amplification of catch bond lifetimes.
In addition to analyzing conformational extensions (Fig. 2C), bond lifetime measurements as a function of force were performed using wild-type (WT), CβFG loop-deleted (ΔFG) (18, 19) and H57 Fab-stabilized TCRαβ heterodimers paired with pMHC molecules (Fig. 2D). Note that H57 binds the CβFG loop (16). The MHC class I H-2 Kb molecules were complexed with the strong agonist peptide VSV8, the weak agonist L4 containing a single valine-to-leucine substitution at the p4 position of VSV8, or the unrelated Sendai virus nonamer peptide SEV9. SEV9 binds to Kb with comparable affinity to VSV8 but is unable to activate N15TCR-expressing T cells (20). SM measurements for the WT-TCRαβ showed catch bond behavior with peak lifetimes at ∼15 pN for VSV8, a weaker catch bond character for L4, and a slip bond for SEV9 (Fig. 2D). ΔFG shows significantly reduced bond lifetime, shifting the maximal lifetime to a lower force of ∼10 pN (Fig. 2D), and lessens the ability of the TCR to discriminate different ligands bound to the same MHC molecule (see below). In contrast, stabilizing the CβFG loop via H57 Fab ligation amplifies catch bond lifetimes (Fig. 2D). By comparison, the Cα-binding antibody H28 only modestly extends bond lifetime (SI Appendix, Fig. S1).
Conformational Transition Is Linked to the FG Loop.
Before rupture, we frequently observed a signature conformational transition spanning 8–15 nm (Fig. 2C). The upward transition represents an extension of the system incorporating rotations of domains, extensions, and potential unfolding. The transition is coupled to the state of the CβFG loop in several respects. First, removing the CβFG loop weakens an energetic barrier, as TCRαβΔFG transitions frequently occurred during the ramp phase (compare Fig. 2C, WT vs. ΔFG). Second, stabilizing the CβFG loop with H57 Fab virtually abolished the transition (Fig. 2C, H57), suggesting that this loop region controls access to the transition. Third, ΔFG altered the displacement magnitude, described below.
Single-Molecule Measurements on Isolated Single Cells.
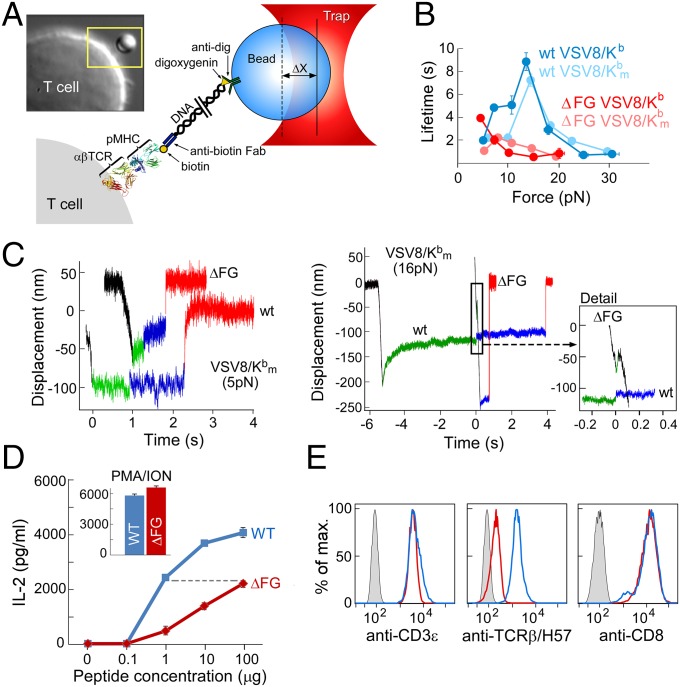
Single-molecule measurements on isolated single cells (SMSC), the second assay, corroborate the SM observations (Fig. 3). Single tethers on single cells were constructed using anti-digoxygenin beads that linked a digoxygenin-adducted 3,500-bp DNA molecule covalently bound to the thiol group in the hinge domain of a biotin-specific antibody that binds single biotin-labeled pMHC molecules (Fig. 3A). SMSC studies were performed by trapping and calibrating a bead, positioning cells to facilitate tether formation, displacing cells to a fixed load, and recording bead position until rupture, as with SM assay. Although the cell system is much more compliant due to the cellular connectivity of the load pathway, the measurements recapitulate a more natural state of the αβTCR–pMHC complex. As with SM measurements, catch bonds were seen peaking at ∼15 pN but with generally longer lifetimes (Fig. 3B). Dramatic reduction in bond lifetime was seen for N15TCRαβΔFG cells. Despite the gradual creep in position and other displacements as a consequence of membrane compliance and cytoskeletal attachment in single-cell measurements, we were able to identify records displaying the signature conformational transitions found in SM studies (Fig. 3C). The N15αβΔFG cells show early transition including some during the ramp phase (Fig. 3C). This difference is functionally correlated with a 100-fold reduction in responsiveness to the cognate VSV8 peptide as judged by IL-2 production when stimulated through the TCR, despite comparable surface TCR and coreceptor levels as assessed by anti-CD3ε and anti-CD8α mAbs (Fig. 3 D and E).
Fig. 3.
Single-cell studies. (A) Cartoon showing the SMSC assay along with a photomicrograph of a T cell with tethered bead in the yellow box. (B) Lifetime-force plots comparing N15WT and N15ΔFG T-cell ligation with VSV8/Kb and VSV8/Kbm, a mutant pMHC lacking the ability to bind CD8, show catch bonds peaking at 15 pN. (C) Representative traces at low force (5 pN) and high force (16 pN) comparing WT to ΔFG for VSV8/Kbm, where the structural transition is clearly visible. ΔFG traces at 16 pN exhibit transitions early on, here during the loading phase of the trace. (D) IL-2 production requires a 2-log-greater VSV8 peptide concentration for ΔFG than WT despite comparable capacity to produce equivalent maximal IL-2 production triggered by phorbal myristate acetate (PMA) plus ionomycin (ION). (E) Flow cytometry confirms similar CD3ε and CD8 density and lack of FG loop seen in H57-binding assay. Color code is as in D with gray curve showing background staining.
CD8αβ is an important T-cell surface molecule, absent in the SM assay, that functions as a coreceptor by associating with the αβTCR–pMHC complex via binding to the membrane proximal α3 domain of H-2Kb (reviewed in ref. 1). To measure the influence of CD8, we performed our assay on cells using a mutant pMHC (VSV8/Kbm) unable to bind CD8αα and CD8αβ dimers (described below). As shown in Fig. 3B, a general reduction in bond lifetime was observed, shifting the curve closer to SM and exposing the TCRαβΔFG as forming a weak catch bond. Much of the “slip bond” character seen in the ΔFG native system can be attributed to CD8 binding, which masks the diminished pMHC interaction.
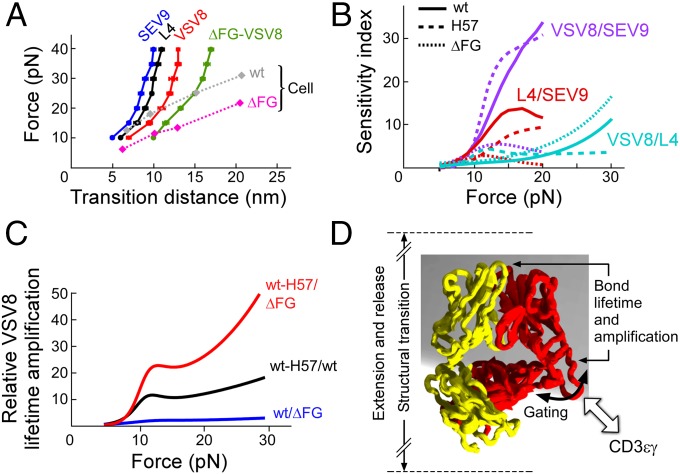
When comparing transitions, plots of magnitude vs. force (Fig. 4A) demonstrate that greater transition displacements are seen for agonists than for nonagonists and in SMSC vs. SM systems. Analysis of the relative response to agonists and nonagonists by comparing ratios of lifetimes for pairs of peptides bound to the same MHC, sensitivity index plots, reveal a peak for VSV8/SEV9 at 15 pN but minimal, if any, discrimination at low loads (Fig. 4B). Plots comparing ratios of lifetimes for VSV8 demonstrate greatest amplification for the stabilized CβFG loop region and with increasing force (Fig. 4C).
Fig. 4.
Force-dependent structural transitions, ligand sensitivity, amplification factors and an αβTCR model. (A) The major structural transition distance, measured as the difference in bead position before and after the transition, is plotted vs. force for WT-VSV8 cognate peptide (red), -L4 partial agonist (black), -SEV9 irrelevant ligand (blue), and ΔFG-VSV8 (green) showing a ligand dependence with greater ligand potency exhibiting larger structural transition distances in SM systems. Force-transition distance plots for cell systems; SMSC (diamonds with dotted lines) exhibit greater displacement with VSV8 for ΔFG than for WT and are generally larger than the SM transitions. (B) Sensitivity plots comparing lifetime ratios of various antigens for WT (solid), H57 (dashed), and ΔFG (dotted). Plots were constructed from fits in Fig. 2. (C) Amplification factors associated with lifetime enhancement for VSV8 binding including ratios of WT-H57/ΔFG (red), WT-H57/WT (black), and WT/ΔFG (blue). Amplification factors rise abruptly at 10 pN, are greatest with stabilization of the FG loop, and increase generally as a function of force. (D) Model for force-induced motions and gating associated with the CβFG loop region. The FG loop region couples to the binding interface strength and conformational change magnitude. Associations with molecules such as CD3εγ may dramatically stabilize the CβFG loop, influencing bond lifetime and force transfer response of the loaded αβTCR–pMHC complex system.
Discussion
Using both SM and SMSC assays, we have compared the strength of pMHC interactions with WT-TCRαβ, TCRαβΔFG, and H57 Fab-stabilized WT-TCRαβ. We have found strong evidence for allostery in that the state of the CβFG loop region dramatically modulates the strength of the TCRαβ–pMHC bond. Moreover, we observe a mechanical extension, the displacement of which correlates with ligand potency and the strength of which correlates with the CβFG loop region structure. Single-molecule records are consistent with a model where an unloaded compact TCR binds weakly, a loaded compact TCR binds strongly, and a stabilized CβFG loop region further strengthens binding, whereas release occurs through an extended TCRαβ heterodimer. The adjacent CD3εγ ectodomains may stabilize the CβFG loop region, prolonging bond lifetime under force in SMSC relative to SM experiments as detailed below. Our model implies a mechanism whereby the CβFG loop enhances mechanosensor action through force-driven gating of initial access and stabilization of productive pMHC interactions but release of unproductive interactions, thereby controlling catch bond strength and bond lifetime (Fig. 4D).
These data confirm that the αβTCR is a mechanosensor activated by pN forces upon pMHC ligation. More importantly, they show that the CβFG loop region allosterically controls the V domain module’s catch bond lifetime and peptide discrimination via force-driven conformational transitions. By contrast, CD8 neither contributes to catch bond formation in mature αβT cells as shown here nor is a mechanosensor as revealed by earlier data showing an inability of CD8-directed force application to stimulate T-cell activation (11).
The force-induced structural transition observed in SM experiments is large, being on the order of ∼120 Å (Fig. 4A). That said, ∼40 Å of transition is observed differentially between VSV8/Kb (agonist) vs. SEV9/Kb (irrelevant) pMHC ligation of the N15αβTCR heterodimer (Fig. 4A). This distance is more than the ∼15 Å permitted by simple extension of the Vβ-Cβ connector. In fact, in SM assays, displacements for TCRαβΔFG were greater than WT-TCRαβ (Fig. 4A). Therefore, additional rearrangements, outside of the FG loop, in the TCRαβ heterodimer–pMHC complex must exist. These might include extension of the long (>20-aa residues) connecting peptides between Cβ and the transmembrane surrogate (LZ) and Cα and LZ or possibly even sliding of the Vβ and Vα domains relative to one another at the hydrophobic interface of the V module. Major motion within individual V and C domains of the TCR heterodimer per se is unlikely, given the existence of intradomain disulfide bonds in each (Fig. 1B), but the A and/or G strands nonetheless might be flayed via force. Although our focus has been on the large transition, conformational change was also observed featuring reversible, low-amplitude, 1- to 3-nm motions of the baseline (Fig. 3C, WT), suggesting reconfigurations of the αβTCR–pMHC complex. Single-molecule FRET analysis with fluorophores placed in individual domains can more precisely define the nature of domain extension and twisting in future investigation.
Our prior studies (18, 19) revealed that the CβFG loop facilitates thymocyte development at both CD4−CD8− double negative and CD4+CD8+ double-positive stages. The former requires the pre-TCR, a molecular complex involving the same β and CD3 subunits but with the β-chain ectodomain stalk disulfide linked to that of pTα, a surrogate α-chain lacking a V domain. Given the current data, it is tempting to speculate that the CβFG loop-regulated transition observed for the TCRαβ heterodimer may be applicable to the pre-TCR, but in even greater magnitude in the absence of a Vα pairing constraint. The suggestion that pMHC or other self-ligands could interact with the unpaired Vβ domain (21) is consistent with this possibility and worthy of further investigation.
The Cβ FG loop evolved in mammals at the same time as molecular speciation of CD3γ and CD3δ genes from a single CD3 precursor gene among Gnathostomata, a group of vertebrates with adaptive immunity involving a recombinatorial system (VDJ) (17). The sensitivity of allosteric regulation makes this adaptation highly efficient for antigen recognition. That requirement may be less critical for recognition by γδTCRs, given the potential for high copy numbers of their less diverse stimulating ligands on APCs (22, 23). The distinctive CD3εγ topology adjacent to the CβFG loop can further maximize antigen activation by facilitating force transfer from the TCRαβ heterodimer to adjacent CD3εγ (17, 24, 25). Perhaps force-mediated structural transitions enrich for conformers with the greatest intersubunit ectodomain complementarity, thereby favoring ligation-mediated signaling. That H57Fab prolongs TCR–pMHC bond lifetime (Fig. 2H) yet blocks pMHC-triggered T-cell responses (11) is consistent with such a notion. How CD3εγ may associate with pretransition, intermediate, or posttransition states is unclear. Pretransition binding of CD3 to a compact state might extend the lifetime of the αβTCR–pMHC bond. During the transition, the bound CD3 heterodimer may participate in the mechanical change, being pushed and pulled to expose sequestered CD3 cytoplasmic tail ITAMs. Posttransition binding to an extended TCR state may also modulate CD3 binding in a positive and/or negative sense in ways that need to be defined.
Mechanotransduction is obviously critical for αβTCR function, as evident from the above studies. Understanding the structural consequences of pN load will be important to elucidate T-lineage cell biology related to cognate recognition of antigen as well as thymocyte development. Mechanical influence has been established in many systems including stem cell differentiation (26), cell migration and rolling (27), focal adhesion development (28), and integrin adhesion (29). Single-molecule methods have been pioneered largely through the study of biological motors using classical assays such as our SM system (30), yet there is great opportunity for using single-molecule methods to probe machinery on or within intact cells. The approach presented here bridges the classic isolated single-molecule assays to the more biologically relevant single molecule in a single-cell system. The SMSC system is ideally suited for in situ study of surface membrane-embedded receptors and represents an evolution of single-molecule technology.
Our studies begin to address how it is possible that αβTCRs, whose affinity in the absence of load (i.e., in solution) is extremely weak, can nevertheless mediate exquisite sensitivity and specificity in a noisy chemical environment on the antigen-presenting cell surface. There, many identically sized peptides are bound to the same type of MHC molecule, forming an array of ∼100,000 different pMHC complexes. The challenge for any T lymphocyte is to use its clonally endowed 20,000 αβTCR surface copies to rapidly scan this collection of diverse pMHCs on each cell where a dozen or fewer relevant foreign peptides may be embedded. In this context, load-induced transitions tune αβTCR–pMHC bond lifetime with high specificity. In particular, maximal nonlinear dynamic bond lifetime is associated with TCRαβ structural transitions that readily distinguish among peptides, even when differences compose a single residue (VSV8 vs. L4). On the other hand, without force, discrimination is minimal. Hence, during immune surveillance as an αβTCR is gaffed (i.e., hooked) by its specific pMHC, force is generated, bond lifetime extends, and a structural transition is induced in TCRαβ. These features associated with catch bond formation likely are sufficient to promote further αβTCR quaternary change, CD3 heterodimer interactions, and transmembrane and cytoplasmic tail segment alterations to foster downstream signaling events (1, 11, 31). A small number of those productive interactions in time and space will result in quantal T-cell activation that can be further sustained at the immunological synapse (32, 33). Unlike the cognate peptide, unrelated or variant peptides will not induce either these transitions or extend bond lifetime sufficient to promulgate a signaling event. Remarkably, then, the TCR is able to detect very few side chains of a peptide in the MHC groove even though the majority of recognition surface contacts are shared in common with other pMHC complexes.
That the αβTCR physically goes through a mechanical transition that prolongs the αβTCR–pMHC bond lifetime has a number of immediate consequences. First, the extension represents a previously unknown state. As in physical changes observed with integrin αβ heterodimers, affinities between compact and extended states can be very different. Interestingly, the mechanical action of the αβTCR appears to work with a logic opposite to that of the integrins because in the TCR the compact form binds strongly whereas the extended form binds weakly. Leading from an “on state” as opposed to an “off state,” as in integrins where ligands are plentiful, may be a consequence of the αβTCR seeking out rare binding events. Second, the ligand-dependent displacement seen here may differentially couple to mechanisms that critically depend on distance such as TCR-APC intercellular membrane proximity. Third, the mechanical transition in the αβTCR–pMHC system is regulated via the FG loop region, as observed by H57 Fab stabilization, which may also be occurring through the physiological CD3 associations noted above. This partitioning of function allows the mechanism of discrimination to be conserved within the constant region of the TCR while maintaining near-infinite variability in the ligand-recognition elements in the V-domain module. Fourth, kinetic proofreading mechanisms require working outside of thermal equilibrium and are thus generally energy intensive (31, 34). Mechanical work through a mechanosensor mechanism can drive such nonequilibrium signaling events, sustaining triggering during immune surveillance and at the immunological synapse while using forces derived via concomitant cell movements.
Materials and Methods
TCRαβ Production and Peptide and Protein Preparation.
TCR N15α and β-subunits were fused to basic and acidic LZ components, respectively, as previously described (35). They were then separately cloned into the eukaryotic expression vector containing human cytomegalovirus promotor designed for expression of antibody γ-chain (36) using AgeI/SalI to insert each subunit in-frame, retaining the vector signal sequence while adding a stop codon immediately following the LZ motif. Both WT-TCRα- and β-subunits were expressed simultaneously after transient cotransfection in Freestyle 293F cells (Life Technologies) according to the manufacturer’s recommendations. A second TCRβ construct wherein the Cβ FG loop was excised as previously described (18, 19), termed N15βΔFG, was used for cotransfection with WT-TCRα to create N15αβΔFG. Proteins were purified from cell supernatants essentially as described (35). Although yields were lower for the N15αβΔFG than for N15αβWT, we were able to confirm purification of the N15αβΔFG through SDS/PAGE separation, band excision, trypsin digestion, and identification of both N15α and N15β subunit peptides via electrospray mass spectrometry. Singly biotinylated pMHC were purified (37) and biotinylated (38) as described. Peptides were synthesized by United Biosystems. The monoclonal anti-Cβ antibody H57 was produced as described (35), and Fab fragments were generated and purified using a Pierce Fab Micro Preparation Kit (Thermo Scientific) according to the manufacturer’s directions.
Single-Molecule Assay.
For single-molecule measurements, a complex of N15αβTCR and pMHC was tethered between a single trapped bead and glass surface. The overall assay geometry included a N15TCRαβ heterodimer fused to a leucine zipper at the C terminus. A 1,010-bp DNA molecule with anti-leucine zipper antibody (2H11) on one 5′ end and a digoxygenin on the opposite 5′ end, described below, was used to attach the N15TCRαβ heterodimer to an anti-digoxygenin–coated bead. The DNA served as a spacer between the bead and N15TCRαβ. A biotinylated pMHC complex, VSV8/Kb, was immobilized on a PEG functionalized coverglass surface using biotin–streptavidin chemistry. The schematic diagram of our single-molecule assay is shown in Fig. 2.
The DNA linker was constructed by PCR of a 1,010-bp region of the M13mp18 plasmid with 5′ primers of an amino group and digoxigenin. The 5′ amino group was cross-linked to 2H11 using carbodiimide cross-linker strategy (EDC chemistry). A 10-μL flow cell was prepared using two pieces of double-sided sticky tape. Cover-glass surfaces were coated with functionalized PEG silane 99% mixed with 1% biotin–PEG silane similar to previously described work (39). Streptavidin (1 mg/mL) was introduced into the flow cell, incubated for 10 min, and washed with PBS buffer. A total of 20 μL of biotinylated pMHC (1 μg/1 μL) was introduced and incubated for 30 min and washed again with PBS buffer. During this time period, N15TCRαβ (100 ng/1 μL) and 2H11-DNA-digoxigenin (200 ng/μL), +/−H57 or H28 mAb (1mg/mL), were mixed, added to the flow cell, and incubated for 30 min. After washing with 30 μL of PBS buffer, anti-digoxygenin–coated (Roche Diagnostics) polysterene beads (Spherotech Inc.) were introduced followed by a 30-μL PBS buffer wash. The flow cell was sealed with nail polish and loaded on the optical-trapping instrument for measurements. Tethers were found by eye, centered using an automated procedure (40), and trapped with typical stiffness of ∼0.25 pN/nm. Bead positions were sampled at 3 kHz, antialias-filtered at 1.5 kHz, and recorded until rupture. Control experiments where N15αβTCR was excluded from the assay resulted in no tethers.
Single-Cell Assay.
Tethers for SMSC assays were constructed using anti-digoxygenin–coated polystyrene beads (1.0 μm in diameter, Spherotech Inc.) to tether the digoxigenin-labeled, 3,500-bp DNA at one end. At the other end, a sulfo-SMCC (Pierce) reaction was used to bind the free thiol group of the half anti-biotin antibody (see SI Appendix). Finally, biotin-labeled pMHC was linked to the half anti-biotin antibody. Specifically, 0.5 μL 0.25% wt/vol anti-digoxygenin beads were washed two times by PBS buffer (Cellgro) containing 0.02% Tween 20 (PBST) and were incubated with 10 μL of half–anti-biotin functionalized DNA (∼65 ng/μL). The bead surface was saturated with these 3,500-bp DNA tethers to reduce nonspecific binding between the bead and cell surface. After 1 h of incubation, the DNA-labeled bead was washed three times with PBST buffer and then resuspended in a solution containing 5 μL of 20 pM biotinylated pMHC in PBS buffer for 1 h incubation. After two washes with PBST buffer, the bead slurry was diluted 200-fold for bond lifetime measurements. All of the incubations were performed at room temperature.
Cells were cultured in selection medium [complete RPMI medium 1640 (Life Technology) plus 0.1 mg/mL G418 (Life Technology) and 2 mg/mL hygromycin B (Life Technology)] and reached the log phase before the experiments. Cells were washed one time with PBS and then kept in colorless complete RMPI 1640 medium (Life Technology) at 2 million/mL; 20 μL of cells were introduced into the flow chamber, and cells were bound to a poly-l-lysine coverslip prepared as described previously (11). The coverslip surface was further blocked by 1 mg/mL casein (Sigma-Aldrich) in PBS buffer (11). After a 10-min incubation at 37 °C, the pMHC-DNA bead slurry (20 μL) was introduced into the flow cell. The tether-functionalized bead was trapped and brought in the vicinity of N15αβWT or N15αβΔFG cells to form a stable tether. Tether formation was verified and then loaded by displacing the cell relative to the fixed trap using fine motions of the piezo stage. Bead positions were sampled at 3 kHz, antialias-filtered at 1.5 kHz, and recorded until complete rupture. Assays were performed at 37 °C.
Supplementary Material
Acknowledgments
We thank the NIH Tetramer Core Facility at Emory University and Cheng Zhu for providing the VSV8/H2-Kb-α2A2 tetramer. This work is supported by NIH Grants R01AI100643, R01AI37581, and P01GM047467. D.K.D. is a George Russell Chambers postdoctoral fellow. Flow cytometry experiments were performed in the Vanderbilt University Medical Center Flow Cytometry Shared Resource supported by The Vanderbilt-Ingram Cancer Center (NIH Grant CA68485) and the Vanderbilt Digestive Disease Research Center (NIH Grant DK058404).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424829112/-/DCSupplemental.
References
- 1.Wang JH, Reinherz EL. The structural basis of αβ T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunol Rev. 2012;250(1):102–119. doi: 10.1111/j.1600-065X.2012.01161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin L, Scott-Browne J, Kappler JW, Gapin L, Marrack P. T cells and their eons-old obsession with MHC. Immunol Rev. 2012;250(1):49–60. doi: 10.1111/imr.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 2008;135(4):702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aivazian D, Stern LJ. Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nat Struct Biol. 2000;7(11):1023–1026. doi: 10.1038/80930. [DOI] [PubMed] [Google Scholar]
- 5.Nika K, et al. Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity. 2010;32(6):766–777. doi: 10.1016/j.immuni.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Au-Yeung BB, et al. The structure, regulation, and function of ZAP-70. Immunol Rev. 2009;228(1):41–57. doi: 10.1111/j.1600-065X.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- 7.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12(11):749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Man K, et al. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat Immunol. 2013;14(11):1155–1165. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- 9.von Boehmer H, et al. Thymic selection revisited: How essential is it? Immunol Rev. 2003;191:62–78. doi: 10.1034/j.1600-065x.2003.00010.x. [DOI] [PubMed] [Google Scholar]
- 10.Kato L, et al. An evolutionary view of the mechanism for immune and genome diversity. J Immunol. 2012;188(8):3559–3566. doi: 10.4049/jimmunol.1102397. [DOI] [PubMed] [Google Scholar]
- 11.Kim ST, et al. The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009;284(45):31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li YC, et al. Cutting Edge: Mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J Immunol. 2010;184(11):5959–5963. doi: 10.4049/jimmunol.0900775. [DOI] [PubMed] [Google Scholar]
- 13.Husson J, Chemin K, Bohineust A, Hivroz C, Henry N. Force generation upon T cell receptor engagement. PLoS ONE. 2011;6(5):e19680. doi: 10.1371/journal.pone.0019680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC. Mechanosensing in T lymphocyte activation. Biophys J. 2012;102(2):L5–L7. doi: 10.1016/j.bpj.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell. 2014;157(2):357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody. EMBO J. 1998;17(1):10–26. doi: 10.1093/emboj/17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim ST, et al. Distinctive CD3 heterodimeric ectodomain topologies maximize antigen-triggered activation of alpha beta T cell receptors. J Immunol. 2010;185(5):2951–2959. doi: 10.4049/jimmunol.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasada T, et al. Involvement of the TCR Cbeta FG loop in thymic selection and T cell function. J Exp Med. 2002;195(11):1419–1431. doi: 10.1084/jem.20020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Touma M, et al. The TCR C beta FG loop regulates alpha beta T cell development. J Immunol. 2006;176(11):6812–6823. doi: 10.4049/jimmunol.176.11.6812. [DOI] [PubMed] [Google Scholar]
- 20.Sasada T, Ghendler Y, Wang JH, Reinherz EL. Thymic selection is influenced by subtle structural variation involving the p4 residue of an MHC class I-bound peptide. Eur J Immunol. 2000;30(5):1281–1289. doi: 10.1002/(SICI)1521-4141(200005)30:5<1281::AID-IMMU1281>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 21.Zhou B, et al. A conserved hydrophobic patch on Vβ domains revealed by TCRβ chain crystal structures: Implications for pre-TCR dimerization. Front Immunol. 2011;2:5. doi: 10.3389/fimmu.2011.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luoma AM, et al. Crystal structure of Vδ1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human γδ T cells. Immunity. 2013;39(6):1032–1042. doi: 10.1016/j.immuni.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chodaczek G, Papanna V, Zal MA, Zal T. Body-barrier surveillance by epidermal γδ TCRs. Nat Immunol. 2012;13(3):272–282. doi: 10.1038/ni.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun ZY, et al. Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling. Proc Natl Acad Sci USA. 2004;101(48):16867–16872. doi: 10.1073/pnas.0407576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun ZJ, Kim KS, Wagner G, Reinherz EL. Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer. Cell. 2001;105(7):913–923. doi: 10.1016/s0092-8674(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 26.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 27.Alon R, Hammer DA, Springer TA. Lifetime of the P-selectin-carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature. 1995;374(6522):539–542. doi: 10.1038/374539a0. [DOI] [PubMed] [Google Scholar]
- 28.del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323(5914):638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong F, García AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185(7):1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Block SM. Real engines of creation. Nature. 1997;386(6622):217–219. doi: 10.1038/386217a0. [DOI] [PubMed] [Google Scholar]
- 31.Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol. 2014;15(9):798–807. doi: 10.1038/ni.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4(6):565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 33.O’Donoghue GP, Pielak RM, Smoligovets AA, Lin JJ, Groves JT. Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. eLife. 2013;2:e00778. doi: 10.7554/eLife.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci USA. 1995;92(11):5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, et al. Crystallization of a deglycosylated T cell receptor (TCR) complexed with an anti-TCR Fab fragment. J Biol Chem. 1996;271(52):33639–33646. doi: 10.1074/jbc.271.52.33639. [DOI] [PubMed] [Google Scholar]
- 36.Tiller T, Busse CE, Wardemann H. Cloning and expression of murine Ig genes from single B cells. J Immunol Methods. 2009;350(1-2):183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 37.Kern PS, et al. Structural basis of CD8 coreceptor function revealed by crystallographic analysis of a murine CD8alphaalpha ectodomain fragment in complex with H-2Kb. Immunity. 1998;9(4):519–530. doi: 10.1016/s1074-7613(00)80635-4. [DOI] [PubMed] [Google Scholar]
- 38.Moody AM, Xiong Y, Chang HC, Reinherz EL. The CD8alphabeta co-receptor on double-positive thymocytes binds with differing affinities to the products of distinct class I MHC loci. Eur J Immunol. 2001;31(9):2791–2799. doi: 10.1002/1521-4141(200109)31:9<2791::aid-immu2791>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 39.Shin Y, et al. Single-molecule denaturation and degradation of proteins by the AAA+ ClpXP protease. Proc Natl Acad Sci USA. 2009;106(46):19340–19345. doi: 10.1073/pnas.0910484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang MJ, Asbury CL, Shaevitz JW, Block SM. An automated two-dimensional optical force clamp for single molecule studies. Biophys J. 2002;83(1):491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.